Abstract
A relationship between bitter and fat taste sensitivity, CD36 rs1761667 and TAS2R38 has been demonstrated. However, research is scarce and does not take diet into account. This study aimed to explore associations between genetics, fat and bitter taste sensitivity and dietary fat intake in healthy UK adults. A cross-sectional study was carried out on 88 Caucasian participants (49 females and 39 males aged 35 ± 1 years; body mass index 24.9 ± 0.5 kg/m2). Bitter taste sensitivity was assessed using phenylthiocarbamide (PTC) impregnated strips and the general Labeled Magnitude Scale. Fat taste sensitivity was assessed by the Ascending Forced Choice Triangle Procedure and dietary intake with a semi-quantitative food frequency questionnaire. Genotyping for rs713598, rs1726866, rs10246939, and rs1761667 was performed. Participants with TAS2R38 PAV/PAV diplotype perceived PTC strips as more bitter than groups carrying AVI haplotypes (AVI/AVI, P = 1 × 10−6; AVI/AAV, P = 0.029). CD36 rs1761667 was associated with fat taste sensitivity (P = 0.008). A negative correlation between bitter taste sensitivity and saturated fat intake was observed (rs = −0.256, P = 0.016). When combining the CD36 genotypes and TAS2R38 diplotypes into one variable, participants carrying both TAS2R38 AVI haplotype and CD36 A allele had a higher intake of saturated fat compared to carriers of CD36 GG genotype or TAS2R38 PAV/PAV and PAV/AAV diplotypes (13.8 ± 0.3 vs. 12.6 ± 0.5%TEI, P = 0.047) warranting further exploration in a larger cohort.
Keywords: diet, rs1761667, CD36, TAS2R38, taste perception
Introduction
Taste sensitivity is an important factor in dietary habit development (Karmous et al. 2018). The 5 defined human tastes are sweet, sour, bitter, salty, and umami (Ikeda 1909), with a potential sixth taste, fat taste (“oleogustus”) recognized recently (Mattes 2010). The consumption of large amounts of dietary fat constitutes an unhealthy dietary pattern (World Health Organisation [WHO] 2020). Differing taste sensitivity thresholds, which can impact dietary fat consumption, may influence this unhealthy dietary pattern (Duffy and Bartoshuk 2000; Graham et al. 2021). Research has identified genetic predisposition to all 6 tastes (Melis et al. 2020), although these have scarcely been studied together.
A wealth of research has reported a clear disparity in the ability to detect bitter compounds such as phenylthiocarbamide (PTC) and 6-n-propylthiouracil (PROP); a disparity which may be affected by genetics. More specifically, bitter taste sensitivity follows a bimodal distribution, with distinct phenotypes being either non-taster or taster.
To date, various candidate genes have been associated with PROP taste sensitivity, such as the taste receptors from the taste receptor 2 family and the gustin gene, carbonic anhydrase VI, (CA6) (Melis et al. 2013; Roura et al. 2015). The bitter taste receptor 2 member 38 (TAS2R38) is the most researched receptor to date regarding PROP or PTC taste sensitivity. It contains 3 coding single-nucleotide polymorphisms (SNPs): rs713598 (Pro49Ala), rs1726866 (Ala262Val), and rs10246939 (Val296Ile). These may explain more than 70% of bimodal distribution in PTC taste sensitivity (Kim et al. 2003; Risso et al. 2016). They also create common taster Pro-Ala-Val (PAV) and non-taster Ala-Val-Ile (AVI) haplotypes, observed in over 90% of the Caucasian population (Kim et al. 2005). In addition, rare haplotypes such as Ala-Ala-Val (AAV), Ala-Ala-Ile (AAI), Pro-Ala-Ile (PAI), and Pro-Val-Ile (PVI) have been identified and may be associated with intermediate sensitivities to PTC and PROP (Tepper et al. 2008; Risso et al. 2016).
Research on genetic determinants of PROP/PTC taste sensitivity has mostly been conducted in Caucasian populations (North Americans or Europeans) that are more likely to be carriers of the non-taster TAS2R38 AVI haplotype compared to African or Asian populations (Risso et al. 2016). Consequently, Caucasians have also been identified as having lower PROP taste sensitivity than the 2 above-mentioned populations (Williams et al. 2016; Yang et al. 2020).
Regarding dietary intake, lower bitter taste sensitivity has been associated with higher acceptance and intake of foods with a bitter taste (brassica vegetables, spinach, coffee) (Akella et al. 1997; Drewnowski et al. 1998, 1999), as well as a higher preference for sweet and fatty tasting foods (Duffy and Bartoshuk 2000), however, these findings are not consistent across studies (Timpson et al. 2005; O’Brien et al. 2013). Nevertheless, the association between bitter taste sensitivity and intake of foods other than those containing bitter tasting compounds, suggests an interaction with other taste modalities. Considering a larger proportion of bitter non-taster genotypes and phenotypes in Caucasians and the fact these may be associated with diets high in sugar and fat, further research is warranted in this population.
In addition to the above, genetic variants in fat taste sensitivity (FTS) have been reported. There have been 2 candidate genes of focus within human research; the cluster of difference 36 (CD36) and G-protein coupled receptor 120 (GPR120) (Daoudi et al. 2015; Costanzo et al. 2019). There is significant evidence of a link between variants within CD36 and FTS, specifically the rs1761667 (A/G) SNP. This has been associated with FTS (Pepino et al. 2012; Daoudi et al. 2015; Sayed et al. 2015) and dietary fat intake (Pepino et al. 2012; Pioltine et al. 2016; Ramos-Lopez et al. 2016; Fujii et al. 2019). The CD36 receptor, a membrane protein belonging to the class B scavenger receptor family located in taste bud cells, has been shown to bind to varying concentrations of saturated and unsaturated long-chain fatty acids (LCFA) (Besnard et al. 2016). To date, it is the only defined fat receptor with a high affinity to LCFA (Khan et al. 2020). Individuals with the A-allele have demonstrated reduced protein levels (Ghosh et al. 2011; Love-Gregory and Abumrad 2011), and therefore have a higher fat detection threshold (hyposensitive) and consequently cannot taste fat as successfully (Melis et al. 2015; Sayed et al. 2015). These individuals are likely to consume higher quantities of foods containing fatty acids, potentially leading to weight gain (Besnard et al. 2016), although there is paucity in research and what is available is largely heterogeneous (Tucker et al. 2017).
A relationship between bitter and fat taste may be apparent. Prior to the discovery of fat taste and associated receptors, Tepper and Nurse (1997) described PROP tasters to have a greater ability for oral texture perception through a greater density of trigeminal fibers, thus, a better ability to detect fat. Since this, a relationship between PROP tasters and preference for fat has been demonstrated (Tepper and Nurse 2006; Hayes and Duffy 2007). More recently, and in light of this, the CD36 rs1761667 SNP has been investigated together with PROP taster status and TAS2R38 haplotypes (Sollai et al. 2019). Although results are consistent regarding the association between fat and bitter taste by both CD36 rs1761667 and bitter taste TAS2R38 haplotypes, research is scarce and is yet to be undertaken in a healthy UK cohort comprehensively assessing whether genetic disparities impact dietary intake, alongside taste sensitivity. Therefore, the aim of the current study was to explore the associations between genetics, fat and bitter taste sensitivity and dietary fat intake in healthy UK adults.
