Abstract
In this study, two different approaches were applied in the analysis of the GAA gene. One was analyzed based on patients with Pompe disease, and the other was analyzed based on GAA genomic data from unaffected carriers in a general population genetic database. For this, GAA variants in Korean and Japanese patients reported in previous studies and in patients reported in the Pompe disease GAA variant database were analyzed as a model. In addition, GAA variants in the Korean Reference Genome Database (KRGDB), the Japanese Multi Omics Reference Panel (jMorp), and the Genome Aggregation Database (gnomAD) were analyzed. Overall, approximately 50% of the pathogenic or likely pathogenic variants (PLPVs) found in unaffected carriers were also found in real patients with Pompe disease (Koreans, 57.1%; Japanese, 46.2%). In addition, there was a moderate positive correlation (Spearman’s correlation coefficient of 0.45–0.69) between the proportion of certain PLPVs in patients and the minor allele frequency of their variants in a general population database. Based on the analysis of general population databases, the total carrier frequency for Pompe disease in Koreans and Japanese was estimated to be 1.7% and 0.7%, respectively, and the predicted genetic prevalence was 1:13,657 and 1:78,013, respectively.
Keywords: Pompe disease, GAA gene, general population database, carrier frequency, genetic prevalence
1. Introduction
Pompe disease, or glycogen storage disease type II (MIM #232300), is a monogenic autosomal recessive disorder caused by deficiency of lysosomal alpha-glucosidase (GAA). This deficiency results in the accumulation of lysosomal glycogen in various body tissues, especially in cardiac and skeletal muscles [1,2,3]. Pompe patients who develop hypertrophic cardiomyopathy and general muscle weakness within the first year of life are classified as having classic infantile Pompe disease. Without enzyme replacement therapy (ERT), classic infantile Pompe disease is typically fatal within the first year of life. Nonclassic Pompe disease (or late-onset Pompe disease (LOPD) or childhood or adult onset Pompe disease) [3] is associated with a slowly progressive weakness of proximal muscles and respiratory dysfunction. Patients with nonclassic Pompe disease either develop symptoms without cardiac involvement before 1 year of life or develop symptoms after the first year of life. Given the benefits of early diagnosis and treatment with ERT, Pompe disease was included in the recommended uniform screening panel (the newborn screening program, NBS) in the USA [4].
In general, a research on rare diseases is conducted with the clinical and genetic information of patients. However, a huge amount of genetic information has been released to public databases, allowing us to think of new approaches to genetic diseases. Theoretically, the prevalence of a specific Mendelian disease is estimated by analyzing the proportion of unaffected carriers (carrier frequency) with the genomic information in the general population. In this study, two approaches were applied in the analysis of Pompe disease. One was based on the literature review of patients with Pompe disease reported, and the other was based on the genomic information from the general population. For these, the GAA gene in patients and the general population was analyzed.
2. Materials and Methods
2.1. Analysis Workflow
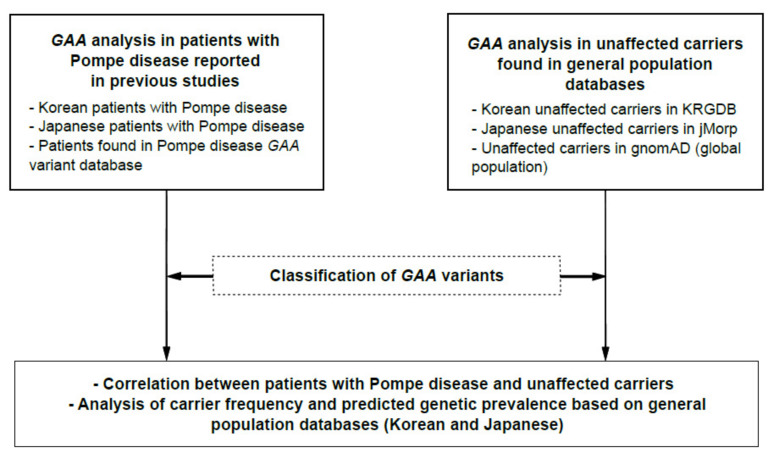
The entire analysis workflow for the two approaches is presented in Figure 1. A literature search for Korean and Japanese patients with Pompe disease was conducted, and the causative GAA variants in the patients were analyzed. In this study, newborn cases without specific symptoms or signs were excluded from the analysis. For the GAA analysis in unaffected carriers, the GAA gene from both Korean and Japanese general population databases was analyzed. Recently, a database containing Korean genomic information was released, called the Korean Reference Genome Database (KRGDB, http://coda.nih.go.kr/coda/KRGDB/index.jsp, accessed on 8 February 2021), which contains 1722 Korean genomic data [5]. In the present study, GAA genetic variants found in KRGDB (30× coverage group, 1465 individuals) were analyzed. In addition, the Japanese Multi Omics Reference Panel (jMorp, https://jmorp.megabank.tohoku.ac.jp/202102/variants, accessed on 16 March 2021) was used to analyze GAA variants in the Japanese general population [6,7]. To date, the jMorp database contains the genomic data (whole-genome sequencing data) of 8380 Japanese individuals.
Figure 1.
Analysis workflow in this study. The Pompe disease GAA variant database (http://www.pompevariantdatabase.nl/) was accessed on 16 March 2021, KRGDB (http://coda.nih.go.kr/coda/KRGDB/index.jsp) was accessed on 8 February 2021, jMorp (https://jmorp.megabank.tohoku.ac.jp/202102/variants) was accessed on 16 March 2021, and gnomAD (https://gnomad.broadinstitute.org/) was accessed on 17 March 2021.
