Abstract
Ovarian failure (OF) is a common cause of infertility usually diagnosed as idiopathic, with genetic causes accounting for 10–25% of cases. Whole-exome sequencing (WES) may enable identifying contributing genes and variant profiles to stratify the population into subtypes of OF. This study sought to identify a blood-based gene variant profile using accumulation of rare variants to promote precision medicine in fertility preservation programs. A case–control (n = 118, n = 32, respectively) WES study was performed in which only non-synonymous rare variants <5% minor allele frequency (MAF; in the IGSR) and coverage ≥ 100× were considered. A profile of 66 variants of uncertain significance was used for training an unsupervised machine learning model to separate cases from controls (97.2% sensitivity, 99.2% specificity) and stratify the population into two subtypes of OF (A and B) (93.31% sensitivity, 96.67% specificity). Model testing within the IGSR female population predicted 0.5% of women as subtype A and 2.4% as subtype B. This is the first study linking OF to the accumulation of rare variants and generates a new potential taxonomy supporting application of this approach for precision medicine in fertility preservation.
Keywords: ovarian failure, whole exome sequencing, single nucleotide variant, infertility, precision medicine, prediction model, genomic taxonomy, genome variant profile, personalized medicine, ovary
1. Introduction
Ovarian failure (OF) is characterised by accelerated attrition of the ovarian follicle reserve, amenorrhea, dramatic hypoestrogenism, and elevated gonadotropin levels, but these manifestations differ depending on aetiology [1,2,3]. OF may result from genetic (familial or sporadic), cytogenetic, environmental, iatrogenic, autoimmune, or metabolic disorders; genetic causes account for about 10–25% of cases [4,5], and autoimmune conditions account for 4–30% of cases [6]. Though women with OF can achieve pregnancy [7,8], OF usually presents as infertility because the ovarian reserve is nearly or completely exhausted [9]. OF is often diagnosed as idiopathic, and research is needed to better define the origins of OF and identify risk factors to aid in early diagnosis and inform treatment measures [10,11,12].
Anti-Müllerian hormone (AMH) and follicle stimulating hormone (FSH) measurements aid OF diagnosis [13,14]. However, both are limited as predictive biomarkers because modest increases or decreases in AMH are difficult to detect and do not characterise subtypes of OF; meanwhile, FSH has less sensitivity than AMH and depends on the day of the cycle in which the sample is obtained [15,16,17]. Intriguingly, as for other conditions [18], high-throughput genomics data may help discern subtypes and stages of OF.
Heritability plays a clear role in OF, and studies of familial OF had shed light on genetic aspects of the condition [19,20,21]. While familial studies focus on detecting causative variants in one or few genes in one family, population studies identify variants shared by individuals independent of familial relationship and inheritance [22,23,24]. Next-generation sequencing (NSG) based on whole-exome sequencing (WES) characterises known and unknown variation within gene-coding regions in each studied sample, significantly improving the power of previous studies focused on discovery of variants in the population [19,24,25,26,27].
Despite advantages of WES, the large degree of genetic variation creates challenges in identifying meaningful changes. Therefore, strategies are needed to identify candidate variants that can be prioritised by predicted protein defects, frequency in the population, and evidence of evolutionary pressure including negative selection [22,28,29,30,31,32]. Existing studies often lack (1) negative controls (i.e., age-matched individuals without OF); (2) characterisation of genetic variation identified in genes not known to be associated with the phenotype of interest; and (3) implementation of machine learning strategies such as machine learning algorithms that provide a more comprehensive picture, as is required in reproductive precision medicine [18]. Manipulation of this information by machine learning algorithms, can be applied to stratify populations into disease subtypes based on an additive model considering the presence or absence of DNA variants [33,34,35]. Thus, we performed a WES-based case–control study to describe genomic profiles based on multivariant models considering the presence or absence of DNA variants (single-nucleotide variants (SNVs)) as preventive screening to identify women at risk of OF. This predictive model will inform precision medicine in fertility preservation programs.
2. Materials and Methods
2.1. Participants and Inclusion Criteria
A WES-based case–control study was conducted between 2017 and 2019 at infertility clinics in collaboration with a genetic diagnosis and reproductive medicine research department. The study recruited 118 women diagnosed with OF and 32 women as controls from Spain. Women exhibiting amenorrhea for >6 months with AMH values < 0.3 ng/mL, FSH values > 20 IU, and <5 follicles upon antral follicle count (AFC) via transvaginal ultrasound were classified as OF. Controls had AMH values > 1.5 ng/mL, FSH < 10 IU, and AFC > 10. Clinical outcome (live birth (LB)) for both groups was determined. All patients were <40 years old at the time of recruitment and were selected following clinical criteria with idiopathic disease and normal karyotypes, no FRM1 permutations, and no history of pelvic surgery, radiotherapy, chemotherapy, or autoimmune disorders. The International Genome Sample Resource (IGSR) database, which contains genomic variant information including allele frequencies, normal genomic variability, and ethnicity from 1271 healthy female individuals (OF was not considered an exclusion criterion), was used as a pseudo-control population to optimise the study complementing control population [36,37,38]. The Shapiro–Wilk test was used to check normality of clinical variables, hormone levels, age, body mass index, AFC, and LB, while the Wilcoxon and Fisher tests were used to evaluate clinical ranges of AMH and presence or absence of variants, respectively, between cases and controls [39,40,41]. The study was approved by the institutional review board of the Instituto Valenciano de Infertilidad and Hospital La Fe (1709-PAM-090-PR).
2.2. Pre-Processing, NGS, and Variant Calling
Peripheral blood genomic DNA was isolated (Maxwell 16 lev blood DNA, Promega, Madison, WI, USA) and quantified using fluorescence spectroscopy (Qubit). Absorbance readings with Nanodrop confirmed the purity of DNA, with all samples yielding a 260:280 ratio of >1.8. DNA integrity was evaluated using TapeStation (Agilent Technologies, Santa Clara, CA, USA), and the DNA integrity number (DIN) was >7 (recommended threshold for NGS library prep strategies) for each sample. WES was performed on all DNA samples using SureSelect Clinical Research Exome V2 (Agilent Technologies) and Illumina sequencers (MiSeq or NextSeq). Reads from the 18,311 sequenced genes were aligned to the human reference genome (hg19) using the Burrows-Wheeler algorithm (version 0.7.17) mapper [42]. Subsequent variant calling was performed using GATK software (version 3.6.0) following the standard pipeline the standard pipeline recommended by the developers of the software [43]. Variants were annotated using SnpEff software (version 4.3) [44] to obtain information on which position of the genome is affected by the variant, including if it is a protein coding sequence, which gene and in what position of said gene is located, and the biological consequences expected from the changes (e.g., if the variant disrupts the triplet reading frame of the DNA, is called a Frameshift variant). Furthermore, SnpEff retrieves information from the IGSR that allow the user to know if a certain variant is registered or not in the database [44].
2.3. Variant Filtering, Processing, and Prioritisation
Variants were filtered based on several criteria (further detailed in Supplemental Figure S1):
A moderate or deleterious effect on protein coding sequence (according to SnpEff annotations). Moderate effect included Missense variants, UTR (5′ + 3′) and splice (acceptor or donor) variants; while deleterious effects included Frameshift, Nosense (stop codon gain/loss) variants, protein to protein contact modifier variants, structural interaction modifier variants and disruptive inframe variants.
Variants absent from the IGSR database were retained for downstream analysis. Meanwhile, variants present in the IGSR were only kept if their minor allele frequency (MAF) was lower than <0.05, based on the premise that purifying selection decreases allele frequency of variants that confer less fitness.
Passage of quality criteria for coverage (>100×) as well as several parameters evaluated by GATK: Genotype Quality (GQ), which evaluates the confidence of the genotype attributed to a patient for a certain variant (homozygous for the reference allele, heterozygous or homozygous for the disease-associated allele); Position depth (DP) or total number of reads detected at a given position of the genome; Allele Depth (AD), the number of reads for the variant in that position. Further information is annexed with Supplementary Figure S1.
After applying these filters, remaining variants were used to (i) identify variants with significant differences in frequency between controls and cases and (ii) classify variants if they were present in at least 10% of cases and completely absent from controls. This last point was based on the premise that variants found only in one individual are related to individual variation while variants shared by a subgroup have a higher probability of being biomarkers related to the disease. Prioritized variants in this step were researched to find possible links with fertility. In addition to IGSR, GnomAD, and dbSNP databases were consulted to determine if identified variants were already reported [45,46], and Genecards, Uniprot, and Gene Ontology databases were used to annotate gene function [47,48,49]. Case and control variant frequencies were tested by Fisher test in the R environment (version 3.4.4, 15 March 2018) [50], while variant processing and prioritisation were done in the Python environment (version 3.5.2, 26 July 2016) [51].