Materials and methods
Study design and participants
The participants were healthy Caucasian adults aged 18–65 years and living in the United Kingdom. Participants were recruited via word of mouth and internet postings. Exclusion criteria were pregnancy, breastfeeding, chronic medical conditions, food allergies, smoking, lactose intolerance, and intake of any medication that may affect taste perception.
At baseline visit, anthropometric measurements including weight (kg), height (m), and waist circumference (cm) were recorded by the research team. Participants provided a 2 mL saliva sample for genotyping and took part in bitter and FTS tests. Participants were asked to refrain from consumption of any food or drink for 1 h prior to testing. All participants provided demographic information and completed a food frequency questionnaire (FFQ) administered online (Google Forms).
All procedures involving human participants were approved by the St Mary’s and Oxford Brookes University Ethics Committees. Written informed consent was obtained from each participant before the baseline data collection, stating they can withdraw from the study at any point. This study is registered as Genetics of Bitter and Fat Taste at ClinicalTrials.gov NCT04038281.
Demographic information
Self-reported demographic data (age, sex, ethnicity, income, occupation, and education level) were collected using an online questionnaire (Google Forms).
Anthropometric measurements
Height (m) (Free Standing Height Measure, SECA GmbH & Co.) and weight (kg) (Portable Scale MS-4203, Marsden Weighing Group) were recorded by the research team to the second decimal place. Body mass index (BMI) was calculated using the equation: weight (kg)/height (m2) (World Health Organization 2018).
Bitter taste sensitivity
The participants rated the intensity of PTC impregnated strip (EISCO labs, Product FSC1031) using the general Labeled Magnitude Scale (gLMS). The gLMS weighted scale labels were: “no sensation” (0), “barely detectable” (1.4), “weak” (6), “moderate” (17), “strong” (35), “very strong” (53), and “the strongest imaginable sensation of any kind” (100) (Roura et al. 2015). Before rating the intensity of the PTC strip, participants were instructed to remember the strongest sensation of any kind they had experienced or the strongest sensation they could imagine happening to them. They were explained these would be deemed as the strongest sensations of any kind on the gLMS scale (Hayes et al. 2013). This was used to guide participants when rating the PTC intensity.
Fat taste sensitivity
The Oral Fatty Acid Threshold Assessment and Ascending Forced Choice Triangle Procedure was carried out to determine each participant’s oleic acid (C18:1) detection threshold (FTS). The method used, and standard operating procedure followed, is described in full in Haryono et al. (2014). Briefly, each participant was presented with 3 cups (30 mL UTH-milk based vehicles) in a random order, 2 controls (oleic-) and 1 containing oleic acid (oleic+; 0.02, 0.06, 1, 1.4, 2, 2.8, 3.8, 5, 6.4, 8, 9.8, 12, 20 mM). A participant was required to select the oleic+ solution correctly 3 times at the same concentration to define their threshold. If they were incorrect at any point, a further 3 cups were presented, one containing the higher oleic+ concentration and 2 oleic- solutions. Participants were categorized by their FTS result: hypersensitive tasters have a FTS below 3.8 mM, hyposensitive tasters have a FTS above or equal to 3.8 mM and participants who fail to identify the oleic+ sample at the maximum concentration (20 mM) are defined as non-tasters (excluded from analysis) (Stewart et al. 2011; Haryono et al. 2014).
Testing was conducted on one occasion for each participant. Samples were served at room temperature and presented to participants in individual sections within either the St Mary’s University Nutrition laboratory or Oxford Brookes University sensory laboratory. Red lighting was used to mask visual differences between the samples, nose clips were worn to inhibit olfactory input, textural differences were avoided with the addition of textural agents (gum Arabic and liquid paraffin), and post-ingestive regulation was followed by the sip-and-spit procedure.
Dietary intake
Habitual dietary intake was assessed with a validated semi-quantitative FFQ (EPIC Norflok). The questionnaires were analyzed using the open source, cross-platform tool FFQ EPIC tool for analysis (FETA) (Mulligan et al. 2014) and information on energy and dietary macronutrient intake obtained. More specifically, total carbohydrate, total fat, monounsaturated fatty acid (MUFA), polyunsaturated fatty acid (PUFA), saturated fatty acid (SFA), and total protein were quantified. Intakes of macronutrients were converted into percentage of total energy intake (%TEI) for analyses.
Single-nucleotide polymorphism genotyping
From each participant, a 2 mL saliva sample was collected (SalivaGene Collection Module II; Stratec Molecular GmbH). A stabilizer provided by the manufacturer was added to the saliva sample which was then kept at −20 °C until DNA was isolated. Genomic DNA was isolated using a PSP Saliva-Gene 17 DNA Kit 1011 (Stratec Molecular GmbH) in agreement with the manufacturer procedures. Quality and quantity of the DNA were measured using spectroscopy (Nanodrop, Thermo Fisher). Genotyping was then performed using predesigned TaqMan SNP genotyping assays for the SNPs: rs1761667, rs713598, rs1726866, and rs10246939 and the StepOnePlus thermocycler (Applied Biosystems) with 2 technical replicates for each sample. The PCR amplification was then completed under the conditions stated by the manufacturer. TAS2R38 haplotypes, defined by rs713598, rs1726866, and rs10246939, were determined using Haploview software (Barrett et al. 2005).
Statistical analyses
Hardy Weinberg equilibrium was assessed for all SNPs using Chi-square goodness of fit test. Continuous variables are presented as mean ± standard error of the mean (SEM) or median (interquartile range) and were tested for normality with Shapiro-Wilk test. Categorical variables are presented as absolute (relative) frequencies. Differences in anthropometry, genotype frequencies, bitter and fat taste sensitivity, and dietary intake between males and females were tested with an independent samples t-test (with Levene’s test for equality of variance), Mann–Whitney U or Fisher’s Exact test, where appropriate. Individuals who failed to identify the oleic+ solution at 20 mM were defined as non-tasters, therefore have no measurable threshold and were excluded from further analyses on FTS and measurements of dietary intake by CD36 genotypes, in line with others (Burgess et al. 2018).
Spearman’s correlation was used to explore the associations between bitter and fat taste sensitivity as continuous variables. Kruskal–Wallis H tests were used to test the difference in bitter taste sensitivity between TAS2R38 diplotype groups and CD36 genotypes. Bonferroni adjustment was considered for pairwise comparisons. Mann–Whitney U test was used to analyze the differences in bitter taste sensitivity according to the TAS2R38 rs713598, rs1726866, and rs10246939. Genotypes were dichotomized into carriers of non-taster (Ala, Val, Ile) and homozygous taster alleles (Pro, Ala, Val). Chi-square or Fisher’s Exact test, where appropriate, were used to assess the associations between CD36 genotypes (AA, AG and GG, and AA/AG and GG), TAS2R38 diplotypes and FTS categories, and to assess the associations between CD36 genotypes (AA, AG and GG, and AA/AG and GG) and TAS2R38 diplotypes. Kruskal–Wallis H test was used to explore the difference in fat taste threshold (mM) between TAS2R38 diplotypes and CD36 genotypes with Bonferroni adjustment for multiple comparisons.