In order to compare GAA variants between general databases, excluding common variants, GAA variants with a minor allele frequency (MAF) < 1% in East Asians in the Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/, accessed on 17 March 2021, search by genomic region: chr17:78,075,380-78,093,680 (GRCh37/hg19)) [8] were compared with those in KRGDB and JMorp. For a comparison between patients with Pompe disease and the general population, GAA variants in Korean or Japanese patients were compared with those found in KRGDB or JMorp. In addition, GAA variants in the Pompe disease GAA variant database [9] (http://www.pompevariantdatabase.nl/, accessed on 16 March 2021) were compared with those in the general population (global) in gnomAD (https://gnomad.broadinstitute.org/, accessed on 17 March 2021). A Venn diagram for comparative analysis used InteractiVenn [10] (Figure 2). A correlation between the proportions of certain PLPVs among all PLPVs found in total patients considering the frequency of detection (for simplicity, the proportions of certain PLPVs) and the MAF of those variants in a general population database was analyzed using Spearman’s rank correlation analysis. To determine the clinical severity of Pompe disease per specific GAA variant, information provided by the Pompe disease GAA variant database was used [9] (http://www.pompevariantdatabase.nl/, accessed on 16 March 2021).
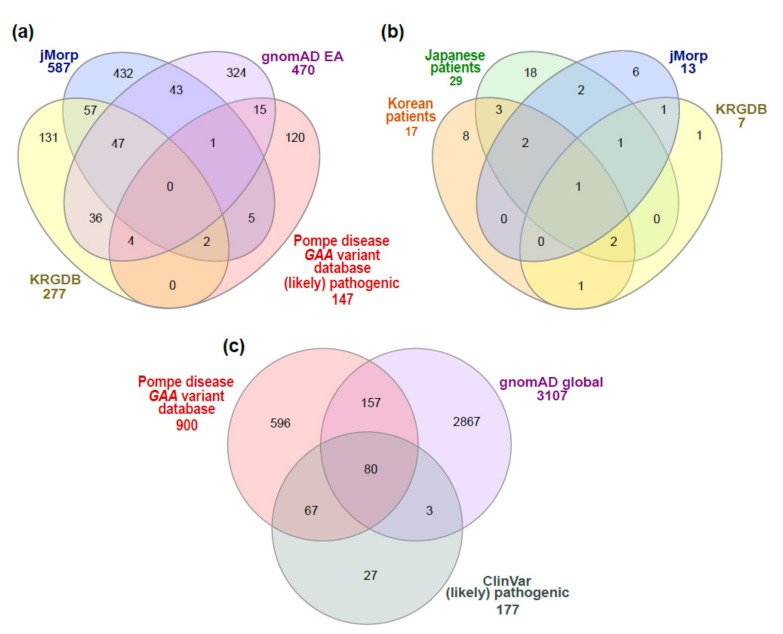
Figure 2.
GAA variants found in patients with Pompe disease or in general population databases. (a) Number of GAA variants with MAF < 1% in general population databases (KRGDB, jMorp, and gnomAD) (East Asian) and number of (likely) pathogenic variants (with a review status of ≥2 gold stars in ClinVar) in the Pompe disease GAA variant database; (b) comparison of (likely) pathogenic variants found in Korean or Japanese patients, KRGDB, and jMorp; and (c) number of GAA variants in the Pompe disease GAA variant database, number of GAA variant with MAF < 1% in gnomAD (global), and number of (likely) pathogenic variants with a review status of ≥2 gold stars in ClinVar.
2.2. GAA Variant Classification
All GAA variants were analyzed based on NM_000152.5 (NP_000143.2) and described following the Human Genome Variation Society (HGVS) variant nomenclature standards ((http://varnomen.hgvs.org/, accessed on 17 March 2021). The GAA variants described in an incorrect nomenclature, which were reported in the previous literature, were not included in this study. The GAA variants in KRGDB, jMorp, and previous literature on Korean or Japanese patients with Pompe disease were classified or reclassified according to the 2015 American College of Medical Genetics and Genomics and the Association for Molecular Pathology standards and guidelines (2015 ACMG/AMP guidelines) [11] and specifications by a ClinGen lysosomal storage disorders expert panel (https://clinicalgenome.org/affiliation/50009/, accessed on 20 March 2021). Briefly, the PVS1, PS1, PS3, PM2, PM5, and PP4 ACMG/AMP variant criteria by the ClinGen lysosomal storage disorders expert panel (https://clinicalgenome.org/affiliation/50009/, accessed on 20 March 2021) were applied. The PM3 criterion was applied following a general recommendation by the Sequence Variant Interpretation Working Group (https://clinicalgenome.org/working-groups/sequence-variant-interpretation/, accessed on 20 March 2021); that is, each proband was given point values considering the direction of avoiding circular logic and combined values, and then the strength level for PM3 was determined. For the PP3 criterion, REVEL (>0.75 for missense variants) [12,13], MutationTaster [14], MaxEntScan (for predicted impact on splicing) [15], and spliceAI (for the predicted impact on splicing) [16] were used. For checking critical functional domains (catalytic barrel and active site) when applying the PVS1 ACMG/AMP variant criterion, Pfam (https://pfam.xfam.org/, accessed on 20 March 2021), InterPro (https://www.ebi.ac.uk/interpro/, accessed on 20 March 2021), and UniProt (https://www.uniprot.org/, accessed on 20 March 2021) were used. Among the GAA variants reported in the Pompe disease GAA variant database, variants reported as pathogenic or likely pathogenic variants (PLPVs) with a review status of ≥2 gold stars in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, assessed on 27 April 2021) were classified as GAA PLPVs.
2.3. Analysis of Carrier Frequency and Predicted Genetic Prevalence
The carrier frequency (CF) and predicted genetic prevalence (pGP) were analyzed based on the heterozygous PLPVs. Neither the KRGDB nor the jMorp database provides information about homozygous variants. Thus (likely) pathogenic variants found in these databases were considered heterozygous variants because the general population assumes that there are no rare diseases. The CF and pGP were calculated as previously described [8,17].