2.4. Patient Stratification and Ovarian Failure Subtypes Prediction
Prioritised variants were used to stratify the study population based on patient genotype (homozygous for the reference allele, heterozygous or homozygous for the disease-associated allele). In order to find different subtypes of ovarian failure in the study population, unsupervised hierarchical clustering—with similarity and genomic distance values calculated with the Jaccard coefficient [52]—was used to group patients based on genomic variant profile. Jaccard’s formula is J (X, Y) = |X∩Y|/|X∪Y| where X is the genotype profile of the variants of one patient, while Y is the same for another patient. Calculating the similarity coefficient with Jaccard allow grouping together patients that share not only the same variants but also the same genotype for said variants. Optimal clustering was achieved with Ball and Hartigan indexes implemented in Nbclust (version 3.0) in R [53], which increase differences inter-cluster while trying to maximize similarities intra-cluster; this is, trying to find clusters of patients that are really similar by the genomic profile while separating them as much as possible from other patients that do not share similar profiles. Using the subtypes of ovarian failure generated and to generate a model capable of distinguish patients with OF, a Random Forest algorithm was trained with a 10-fold cross-validation (CV) 100 times in WEKA platform software (version 3.8.2, 22 December 2017) [54], WEKA by default stratified the folds maintaining the proportion of cases and controls (79%/21%). With Random Forest, the model created 500 forest each iteration and selected the best consensual tree [55,56]. Random forest also assigned a score to each variant based on the mean decrease in impurity (MDI), which represents how informative each variant is for stratifying the population according to genotype of said variant [57]. Key variants in the stratification were analysed to look for potential relations with fertility. Finally, to ascertain if the genomic variant profile found in our population is detected in an independent population and ensure that a determined subtype is reproducible, the female population from the IGSR database (n = 1271) [36] was evaluated using our predictive model.
3. Results
3.1. Clinical Characterisation and Sequencing Quality of the Study Population
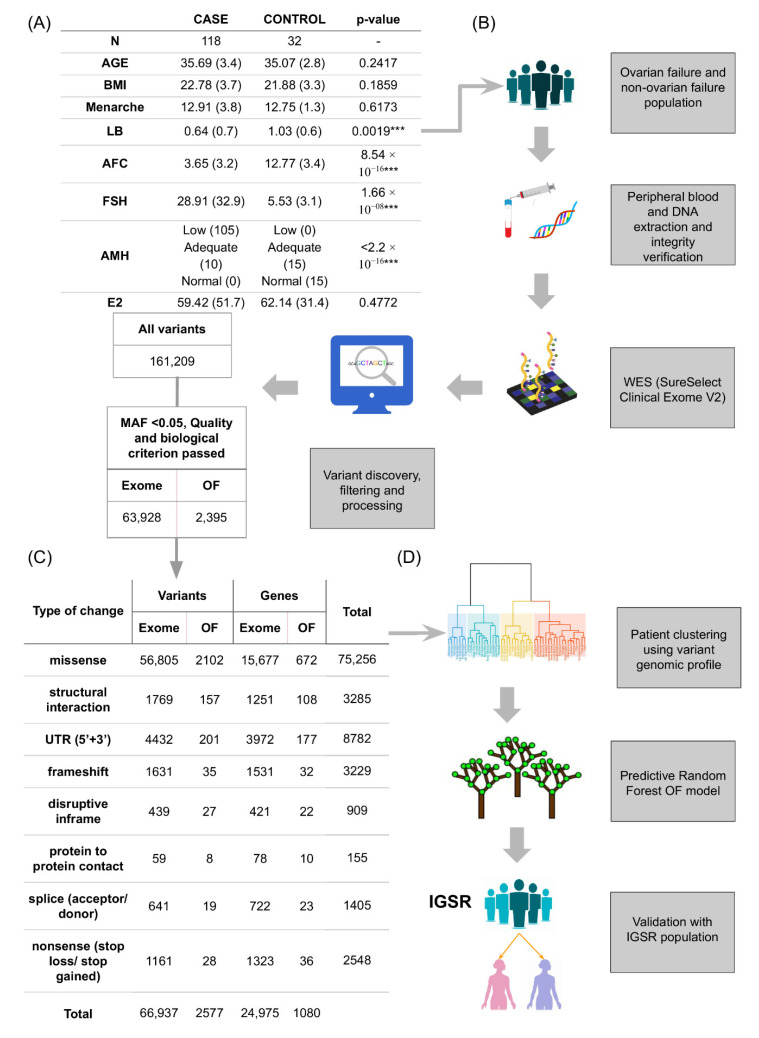
High-depth exome sequencing data were achieved with an average > 100× for 15,903 genes (86.85% of the studied 18,311 genes) and >25× for 16,773 genes (91.66% of the studied 16,773 genes) among 118 patients with OF and 32 controls. Phenotypically characterised cases and controls had significantly different mean values of AMH, AFC, FSH, and LB (p < 0.01), highlighting the clinical differences associated with OF related to controls (Figure 1A). For genomic study of SNVs associated with OF, we focused on variants absent in the IGSR with a low MAF (<0.05) in the IGSR population and changes predicted to cause a moderate to deleterious effect at the protein level. With these criteria, we ensured that variants were under pressure of purifying selection.
Figure 1.
Study design and variant prioritisation. (A) Population demographics and clinical data. Means and standard deviations (in brackets) are shown. For age, body mass index (BMI), age at menarche, live birth (LB), antral follicle count (AFC), follicle stimulating hormone (FSH), and oestradiol (E2) contrasts. The Shapiro–Wilk test was used to check normality and the Wilcoxon test to evaluate differences between cases and controls. The Fisher test was used to evaluate differences between cases and controls for anti-Müllerian hormone (AMH) levels according to Reference Laboratory hormonal ranges (low: <0.68 ng/mL; adequate: 0.68–2.27 ng/mL; normal: >2.27 ng/mL) (*** p < 0.01). (B) Pipeline for filtering variants. Women diagnosed with ovarian failure (OF) were recruited as cases (n = 118) and those without ovarian failure as controls (n = 32). Whole-exome sequencing (WES) of DNA from peripheral blood was performed in all samples using SureSelect Clinical Research Exome V2 (Agilent Technologies) and Illumina sequencing (Miseq or Nextseq). Variant calling was performed using GATK software. (C) Prioritised variants. Variants that passed quality and biological criteria (minor allele frequency (MAF), type of change at protein level, sequencing parameters) are shown for targeted analysis of genes previously associated with ovarian physiology and from whole exome analysis. Number of variants and genes affected for each predicted change in protein function are also represented. (D) Pipeline of the predictive OF model. A random forest predictive model is built using the prioritised variants and then validated with the pseudo-control population of the International Genome Sample Resource (IGSR) (n = 1271).
3.2. Genomic Variation Hypotheses
Gene-targeted and non-targeted hypotheses related to ovarian failure were developed from the 161,209 variants identified in the 18,311-gene panel (Figure 1B). In the gene-targeted approach, we focused on finding more variants in genes previously associated with ovarian physiology; for the non-targeted approach, the whole gene panel was considered to identify main variants in novel genes not previously associated with OF. There were 2395 variants in genes associated with ovarian physiology identified in the targeted approach, and 63,928 variants considered candidate disease-associated alleles in the non-targeted approach (Figure 1C); 57,866 synonymous variants were excluded because no effect was predicted. However, contrasting the proportions of variants found in targeted genes to the proportions of variants found in remaining genes of the exome (non-targeted) revealed that targeted genes related to ovarian physiology accumulated fewer variants (Fisher test, p < 2.2 × 10−16, odds ratio = 1.2), so the targeted hypothesis was discarded. Variants found in the whole exome were classified by the predicted type of change in the coding sequence. The 63,928 variants showed moderate to deleterious effects, with most changes being missense in the UTR or in structural interaction and frameshifts (Figure 1C). The rest of the experimental design covered in the following points is shown in Figure 1D.