Spearman’s correlation was used to assess the associations between dietary fat intake (total, MUFA, PUFA, and SFA) and bitter taste sensitivity, and FTS. One-way analysis of variance (ANOVA) or Kruskal–Wallis H, were appropriate, were used to test for differences in dietary intake between TAS2R38 diplotype groups, and between rs1761667 genotypes (AA, AG, and GG). Independent samples t-test (with Levene’s test for equality of variance) or Mann–Whitney U test, where appropriate, were used to test for differences in dietary intake between rs713598, rs1726866, rs10246939 (carriers of the non-taster and homozygous taster allele), and rs1761667 genotypes (AA/AG and GG) as well as a variable combined of CD36 genotypes and TAS2R38 diplotypes (Non-tasters: participants carrying both TAS2R38 AVI haplotype and CD36 A allele vs. Tasters: carriers of CD36 GG genotype or TAS2R38 PAV/PAV and PAV/AAV diplotypes). Participants with AVI/PAV diplotype were grouped with non-tasters considering that larger proportion of our study population carrying this diplotype was deemed a non-taster using the classification by Roura et al. (2015) explained below. Finally, 2-way ANOVA was conducted to explore the interaction between fat and bitter taster categories on dietary fat intake (total fat, MUFA, PUFA, and SFA). For this purpose, PTC ratings were used to categorize the participants into 3 distinct taster groups. The cut-off criteria were: hyposensitive taster (non-taster) ≤15.5, normal taster >15.5, and hypersensitive taster ≥51 (Roura et al. 2015). Considering a low number of hypersensitive tasters, these were excluded from the analysis. Bonferroni adjustment was used for multiple comparisons.
SPSS was used throughout (IBM Corp. Released 2016. IBM SPSS Statistics for Windows, Version 24.0. IBM Corp.). All tests were 2-tailed, with P < 0.05 considered statistically significant.
Results
Participant characteristics
Participant characteristics are shown in Table 1. Participants were healthy Caucasians (49 females (56%) and 39 males (44%)) with mean age 35 ± 1 years and BMI 24.9 ± 0.5 kg/m2. There were no differences in any of the presented variables or genotype frequencies according to sex, therefore males and females were combined in all analyses (data not shown). No differences in BMI were found between genotypes/diplotypes or bitter and fat taster categories (data not shown). Genotype/diplotype frequency of fat non-tasters can be found in Supplementary Table 1.
Table 1.
Participant characteristics (n = 88, n = 69)
| All participants (n = 88) | Fat-tasters only* (n = 69) | |
|---|---|---|
| Age (years) | 35 ± 1 | 34.7 ± 1.7 |
| BMI (kg/m2) | 24.9 ± 0.5 | 25.0 ± 0.6 |
| 18.5–24.9 kg/m2n (%) | 49 (56) | 41 (59) |
| ≥25.0 kg/m2n (%) | 39 (44) | 28 (41) |
| Sex n (%) | ||
| Female | 49 (56) | 40 (58) |
| Male | 39 (44) | 29 (42) |
| Bitter taste intensity rating m (IQR) | 6 (18.5) | 8 (27.5) |
| Fat taste category n (%) | ||
| Hyposensitive | 42 (48) | 42 (48) |
| Hypersensitive | 27 (31) | 27 (31) |
| Non-taster | 19 (21) | - |
| Energy (kcal) | 1656 ± 79 | 1709 ± 94 |
| Carbohydrate (%TEI) | 43.6 ± 0.8 | 44.4 ± 0.9 |
| Protein (%TEI) | 19.3 ± 0.3 | 19.2 ± 0.4 |
| Total fat (%TEI) | 37.5 ± 0.6 | 37.2 ± 0.7 |
| MUFA (%TEI) | 14.1 ± 0.3 | 14.1 ± 0.4 |
| PUFA (%TEI) | 6.6 ± 0.2 | 6.7 ± 0.3 |
| SFA (%TEI) | 13.3 ± 0.2 | 13.1 ± 0.3 |
Data presented as mean ± SEM, median (IQR), or absolute (relative) frequencies.
*Participants with a defined fat taste threshold.
The TAS2R38 and CD36 SNPs were in Hardy Weinberg equilibrium (P = 0.825, P = 0.573, P = 0.573, and P = 0.217 for the rs713598, rs1726866, rs10246939, and rs1761667, respectively). Haplotype frequencies of TAS2R38 in the study population were the following: AVI (53%), PAV (42%), and AAV (5%) and allele frequencies of CD36 rs1761667 were A (61%) and G (39%).
Taste sensitivity and genetics
There was no correlation between fat and bitter taste sensitivity (rs = 0.038, P = 0.758, data not shown, n = 69).
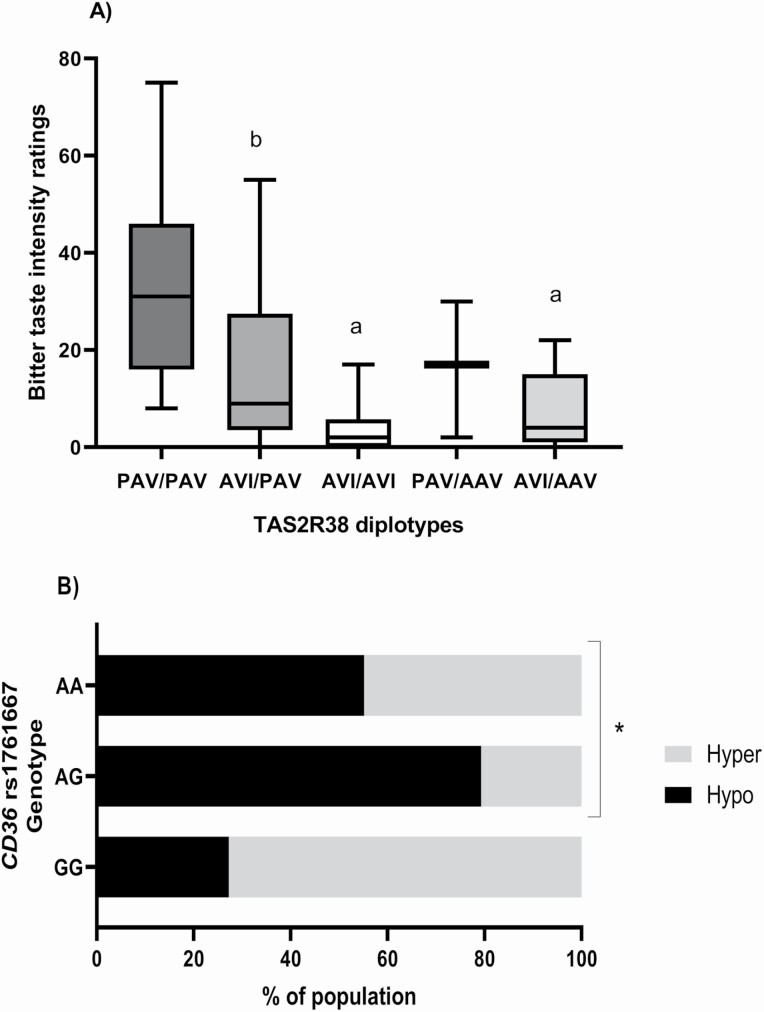
As shown in Figure 1, participants carrying PAV/PAV diplotype had higher median ratings of PTC intensity (median [IQR] 31 [30]) compared to participants with AVI haplotype (AVI/AVI, median [IQR] 2 [6] P = 1 × 10−6; AVI/AAV, median [IQR] 4 [14], P = 0.029, n = 88). Similarly, those classified as AVI/PAV had higher PTC ratings (median [IQR] 9 [24]) than those homozygous for AVI haplotype (P = 0.002, n = 88). Carriers of non-taster alleles for rs713598 (Ala), rs1726866 (Val), and rs10246939 (Ile) had lower ratings of bitterness compared to those homozygous for the taster alleles (Pro, Ala, and Val, data not shown).