3. Results
3.1. GAA Variants Found in Patients with Pompe Disease or General Population Databases
The GAA variants in Korean or Japanese patients with Pompe disease reported in previous studies are described in Table 1. A total of 10 studies evaluating the GAA variants in Korean patients with Pompe disease were reviewed [18,19,20,21,22,23,24,25,26,27]. To date, 17 different PLPVs (total of 59 PLPV alleles) in GAA have been reported in 32 Korean patients with Pompe disease (Table 1). GAA variants classified as variants of uncertain significance (VUS) because of insufficient pathogenic evidence (c.1669A>T (p.Ile557Phe) and c.2132C>G (p.Thr711Arg) reported by Kim EH et al. [27]) or GAA variants described in an incorrect nomenclature were excluded in this study. A total of 17 studies on Japanese patients with Pompe disease were reviewed [28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44], and 29 different GAA PLPVs (total of 130 PLPV alleles) were reported in 76 Japanese patients with Pompe disease. Of the GAA variants reported in 17 Japanese studies, 11 were classified as VUS and one was classified as benign, which were excluded from this analysis.
Table 1.
Pathogenic or likely pathogenic variants found in Korean or Japanese patients with Pompe disease or in general population databases.
| Variant | Korean Patients | Japanese Patients | General Population Databases, MAF | |||||
|---|---|---|---|---|---|---|---|---|
| Allele Count | [Ref] (HT/HM) | Allele Count | [Ref] (HT/HM) | KRGDB 1 | jMorp 2 | gnomAD 3, East Asian | gnomAD 3, Global | |
| c.-32-13T>G | 0 | 0 | 0.000342 | 0 | 0.000207 | 0.003401 | ||
| c.2T>C | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0 | |
| c.118C>T (p.Arg40 *) | 1 | [20] (1/0) | 1 | [28] (1/0) | 0 | 0 | 0.000050 | 0.000014 |
| c.169C>T (p.Gln57 *) | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0 | |
| c.307T>C (p.Cys103Arg) | 0 | 0 | 0 | 0.000060 | 0 | 0 | ||
| c.309C>A (p.Cys103 *) | 0 | 2 | [29] (0/1) | 0 | 0 | 0 | 0 | |
| c.483dup (p.Lys162Glnfs*15) | 0 | 2 | [30] (2/0) | 0 | 0 | 0 | 0 | |
| c.546G>A (p.Thr182=) | 0 | 0 | 0 | 0.000179 | 0.000102 | 0.000030 | ||
| c.546G>T (p.Thr182=) | 2 | [22] (1/0), [25] (1/0) | 27 | [28] (6/5), [34] (0/2), [35] (2/1), [36] (1/0), [37] (0/1) | 0 | 0.000119 | 0 | 0 |
| c.547-1G>C | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0 | |
| c.569G>A (p.Arg190His) | 0 | 1 | [28] (1/0) | 0 | 0.000060 | 0 | 0.000016 | |
| c.655G>A (p.Gly219Arg) | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0.000018 | |
| c.670C>T (p.Arg224Trp) | 0 | 1 | [31] (1/0) | 0 | 0 | 0 | 0.000022 | |
| (c.752C>T; c.761C>T) ((p.Ser251Leu; p.Ser254Leu) | 0 | 2 | [28] (1/0) | 0.005476 | 0.001850 | 0.002759 | 0.000195 | |
| c.756_757insT (p.Pro253Serfs*77) | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0 | |
| c.796C>T (p.Pro266Ser) | 1 | [26] (1/0) | 2 | [28] (2/0) | 0 | 0 | 0 | 0 |
| c.841C>T (p.Arg281Trp) | 0 | 0 | 0 | 0.000060 | 0 | 0.000205 | ||
| c.875A>G (p.Tyr292Cys) | 4 | [18] (1/0), [19] (1/0), [21] (1/0), [23] (1/0) | 0 | 0 | 0 | 0 | 0.000008 | |
| c.1156C>T (p.Gln386 *) | 1 | [23] (1/0) | 0 | 0 | 0 | 0 | 0 | |
| c.1225dup (p.Asp409Glyfs*97) | 1 | [20] (1/0) | 0 | 0 | 0 | 0 | 0 | |
| c.1309C>T (p.Arg437Cys) | 3 | [18] (1/0), [19] (1/0), [25] (1/0) | 12 | [28] (6/0), [33] (2/0), [38] (1/0), [39] (0/1), [40] (1/0) | 0 | 0.000060 | 0 | 0.000008 |
| c.1316T>A (p.Met439Lys) | 14 | [18] (4/0), [19] (1/0), [20] (1/0), [22] (1/0), [23] (1/0), [24] (1/0), [25] (3/0), [26] (2/0) | 3 | [28] (0/1), [35] (1/0) | 0.001027 | 0 | 0.000384 | 0.000028 |
| c.1322_1326+9del | 2 | [18] (1/0), [19] (1/0) | 0 | 0 | 0 | 0 | 0 | |
| c.1447G>A (p.Gly483Arg) | 0 | 0 | 0 | 0.000060 | 0 | 0.000008 | ||
| c.1579_1580del (p.Arg527Glyfs*3) | 2 | [19] (1/0), [20] (1/0) | 0 | 0 | 0 | 0 | 0.000004 | |
| c.1582_1583del (p.Gly528Leufs*2) | 1 | [18] (1/0) | 0 | 0 | 0 | 0 | 0 | |
| c.1585_1586delinsGT (p.Ser529Val) | 0 | 7 | [32] (2/2), [41] (1/0) | 0 | 0 | 0 | 0 | |
| c.1696T>C (p.Ser566Pro) | 0 | 4 | [28] (2/0), [30] (2/0) | 0 | 0 | 0 | 0 | |
| c.1735G>A (p.Glu579Lys) | 0 | 2 | [28] (1/0), [42] (1/0) | 0 | 0 | 0 | 0.000007 | |
| c.1798C>T (p.Arg600Cys) | 0 | 20 | [28] (7/0), [31] (1/0), [32] (7/1), [35] (2/0), [36] (1/0) | 0 | 0.000239 | 0 | 0.000004 | |