3.3. A Genomic Variant Profile Predictive of OF
A significant difference (p < 0.01) in the distributions of allele frequencies between cases and controls was observed in 116 of the 63,928 candidate variants (Supplementary Table S1). Interestingly, only one of them, the missense variant c.902C > G (p.Ala301Gly) inducing an alanine to glycine change, was identified in the macrophage stimulating 1-like (MST1L) gene in 14 controls and four cases with a significant difference in proportions (FDR = 0.03, odds ratio = 16.6) (highlighted in Supplementary Table S1). Given the unique intra-variability of each individual and that finding variants shared by several individuals is complex, to ensure that the accumulation of variants was predictive of OF, we identified variants shared by at least 10% of cases and not present in controls. The 66 variants of uncertain significance (VUS) absent in controls with high case prevalence in >10% of cases affected 62 genes (Supplementary Table S2). Important variants by their prevalence in cases are shown in Table 1. One variant, affecting the mucin 6 (MUC6) gene (c.5297C > T; p.Thr1766Ile), was identified in 26% (i.e., 31 of 118) of patients with OF. An additional variant, c.715G > A (p.Ala239Thr), affecting the ankyrin repeat domain 20 family member A4 (ANKRD20A4), was absent in IGSR, dbSNP, and GnomAD databases. In addition to the MUC6 variant, five variants were shared by >20 cases: c.529A > G (p.Ser177Gly) affecting bromodomain and PHD finger containing 3 (BRPF3) in 22 cases, c.1435G > A (p.Ala479Thr) affecting adaptor related protein complex 5 subunit mu 1 (AP5M1) in 22 cases, c.880A > T (p.Met294Leu) affecting cysteine rich secretory protein LCCL domain containing 2 (CRISPLD2) in 21 cases, c.692C > G (p.Ala237Gly) affecting galactosamine (N-acetyl)-6-sulfatase (GALNS) in 20 cases, and c.539C > G (p.Thr180Ser) affecting mini chromosome maintenance complex component 5 (MCM5) in 20 cases (highlighted in Table 1). Three of the 66 variants affected three genes previously associated with infertility: variant c.181G > C (p.Ala60Pro) of mutS homolog 3 (MSH3) in 15 cases, c.1534G > A (p.Val512Ile) in gamma-glutamyltransferase 1 (GGT1) in 14 cases, and c.782G > A (p.Arg261Gln) of aquaporin 8 (AQP8) in 13 cases (as noted in Table 1).
Table 1.
Novel variants predictive of ovarian failure. Chromosomal and genomic positions of the variants, as well as bases and amino acid changes at the indicated number in the sequences, dbSNP IDs (if known), type of changes, amino acid class, polarity and charge changes, genes affected, number of cases affected by the variant, coverage (mean and standard deviation), and accession numbers. Variants were present in at least 10% of cases (12 women out of 118) and no controls. Gene Ontology and Genecards databases were consulted to annotate gene function. 1 Top 6 variants shared by >20 cases. 2 Top variants valued by random forest algorithm to stratify the population. 3 The three variants found affecting genes already related to ovarian failure in the literature. 4 Variant absent in International Genome Sample Resource, dbSNP, and GnomAD databases.
| Chromosome and Position | Change at Sequence and Aa Level | Rs | Type of Change | Amino Acid Class, Polarity, and Charge Change | Gene | Function | N Cases Affected | Coverage | Accession Number |
|---|---|---|---|---|---|---|---|---|---|
| 2; 84897501 2 | c.6356A > G, p.Tyr2119Cys | rs17025409 | Missense variant | Aromatic polar neutral > sulfuric nonpolar neutral | DNAH6 | Microtubule activity | 17 | 146 (18.08) |
NM_001370.1 |
| 2; 84932720 2 | c.8576A > G, p.Lys2859Arg | rs61750773 | Missense variant | Basic polar positive > basic polar positive | DNAH6 | Microtubule activity | 19 | 146 (20.02) |
NM_001370.1 |
| 2; 85059227 2 | c.1034C > T, p.Arg345His | rs61744273 | Missense variant | Basic polar positive > basic aromatic polar positive-neutral | TRABD2A | Negative regulation of WNT signalling pathway | 18 | 287 (42.23) |
NM_001277053.1 |
| 5; 79950724 3 | c.181G > C, p.Ala60Pro | rs2001675 | Missense variant | Aliphatic nonpolar neutral > cyclic nonpolar neutral | MSH3 | DNA repair | 15 | 145 (57.73) |
NM_002439.4 |
| 6; 36168628 1 | c.529A > G, p.Ser177Gly | rs45504893 | Missense variant | Hydroxylic polar neutral > aliphatic nonpolar neutral | BRPF3 | Chromatin organisation | 22 | 320 (44.1) |
NM_015695.2 |
| 9; 69391207 4 | c.715G > A, p.Ala239Thr | Missense variant | Aliphatic nonpolar neutral > hydroxylic polar neutral | ANKRD20A4 | Unknown | 13 | 125 (8.03) |
NM_001098805.1 | |
| 11; 1017471 2 | c.5330G > A, p.Gly1777Asp | Missense variant | Aliphatic nonpolar neutral > acid acidic polar negative | MUC6 | Cytoprotection of epithelial surfaces | 13 | 448 (194.38) |
NM_005961.2 | |
| 11; 1017504 1,2 | c.5297C > T, p.Thr1766Ile | Missense variant | Hydroxylic polar neutral > aliphatic nonpolar neutral | MUC6 | Cytoprotection of epithelial surfaces | 31 | 448 (147.15) |
NM_005961.2 | |
| 14; 57755564 1 | c.1435G > A, p.Ala479Thr | rs35759976 | Missense variant | Aliphatic nonpolar neutral > hydroxylic polar neutral | AP5M1 | Apoptosis | 22 | 179 (14.19) |
NM_018229.3 |
| 16; 25239809 2,3 | c.782G > A, p.Arg261Gln | rs111840156 | Missense variant | Basic polar positive > amide polar neutral | AQP8 | Cellular response to cAMP | 13 | 265 (24.69) |
NM_001169.2 |
| 16; 84902483 1 | c.880A > T, p.Met294Leu | rs72799568 | Missense variant | Sulfuric nonpolar neutral > aliphatic nonpolar neutral | CRISPLD2 | Extracellular matrix assembly | 21 | 247 (145.71) |
NM_031476.3 |
| 16; 88902199 1 | c.692C > G, p.Ala237Gly | rs34745339 | Structural interaction variant, missense variant | Aliphatic nonpolar neutral > aliphatic nonpolar neutral | GALNS | Degradation of glycosaminoglycans | 20 | 214 (33.02) |
NM_000512.4 |
| 17; 71232990 2 | c.1369C > A, p.Arg457Ser | rs61729639 | Missense variant | Basic polar positive > hydroxylic polar neutral | SPEP1 | Unknown | 14 | 180 (38.16) |
NM_001288771.1 |
| 22; 17450929 2 | c.841G > A, p.Ala281Thr | rs61741409 | Missense variant | Aliphatic nonpolar neutral > hydroxylic polar neutral | GAB4 | Unknown | 14 | 236 (36.89) |
NM_001037814.1 |
| 22; 17450952 2 | c.818T > C, p.Leu273Pro | rs11703655 | Missense variant | Aliphatic nonpolar neutral > cyclic nonpolar neutral | GAB4 | Unknown | 14 | 236 (43.66) |
NM_001037814.1 |
| 22; 25024326 3 | c.1534G > A, p.Val512Ile | Structural interaction variant, missense variant | Aliphatic nonpolar neutral > aliphatic nonpolar neutral | GGT1 | Proteolysis | 14 | 112 (17.48 |
NM_013430.2 | |
| 22; 35802661 1 | c.539C > G, p.Thr180Ser | rs2307340 | Missense variant | Hydroxylic polar neutral > hydroxylic polar neutral | MCM5 | DNA replication initiation | 20 | 309 (92.27) |
NM_006739.3 |
3.4. A New Genomic Taxonomy of OF
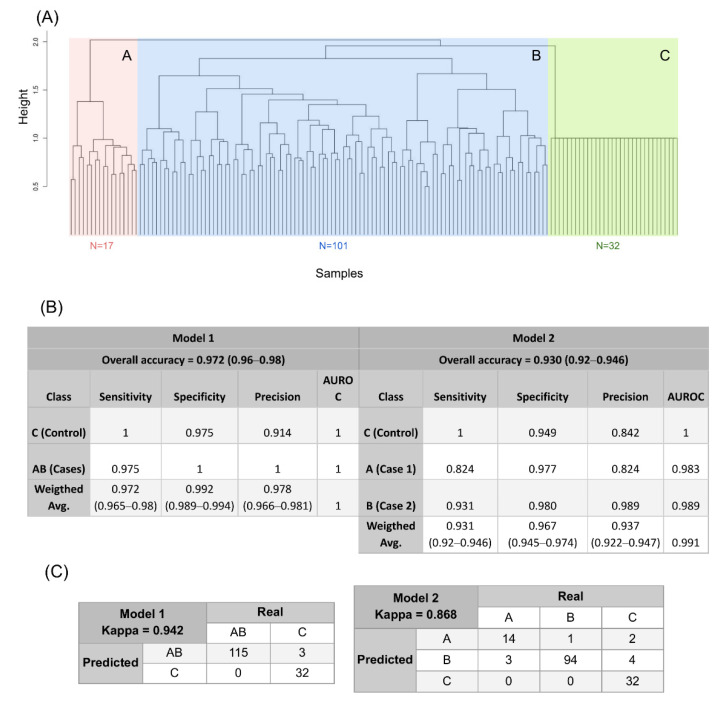
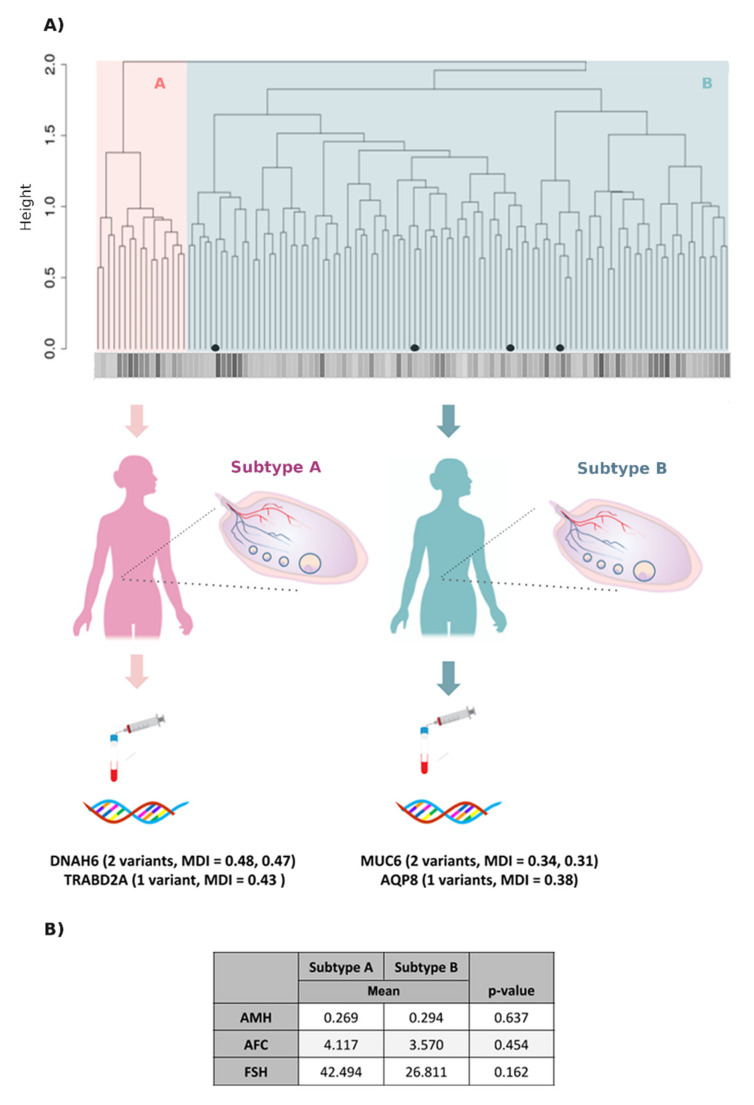