Figure 1.
Genetics and taste sensitivity. (A) TAS2R38 diplotypes and bitter taste sensitivity; total n = 88, PAV/PAV = 13, AVI/PAV = 41, AVI/AVI = 24, PAV/AAV = 3, AVI/AAV = 7, a: different than PAV/PAV (AVI/AVI, P = 1 × 10-6; AVI/AAV, P = 0.029), b: different than AVI/AVI (P = 0.002). Line represents the median and whiskers min and max values, Kruskal–Wallis H test with Bonferroni adjusted P values. (B) CD36 rs1761667 and fat taste sensitivity, total n = 69, AA = 29, AG = 29, GG = 11, *P = 0.008, Fischer’s Exact test.
The CD36 rs1761667 was associated with FTS (P = 0.008, n = 69) when analyzed as 3 genotype groups (AA, AG, and GG). Here, a larger proportion of hyposensitive tasters had the AG genotype (55%), this remained significant after Bonferroni corrections were applied (Figure 1). For exploratory purposes only, non-tasters were included in further analysis, results were consistent (P = 0.033; Supplementary Figure 1), however this was no longer significant after Bonferroni correction applied. When genotypes were combined by variant allele (AA/AG, and GG), a larger percentage of participants carrying the A allele (67.2%) were classified as hyposensitive tasters compared to those homozygous for the G allele (P = 0.013, n = 69, data not shown). Similar was observed when fat taste threshold was treated as a continuous variable (Supplementary Table 3).
There was no association between TAS2R38 diplotypes and CD36 rs1761667 (P = 0.622, 0.963, respectively for AA, AG and GG, and AA/AG and GG, n = 88). There was also no difference in PTC ratings of bitterness according to CD36 rs1761667 genotypes (P = 0.782, 1.000, respectively for AA, AG and GG, and AA/AG and GG, n = 88) or TAS2R38 diplotypes and fat taste categories (P = 0.384, n = 69). There were no differences in fat taste threshold between TAS2R38 diplotypes (Supplementary Table 3).
Associations between genetics, taste sensitivity, and diet
As shown in Figure 2, the ratings of PTC intensity were negatively correlated with SFA (%TEI) (rs = −0.256, P = 0.016, n = 88). There were no correlations between bitter taste sensitivity, total fat, MUFA, and PUFA intakes. When excluding participants carrying AVI/AVI diplotype, there was no correlation between PTC bitter taste intensity and dietary fat intake (rs = −0.229, P = 0.069; rs = −0.199; P = 0.115; rs = −0.184; P = 0.145; rs = −0.166; P = 0.191 for total fat, MUFA, PUFA, and SFA, respectively). Similarly, there were no correlations between fat taste threshold and any of the presented variables (Figure 3, n = 69). SFA (%TEI) and total fat (%TEI) (rs = 0.656, P = 3.9 × 10−12) and total fat (%TEI) and energy intake (kcal) (rs = 0.225, P = 0.035) were positively correlated in the total cohort (data not shown).
Figure 2.
The correlations between bitter taste sensitivity (PTC intensity rating) and dietary fat intake (n = 88). Spearman’s correlation.
Figure 3.
The correlations between fat taste threshold and dietary fat intake (n = 69). Oleic acid concentrations/fat taste threshold was: 0.02, 0.06, 1, 1.4, 2, 2.8, 3.8, 5, 6.4, 8, 9.8, 12, and 20 mM. Spearman’s correlation.
There were no differences in energy and macronutrient intakes according to TAS2R38 diplotypes (Table 2, n = 88) or CD36 rs1761667 (Table 3, n = 69). Similar findings were observed when rare diplotypes AVI/AAV and PAV/AAV were excluded from the analyses (data not shown, n = 78). When analyzing individual TAS2R38 SNPs, there was a significant difference in SFA between rs1726866 and rs10246939 genotypes. Those carrying the non-taster allele for both SNPs (Val and Ile) had higher intake compared to participants homozygous for the taster allele (Ala and Val), both 13.6 ± 0.3 versus 12.1 ± 0.6 %TEI, P = 0.032, n = 88. When combining the CD36 genotypes and TAS2R38 diplotypes into one variable, participants carrying both TAS2R38 AVI haplotype and CD36 A allele had a higher SFA intake compared to carriers of CD36 GG genotype or TAS2R38 PAV/PAV and PAV/AAV diplotypes (13.8 ± 0.3 vs. 12.6 ± 0.5 %TEI, P = 0.047, Supplementary Table 2, n = 88). Similar was observed when only TAS2R38 combined diplotypes were compared (AVI/AVI, AVI/AAV, AVI/PAV vs. PAV/PAV, PAV/AAV, data not shown).
Table 2.
Energy and macronutrient intakes according to TAS2R38 diplotypes (n = 88)
| PAV/PAV (n = 13) | AVI/PAV (n = 41) | AVI/AVI (n = 24) | PAV/AAV (n = 3) | AVI/AAV (n = 7) | P-value | |
|---|---|---|---|---|---|---|
| Energy (kcal) | 1794 ± 224 | 1683 ± 137 | 1512 ± 89 | 1458 ± 100 | 1814 ± 289 | 0.666 |
| Protein (%TEI) | 20.4 ± 0.7 | 19.2 ± 0.6 | 19.0 ± 0.7 | 20.7 ± 2 | 18.9 ± 1.8 | 0.735 |
| CHO (%TEI) | 45. 1 ± 1.5 | 43.2 ± 1.3 | 42.3 ± 1.6 | 48.3 ± 4.5 | 44.9 ± 2.9 | 0.631 |
| Total fat (%TEI) | 35.2 ± 1.4 | 37.9 ± 1.1 | 38.4 ± 1.3 | 34.0 ± 4.2 | 37.4 ± 1.3 | 0.493 |
| SFA (%TEI) | 12.5 ± 0.7 | 13.5 ± 0.4 | 14.2 ± 0.6 | 10.0 ± 0.6 | 12.9 ± 0.6 | 0.082 |
| MUFA (%TEI) | 13.2 ± 0.6 | 14.4 ± 0.5 | 14.4 ± 0.6 | 14.0 ± 2.5 | 14.4 ± 0.5 | 0.613 |
| PUFA (%TEI) | 6.2 ± 0.3 | 6.8 ± 0.4 | 6.6. ± 0.3 | 6.7 ± 0.7 | 6.6. ± 0.5 | 0.928 |
Data presented as mean ± SEM (one-way ANOVA).
CHO, carbohydrate; TAS2R38, Taste 2 receptor member 38.
Table 3.