| c.1822C>T (p.Arg608 *) | 6 | [18] (2/0), [19] (2/0), [22] (1/0), [24] (1/0) | 4 | [28] (1/1), [35] (1/0) | 0 | 0 | 0.000051 | 0.000018 |
| c.1826dup (p.Tyr609 *) | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0.000008 | |
| c.1857C>G (p.Ser619Arg) | 7 | [20] (1/0), [21] (1/0), [23] (2/0), [24] (1/0), [25] (1/0), [26] (1/0) | 15 | [28] (4/3), [31] (0/1), [33] (0/1), [42] (1/0) | 0.000342 | 0.000418 | 0 | 0 |
| c.1935C>A (p.Asp645Glu) | 0 | 3 | [32] (1/1) | 0 | 0 | 0.001729 | 0.000124 | |
| c.1979G>A (p.Arg660His) | 0 | 2 | [31] (2/0) | 0 | 0 | 0 | 0.000037 | |
| c.2014C>T (p.Arg672Trp) | 0 | 0 | 0 | 0.000060 | 0 | 0.000008 | ||
| c.2015G>A (p.Arg672Gln) | 5 | [20] (1/0), [24] (2/0), [25] (1/0), [26] (1/0) | 8 | [32] (2/2), [43] (0/1) | 0.000343 | 0 | 0.000111 | 0.000021 |
| c.2171C>A (p.Ala724Asp) | 3 | [18] (1/0), [19] (1/0), [25] (1/0) | 0 | 0 | 0 | 0 | 0 | |
| c.2177C>G (p.Pro726Arg) | 0 | 2 | [28] (1/0), [40] (1/0) | 0 | 0 | 0 | 0 | |
| c.2297A>G (p.Tyr766Cys) | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0.000025 | |
| c.2238G>C (p.Trp746Cys) | 5 | [19] (1/0), [22] (1/0), [24] (1/0), [25] (2/0) | 0 | 0.000685 | 0 | 0.000351 | 0.000308 | |
| c.2238G>T (p.Trp746Cys) | 0 | 0 | 0 | 0.000119 | 0 | 0 | ||
| c.2326C>T (p.Gln776 *) | 0 | 2 | [33] (2/0) | 0 | 0 | 0 | 0 | |
| c.2407_2413del (p.Gln803 *) | 1 | [23] (1/0) | 0 | 0 | 0 | 0 | 0 | |
| c.2481+1G>A | 0 | 1 | [28] (1/0) | 0 | 0 | 0 | 0 | |
| c.2647-7G>A | 0 | 0 | 0.000342 | 0.000298 | 0 | 0.000018 | ||
1 The Korean Reference Genome Database (KRGDB, http://coda.nih.go.kr/coda/KRGDB/index.jsp, accessed on 8 February 2021). 2 The Japanese Multi Omics Reference Panel (jMorp, https://jmorp.megabank.tohoku.ac.jp/202102/variants, accessed on 16 March 2021). 3 The Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/, accessed on 17 March 2021). *, stop codon; Ref, references; HT, heterozygous allele count; HM, homozygous allele count; MAF, minor allele frequency.
There were 277 GAA variants with MAF < 1% in KRGDB, 587 variants in jMorp, and 470 variants in the East Asian population in gnomAD (Figure 2a). Of those, 47 variants were included in three databases. In addition, there were 7 GAA PLPVs in KRGDB and 13 PLPVs in jMorp (1 suspicious PLPV (>100 bp indel) was excluded) (Table 1, Figure 2b). Most of the (likely) pathogenic variants (with a review status of ≥2 gold stars in ClinVar) reported in the Pompe disease GAA variant database were not found in East Asian general population databases, such as KRGDB, JMorp, and gnomAD East Asian (81.6% (120/147), Figure 2a).
Overall, a total of 46 different variants from previous Korean or Japanese studies, KRGDB, or jMorp were classified into PLPVs (Table 1). Of those, 4 PLPVs were in both Korean patients and KRGDB, 6 PLPVs were in both Japanese patients and jMorp, and only 1 PLPV (c.1857C>G, p.Ser619Arg) was found in all Korean and Japanese patients, KRGDB, and jMorp (Figure 2b). Of the 46 PLPVs, there were 21 (likely) pathogenic variants with a review status of 2 or more gold stars (2 or 3) in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, assessed on 27 April 2021), and the other 25 were (likely) pathogenic variants with a review status of <2 gold stars (0 or 1), variants of uncertain significance, variants with conflicting interpretations of pathogenicity, or absent in ClinVar. The ACMG evidence codes for the other 25 variants are described in Table S1.
Of the 900 GAA variants reported in the Pompe disease GAA variant database, 147 variants were classified as PLPVs with a review status of ≥2 gold stars in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, assessed on 27 April 2021) (Figure 2c). Among those, 80 PLPVs were found in gnomAD.
3.2. Correlation between Patients with Pompe Disease and Unaffected Carriers
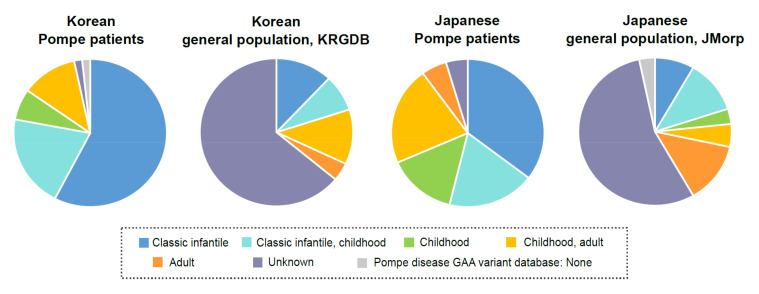
It was found that the overall distribution of clinical severity associated with GAA PLPVs detected in patients with Pompe disease and those in unaffected carriers differed (Figure 3). Especially, more GAA PLPVs associated with classic infantile Pompe disease were found in patients with Pompe disease than in unaffected carriers.