Based on the 66 variants present in >10% of OF cases and absent in controls, the clustering based on the genomic variant profile distinguished two main subtypes of OF (subtypes A and B) distinct from controls (C) (Figure 2A). Based on genomic distance, subtype B was more similar to controls than A. The predictive value of the 66 variant profiles distinguished OF cases (A, B) and controls (C) with an average of 97.2% (ranging 0.96–0.98) through the 100 interactions of the Random Forest model, an average sensitivity of 97.2% (ranging 0.965–0.98) an average specificity of 99.2% (ranging 0.989–0.994) (Figure 2B, left). Major variant contributors to the stratification were SPEP1 and GAB4 missense variants (c.1369C > A, p.Arg457Ser and c.818T > C, p.Leu273Pro, with MDI 0.2 and 0.15, respectively). The model also distinguished two genomic subtypes of OF (A and B) with an average of 93.3% accuracy (ranging 0.92–0.946) through the 100 iterations of the model, an average of 93.31% (ranging 0.92–0.946) sensitivity, and an average of 96.57% specificity (ranging 0.945–0.974) (Figure 2B, right); 14.4% of OF patients were classified as type A and 85.6% as type B, and three patients were incorrectly classified between cases and controls (Figure 2C, left) and an average of 10 patients were incorrectly classified as the other subtype or control when comparing subtypes (Figure 2C, right). Further, there were no clinical differences of significance in mean values for AMH, FSH, or AFC between subtypes A and B, so the difference was only at the genetic level (Figure 3B). The number of disease-associated variants (n = 66) accumulated by each patient ranged 1–15, with most accumulating nine variants (n = 17) (Supplementary Figures S2 and S3A). Additionally, the MST1L variant c.902C > G (p.Ala301Gly) was confined to subtype B (highlighted in Figure 3A). Genomic characterisation of the subtypes revealed three variants characteristic of subtype A; two affecting the dynein axonemal heavy chain 6 (DNAH6) gene (c.6356A > G, p.Tyr2119Cys and c.8576A > G, p.Lys2859Arg, MDI 0.48 and 0.47, respectively), and one affecting traB domain containing 2A (TRABD2A) (c.1034G > A, p.Arg345His, MDI 0.43); also identified were two previously mentioned variants (AQP8 and MUC6 (MDI 0.38 and 0.31, respectively)) and a second MUC6 variant (c.5330G > A, p.Gly1777Asp, MDI 0.34) characteristic of subtype B (Figure 3A, bottom) (as noted in Table 1).
Figure 2.
Genomic taxonomy of ovarian failure. (A) Dendrogram obtained from unsupervised clustering of case and control individuals. Three clusters are distinguished: clusters A (red, n = 17) and B (blue, n = 101) group case individuals into two distinct genomic profiles, while cluster C (green, n = 32) contains all control individuals. Genomic distance is represented by height for all groups, with a greater height indicating a larger difference between groups. (B) Prediction performance parameters. Parameters were obtained after executing a random forest algorithm 100 times, with 500 trees created in each iteration with 10-fold stratified cross-validation. Parameters are shown for model 1 (left) and model 2 (right), with the corresponding values of accuracy, sensitivity, specificity, precision, and ROC area obtained for each class and a weighted average in total. Kappa statistic for each model also is shown. (C) Prediction performance confusion matrices. A matrix is shown for model 1 (left), where controls (group C) were distinguished from cases (groups A, B). All 32 controls were correctly classified in cluster C, but three cases were misclassified. A matrix for model 2 (right) distinguishing the two genomic profiles for ovarian failure (groups A and B), with all controls correctly classified (group C) and 10 cases incorrectly classified as either controls or the other subtype. AUROC = area under ROC curve.
Figure 3.
Characterisation of the two genomic subtypes of ovarian failure. (A) Dendrogram analysis of both subtypes. Genomic profiles of subtypes A (red cluster, n = 17) and B (blue cluster, n = 101) obtained from unsupervised clustering were examined for the number of variants accumulated in each individual (represented in greyscale). Macrophage stimulating 1 like variant was found in four cases, all in the B subtype (black point in dendrogram). Mean decrease in impurity (MDI) scores assigned by the random forest algorithm was evaluated, and two genomic variants for dynein axonemal heavy chain 6 (DNAH6) and one for TraB domain containing 2A (TRABD2A) genes were the most characteristic of subtype A. Meanwhile, two variants for mucin 6 (MUC6) and one for aquaporin 8 (AQP8) were the most characteristic of subtype B (bottom). (B) Clinical comparison of subtypes A and B. Shapiro and Wilcoxon tests were applied to contrast the mean antral follicle counts (AFC) and levels of follicle-stimulating hormone (FSH) and anti-Müllerian hormone (AMH). No statistical differences were found.
3.5. Testing the Genomic Predictive Model in the IGSR Population
To determine whether the model could predict subtypes of OF in an independent population with unknown fertility, we tested our model in the female IGSR population (n = 1271). A prediction score of 0–1 was associated with each individual to determine diagnostic power, and only individuals with a predictive score ≥ 0.9 were considered predicted to the specific subtype. In the IGSR population, 7/1271 (0.5%) women were predicted as subtype A, while 31/1271 (2.4%) were considered subtype B.
4. Discussion
We describe the first predictive model of OF based on a genomic variant profile obtained through blood WES and machine learning algorithms. The model was effective in identifying and stratifying patients into two subtypes of OF (A and B), considering a pattern of 66 variants rather than individual variant effects. Subtypes of OF were tested in an independent IGSR population of 1271 women—only 0.5% of women were predicted as subtype A, a feasible proportion considering that OF prevalence is estimated at 1% in women < 40 years old [1,2,3]. We believe that the 2.4% of women predicted as subtype B may be overestimated given that our model was constructed under the assumption that the 66 variants are absent from controls and our design only considered 32 controls. However, the IGSR population introduced a higher population variability that could contribute to false positives in subtype B, as this profile was more similar to controls than subtype A. Additionally, other phenomena influencing disease prevalence such as penetrance or expressivity together with environmental factors could affect the final phenotype. Thus, subtype A should be considered the most distinguishable from controls, with likely fewer false positives and with the most potential to be useful in fertility preservation programs. However, further prospective studies are needed to evaluate the prediction ability of this model in relation to clinical phenotype.
