Abstract
Background
Pseudomonas aeruginosa is of great concern among MDR bacteria and rapid and reliable in vitro antibiotic susceptibility testing methods are extremely necessary. Colistin is, in many cases, among the limited useful alternatives for these isolates. Unfortunately, only a few reliable in vitro methods are validated for testing susceptibility to colistin. Although EUCAST and CLSI recommend broth microdilution (BMD) as the standard method for antibiotic susceptibility testing, this method is not routinely performed in microbiology laboratories. However, some commercial products based upon BMD have tested well and offer consistent results.
Objectives
To evaluate the performance of the colorimetric Rapid Polymyxin Pseudomonas Test (RPPT) (ELITech Microbiology, France).
Methods
Eighty-seven clinical P. aeruginosa strains, prospectively collected in two microbiology laboratories exhibiting either susceptibility or various degrees of multidrug resistance, including to colistin, were used. Different susceptibility testing methods were simultaneously performed and compared with reference BMD and interpreted using 2020 EUCAST criteria.
Results
Results indicate an essential agreement (EA) of 97.7% for RPPT while the other tests did not reach 90% of EA [66.7% MicroScan, 63.2% Etest (bioMérieux, France) and 60.9% other MIC Test Strips (MTS, Liofilchem, Italy)]. The categorical agreement was 98.9% for RPPT, 87.4% for MTS, 85.1% for Etest and 64.4% for MicroScan.
Conclusions
The RPPT was able to accurately detect both colistin-susceptible and -resistant isolates within 4 h, offering a rapid alternative for a prompt decision about the inclusion of this antibiotic in a patient’s treatment.
Introduction
MDR microorganisms have increased in recent years, mainly among clinically important Gram-negative bacilli, as a result of which few therapeutic alternatives are left. Because of this, polymyxins, and more specifically colistin, regained their place as an option for treatment use.1
In vitro susceptibility testing of colistin is problematic due to certain chemical characteristics of the antibiotic, mainly its net cationic nature and its high molecular weight hindering its agar diffusion. Moreover, heteroresistance to colistin is a frequent behaviour observed in many bacterial populations.2 Taking all this into account, in vitro results and the resulting susceptibility status have a strong impact in clinical and stewardship decisions, impacting colistin’s clinical use. The reference method, according to EUCAST and CLSI is broth microdilution (BMD) following the ISO standard 20776-1. This method is laborious for daily laboratory practice and results are read after 18–20 h, thus reinforcing the need for other reliable and, if possible, rapid methods.
A comparative study was recently published in order to evaluate seven commercially available products for colistin antimicrobial susceptibility testing (AST).3 In that study, five commercial BMD products and two gradient strip tests were compared with the reference method. The group concluded that commercial broth microdilution methods generally performed well, however, the performance of the two gradient tests was unacceptable. They advised laboratories not to trust colistin gradient tests or disc diffusion and recommended the use of broth microdilution-based methods.3 After this work, a warning from EUCAST has been issued about commercial methods to test colistin.4
The Rapid Polymyxin™ Pseudomonas test (RPPT) (ELITech Microbiology, France) is a liquid colorimetric method with freeze-dried colistin (2, 4 and 8 mg/L concentration in each well) that also contains a glucose culture medium and bromocresol purple as a pH indicator. If the isolate is able to grow in a defined colistin concentration, a colour change (green to purple) is evidenced. Results can be read less than 4 h after inoculation. A positive and a negative control are included.5 The objective of this study was to evaluate the performance of the colorimetric RPPT to confirm the colistin susceptibility status in a prospectively collected Pseudomonas aeruginosa isolates exhibiting either susceptibility or various degrees of multidrug resistance, including to colistin, in two clinical microbiology laboratories in comparison with different AST methods.6
Materials and methods
AST was performed on 75 prospectively collected P. aeruginosa isolates that were recovered in routine daily work at the Microbiology Department, Ramón y Cajal University Hospital in Madrid (Spain) in the last quarter of 2018 and 12 isolates recovered in a contemporary period at the Microbiology Department of the Son Espases University Hospital in Palma de Mallorca (Spain). A total of 93.1% (n = 81) isolates came from clinical samples (respiratory samples, 33; urine, 23; organic fluid, 8; abscess, 7; blood, 5; exudate, 2; and prosthesis, wound and biopsy, 1 each) and 6.9% (n = 6) from rectal colonization. Among the group of 75 isolates, 54 exhibited various degrees of resistance: 43 MDR, 8 XDR, and 3 pandrug resistant (PDR) according to the results obtained with the routinely used automated AST system MicroScan WalkAway plus (Beckman, West Sacramento, CA, USA), using the Negative Combo Panel 58 (NC58) for non-fermenting microorganisms. The 33 non-MDR isolates were included for comparison purposes when the analysis of the results was performed.