Energy and macronutrient intakes according to CD36 rs1761667 (n = 69)
| AA (n = 29) | AG (n = 29) | GG (n = 11) | AA/AG (n = 58) | P-value* | P-value** | |
|---|---|---|---|---|---|---|
| Energy (kcal) | 1803 ± 152 | 1631 ± 156 | 1670 ± 150 | 1717 ± 109 | 0.416 | 0.611 |
| Protein (%TEI) | 19.1 ± 0.6 | 19.3 ± 0.7 | 19.1 ± 0.9 | 19.2 ± 2.2 | 0.952 | 0.786 |
| CHO (%TEI) | 44.7 ± 1.4 | 43.9 ± 1.3 | 45.2 ± 2.2 | 44.3 ± 0.9 | 0.846 | 0.703 |
| Total fat (%TEI) | 36.9 ± 1.3 | 37.9 ± 1.0 | 36.0 ± 1.7 | 37.4 ± 1.7 | 0.453 | 0.458 |
| SFA (%TEI) | 12.5 ± 0.5 | 13.8 ± 0.4 | 12.9 ± 1.0 | 13.2 ± 0.3 | 0.156 | 0.762 |
| MUFA (%TEI) | 14.2 ± 0.5 | 14.2 ± 0.6 | 13.7 ± 0.7 | 14.2 ± 0.3 | 0.869 | 0.811 |
| PUFA (%TEI) | 7.0 ± 0.5 | 6.5 ± 0.5 | 6.0 ± 0.3 | 6.8 ± 0.3 | 0.514 | 0.312 |
Data presented as mean ± SEM. Kruskal–Wallis, Independent T-test or Mann–Whitney U test where appropriate.
CHO, carbohydrate.
*P-value difference in diet between 3 genotypes (AA, AG, and GG), **P-value difference in diet between two genotypes (AA/AG and GG).
Finally, results of the 2-way ANOVA showed no interaction between fat (hypo and hyper) and bitter (non-taster and taster) taster categories on total fat (P = 0.111), MUFA (P = 0.474), PUFA (P = 0.220), and SFA (P = 0.218). There were also no main effects of bitter taste category on total fat (P = 0.311), MUFA (P = 0.457), PUFA (P = 0.688), and SFA (P = 0.224). Similarly, there were no main effects of fat taste category on total fat (P = 0.186), MUFA (P = 0.406), PUFA (P = 0.145), and SFA (P = 0.702, Figure 4).
Figure 4.
Difference in dietary fat intake according to bitter taster status in (A) fat hypersensitive taster (total n = 27, bitter taster = 9, bitter non-taster = 18) and (B) fat hyposensitive taster group (total n = 40, bitter taster = 14, bitter non-taster = 26). Error bars represent ± SEM.
Discussion
The aim of this study was to explore the associations between genetics, taste sensitivity (bitter and fat), and dietary fat intake in healthy UK adults. We have demonstrated a difference in bitter taste sensitivity between TAS2R38 diplotypes and an association between CD36 rs1761667 and FTS. We did not find an association between TAS2R38 and FTS, and CD36 rs1761667 and bitter taste sensitivity. When analyzing dietary intake, although there was no association between either TAS2R38 diplotypes or CD36 rs1761667 and dietary intake, we did observe a difference in SFA according to TAS2R38 rs1726866 and rs10246939 genotypes and a negative correlation between bitter taste sensitivity and SFA. Finally, we did not observe an interaction between bitter and fat taste phenotypes on dietary fat intake. However, when combining the CD36 genotypes and TAS2R38 diplotypes into one variable, participants carrying both TAS2R38 AVI haplotype and CD36 A allele had a higher intake of saturated fat compared to carriers of CD36 GG genotype or TAS2R38 PAV/PAV and PAV/AAV diplotypes.
The associations between TAS2R38, bitter taste, and diet
We observed differences in the PTC ratings of bitterness according to TAS238 diplotype groups. Participants with PAV/PAV diplotype had higher ratings than those carrying AVI haplotype and participants classified as AVI/PAV had higher ratings than those homozygous for AVI haplotype. This is in line with previous research where AVI haplotype was associated with bitter non-taster and PAV with a bitter taster phenotype (Bufe et al. 2005; Kim et al. 2005; Tepper 2008).
In addition to the associations between genetics and taste perception, we also observed an inverse association between bitter taste sensitivity and SFA. Moreover, SFA was positively associated with total fat intake in our study population. This negative association between bitter taste sensitivity and dietary fat intake is in line with previous research reporting higher preference and intake of dietary fat in bitter non-tasters compared to tasters (Tepper and Nurse 1998; Duffy 2004; Choi and Chan 2015). Considering that total fat intake was positively associated with energy intake, a higher intake of SFA may be an indicator of a more energy dense pattern of dietary intake. Since we did not explore dietary patterns, this warrants further research in a similar study population.
The mechanism behind the association between bitter taste sensitivity and dietary fat intake is not entirely clear. It may be that interaction between bitter and fat taste perception exists and this will be discussed later. Considering that the correlation between bitter taste sensitivity and SFA was no longer significant once participants with AVI/AVI diplotype were excluded, this association appears to be driven by genetic predisposition. In this sense, TAS2R38 is expressed in the gastrointestinal tract where it may regulate the release of satiety hormones and influence the postprandial response to nutrients (Rozengurt 2006; Dotson et al. 2010). We observed a higher intake of SFA in carriers of the non-taster alleles (Val and Ile) for the rs1726866 and rs10246939 compared to those homozygous for the taster allele (Ala and Val). Similar was observed when TAS2R38 diplotyes were combined into carriers of the non-taster AVI haplotype and compared to those carrying PAV/PAV or PAV/AAV diplotype. The fact that we did not observe a similar difference in SFA when TAS2R38 diplotypes were analyzed as separate groups may be due to a smaller sub-group sample size when splitting participants into these; this warrants further investigation in a larger sample size study. Interestingly, Dotson et al. (2010) observed an increased eating disinhibition in carriers of the rs1726866 Val, non-taster, allele in their population of Amish women. The authors did not explore dietary intake, however the associations between saturated, total fat and energy intake in our study population suggest that TAS2R38 may be associated with both eating behavior, such as eating disinhibition, and a more energy dense dietary pattern. There are number of proposed mechanisms including impaired release of satiety hormones (glucagon-like peptide 1 [GLP-1], insulin) and increased levels of leptin in carriers of the non-taster alleles that warrant further investigation.
Besides the potential effects of TAS2R38 intestinal expression on hormone signaling, genetic variations in the CA6 gene may provide an explanation for the association between bitter taste sensitivity and dietary fat intake. Lower fat intake, as %TEI, was observed in UK individuals carrying the AA genotype of the CA6 rs2274333 compared to heterozygous AG individuals (Shen et al. 2017). This genotype has been associated with greater bitter taste sensitivity (i.e., PROP super-taster status), through greater fungiform papillae density in AA genotypes compared to homozygous GG genotypes (Melis et al. 2013). Considering that greater fungiform papillae density has also been associated with improved FTS (Zhou et al. 2020), there may be an interaction between TAS2R38 and CA6 on dietary fat intake in our study population. These interactions require further research in a similar study population.
Finally, due to the cross-sectional nature of the present study, it is not possible to determine the direction of the association between bitter taste sensitivity and dietary fat intake. Besides the possibility that lower bitter taste sensitivity leads to a higher fat intake, the opposite may also be correct. Jeon et al. (2008) suggested that a low-cholesterol diet, likely low in saturated fat, increases the sensitivity of intestinal bitter taste signaling system making the gut more responsive to the presence of bitter tasting compounds. Further intervention studies are, therefore, warranted to explore the cause-and-effect relationship between bitter taste and dietary fat intake.