Prior exome-sequencing studies seeking to identify new variants associated with OF [22,30,58] focused on established genes associated with OF; in our study, DNA sequences in genes related to ovarian physiology exhibited proportionately fewer variants than remaining genes in the exome. Further, prior studies identified variants shared by a few patients with OF and did not include controls [22,29,31,32,59,60]. The prioritisation criteria used in our study ensure that variants are rare and are likely under purifying selective pressure based on the potential cumulative adverse effects of the variants on genetic fitness of the OF population at a functional level [25,61]. In contrast to GWAS studies that use populations of thousands [62,63,64,65], our study had a modest sample size but was larger than similar studies in Europe or the USA [22,32,66,67]. The efficacy of the contrast of proportions approach was lower than GWAS studies, as expected, although we did identify 116 disease-associated variants with p < 0.01 and one variant with an adjusted p < 0.05 that was over-represented in controls. This adjusted variant affected MST1L, which encodes a protein with serine-type endopeptidase activity but no other known functions [47,48]. Overrepresentation of the MSTL1 variant in controls may protect against OF, suggesting an advantage conferred by the G allele, but this requires further research because the number of controls in this study was small.
The fingerprint or genomic intra-variability of each individual presents a challenge in variant profiling [68]. A presence > 10% (i.e., 12 out of 118 OF patients) was deemed necessary to identify variants fixed in the OF population, and we identified 66 VUS in genes not previously associated with OF matching these criteria. In addition to identifying variants in genes not previously associated with fertility, we identified three variants affecting genes previously associated with OF: MSH3, GGT1, and AQP8. MSH3, part of the post-replication DNA repair system, is required for fertility; mice lacking Mshl3 are sterile and their oocytes fail to complete meiosis I [69]. Ggt1 knock-out mice are infertile, lack antral follicle development, and do not respond to external gonadotropins [70]. AQP8 plays an important role in the apoptosis of granulosa cells, and mice lacking Aqp8 develop mature follicles and are more fertile than wild-type mice [71]; therefore, further research is needed to determine how the AQP8 variants impact OF.
The top six variants shared by patients with OF that were most representative of our population occurred in MUC6, BRPF3, AP5M1, CRISPL2, GALNS, and MCM5. A variant in MUC6 was found in 31 cases (25% of study population). MUC6 encodes a protein associated with protecting epithelial surfaces against chemical agents [72] and is related to ovarian tumours in mice [73]. Given that OF may have an environmental aetiology [11,20,74], we hypothesise that it may protect the ovary from environmental pollution and chemotoxicity and its variants may confer differential sensitivity to environmental agents. Another variant shared by 22 patients (20% of population) is within BRPF3 and is associated with reorganisation of chromatin and acetylation of histone H3K14, which is needed for efficient activation of DNA replication [75]. Chromatin organisation and DNA replication are imperative during follicular development [19,76], suggesting a role of BRPF3 in oocyte maturation and fertility. Further, an AP5M1 variant was found in 22 women. AP5M1 induces apoptosis in cervical carcinoma cells [77] and may play a similar role in primordial follicle death and premature loss of ovarian reserve. Twenty-one cases had a variant in CRISPL2, which promotes extracellular matrix assembly. The bovine CRISPL2 homolog is upregulated in granulosa cells in ovulatory follicles and could play an important role in human fertility [78]. GALNS was affected in 20 patients, participates in degradation of glycosaminoglycans, and is highly expressed in the ovary [79]. Finally, 20 patients had a variant affecting MCM5, which encodes a protein that is part of a molecular complex involved in DNA replication. Alterations in members of the same gene family, MCM8 and MCM9, affect DNA repair and cause OF [80,81,82].
Precision medicine describes new disease stages and treatment targets based on genomic profiles [18]. A profile based on the 66 VUS identified in this study distinguished OF from controls with 97.2% accuracy and stratified the OF population into two different groups with 93% accuracy, 93.31% sensitivity, and 96.67% specificity. Three patients in subtype A and six in subtype B failed to classify as their actual subtype. Subtype B was closer to the control group and contained all four cases sharing the MST1L variant. These results suggest two genomic subtypes of OF, one with a specific genetic profile (A) and another (B) genetically distinct but closer to our control group. Two variants, one in SPEP1 and the other in GAB4, were the most informative in case vs. control classification. Little is known, however, about the functions of both genes. Three gene variants were the most informative for classifying patients into subtype A (one in TRABD2 affecting 18 patients, and two in DNHA6 affecting 17 and 19 patients) and three into subtype B (AQP8 and MUC6 variants, and an additional MUC6 variant affecting 13 patients). TRABD2A is a metalloprotease that acts as a negative regulator of WNT signalling [83] by cleaving WNT3A, which is needed in synergy with R-spondin2 for follicular development in mice [84]. TRABD1A also cleaves WNT5A, a protein that decreases ovulation and increases follicular atresia. WNT5A is a physiologic inhibitor of gonadotropin signalling in humans [85], and female Wnt5a knockout mice are subfertile [86]. DNAH6 belongs to the dynein family of genes and encodes part of the microtubule-associated motor protein complex. Other dynein family members, DNAH5 and DNAH1, are associated with infertility [87,88]. Mutations in DNAH6 cause primary ciliary dyskinesia and Huntington’s disease, both of which are associated with infertility [89,90].
Although we purport functional roles for the genes involved in the genomic variant profile, this does not imply causation in reference to OF. Not all variants of the profile need to have a direct link with the pathology; these variants could be biomarkers without an implication for fertility. Importantly, the subtypes we described were distinguishable only by their genetic profiles and not by clinical parameters (including FSH), suggesting that they are detectable with the molecular deepness of genomic profiling. This highlights that a deeper understanding of the variant profile could change OF taxonomy and molecular classification. Indeed, this is interesting because the clinical criteria do not distinguish aetiologies or subtypes of OF. Subtype A could be easier to distinguish from controls for preventive detection in women at risk to experience infertility who can be identified only from genomic information. Whether these subtypes have different aetiologies or confer other clinical implications requires further studies.
We distinguished a genomic variant profile between OF cases and controls that revealed two subtypes of OF. While the inheritance and causative nature of the variants are not known, the distinct variant profiles serve as building blocks for a predictive model to detect subtype A in the general population and offer a promising first step toward using genomic and personalised medicine to predict OF in fertility preservation programs. We acknowledge that the good predictive value of our model depends on absence of the 66 variants for proper prediction; thus, as more individuals are tested, the performance of the model may decrease, such as overestimating the number of women with subtype B. Nonetheless, we believe this distinct genomic profile is capable of predicting OF in the general population, especially for subtype A. Clinical follow-up studies and prediction models testing in an independent population will be required to ascertain significance of the identified subtypes and overcome clinical and technical limitations of this study. Nevertheless, and given our sample size, we trust that the cross-validation models here developed avoids overfitting by using all samples in both training and testing phases.
5. Conclusions
We described the genomic profile of 66 VUS not previously associated with OF. The variant profile was used to create a predictive model capable of identifying OF individuals and classifying them into two genomic subtypes (A and B) with high accuracy, specificity, and sensitivity. One subtype was predicted accurately in a feasible proportion of the IGSR cohort as a surrogate of the general population. Thus, the identified variants may help establish a variant profile as a preventive biomarker in fertility preservation programs as a minimally invasive test in blood samples. Further prospective studies in an independent population are needed to determine reproducibility of the model and evaluate preventive potential of the two genetic subtypes in clinical practice.