The identification of isolates was confirmed by MALDI-TOF MS using the Microflex LT/SH Smart System with the MALDI Biotyper software (Bruker Daltonics, Bremen, Germany). Colistin reference MICs were determined by BMD in accordance with the ISO standard 20776-1:2019 recommendations7 on 96 U-well polystyrene Microtiter™ plates (Thermo Fisher Scientific, Cleveland, OH, USA) with 2-fold dilutions ranging from 128 to 0.125 mg/L, using colistin sulphate (Sigma–Aldrich, St Louis, MI, USA) and BBL™ cation-adjusted Mueller–Hinton II broth (Becton Dickinson, Sparks, MD, USA) freshly prepared. The Rapid Polymyxin Pseudomonas test was performed in all isolates according to the instructions of the manufacturer. Briefly, this method consists in prepare a standardized suspension of each isolate in a specific medium; 100 µL of this suspension is carefully placed in the wells of the kit, which contain known amounts of colistin, to obtain final concentrations of 0 (growth control), 2, 4 and 8 mg/L. The negative control well is inoculated with a suspension of NaCl only. The reading of results is done after 3 or 4 h of incubation at 37 °C. A valid result is obtained when a colour shift is observed in the growth control well, and no colour shift in the negative control well. The result is interpreted as an MIC for colistin. Two commercial MIC gradient tests, Etest® (bioMérieux, Marcy-l’Étoile, France) and Liofilchem® MIC Test Strips (MTS) (Liofilchem, Roseto degli Abruzzi, Italy) were included (MIC values that fell between standard 2-fold dilutions were rounded up to the next upper 2-fold value before categorization).
The interpretation of the MIC values was performed according to EUCAST version 10 [susceptible ≤2 mg/L; resistant >2 mg/L; area of technical uncertainty (ATU) 4 mg/L].8 All tests were conducted in parallel using the same inoculum. P. aeruginosa ATCC 27853, Escherichia coli ATCC 25922 and the colistin resistant E. coli NCTC 13846 were included as quality controls in all tests. Essential agreement (EA, MICs within ± 1 dilution of reference MICs) and categorical agreement (CA, number of tests with correct susceptibility categorization) as well as AST errors [major (ME) and very major (VME) errors] were calculated. Error levels were expressed as percentages.9 Minimal EA between techniques was established in 90%.10 Isolates with VMEs were retested by broth microdilution to ensure an accurate and reproducible reference MIC result. Due to the diverse antibiotic concentrations included in the commercial tests used for comparative purposes, it was not possible to determine the exact differences in MICs between the reference method and such tests when MIC values were in the lower or upper limit of the studied concentration range and, consequently, were included in the EA. Cohen’s κ coefficient for agreement between the reference and the comparative methods was calculated and values were interpreted according to the Landis and Koch classification.11
Results