The associations between CD36 rs1761667, fat taste, and diet
Furthermore, we observed that the A allele of CD36 rs1761667 was associated with FTS, specifically a larger percentage of participants carrying the A allele were classified as hyposensitive tasters. This is in line with previous research (Keller et al. 2012; Pepino et al. 2012; Sayed and Khan 2015; Melis et al. 2015; Mrizak et al. 2015; Burgess et al. 2018; Chmurzynska et al. 2020) and supports that LCFA evoke calcium signaling in gustatory cells expressing CD36 (El-Yassimi et al. 2008), and that lower protein levels may be related to the A allele hindering ability to detect fat. To date, although research is supportive toward an association between rs1761667 and FTS findings are largely heterogeneous, specifically, regarding ethnicity, which has been shown to modify responses to taste sensitivity (El-Sohemy et al. 2007). Only Melis et al. (2015); Burgess et al. (2018); and Sollai et al. (2019) investigated a Caucasian cohort, similar to ours. Chmurzynska et al. (2020) state recruitment was carried out in Poland, but otherwise does not specify ethnicity of participants. Our results corroborate Melis et al. (2015), and Sollai et al. (2019) but contrast, Burgess et al. (2018) who reported no association between rs1761667 genotype and FTS or perception of fat in the Caucasian sub-group. Results may differ to ours due to Burgess et al. (2018) having a lower sample size (n = 36) than us (n = 69) and Melis et al. (2015) (n = 64), and thus may have resulted in a type II error. Overall, it is evident the CD36 rs1761667 A-allele may hinder ability to detect fat in Caucasian participants, although research is scarce. Here, it is important to state that other factors may lead to differing taste sensitivity levels alongside rs1761667 genotype. This includes both mechanistic factors, for example rs1527483, another SNP on the CD36 gene that has been associated with instantaneous orosensory fat taste sensitivity (Plesník et al. 2018), and interactions between FTS and other tastes, which will be discussed below. Further, an additional factor to consider are fat non-tasters, despite constituting a comparatively small percentage of the population it is unclear whether this sub-population are associated with the same genetic pattern demonstrated by us and others (Keller et al. 2012; Pepino et al. 2012; Amira Sayed and Khan 2015; Melis et al. 2015; Mrizak et al. 2015; Burgess et al. 2018; Chmurzynska et al. 2020). Such genotypic conclusions cannot yet be drawn since many excluded non-tasters from their analysis (Burgess et al. 2018; Karmous et al. 2018; Bajit et al. 2020; Melis et al. 2020) due to no measurable threshold when undertaking the forced choice triangle method and a small sub-sample. We have included data for the fat non-tasters in our genetic analysis to aid future research comparisons.
It has been stated that a reduced ability to taste fat may lead to greater consumption (Besnard et al. 2016). Despite the association found between CD36 rs1761667 and FTS, we did not observe a difference in dietary intake (total energy, carbohydrate, protein, fat, MUFA, PUFA, or SFA). Our findings may be influenced by the majority of our population carrying at least one A allele (81%). To our knowledge, only Graham et al. (2021) and our study assess rs1761667 genotype and dietary intake on a solely Caucasian healthy cohort. Similarly, Pepino et al. (2012) reported no association between genotype and diet, using a mixture of Caucasian and African American (n = 21) participants. Others have reported that the rs1761667 A allele is associated with a higher dietary fat intake. For example, Ramos-Lopez et al. (2016) reported that in participants with chronic hepatitis C the AA genotype is associated with a higher total fat intake (%TEI) and higher SFA (%TEI) (P < 0.05), using a 3-day dietary food record. No differences between MUFA and PUFA were found. Similarly, Fujii et al. (2019), using Japanese (n = 495) participants demonstrated the AA genotype was significantly associated with higher total fat, SFA, MUFA, PUFA, omega-3, and omega-6 intake (P < 0.05), using a short FFQ. In contrast to this, and contradicting mechanisms associated to the A allele causing a reduced protein expression (Melis et al. 2017). Pioltine et al. (2016) reported the A allele was associated with a decreased intake of total fat (g/day), PUFA and MUFA (% kcal and g/day), fatty foods (portion and g/day), and vegetable oils (mL/day) in Brazilian children and adolescents with obesity, using two 24-h dietary recalls. It is evident that research regarding dietary intake and rs1761667 genotype is highly heterogeneous, preventing any clear conclusion from being drawn. This warrants further research in an ethnically homogenous, healthy cohort of adults or children, similar to our own, with consistent dietary collection methods.
Potential interactions between fat and bitter taste
In our study population, bitter and FTS were not correlated. Also, we found neither an association between TAS2R38 diplotypes and FTS nor CD36 rs1761667 and bitter taste sensitivity. Our findings contradict other research, reporting an association between the two tastes (Melis et al. 2015; Sollai et al. 2019). Melis et al. (2015), using 64 Italian participants, displayed that perception of fatty acids was associated with rs1761667 CD36 and that AVI/AVI participants exhibited a 5-fold higher oleic acid threshold than their PAV/PAV counterparts. Later, Sollai et al. (2019), reported similar results but using electrophysiological recordings from the tongue in response to oleic acid in a sample of 35 Italian adults. Similar results have been reported by Karmous et al. (2018), who also displayed a correlation between fat and bitter taste, however in a non-Caucasian (Tunisian) population and by Melis et al. (2020) in patients with inflammatory bowel disease. The fact we did not observe similar associations may be attributed to our study population being UK based and having different allele and haplotype frequencies compared to populations such as Tunisians explored by Karmous et al. (2018). Furthermore, differences in methods of taste sensitivity measurement between studies may also explain discrepancies in results.
None of the aforementioned studies explored the dietary intake of participants. In this sense, we observed an association between genetic predisposition to bitter taste, bitter taste sensitivity and dietary fat intake, where non-tasters have a higher intake of SFA than tasters. Although we did not observe an interaction between bitter and fat taste categories on dietary fat intake, we observed a higher intake of SFA in participants carrying both non-taster CD36 allele (A) and TAS2R38 haplotype (AVI) compared to those carrying either taster CD36 genotype (GG) or TAS2338 haplotype (PAV/PAV and PAV/AAV). This may suggest that genetic predisposition to hyposensitivity to both fat and bitter taste leads to an increased dietary fat intake, and supports previously observed interactions between the two tastes (Melis et al. 2015, 2020; Karmous et al. 2018; Sollai et al. 2019). It may also corroborate proposed mechanisms whereby TAS2R38 may be involved in the textural perception of fat, whereas CD36 may determine the chemosensory detection of fat (Keller 2012). Considering that higher SFA intake was also observed in carriers of TAS2R38 haplotype (AVI) compared to those carrying PAV/PAV or PAV/AAV diplotypes it may be that TAS2R38 is driving these differences. Due to the small sample in our study, we were not able to determine exact contribution of TAS2R38 diplotypes and CD36 rs1761667 in explaining SFA using regression analysis. These results should therefore be considered hypothesis generating and replicated in a larger cohort.
Strengths and limitations
Besides the fact we comprehensively investigated the associations between genetics, taste, and diet, a strength of this study is an ethnically homogenous population enabling a more valid interpretation of genetic association results. However, our population was not homogenous regarding sex. Barragán et al. (2018) reported that sex differences exist in ability to taste. There were no differences between sexes found in any of the variables tested; however, future research should endeavor to recruit a sex specific cohort or have a sample large enough for sex-specific analyses. Our sample size, although in line with other published research (Melis et al. 2015, 2020; Karmous et al. 2018; Sollai et al. 2019), was low regarding subgroup analysis (Grimaldi et al. 2017). This limits the conclusions that can be drawn and results should be replicated in a larger sample size study.