Acknowledgments
The authors thank the study participants and IVI-RMA Pamplona, IVI Foundation, the University of Valencia, the University of Navarra, CIMA LAB Diagnostics, the Government of Navarra, and FEDER for their support. They also thank Fresh Eyes Editing, LLC team, especially Sheila Cherry, for professional assistance in the preparation of this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jpm11070609/s1, Figure S1: Pipeline for filtering genomic variants. A first filter was made to retain variants affecting protein coding sequences. From these, the International Genome Sample Resource (IGSR) database, which contains variants of >3000 healthy individuals, was consulted. Variants found in the database with a minor allele frequency (MAF) < 0.05 were kept. Variants absent from the database also were retained. Two criteria were applied to this set of variants. Quality criteria values were based on confidence of a genotype attributed to a specific sample, or genotype quality (GQ); total amount of reads for a given position, or position depth (DP); and number of reads for the given variant detected in the sequencing, or allele depth (AD). In addition, AD should account for ≥20% of position depth. Biological criteria were based on the predicted protein-level effect of the variant, which could range from deleterious to moderate. Figure S2: Frequency of ovarian failure variants accumulated in each patient. Number of patients is presented on the Y-axis, and number of presenting variants from the genomic profile of 66 variants associated with ovarian failure on the X-axis of the histogram. Most patients (17) shared 9 variants. The maximum number of shared variants was 15, seen in 4 patients. Figure S3: DNAH6 and TRABD2A variants. Screenshots are shown for 3 variant s (2 for DNAH6, 1 for TRABD2A), highlighting the genome position (left corner, in green) and the presence of the variant in all corresponding case samples in the respective gene (highlighted in the centre of the picture, in green). Table S1: Significant variants found after using contrast of proportions. Fisher test identified 116 significant variants (p < 0.01). Chromosome, position, change at sequence and amino acid level, type of change, genes affected, and dbSNP identifier, if known, are shown. 1 = Significant variant after adjusting p-values for FDR (adj. < 0.05). Table S2: Novel sixty-six variants predictive of ovarian failure. Chromosomal and genomic positions of the variants, as well as bases and amino acid changes at the indicated number in the sequences, dbSNP IDs (if known), type of changes, amino acid class, polarity and charge changes, genes affected, number of cases affected by the variant, and coverage and accession numbers. Variants were present in at least 10% of cases (12 women out of 118) and no controls. Gene Ontology and Genecards databases were consulted to annotate gene function. 1 Top 6 variants shared by >20 cases. 2 Top variants valued by random forest algorithm to stratify the population. 3 The three variants found affecting genes already related to ovarian failure in the literature. 4 Variant absent in International Genome Sample Resource, dbSNP, and GnomAD databases.
Author Contributions
The research idea was conceived by P.D.-G. and J.R.; P.R. and J.R. supported the study, and P.D.-G. coordinated and supervised the study. Clinical criteria and patient recruitment were managed by P.R. and J.R. and primarily performed by B.M.-M. with the help of M.R. Clinical follow-up was done mainly by B.M.-M. and supervised by P.R. with the help of M.R. Study design was determined by P.D.-G. with the help of P.R. and G.A.-A. as well as A.P.-G. in the design of sequencing strategies. Sample pre-processing, DNA extraction, and exome sequencing were coordinated by G.A.-A. and A.P.-G. The bioinformatics exome pre-processing pipeline was designed and implemented by F.J.G.-A. and supervised by G.A.-A. and P.D.-G. with the help of A.A. and I.H.-C., and P.S.-L. designed the bioinformatic downstream analysis. A.A. and I.H.-C. implemented the prediction models with the help of P.S.-L. and were supervised by P.D.-G. Variant prioritisation and genomic analysis was mainly done by A.A. and I.H.-C. with the help of P.S.-L. and supervised by P.D.-G. Data interpretation and visualisation were mainly done by I.H.-C. and P.D.-G. with the help of B.M.-M. and P.R. Tables and figures were designed by I.H.-C. with the help of P.S.-L. and supervised by P.D.-G. The manuscript was written by I.H.-C. and P.D.-G. and was supervised by all co-authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the IVI-RMA IVI Foundation, Valencia, and by IVI-RMA global Pamplona, Spain (1709-PAM-090-PR). Research was co-financed by Navarra Government and European Regional Development funds (p.o FEDER 2014-2020): 0011-1365-2017-000265. Ismael Henarejos-Castillo is supported by the ACIF/2019/148 predoctoral program fellowship from the Conselleria de Educacion Investigacion Cultura y Deporte, Generalitat de Valencia, Spain. Begoña Martinez-Montoro is co-funded by the predoctoral program “doctorados industriales 2019–2021” from the Navarra Government 0011-1408-2018-000011.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the institutional review board of the Instituto Valenciano de Infertilidad and Hospital La Fe (1709-PAM-090-PR).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors report no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tucker E., Grover S.R., Bachelot A., Touraine P., Sinclair A.H. Premature Ovarian Insufficiency: New Perspectives on Genetic Cause and Phenotypic Spectrum. Endocr. Rev. 2016;37:609–635. doi: 10.1210/er.2016-1047. [DOI] [PubMed] [Google Scholar]
- 2.Coulam C.B., Adamson S.C., Annegers J.F. Incidence of Premature Ovarian Failure. Obstet. Gynecol. Surv. 1987;42:182–183. doi: 10.1097/00006254-198703000-00020. [DOI] [PubMed] [Google Scholar]
- 3.Salvador-Carulla L., Bertelli M., Martinez-Leal R. The road to 11th edition of the International Classification of Diseases: Trajectories of scientific consensus and contested science in the classification of intellectual disability/intellectual developmental disorders. Curr. Opin. Psychiatry. 2018;31:79–87. doi: 10.1097/YCO.0000000000000396. [DOI] [PubMed] [Google Scholar]
- 4.Torrealday S., Kodaman P., Pal L. Premature Ovarian Insufficiency—An update on recent advances in understanding and management. F1000Research. 2017;6:2069. doi: 10.12688/f1000research.11948.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudnicka E., Kruszewska J., Klicka K., Kowalczyk J., Grymowicz M., Skórska J., Pięta W., Smolarczyk R. Premature ovarian insufficiency—Aetiopathology, epidemiology, and diagnostic evaluation. Menopausal Rev. 2018;17:105–108. doi: 10.5114/pm.2018.78550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirshenbaum M., Orvieto R. Premature ovarian insufficiency (POI) and autoimmunity—An update appraisal. J. Assist. Reprod. Genet. 2019;36:2207–2215. doi: 10.1007/s10815-019-01572-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calik-Ksepka A., Grymowicz M., Bronkiewicz W., Urban A., Mierzejewski K., Rudnicka E., Smolarczyk R. Spontaneous pregnancy in a patient with premature ovarian insufficiency—Case report. Menopausal Rev. 2018;17:139–140. doi: 10.5114/pm.2018.78560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J.L., Chabbert-Buffet N., Darai E. Diminished ovarian reserve, premature ovarian failure, poor ovarian responder—A plea for universal definitions. J. Assist. Reprod. Genet. 2015;32:1709–1712. doi: 10.1007/s10815-015-0595-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pastore L.M., Christianson M.S., Stelling J., Kearns W.G., Segars J.H. Reproductive ovarian testing and the alphabet soup of diagnoses: DOR, POI, POF, POR, and FOR. J. Assist. Reprod. Genet. 2018;35:17–23. doi: 10.1007/s10815-017-1058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora P., Polson D.W. Diagnosis and management of premature ovarian failure. Obstet. Gynaecol. 2011;13:67–72. doi: 10.1576/toag.13.2.67.27648. [DOI] [Google Scholar]