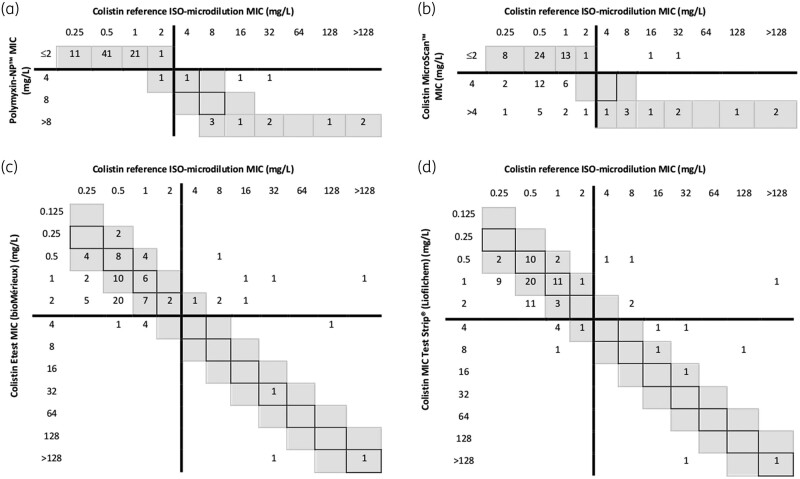
Colistin reference BMD MICs for the 87 P. aeruginosa isolates ranged from 0.25 to >128 mg/L. Colistin resistance was observed in 12 isolates: 2 MDR, 1 XDR and 1 PDR (all from respiratory samples) and 8 non-MDR (3 from urine, 4 from respiratory samples and 1 from organic fluid). In only one non-MDR isolate, the colistin BMD MIC value corresponded to the ATU category. The correlation of resistance MIC values obtained with the reference BMD was good for the RPPT but poor for automated microdilution and MIC gradient tests results (Figure 1).
Figure 1.
Correlation between the reference broth microdilution method results and (a) Rapid Polymyxin Pseudomonas test, (b) MicroScan WalkAway plus using NC58, (c) colistin Etest and (d) colistin MIC Test Strip for 87 P. aeruginosa isolates. MICs within essential agreement (within ± 1 dilution of reference MICs) are highlighted in grey and MICs identical to reference MICs are within boxes. EUCAST breakpoints (susceptible ≤2, resistant >2 mg/L) are shown as lines.
Overall, an EA of 97.7% was obtained for RPPT while the other tests did not reach the minimum 90% of EA required (66.7% for MicroScan, 63.2% for Etest and 60.9% for MTS).9 The CA was 98.9% for RPPT, 87.4% for MTS, 85.1% for Etest and 64.4% for MicroScan. Considering error classes, a 38.7% rate of MEs (false resistant results) was observed when the isolates were tested with MicroScan, 8.0% with MTS, 6.7% with Etest and 1.3% with RPPT. A 66.7% rate of VMEs (false susceptible results) was observed when the isolates were tested by Etest, 41.7% when using MTS and 16.7% with MicroScan. The acceptable inter-method error percentage of VMEs and MEs is ≤1.5% and ≤3%, respectively.12 The κ coefficient for the different methods when compared with the reference BMD was: 0.953 (almost perfect) for RPPT, 0.486 (moderate) for MTS, 0.230 (fair) for Etest and 0.298 (fair) for MicroScan.
When considering different levels of colistin resistance, i.e. non-MDR, MDR, XDR and PDR, an EA of 93.9% was obtained for RPPT with the non-MDR isolates, conversely, the worst results for this group (lowest CA) were obtained with MicroScan (24.2%), which had a tendency to overestimate MIC values (Table 1). For MDR isolates, EA was 100% for RPPT, 90.7% for MicroScan, 72.1% for MTS and 53.5% for Etest; CA was 97.7% for RPPT, 97.7% for Etest and 90.7% for both MicroScan and MTS while MEs were found with MicroScan (9.8%), MTS (7.3%) and both RPPT and Etest (2.4%); VMEs were observed only with the two gradient tests (50%). In XDR isolates, EA was 100% for RPPT and MicroScan and 50% for Etest and MTS. CA was 100% for RPPT and MicroScan and 87.5% for Etest and MTS. MEs were only found with gradient tests (14.3%). No VMEs were found in this category. Finally, in PDR isolates, EA was 100% for RPPT and 33.3% for MicroScan, Etest and MTS. CA was 100% for RPPT, Etest and MTS and 33.3% for MicroScan. A 100% rate of MEs was detected with MicroScan. No VMEs were found in this category.
Table 1.