Figure 3.
Overall distribution of clinical severity associated with GAA (likely) pathogenic variants detected in Korean or Japanese patients with Pompe disease and those found in unaffected carriers.
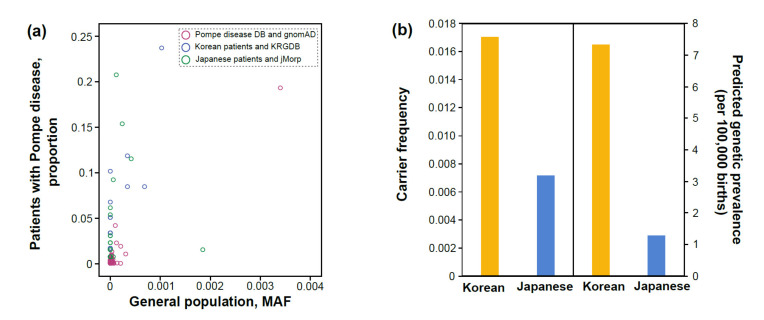
Spearman’s correlation coefficient between the proportion of certain PLPVs among all PLPVs found in total Korean patients (for simplicity, the proportion of certain PLPVs) and the MAF of their variants in KRGDB was 0.69 (p = 0.002), and it was 0.45 (p = 0.014) for Japanese patients. In addition, Spearman’s correlation coefficient between the proportion of certain PLPVs in the Pompe disease GAA variant database and their MAF in gnomAD (global) was 0.54 (p = 2.64 × 10−12) (Figure 4a).
Figure 4.
(a) Scatterplot of the proportion of certain (likely) pathogenic variants among all (likely) pathogenic variants found in total patients considering the frequency of detection (Y) and the minor allele frequency of those variants in a general population database (X). Purple line circles for patients found in the Pompe disease GAA variant database (Pompe disease DB), blue line circles for Korean patients, and green line circles for Japanese patients; (b) carrier frequency and predicted genetic prevalence for Pompe disease in Koreans and Japanese.
3.3. Carrier Frequency and Predicted Genetic Prevalence Based on General Population Databases
The total CF for Pompe disease in Koreans was estimated to be 1.7%, and the pGP was 1:13,657 (7.32 per 100,000 births) based on KRGDB (Figure 4b). In addition, the CF for Pompe disease in Japanese was predicted to be 0.7%, and the pGP was 1:78,013 (1.28 per 100,000 births) based on jMorp (Figure 4b).
4. Discussion
The main questions in this study are how GAA variants detected in Pompe patients are related to those in unaffected carriers and, on the contrary, how genomic information from the healthy population reflects the likelihood of developing Pompe disease. Two aspects can be considered to analyze how much GAA PLPVs found in patients and unaffected carriers have in common. One is to consider the qualitative aspect and to analyze how identical the GAA PLPVs between two groups are. The other is the quantitative aspect, which is whether certain GAA PLPVs frequently found in patients with Pompe disease are also found at a high frequency in unaffected carriers. In this study, Koreans and Japanese and a wider range of ethnic groups were independently analyzed to identify questions related to Pompe disease and associated GAA variants.
Of the 17 different PLPVs detected in Korean patients with Pompe disease, 23.5% (4 PLPVs) were found in unaffected Korean carriers in KRGDB. In addition, 20.7% (6/29) of the PLPVs detected in Japanese patients were found in unaffected Japanese carriers in JMorp (Figure 2b and Table 1). Among the PLPVs detected in Korean or Japanese patients with Pompe disease, certain PLPVs were not found in any general population databases, such as KRGDB, jMorp, and gnomAD (e.g., c.796C>T (p.Pro266Ser), c.2171C>A (p.Ala724Asp), c.1585_1586delinsGT (p.Ser529Val), c.1696T>C (p.Ser566Pro)). This means that there are GAA variants that are enriched especially in patients, which contribute to the development of Pompe disease. In contrast, about 50% of the PLPVs in unaffected carriers are also found in real patients with Pompe disease (Koreans, 57.1%; Japanese, 46.2%) (Figure 2b). When considering GAA PLPVs found in both patients and unaffected carriers, the sum of the proportion of these PLPVs (among all PLPVs found in total patients) in patients occupied up to 50%–60% (52.5% in Korean patients and 59.2% in Japanese patients).
In addition, there was a moderate positive correlation (Spearman’s correlation coefficient of 0.45–0.69) between the proportion of certain PLPVs in patients and the MAF of their variants in a general population database in each of the three independent analyses. However, not all cases where PLPVs were detected in patients with Pompe disease are reported in the literature, so there is a limit to the accuracy of the proportion of certain PLPVs. In this study, Koreans were predicted to have higher CF and pGP than Japanese, and what is interesting is that Spearman’s correlation coefficient in Koreans (0.69) is also higher than in Japanese (0.45).