- 11.Vabre P., Gatimel N., Moreau J., Gayrard V., Picard-Hagen N., Parinaud J., Léandri R. Environmental pollutants, a possible etiology for premature ovarian insufficiency: A narrative review of animal and human data. Environ. Health. 2017;16:37. doi: 10.1186/s12940-017-0242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck-Peccoz P., Persani L. Premature ovarian failure. Orphanet J. Rare Dis. 2006;1:9. doi: 10.1186/1750-1172-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwase A., Nakamura T., Osuka S., Takikawa S., Goto M., Kikkawa F. Anti-Müllerian hormone as a marker of ovarian reserve: What have we learned, and what should we know? Reprod. Med. Biol. 2016;15:127–136. doi: 10.1007/s12522-015-0227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Marca A., Sighinolfi G., Papaleo E., Cagnacci A., Volpe A., Faddy M.J. Prediction of Age at Menopause from Assessment of Ovarian Reserve May Be Improved by Using Body Mass Index and Smoking Status. PLoS ONE. 2013;8:e57005. doi: 10.1371/journal.pone.0057005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alipour F., Rasekhjahromi A., Maalhagh M., Sobhanian S., Hosseinpoor M. Comparison of Specificity and Sensitivity of AMH and FSH in Diagnosis of Premature Ovarian Failure. Dis. Markers. 2015;2015:585604. doi: 10.1155/2015/585604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleicher N., A Kushnir V., Barad D.H. Prospectively assessing risk for premature ovarian senescence in young females: A new paradigm. Reprod. Biol. Endocrinol. 2015;13:34. doi: 10.1186/s12958-015-0026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gleicher N., Weghofer A., Barad D.H. Defining ovarian reserve to better understand ovarian aging. Reprod. Biol. Endocrinol. 2011;9:23. doi: 10.1186/1477-7827-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirnezami R., Nicholson J., Darzi A. Preparing for Precision Medicine. N. Engl. J. Med. 2012;366:489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 19.Huhtaniemi I., Hovatta O., La Marca A., Livera G., Monniaux D., Persani L., Heddar A., Jarzabek K., Laisk-Podar T., Salumets A., et al. Advances in the Molecular Pathophysiology, Genetics, and Treatment of Primary Ovarian Insufficiency. Trends Endocrinol. Metab. 2018;29:400–419. doi: 10.1016/j.tem.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Fortuño C., Labarta E. Genetics of primary ovarian insufficiency: A review. J. Assist. Reprod. Genet. 2014;31:1573–1585. doi: 10.1007/s10815-014-0342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao X., Ke H., Qin Y., Chen Z.-J. Molecular Genetics of Premature Ovarian Insufficiency. Trends Endocrinol. Metab. 2018;29:795–807. doi: 10.1016/j.tem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Patiño L.C., Beau I., Carlosama C., Buitrago J.C., González R., Suárez C.F., Patarroyo M.A., Delemer B., Young J., Binart N., et al. New mutations in non-syndromic primary ovarian insufficiency patients identified via whole-exome sequencing. Hum. Reprod. 2017;32:1512–1520. doi: 10.1093/humrep/dex089. [DOI] [PubMed] [Google Scholar]
- 23.Knauff E.A.H., Franke L., Van Es M.A., Van Den Berg L.H., Van Der Schouw Y.T., Laven J.S.E., Lambalk C.B., Hoek A., Goverde A.J., Christin-Maitre S., et al. Genome-wide association study in premature ovarian failure patients suggests ADAMTS19 as a possible candidate gene. Hum. Reprod. 2009;24:2372–2378. doi: 10.1093/humrep/dep197. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Wang B., Zhang W., Chen B., Luo M., Wang J., Wang X., Cao Y., Kee K. A homozygous NOBOX truncating variant causes defective transcriptional activation and leads to primary ovarian insufficiency. Hum. Reprod. 2017;32:248–255. doi: 10.1093/humrep/dew271. [DOI] [PubMed] [Google Scholar]
- 25.Pasipoularides A. The new era of whole-exome sequencing in congenital heart disease: Brand-new insights into rare pathogenic variants. J. Thorac. Dis. 2018;10:S1923–S1929. doi: 10.21037/jtd.2018.05.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B., Li L., Zhu Y., Zhang W., Wang X., Chen B., Li T., Pan H., Wang J., Kee K., et al. Sequence variants of KHDRBS1 as high penetrance susceptibility risks for primary ovarian insufficiency by mis-regulating mRNA alternative splicing. Hum. Reprod. 2017;32:2138–2146. doi: 10.1093/humrep/dex263. [DOI] [PubMed] [Google Scholar]
- 27.Tucker E.J., Jaillard S., Grover S.R., van den Bergen J., Robevska G., Bell K.M., Sadedin S., Hanna C., Dulon J., Touraine P., et al. TP63-truncating variants cause isolated premature ovarian insufficiency. Hum. Mutat. 2019;40:886–892. doi: 10.1002/humu.23744. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Vinckenbosch N., Tian G., Huerta-Sanchez E., Jiang T., Jiang H., Albrechtsen A., Andersen G., Cao H., Korneliussen T.S., et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat. Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 29.Jaillard S., Sreenivasan R., Beaumont M., Robevska G., Dubourg C., Knarston I.M., Akloul L., van den Bergen J., Odent S., Croft B., et al. Analysis of NR5A1 in 142 patients with premature ovarian insufficiency, diminished ovarian reserve, or unexplained infertility. Maturitas. 2020;131:78–86. doi: 10.1016/j.maturitas.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Liu H., Guo T., Gong Z., Yu Y., Zhang Y., Zhao S., Qin Y. Novel FSHR mutations in Han Chinese women with sporadic premature ovarian insufficiency. Mol. Cell. Endocrinol. 2019;492:110446. doi: 10.1016/j.mce.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q., Li D., Cai B., Chen Q., Li C., Wu Y., Jin L., Wang X., Zhang X., Zhang F. Whole-exome sequencing reveals SALL4 variants in premature ovarian insufficiency: An update on genotype-phenotype correlations. Hum. Genet. 2019;138:83–92. doi: 10.1007/s00439-018-1962-4. [DOI] [PubMed] [Google Scholar]
- 32.Yang X., Touraine P., Desai S., Humphreys G., Jiang H., Yatsenko A., Rajkovic A. Gene variants identified by whole-exome sequencing in 33 French women with premature ovarian insufficiency. J. Assist. Reprod. Genet. 2019;36:39–45. doi: 10.1007/s10815-018-1349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trakadis Y.J., Sardaar S., Chen A., Fulginiti V., Krishnan A. Machine learning in schizophrenia genomics, a case-control study using 5090 exomes. Am. J. Med. Genet. Neuropsychiatr. Genet. 2019;180:103–112. doi: 10.1002/ajmg.b.32638. [DOI] [PubMed] [Google Scholar]
- 34.Ho D.S.W., Schierding W., Wake M., Saffery R., O’Sullivan J. Machine Learning SNP Based Prediction for Precision Medicine. Front. Genet. 2019;10:267. doi: 10.3389/fgene.2019.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valdes G., Luna J., Eaton E., Ii C.B.S., Ungar L.H., Solberg T.D. MediBoost: A Patient Stratification Tool for Interpretable Decision Making in the Era of Precision Medicine. Sci. Rep. 2016;6:37854. doi: 10.1038/srep37854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fairley S., Lowy-Gallego E., Perry E., Flicek P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2020;48:D941–D947. doi: 10.1093/nar/gkz836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudmant P., Rausch T., Gardner E., Handsaker R., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H.-Y., et al. An integrated map of structural variation in 2504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilcoxon F. Probability Tables for Individual Comparisons by Ranking Methods. Biometrics. 1947;3:119–122. doi: 10.2307/3001946. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro S.S., Wilk M.B. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. doi: 10.1093/biomet/52.3-4.591. [DOI] [Google Scholar]
- 41.Fisher R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922;85:87–94. doi: 10.2307/2340521. [DOI] [Google Scholar]
- 42.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Depristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., Del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 49.The UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team . R: A Language and Environment for Statistical Computing. R Core Team; Vienna, Austria: 2017. [Google Scholar]
- 51.van Rossum G., Drake F.L. Python 3 Reference Manual. CreateSpace; Scotts Valley, CA, USA: 2009. [Google Scholar]
- 52.Kosub S. A note on the triangle inequality for the Jaccard distance. Pattern Recognit. Lett. 2019;120:36–38. doi: 10.1016/j.patrec.2018.12.007. [DOI] [Google Scholar]
- 53.Charrad M., Ghazzali N., Boiteau V., Niknafs A. NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J. Stat. Softw. 2014;61:1–36. doi: 10.18637/jss.v061.i06. [DOI] [Google Scholar]
- 54.Witten I.H., Frank E., Hall M.A., Pal C.J. Data Mining: Practical Machine Learning Tools and Techniques. Elsevier Inc.; Amsterdam, The Netherlands: 2016. [Google Scholar]
- 55.Murtagh F., Legendre P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014;31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 56.Chen X., Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99:323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han H., Guo X., Yu H. Variable selection using Mean Decrease Accuracy and Mean Decrease Gini based on Random Forest; Proceedings of the 2016 7th IEEE International Conference on Software Engineering and Service Science (ICSESS); Beijing, China. 26–28 August 2016; pp. 219–224. [Google Scholar]
- 58.Xu K., Chen X., Yang H., Xu Y., He Y., Wang C., Huang H., Liu B., Liu W., Li J., et al. Maternal Sall4 Is Indispensable for Epigenetic Maturation of Mouse Oocytes. J. Biol. Chem. 2017;292:1798–1807. doi: 10.1074/jbc.M116.767061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delcour C., Amazit L., Patino L.C., Magnin F., Fagart J., Delemer B., Young J., Laissue P., Binart N., Beau I. ATG7 and ATG9A loss-of-function variants trigger autophagy impairment and ovarian failure. Genet. Med. 2019;21:930–938. doi: 10.1038/s41436-018-0287-y. [DOI] [PubMed] [Google Scholar]