Performance of four commercial methods when compared with the reference BMD method
| MDR isolates (N = 43) | XDR isolates (N = 8) | PDR isolates (N = 3) | Non-MDR isolates (N = 33) | All isolates (N = 87) | |
|---|---|---|---|---|---|
| (CST R, n = 2) | (CST R, n = 1) | (CST R, n = 1) | (CST R, n = 8) | (CST R, n = 12) | |
| Essential agreement, n (%) | |||||
| RPPT | 43 (100) | 8 (100) | 3 (100) | 31 (93.9) | 85 (97.7) |
| MicroScan | 39 (90.7) | 8 (100) | 1 (33.3) | 8 (24.2) | 58 (66.7) |
| Etest | 23 (53.5) | 4 (50) | 1 (33.3) | 18 (54.5) | 55 (63.2) |
| MTS | 31 (72.1) | 4 (50) | 1 (33.3) | 17 (51.5) | 53 (60.9) |
| Categorical agreement, n (%) | |||||
| RPPT | 42 (97.7) | 8 (100) | 3 (100) | 33 (100) | 86 (98.9) |
| MicroScan | 39 (90.7) | 8 (100) | 1 (33.3) | 8 (24.2) | 56 (64.4) |
| Etest | 42 (97.7) | 7 (87.5) | 3 (100) | 23 (69.7) | 74 (85.1) |
| MTS | 39 (90.7) | 7 (87.5) | 3 (100) | 27 (81.8) | 76 (87.4) |
| Major errors, n (%) | |||||
| RPPT | 1 (2.4) | 0 (0) | 0 (0) | 0 (0) | 1 (1.3) |
| MicroScan | 4 (9.8) | 0 (0) | 2 (100) | 23 (92) | 29 (38.7) |
| Etest | 1 (2.4) | 1 (14.3) | 0 (0) | 3 (12) | 5 (6.7) |
| MTS | 3 (7.3) | 1 (14.3) | 0 (0) | 2 (8) | 6 (8) |
| Very major errors, n (%) | |||||
| RPPT | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| MicroScan | 0 (0) | 0 (0) | 0 (0) | 2 (25) | 2 (16.7) |
| Etest | 1 (50) | 0 (0) | 0 (0) | 7 (87.5) | 8 (66.7) |
| MTS | 1 (50) | 0 (0) | 0 (0) | 4 (50) | 5 (41.7) |
Total P. aeruginosa isolates (n = 87) are split by resistance categories according to the percentages of essential agreement and categorical agreement, and the number of major errors and very major errors.
CST R, colistin-resistant isolates.
The quality control strain results were within expected ranges for all test and methods except for E. coli ATCC 25922 with Etest, for which MICs were below the range in all studies (data not shown).
Discussion
Clinical microbiology laboratories require the implementation of a reliable and, if possible, rapid test to determine colistin susceptibility and to detect colistin-resistant isolates. At present, very few tests are validated that effectively detect colistin resistance apart from the standard ISO BMD, which cannot be performed routinely because it is a laborious and time-consuming method.
The RPPT method has previously been validated in other studies, although these were performed with collections of P. aeruginosa5,13 or with colistin-resistant P. aeruginosa generated through in vitro experiments,14 while our studied isolates had been prospectively collected from clinical samples reflecting the epidemiological overview of our institution.
According to our results, the RPPT performance for detecting the colistin-susceptibility status of P. aeruginosa isolates is promising. This test appears rapid and reliable for laboratory routine testing, being a particularly useful tool in the case of MDR, XDR and PDR isolates for which colistin may be one of the few therapeutic options left.
In daily clinical practice, we determine colistin MICs using MicroScan panels, however, it has been observed that, although this system shows excellent results with Enterobacterales, non-susceptible results for non-fermenters require confirmation by another validated method due to the high rate of false resistance observed.15 The same has been concluded for current MIC gradient strips due the inaccuracy of the results.2
Our study has some limitations. The small number of colistin-resistant P. aeruginosa included in this study can limit definite conclusions about the true performance of the RPPT. In addition, most issues with colistin testing were found among the non-MDR isolates but this does not constitute a relevant problem as the antibiotic is not used for infections caused by these types of isolates. At present, colistin resistance is low in our hospital, even among MDR P. aeruginosa isolates. Nevertheless, the consistency of the results obtained with RPPT when compared with the reference method is strongly promising, thus constituting a reliable alternative method for routine work. It is also important to remark that results are obtained in less than 4 h, which is also an advantage when compared with the standard BMD method that needs an overnight wait for the results.