The incidence of Pompe disease has been estimated to be 1 in 40,000, but varies depending on the geographic region or population [1]. However, the incidence of Pompe disease reported by the NBS is much higher than the estimate [4,8]. Pompe disease has not yet been included in the Korean NBS program. It is important to estimate the incidence or prevalence rate of a disease when considering its inclusion in the newborn screening program. To date, the prevalence or incidence of Pompe disease in Koreans has not been studied. The pGP (1:13,657, 7.32 per 100,000 births) for Korean Pompe disease in this study is comparable to the incidence of 1:16,919 from an NBS program involving 473,738 newborn samples in Taiwan [45]. In this study, the pGP for Pompe disease in Japanese was 1:78,013 (1.28 per 100,000 births). According to a recent study of 103,204 newborns in Japan, the incidence of Pompe disease in Japanese is 1:34,401 (three patients with potential LOPD were identified) [46]. In these three newborns, [c.752C>T; c.761C>T] ([p.Ser251Leu; p.Ser254Leu]) variant was commonly detected, and additionally, c.317G>A (p.Arg106His), c.2003A>G (p.Tyr668Cys), and c.1244C>T (p.Thr415Met) were detected, respectively. According to the 2015 ACMG/AMP guidelines [11] and specifications by a ClinGen lysosomal storage disorders expert panel (https://clinicalgenome.org/affiliation/50009/, accessed on 20 March 2021), the additional three GAA variants are classified as VUS. Therefore, if GAA variants are classified according to the current guidelines and specifications, the incidence of Pompe disease in Japanese might be lower than 1:34,401.
Interestingly, there were differences in the distribution of PLPVs detected in East Asia. The c.1316T>A (p.Met439Lys) variant was the most frequently detected in Korean patients and the second most frequent in the Korean general population, but was not found in other populations in gnomAD. This variant is supposed to be a founder pathogenic variant for Korean Pompe disease. In addition, c.546G>T (p.Thr182=) was the most reported variant in Japanese patients, and none other than the Japanese general population was reported. In addition, c.1316T>A (p.Met439Lys) and c.546G>T (p.Thr182=) were only reported in Korean or Japanese patients in the Pompe disease GAA variant database [9] (http://www.pompevariantdatabase.nl/, last accessed on 27 April 2021). The (c.752C>T; c.761C>T) (p.Ser251Leu; p.Ser254Leu) variant has the highest AF in both KRGDB and jMorp. However, this variant was not identified in Korean patients with Pompe disease and was identified in only one Japanese patient with a homozygous status [28]. This variant has been reported as a common causative variant in Asia, but is mostly identified on the NBS (http://www.pompevariantdatabase.nl/ accessed on 27 April 2021). It is presumed that the clinical severity associated with this variant might be very mild. Therefore, Pompe disease with this variant could not be identified. Additionally, the haplotype frequency (including this variant) for developing Pompe disease might be extremely rare. The c.-32-13T>G variant is the most common pathogenic variant for European Pompe disease [1]. However, this variant was only found in KRGDB and not reported in any Korean or Japanese patients with Pompe disease. The c.2238G>C (p.Trp746Cys) variant was reported as a common pathogenic variant for Pompe disease in mainland China [2]; however, this variant has not been reported in Japanese patients.
5. Conclusions
In this study, two different approaches were made to study Pompe disease. One was to analyze GAA variants based on patients in a traditional way, and the other was to analyze how likely this disease was in the general population. To apply this analysis, the GAA variants found in patients and the general population were interpreted as the same criterion according to the standards/guidelines or specifications for the interpretation of genetic variants, and Pompe disease in Koreans and Japanese was analyzed as a model. In addition, GAA PLPVs (with a review status of ≥2 gold stars in ClinVar) in the Pompe disease GAA variant database and gnomAD were compared.
Although some real PLPVs may have been classified as VUS due to currently insufficient evidence and the accuracy of this analysis is limited because GAA variants in patients with Pompe disease have been analyzed in only those reported in previous studies, the relationship between GAA variants found in patients with Pompe disease and in the general population is predicted to be more than a moderate correlation.
Acknowledgments
The author thanks Jong-Won Kim at the Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, for his valuable comments on this research.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/children8070601/s1: Table S1: Presumed pathogenic or likely pathogenic variants in the GAA gene are found in Korean or Japanese patients or in general population databases.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data analyzed in this study are included in this article and its Supplementary Information files.
Conflicts of Interest
The author declares no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.van der Ploeg A.T., Reuser A.J. Pompe’s disease. Lancet. 2008;372:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 2.Peruzzo P., Pavan E., Dardis A. Molecular genetics of Pompe disease: A comprehensive overview. Ann. Transl. Med. 2019;7:278. doi: 10.21037/atm.2019.04.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gungor D., Reuser A.J. How to describe the clinical spectrum in Pompe disease? Am. J. Med. Genet. A. 2013;161A:399–400. doi: 10.1002/ajmg.a.35662. [DOI] [PubMed] [Google Scholar]