Moreover, in future studies, repeated testing of FTS should be considered. Although some have demonstrated FTS is reproducible (Newman and Keast 2013), others have demonstrated improvement, specifically within the hypersensitive tasters, over time (Tucker and Mattes 2013).
Furthermore, the use of PTC filter strips may result in misclassification of participants into bitter tasters and non-tasters (Lawless 1980). However, more recently, the use of PROP or PTC paper strip has been shown as a valid method to explore genetic predisposition to PTC taste sensitivity (Khataan et al. 2010) and we have used these ratings as a continuous variable in the majority of our analyses. Furthermore, the gLMS may also be more reliable when repeated on multiple occasions (Hayes et al. 2008) and this should be considered in future research. Nevertheless, participants were instructed on the use of the scale, which has been employed in similar studies exploring genetics and bitter taste sensitivity (Yang et al. 2020).
This study explored the associations between PTC taste sensitivity as a proxy for bitter taste sensitivity and TAS2R38 receptor as its determinant. PTC is however, only one of the many bitter tasting compounds and may not be a predictor of general bitter taste sensitivity. There are number of TAS2R bitter taste receptors that are activated by different bitter tasting compounds such as caffeine, quinine, and saccharin requiring further investigation to gain a more comprehensive understanding of bitter taste variability and its effects on dietary intake (Roura et al. 2015).
Lastly, self-reported dietary intake data, collected via validated FFQ, may be prone to misreporting (Shim et al. 2014). However, to improve accuracy, we selected a population specific FFQ (UK) and expressed macronutrients as % TEI which may improve accuracy of comparisons made (Macdiarmid and Blundell 1998). Also, although the FFQ used is a validated method to collect dietary consumption over the previous 12 months and has been calibrated using a 24-h dietary recall, dietary intake may vary over time and FTS has been shown to alter after only weeks of dietary modification (Newman et al. 2016; Costanzo et al. 2019). Therefore, future studies should consider the use of multiple 24-h dietary recalls to collect dietary intake information.
Conclusion
Overall, we confirmed that TAS2R38 haplotypes determine bitter taste sensitivity and CD36 rs1761667 is associated with fat taste sensitivity. Lower sensitivity to bitter taste may also lead to a higher dietary intake of fat. Considering the lack of association between bitter taste sensitivity and SFA when excluding participants carrying TAS2R38 non-taster AVI/AVI diplotype, this appears to be mainly driven by genetic predisposition. Although we did not observe an interaction between bitter and fat taste categories on dietary fat intake, we observed a higher intake of SFA in participants carrying both non-taster CD36 allele (A) and TAS2R38 haplotype (AVI) compared to those carrying either taster CD36 genotype (GG) or TAS2338 diplotype (PAV/PAV and PAV/AAV). This may suggest that genetic predisposition to hyposensitivity to both fat and bitter taste leads to an increased dietary fat intake. Nevertheless, it warrants further research in a larger cohort employing repeated measurements of bitter and FTS and a combination of dietary consumption methods such as FFQ and 24-h recalls.
Supplementary Material
Acknowledgments
We are grateful to the EPIC-Norfolk Study team for the use of the EPICFFQ software. EPIC-Norfolk is supported by programme grants from the Medical Research Council UK (G9502233, G0300128) and Cancer Research UK (C865/A2883).
Funding
The authors did not receive external funding for the research.
Author contributions
C.A.-M.G. and L.P.: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing—original draft, Visualization. E.M.: Investigation, Data Curation. M.B.: Investigation, Data Curation. I.J.E.: Investigation, Data Curation. J.N.K.: Investigation, Data Curation. V.S.: Investigation, Data Curation. Y.W.: Investigation, Data Curation. N.D.: Investigation, Data Curation. N.H.: Investigation, Data Curation. D.B. Investigation, Data Curation. A.K.: Writing—Review and Editing. Y.M.: Conceptualization, Methodology, Validation, Writing—Review and Editing, Supervision. All authors have approved the final article.
Conflict of interest
Y.M. is a shareholder for Nell Health, a lifestyle genotyping company. L.P. is serving on advisory board of DNAfuel LTD.
References
- Akella GD, Henderson SA, Drewnowski A. 1997. Sensory acceptance of Japanese green tea and soy products is linked to genetic sensitivity to 6-n-propylthiouracil. Nutr Cancer. 29(2):146–151. [DOI] [PubMed] [Google Scholar]
- Bajit H, Mohammed OAS, Guennoun Y, Benaich S, Bouaiti E, Belghiti H, Mrabet M, Elfahime EM, El Haloui NE, Saeid N, et al. 2020. Single-nucleotide polymorphism rs1761667 in the CD36 gene is associated with orosensory perception of a fatty acid in obese and normal-weight Moroccan subjects. J Nutr Sci. 9:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán R, Coltell O, Portolés O, Asensio EM, Sorlí JV, Ortega-Azorín C, González JI, Sáiz C, Fernández-Carrión R, Ordovas JM, et al. 2018. Bitter, sweet, salty, sour and umami taste perception decreases with age: sex-specific analysis, modulation by genetic variants and taste-preference associations in 18 to 80 year-old subjects. Nutrients. 10(10):1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J, Fry B, Maller J, Daly M. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21(2):263–265. [DOI] [PubMed] [Google Scholar]
- Besnard P, Passilly-Degrace P, Khan NA. 2016. Taste of fat: a sixth taste modality? Physiol Rev. 96(1):151–176. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. 2005. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 15(4):322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess B, Melis M, Scoular K, Driver M, Schaich KM, Keller KL, Tomassini Barbarossa I, Tepper BJ. 2018. Effects of CD36 genotype on oral perception of oleic acid supplemented safflower oil emulsions in two ethnic groups: a preliminary study. J Food Sci. 83(5):1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmurzynska A, Mlodzik-Czyzewska MA, Galinski G, Malinowska AM, Radziejewska A, Mikołajczyk-Stecyna J, Bulczak E, Wiebe DJ. 2020. Polymorphism of CD36 determines fat discrimination but not intake of high-fat food in 20- to 40-year-old adults. J Nutr. 150(8):2016–2022. [DOI] [PubMed] [Google Scholar]
- Choi SE, Chan J. 2015. Relationship of 6-n-propylthiouracil taste intensity and chili pepper use with body mass index, energy intake, and fat intake within an ethnically diverse population. J Acad Nutr Diet. 115(3):389–396. [DOI] [PubMed] [Google Scholar]
- Costanzo A, Liu D, Nowson C, Duesing K, Archer N, Bowe S, Keast R. 2019. A low-fat diet up-regulates expression of fatty acid taste receptor gene FFAR4 in fungiform papillae in humans: a co-twin randomised controlled trial. Br J Nutr. 122(11):1212–1220. [DOI] [PubMed] [Google Scholar]