- 60.Patiño L.C., Beau I., Morel A., Delemer B., Young J., Binart N., Laissue P. Functional evidence implicating NOTCH2 missense mutations in primary ovarian insufficiency etiology. Hum. Mutat. 2019;40:25–30. doi: 10.1002/humu.23667. [DOI] [PubMed] [Google Scholar]
- 61.Orr H.A. Fitness and its role in evolutionary genetics. Nat. Rev. Genet. 2009;10:531–539. doi: 10.1038/nrg2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin Y., Sun M., You L., Wei D., Sun J., Liang X., Zhang B., Jiang H., Xu J., Chen Z.-J. ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet J. Rare Dis. 2012;7:5. doi: 10.1186/1750-1172-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin Y., Zhao H., Xu J., Shi Y., Li Z., Qiao J., Liu J., Ren C., Chen S., Cao Y., et al. Association of 8q22.3 locus in Chinese Han with idiopathic premature ovarian failure (POF) Hum. Mol. Genet. 2012;21:430–436. doi: 10.1093/hmg/ddr462. [DOI] [PubMed] [Google Scholar]
- 64.Perry J.R.B., Corre T., Esko T., Chasman D.I., Fischer K., Franceschini N., He C., Kutalik Z., Mangino M., Rose L.M., et al. A genome-wide association study of early menopause and the combined impact of identified variants. Hum. Mol. Genet. 2013;22:1465–1472. doi: 10.1093/hmg/dds551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L., Jia P., Wolfinger R.D., Chen X., Zhao Z. Gene set analysis of genome-wide association studies: Methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bramble M., Goldstein E.H., Lipson A., Ngun T., Eskin A., Gosschalk J.E., Roach L., Vashist N., Barseghyan H., Lee E., et al. A novel follicle-stimulating hormone receptor mutation causing primary ovarian failure: A fertility application of whole exome sequencing. Hum. Reprod. 2016;31:905–914. doi: 10.1093/humrep/dew025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Philibert P., Paris F., Lakhal B., Audran F., Gaspari L., Saâd A., Christin-Maitre S., Bouchard P., Sultan C. NR5A1 (SF-1) gene variants in a group of 26 young women with XX primary ovarian insufficiency. Fertil. Steril. 2013;99:484–489. doi: 10.1016/j.fertnstert.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 68.Nichols R.A., Balding D. Effects of population structure on DNA fingerprint analysis in forensic science. Heredity. 1991;66:297–302. doi: 10.1038/hdy.1991.37. [DOI] [PubMed] [Google Scholar]
- 69.Lipkin S.M., Moens P.B., Wang V., Lenzi M., Shanmugarajah D., Gilgeous A., Thomas J., Cheng J., Touchman J.W., Green E.D., et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 70.Kumar T.R., Wiseman A.L., Kala G., Kala S.V., Matzuk M.M., Lieberman M.W. Reproductive Defects in γ-Glutamyl Transpeptidase-Deficient Mice. Endocrinology. 2000;141:4270–4277. doi: 10.1210/endo.141.11.7760. [DOI] [PubMed] [Google Scholar]
- 71.Su W., Guan X., Zhang D., Sun M., Yang L., Yi F., Hao F., Feng X., Ma T. Occurrence of multi-oocyte follicles in aquaporin 8-deficient mice. Reprod. Biol. Endocrinol. 2013;11:88. doi: 10.1186/1477-7827-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xia H.H.-X., Yang Y., Lam S.K., Wong W.M., Leung S.Y., Yuen S.T., Elia G., Wright N.A., Wong B.C.-Y. Aberrant epithelial expression of trefoil family factor 2 and mucin 6 in Helicobacter pylori infected gastric antrum, incisura, and body and its association with antralisation. J. Clin. Pathol. 2004;57:861–866. doi: 10.1136/jcp.2003.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirabayashi K., Yasuda M., Kajiwara H., Itoh J., Miyazawa M., Hirasawa T., Muramatsu T., Murakami M., Mikami M., Osamura R.Y. Alterations in Mucin Expression in Ovarian Mucinous Tumors: Immunohistochemical Analysis of MUC2, MUC5AC, MUC6, and CD10 Expression. Acta Histochem. Cytochem. 2008;41:15–21. doi: 10.1267/ahc.08008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qin Y., Jiao X., Simpson J.L., Chen Z.-J. Genetics of primary ovarian insufficiency: New developments and opportunities. Hum. Reprod. Update. 2015;21:787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Y., Vlassis A., Roques C., LaLonde M., González-Aguilera C., Lambert J., Lee S., Zhao X., Alabert C., Johansen J.V., et al. BRPF 3-HBO 1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016;35:176–192. doi: 10.15252/embj.201591293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swiech L., Kisiel K., Czolowska R., Zientarski M., Borsuk E. Accumulation and dynamics of proteins of the MCM family during mouse oogenesis and the first embryonic cell cycle. Int. J. Dev. Biol. 2007;51:283–295. doi: 10.1387/ijdb.062239ls. [DOI] [PubMed] [Google Scholar]
- 77.Won M., Luo Y., Lee D.-H., Shin E., Suh D.-S., Kim T.-H., Jin H., Bae J. BAX is an essential key mediator of AP5M1-induced apoptosis in cervical carcinoma cells. Biochem. Biophys. Res. Commun. 2019;518:368–373. doi: 10.1016/j.bbrc.2019.08.065. [DOI] [PubMed] [Google Scholar]
- 78.Lussier J.G., Diouf M.N., Lévesque V., Sirois J., Ndiaye K. Gene expression profiling of upregulated mRNAs in granulosa cells of bovine ovulatory follicles following stimulation with hCG. Reprod. Biol. Endocrinol. 2017;15:88. doi: 10.1186/s12958-017-0306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Navani S. The Human Protein Atlas. [(accessed on 6 May 2020)]; Available online: https://www.proteinatlas.org/
- 80.Desai S., Wood-Trageser M., Matic J., Chipkin J., Jiang H., Bachelot A., Dulon J., Sala C., Barbieri C., Cocca M., et al. MCM8 and MCM9 Nucleotide Variants in Women with Primary Ovarian Insufficiency. J. Clin. Endocrinol. Metab. 2017;102:576–582. doi: 10.1210/jc.2016-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dondik Y., Lei Z., Gaskins J., Pagidas K. Minichromosome maintenance complex component 8 and 9 gene expression in the menstrual cycle and unexplained primary ovarian insufficiency. J. Assist. Reprod. Genet. 2019;36:57–64. doi: 10.1007/s10815-018-1325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee K.Y., Im J.-S., Shibata E., Park J., Handa N., Kowalczykowski S.C., Dutta A. MCM8-9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex. Nat. Commun. 2015;6:7744. doi: 10.1038/ncomms8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X., Abreu J.G., Yokota C., MacDonald B.T., Singh S., Coburn K.L.A., Cheong S.-M., Zhang M.M., Ye Q.-Z., Hang H.C., et al. Tiki1 Is Required for Head Formation via Wnt Cleavage-Oxidation and Inactivation. Cell. 2012;149:1565–1577. doi: 10.1016/j.cell.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng Y., Kawamura K., Takae S., Deguchi M., Yang Q., Kuo C., Hsueh A.J.W. Oocyte-derived R-spondin2 promotes ovarian follicle development. FASEB J. 2013;27:2175–2184. doi: 10.1096/fj.12-223412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abedini A., Zamberlam G., Lapointe E., Tourigny C., Boyer A., Paquet M., Hayashi K., Honda H., Kikuchi A., Price C., et al. WNT5a is required for normal ovarian follicle development and antagonizes gonadotropin responsiveness in granulosa cells by suppressing canonical WNT signaling. FASEB J. 2016;30:1534–1547. doi: 10.1096/fj.15-280313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chawengsaksophak K., Svingen T., Ng E.T., Epp T., Spiller C., Clark C., Cooper H., Koopman P. Loss of Wnt5a Disrupts Primordial Germ Cell Migration and Male Sexual Development in Mice1. Biol. Reprod. 2012;86:1–12. doi: 10.1095/biolreprod.111.095232. [DOI] [PubMed] [Google Scholar]
- 87.Ibañez-Tallon I., Gorokhova S., Heintz N. Loss of function of axonemal dynein Mdnah5 causes primary ciliary dyskinesia and hydrocephalus. Hum. Mol. Genet. 2002;11:715–721. doi: 10.1093/hmg/11.6.715. [DOI] [PubMed] [Google Scholar]
- 88.Hu J., Lessard C., Longstaff C., O’Brien M., Palmer K., Reinholdt L., Eppig J., Schimenti J., Handel M.A. ENU-induced mutant allele of Dnah1, ferf1, causes abnormal sperm behavior and fertilization failure in mice. Mol. Reprod. Dev. 2019;86:416–425. doi: 10.1002/mrd.23120. [DOI] [PubMed] [Google Scholar]
- 89.Areal L., Pereira L.P., Ribeiro F., Olmo I.G., Muniz M.R., do Carmo Rodrigues M., Costa P.F., Martins-Silva C., Ferguson S.S.G., Guimarães D.A.M., et al. Role of Dynein Axonemal Heavy Chain 6 Gene Expression as a Possible Biomarker for Huntington’s Disease: A Translational Study. J. Mol. Neurosci. 2017;63:342–348. doi: 10.1007/s12031-017-0984-z. [DOI] [PubMed] [Google Scholar]
- 90.Li Y., Yagi H., Onuoha E.O., Damerla R.R., Francis R., Furutani Y., Tariq M., King S.M., Hendricks G., Cui C., et al. DNAH6 and Its Interactions with PCD Genes in Heterotaxy and Primary Ciliary Dyskinesia. PLoS Genet. 2016;12:e1005821. doi: 10.1371/journal.pgen.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.