Funding
This study was supported by internal funding. Vircell S.L. Spain kindly provided the ELITech Microbiology kits of Rapid Polymyxin Pseudomonas test. C.L.-C. and R.C. are partially supported by Plan Nacional de I + D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0004 and RD16/0016/0011) co-financed by European Development Regional Fund ‘A way to achieve Europe’ (ERDF), Operative Program Intelligent Growth 2014–2020. M.D.-A. was partially supported by the Innovative Medicines Initiative (IMI), European Commission-funded project [iABC grant 115721-2] and Fundación Francisco Soria Melguizo (Madrid, Spain).
Transparency declarations
None to declare.
References
- 1. Vaara M. Polymyxins and their potential next generation as therapeutic antibiotics. Front Microbiol 2019; 10: 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin J, Xu C, Fang R. et al. Resistance and heteroresistance to colistin in Pseudomonas aeruginosa isolates from Wenzhou, China. Antimicrob Agents Chemother 2019; 63: e00556-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Matuschek E, Åhman J, Webster C. et al. Antimicrobial susceptibility testing of colistin - evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect 2018; 24: 865–70. [DOI] [PubMed] [Google Scholar]
- 4. EUCAST. Antimicrobial Susceptibility Testing of Colistin - Problems Detected with Several Commercially Available Products. 2016. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Warnings/Warnings_docs/Warning_-_colistin_AST.pdf.
- 5. Lescat M, Poirel L, Jayol A. et al. Performances of the Rapid Polymyxin Acinetobacter and Pseudomonas tests for colistin susceptibility testing. Microb Drug Resist 2019; 25: 520–3. [DOI] [PubMed] [Google Scholar]
- 6. Magiorakos A-P, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 7. International Organization for Standardization (ISO). ISO 20776-1:2006. Clinical Laboratory Testing and In Vitro Diagnostic Test Systems. Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices. Part 1: Reference Method for Testing the In Vitro Activity of Antimicrobial Agents Against Rapidly Growing Aerobic Bacteria Involved in Infectious Disease. ISO, 2006.
- 8. EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, 2020, Version 10.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf.
- 9. Elder BL, Hansen SA, Kellogg JA, McCurdy BW, et al. ed. Cumitech 31a, Verification and Validation of Procedures in the Clinical Microbiology Laboratory. American Society for Microbiology, 1997. [Google Scholar]
- 10. Humphries RM, Ambler J, Mitchell SL. et al. CLSI Methods Development and Standardization Working Group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol 2018; 56: e01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Landis JR, Koch GG.. The measurement of observer agreement for categorical data. Biometrics 1977; 33: 159–74. [PubMed] [Google Scholar]
- 12. ISO. ISO 20776-2:2007. Clinical Laboratory Testing and In Vitro Diagnostic Test Systems. Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices. Part 2: Evaluation of Performance of Antimicrobial Susceptibility Test Devices. ISO, 2007.
- 13. Sadek M, Tinguely C, Poirel L. et al. Rapid polymyxin/pseudomonas NP test for rapid detection of polymyxin susceptibility/resistance in Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 2020; 39: 1657–62. [DOI] [PubMed] [Google Scholar]
- 14. Javed M, Ueltzhoeffer V, Heinrich M. et al. Colistin susceptibility test evaluation of multiple-resistance-level Pseudomonas aeruginosa isolates generated in a morbidostat device. J Antimicrob Chemother 2018; 73: 3368–74. [DOI] [PubMed] [Google Scholar]
- 15. Jayol A, Nordmann P, André C. et al. Evaluation of three broth microdilution systems to determine colistin susceptibility of Gram-negative bacilli. J Antimicrob Chemother 2018; 73: 1272–8. [DOI] [PubMed] [Google Scholar]