- 4.Bodamer O.A., Scott C.R., Giugliani R., Pompe Disease Newborn Screening Working G. Newborn Screening for Pompe Disease. Pediatrics. 2017;140:S4–S13. doi: 10.1542/peds.2016-0280C. [DOI] [PubMed] [Google Scholar]
- 5.Jung K.S., Hong K.W., Jo H.Y., Choi J., Ban H.J., Cho S.B., Chung M. KRGDB: The large-scale variant database of 1722 Koreans based on whole genome sequencing. Database. 2020;2020 doi: 10.1093/database/baaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tadaka S., Saigusa D., Motoike I.N., Inoue J., Aoki Y., Shirota M., Koshiba S., Yamamoto M., Kinoshita K. jMorp: Japanese Multi Omics Reference Panel. Nucleic Acids Res. 2018;46:D551–D557. doi: 10.1093/nar/gkx978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tadaka S., Hishinuma E., Komaki S., Motoike I.N., Kawashima J., Saigusa D., Inoue J., Takayama J., Okamura Y., Aoki Y., et al. jMorp updates in 2020: Large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 2021;49:D536–D544. doi: 10.1093/nar/gkaa1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park K.S. Carrier frequency and predicted genetic prevalence of Pompe disease based on a general population database. Mol. Genet. Metab. Rep. 2021;27:100734. doi: 10.1016/j.ymgmr.2021.100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nino M.Y., In ‘t Groen S.L.M., Bergsma A.J., van der Beek N., Kroos M., Hoogeveen-Westerveld M., van der Ploeg A.T., Pijnappel W. Extension of the Pompe mutation database by linking disease-associated variants to clinical severity. Hum. Mutat. 2019;40:1954–1967. doi: 10.1002/humu.23854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh R., Oak N., Plon S.E. Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol. 2017;18:225. doi: 10.1186/s13059-017-1353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 15.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 16.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e524. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Hanany M., Rivolta C., Sharon D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc. Natl. Acad. Sci. USA. 2020;117:2710–2716. doi: 10.1073/pnas.1913179117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho A., Kim S.J., Lim B.C., Hwang H., Park J.D., Kim G.B., Jin D.K., Lee J., Ki C.S., Kim K.J., et al. Infantile Pompe disease: Clinical and genetic characteristics with an experience of enzyme replacement therapy. J. Child Neurol. 2012;27:319–324. doi: 10.1177/0883073811420295. [DOI] [PubMed] [Google Scholar]
- 19.Kim M.S., Song A., Im M., Huh J., Kang I.S., Song J., Yang A., Kim J., Kwon E.K., Choi E.J., et al. Clinical and molecular characterization of Korean children with infantile and late-onset Pompe disease: 10 years of experience with enzyme replacement therapy at a single center. Korean J. Pediatr. 2019;62:224–234. doi: 10.3345/kjp.2018.06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko J.M., Park K.S., Kang Y., Nam S.H., Kim Y., Park I., Chae H.W., Lee S.M., Lee K.A., Kim J.W. A New Integrated Newborn Screening Workflow Can Provide a Shortcut to Differential Diagnosis and Confirmation of Inherited Metabolic Diseases. Yonsei Med. J. 2018;59:652–661. doi: 10.3349/ymj.2018.59.5.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee D.H., Qiu W.J., Lee J., Chien Y.H., Hwu W.L. Hypertrophic cardiomyopathy in pompe disease is not limited to the classic infantile-onset phenotype. JIMD Rep. 2014;17:71–75. doi: 10.1007/8904_2014_339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J.H., Shin J.H., Park H.J., Kim S.Z., Jeon Y.M., Kim H.K., Kim D.S., Choi Y.C. Targeted population screening of late onset Pompe disease in unspecified myopathy patients for Korean population. Neuromuscul. Disord. 2017;27:550–556. doi: 10.1016/j.nmd.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Park H.D., Lee D.H., Choi T.Y., Lee Y.K., Lee S.Y., Kim J.W., Ki C.S., Lee Y.W. Three patients with glycogen storage disease type II and the mutational spectrum of GAA in Korean patients. Ann. Clin. Lab. Sci. 2013;43:311–316. [PubMed] [Google Scholar]
- 24.Park H.J., Jang H., Kim J.H., Lee J.H., Shin H.Y., Kim S.M., Park K.D., Yim S.V., Lee J.H., Choi Y.C. Discovery of pathogenic variants in a large Korean cohort of inherited muscular disorders. Clin. Genet. 2017;91:403–410. doi: 10.1111/cge.12826. [DOI] [PubMed] [Google Scholar]
- 25.Park J.S., Kim H.G., Shin J.H., Choi Y.C., Kim D.S. Effect of enzyme replacement therapy in late onset Pompe disease: Open pilot study of 48 weeks follow-up. Neurol. Sci. 2015;36:599–605. doi: 10.1007/s10072-014-2000-5. [DOI] [PubMed] [Google Scholar]
- 26.Park Y.E., Park K.H., Lee C.H., Kim C.M., Kim D.S. Two new missense mutations of GAA in late onset glycogen storage disease type II. J. Neurol. Sci. 2006;251:113–117. doi: 10.1016/j.jns.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Kim E.H., Ko J.M., Lee B.H., Kim G.H., Choi J.H., Yoo H.W. Two patients with atypical infantile Pompe disesase presenting with hypertrophic cardiomyopathy. J. Genet. Med. 2009;6:161–165. [Google Scholar]
- 28.Fukuhara Y., Fuji N., Yamazaki N., Hirakiyama A., Kamioka T., Seo J.H., Mashima R., Kosuga M., Okuyama T. A molecular analysis of the GAA gene and clinical spectrum in 38 patients with Pompe disease in Japan. Mol. Genet. Metab. Rep. 2018;14:3–9. doi: 10.1016/j.ymgmr.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermans M.M., van Leenen D., Kroos M.A., Beesley C.E., Van Der Ploeg A.T., Sakuraba H., Wevers R., Kleijer W., Michelakakis H., Kirk E.P., et al. Twenty-two novel mutations in the lysosomal alpha-glucosidase gene (GAA) underscore the genotype-phenotype correlation in glycogen storage disease type II. Hum. Mutat. 2004;23:47–56. doi: 10.1002/humu.10286. [DOI] [PubMed] [Google Scholar]
- 30.Matsuoka T., Miwa Y., Tajika M., Sawada M., Fujimaki K., Soga T., Tomita H., Uemura S., Nishino I., Fukuda T., et al. Divergent clinical outcomes of alpha-glucosidase enzyme replacement therapy in two siblings with infantile-onset Pompe disease treated in the symptomatic or pre-symptomatic state. Mol. Genet. Metab. Rep. 2016;9:98–105. doi: 10.1016/j.ymgmr.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pipo J.R., Feng J.H., Yamamoto T., Ohsaki Y., Nanba E., Tsujino S., Sakuragawa N., Martiniuk F., Ninomiya H., Oka A., et al. New GAA mutations in Japanese patients with GSDII (Pompe disease) Pediatr. Neurol. 2003;29:284–287. doi: 10.1016/S0887-8994(03)00267-4. [DOI] [PubMed] [Google Scholar]
- 32.Tsujino S., Huie M., Kanazawa N., Sugie H., Goto Y., Kawai M., Nonaka I., Hirschhorn R., Sakuragawa N. Frequent mutations in Japanese patients with acid maltase deficiency. Neuromuscul. Disord. 2000;10:599–603. doi: 10.1016/S0960-8966(00)00142-5. [DOI] [PubMed] [Google Scholar]
- 33.Nabatame S., Taniike M., Sakai N., Kato-Nishimura K., Mohri I., Kagitani-Shimono K., Okinaga T., Tachibana N., Ozono K. Sleep disordered breathing in childhood-onset acid maltase deficiency. Brain Dev. 2009;31:234–239. doi: 10.1016/j.braindev.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Tsuburaya R.S., Monma K., Oya Y., Nakayama T., Fukuda T., Sugie H., Hayashi Y.K., Nonaka I., Nishino I. Acid phosphatase-positive globular inclusions is a good diagnostic marker for two patients with adult-onset Pompe disease lacking disease specific pathology. Neuromuscul. Disord. 2012;22:389–393. doi: 10.1016/j.nmd.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi H., Shimada Y., Ikegami M., Kawai T., Sakurai K., Urashima T., Ijima M., Fujiwara M., Kaneshiro E., Ohashi T., et al. Prognostic factors for the late onset Pompe disease with enzyme replacement therapy: From our experience of 4 cases including an autopsy case. Mol. Genet. Metab. 2010;100:14–19. doi: 10.1016/j.ymgme.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Maimaiti M., Takahashi S., Okajima K., Suzuki N., Ohinata J., Araki A., Tanaka H., Mukai T., Fujieda K. Silent exonic mutation in the acid-alpha-glycosidase gene that causes glycogen storage disease type II by affecting mRNA splicing. J. Hum. Genet. 2009;54:493–496. doi: 10.1038/jhg.2009.66. [DOI] [PubMed] [Google Scholar]
- 37.Hossain M.A., Miyajima T., Akiyama K., Eto Y. A Case of Adult-onset Pompe Disease with Cerebral Stroke and Left Ventricular Hypertrophy. J. Stroke Cerebrovasc. Dis. 2018;27:3046–3052. doi: 10.1016/j.jstrokecerebrovasdis.2018.06.043. [DOI] [PubMed] [Google Scholar]
- 38.Fujimoto S., Manabe Y., Fujii D., Kozai Y., Matsuzono K., Takahashi Y., Narai H., Omori N., Adachi K., Nanba E., et al. A novel mutation of the GAA gene in a patient with adult-onset Pompe disease lacking a disease-specific pathology. Intern. Med. 2013;52:2461–2464. doi: 10.2169/internalmedicine.52.0311. [DOI] [PubMed] [Google Scholar]
- 39.Isayama R., Shiga K., Seo K., Azuma Y., Araki Y., Hamano A., Takezawa H., Kuriyama N., Takezawa N., Mizuno T., et al. Sixty six-month follow-up of muscle power and respiratory function in a case with adult-type Pompe disease treated with enzyme replacement therapy. J. Clin. Neuromuscul. Dis. 2014;15:152–156. doi: 10.1097/CND.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 40.Ishigaki K., Yoshikawa Y., Kuwatsuru R., Oda E., Murakami T., Sato T., Saito T., Umezu R., Osawa M. High-density CT of muscle and liver may allow early diagnosis of childhood-onset Pompe disease. Brain Dev. 2012;34:103–106. doi: 10.1016/j.braindev.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Tsunoda H., Ohshima T., Tohyama J., Sasaki M., Sakuragawa N., Martiniuk F. Acid alpha-glucosidase deficiency: Identification and expression of a missense mutation (S529V) in a Japanese adult phenotype. Hum. Genet. 1996;97:496–499. doi: 10.1007/BF02267074. [DOI] [PubMed] [Google Scholar]
- 42.Ishigaki K., Murakami T., Nakanishi T., Oda E., Sato T., Osawa M. Close monitoring of initial enzyme replacement therapy in a patient with childhood-onset Pompe disease. Brain Dev. 2012;34:98–102. doi: 10.1016/j.braindev.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Huie M.L., Tsujino S., Sklower Brooks S., Engel A., Elias E., Bonthron D.T., Bessley C., Shanske S., DiMauro S., Goto Y.I., et al. Glycogen storage disease type II: Identification of four novel missense mutations (D645N, G648S, R672W, R672Q) and two insertions/deletions in the acid alpha-glucosidase locus of patients of differing phenotype. Biochem. Biophys. Res. Commun. 1998;244:921–927. doi: 10.1006/bbrc.1998.8255. [DOI] [PubMed] [Google Scholar]
- 44.Muraoka T., Murao K., Imachi H., Kikuchi F., Yoshimoto T., Iwama H., Hosokawa H., Nishino I., Fukuda T., Sugie H., et al. Novel mutations in the gene encoding acid alpha-1,4-glucosidase in a patient with late-onset glycogen storage disease type II (Pompe disease) with impaired intelligence. Intern. Med. 2011;50:2987–2991. doi: 10.2169/internalmedicine.50.5563. [DOI] [PubMed] [Google Scholar]
- 45.Chiang S.C., Hwu W.L., Lee N.C., Hsu L.W., Chien Y.H. Algorithm for Pompe disease newborn screening: Results from the Taiwan screening program. Mol. Genet. Metab. 2012;106:281–286. doi: 10.1016/j.ymgme.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Momosaki K., Kido J., Yoshida S., Sugawara K., Miyamoto T., Inoue T., Okumiya T., Matsumoto S., Endo F., Hirose S., et al. Newborn screening for Pompe disease in Japan: Report and literature review of mutations in the GAA gene in Japanese and Asian patients. J. Hum. Genet. 2019;64:741–755. doi: 10.1038/s10038-019-0603-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed in this study are included in this article and its Supplementary Information files.