- Daoudi H, Plesník J, Sayed A, Šerý O, Rouabah A, Rouabah L, Khan NA. 2015. Oral fat sensing and CD36 gene polymorphism in Algerian lean and obese teenagers. Nutrients. 7(11):9096–9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson CD, Shaw HL, Mitchell BD, Munger SD, Steinle NI. 2010. Variation in the gene TAS2R38 is associated with the eating behavior disinhibition in Old Order Amish women. Appetite. 54(1):93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A, Henderson SA, Levine A, Hann C. 1999. Taste and food preferences as predictors of dietary practices in young women. Public Health Nutr. 2(4):513–519. [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Henderson SA, Shore AB, Barratt-Fornell A. 1998. Sensory responses to 6-n-propylthiouracil (PROP) or sucrose solutions and food preferences in young women. Ann N Y Acad Sci. 855:797–801. [DOI] [PubMed] [Google Scholar]
- Duffy VB. 2004. Associations between oral sensation, dietary behaviors and risk of cardiovascular disease (CVD). Appetite. 43(1):5–9. [DOI] [PubMed] [Google Scholar]
- Duffy VB, Bartoshuk LM. 2000. Food acceptance and genetic variation in taste. J Am Diet Assoc. 100(6):647–655. [DOI] [PubMed] [Google Scholar]
- El-Sohemy A, Stewart L, Khataan Ln, Fontaine-Bisson B, Kwong P, Ozsungur S, Cornelis MC. 2007. Nutrigenomics of taste – impact on food preferences and food production. Forum Nutr. 60:176–182. [DOI] [PubMed] [Google Scholar]
- El-Yassimi A, Hichami A, Besnard P, Khan NA. 2008. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J Biol Chem. 283(19):12949–12959. [DOI] [PubMed] [Google Scholar]
- Fujii R, Hishida A, Suzuki K, Imaeda N, Goto C, Hamajima N, Wakai K, Kondo T. 2019. Cluster of differentiation 36 gene polymorphism (rs1761667) is associated with dietary MUFA intake and hypertension in a Japanese population. Br J Nutr. 121(11):1215–1222. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Murugesan G, Chen K, Zhang L, Wang Q, Febbraio M, Anselmo RM, Marchant K, Barnard J, Silverstein RL. 2011. Platelet CD36 surface expression levels affect functional responses to oxidized LDL and are associated with inheritance of specific genetic polymorphisms. Blood. 117(23):6355–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CA-M, Pilic L, King A, Nixon JE, Pipe J, Holton J, Tamba K, Hearne G, Pedlar CR, Lorente-Cebrián S, et al. 2021. Genetic differences in fat taste sensitivity and dietary intake in a UK female cohort. Food Quality and Preference. doi: 10.1016/j.foodqual.2021.104202. [DOI] [Google Scholar]
- Grimaldi KA, van Ommen B, Ordovas JM, Parnell LD, Mathers JC, Bendik I, Brennan L, Celis-Morales C, Cirillo E, Daniel H, et al. 2017. Proposed guidelines to evaluate scientific validity and evidence for genotype-based dietary advice. Genes and Nutrition. 12(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haryono RY, Sprajcer MA, Keast RSJ. 2014. Measuring oral fatty acid thresholds, fat perception, fatty food liking, and papillae density in humans. J Vis Exp. 88:51236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Allen AL, Bennett SM. 2013. Direct comparison of the generalized Visual Analog Scale (gVAS) and general Labeled Magnitude Scale (gLMS). Food Qual Prefer. 28(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Bartoshuk LM, Kidd JR, Duffy VB. 2008. Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem Senses. 33(3):255–265. [DOI] [PubMed] [Google Scholar]
- Hayes JE, Duffy VB. 2007. Revisiting sugar-fat mixtures: sweetness and creaminess vary with phenotypic markers of oral sensation. Chem Senses. 32(3):225–236. [DOI] [PubMed] [Google Scholar]
- Ikeda K. 1909. New seasonings. Journal of the Chemical Society of Tokyo 30:820–836. [Google Scholar]
- Jeon TI, Zhu B, Larson JL, Osborne TF. 2008. SREBP-2 regulates gut peptide secretion through intestinal bitter taste receptor signaling in mice. J Clin Invest. 118(11):3693–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmous I, Plesník J, Khan AS, Šerý O, Abid A, Mankai A, Aouidet A, Khan NA. 2018. Orosensory detection of bitter in fat-taster healthy and obese participants: genetic polymorphism of CD36 and TAS2R38. Clin Nutr. 37(1):313–320. [DOI] [PubMed] [Google Scholar]
- Keller KL. 2012. Genetic influences on oral fat perception and preference: Presented at the symposium “The Taste for Fat: New Discoveries on the Role of Fat in Sensory Perception, Metabolism, Sensory Pleasure and Beyond” held at the Institute of Food Technologists 2011 Annual Meeting, New Orleans, LA, June 12, 2011. J Food Sci. 77(3):S143–S147. [DOI] [PubMed] [Google Scholar]
- Keller KL, Liang LC, Sakimura J, May D, van Belle C, Breen C, Driggin E, Tepper BJ, Lanzano PC, Deng L, et al. 2012. Common variants in the CD36 gene are associated with oral fat perception, fat preferences, and obesity in African Americans. Obesity (Silver Spring). 20(5):1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AS, Keast R, Khan NA. 2020. Progress in lipid research preference for dietary fat : from detection to disease. Prog Lipid Res. 78:101032. [DOI] [PubMed] [Google Scholar]
- Khataan NH, Stewart L, Brenner DM, Cornelis MC, El-Sohemy A. 2010. TAS2R38 genotypes and phenylthiocarbamide bitter taste perception in a population of young adults. J Nutrigenet Nutrigenomics. 2(4–5):251–256. [DOI] [PubMed] [Google Scholar]
- Kim, Un kyung, Jorgenson, E., Coon, H., Leppert, M., Risch, N., & Drayna, D. 2003. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 299(5610):1221–1225. [DOI] [PubMed] [Google Scholar]
- Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. 2005. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Human Mutation. 26(3):199–204. [DOI] [PubMed] [Google Scholar]
- Lawless H. 1980. A comparison of different methods used to assess sensitivity to the taste of phenylthiocarbamide (PTC). Chem Senses. 5(3):247–256. [Google Scholar]
- Love-Gregory L, Abumrad NA. 2011. CD36 genetics and the metabolic complications of obesity. Curr Opin Clin Nutr Metab Care. 14(6):527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdiarmid J, Blundell J. 1998. Assessing dietary intake: who, what and why of under-reporting. Nutr Res Rev. 11(2):231–253. [DOI] [PubMed] [Google Scholar]
- Mattes RD. 2010. Fat taste in humans: is it a primary? In: Montmayeur J-P, le Coutre J, editors. Fat detection: taste, texture, and post ingestive effects. Florida: CRC Press. p. 167–193. https://books.google.co.uk/books?hl=en&lr=&id=jSb7aQxm9UEC&oi=fnd&pg=PA167&dq=mattes+fat+taste+is+it+a+primary&ots=JhFBAO1mjf&sig=L5t_RFCy36IKWRGk_bYEKLo1xEc&redir_esc=y#v=onepage&q=mattes fat taste is it a primary&f=false [PubMed] [Google Scholar]
- Melis M, Atzori E, Cabras S, Zonza A, Calò C, Muroni P, Nieddu M, Padiglia A, Sogos V, Tepper BJ, et al. 2013. The gustin (CA6) gene polymorphism, rs2274333 (A/G), as a mechanistic link between PROP tasting and fungiform taste papilla density and maintenance. PLoS One. 8(9):e74151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis M, Carta G, Pintus S, Pintus P, Piras CA, Murru E, Manca C, Di Marzo V, Banni S, Tomassini Barbarossa I. 2017. Polymorphism rs1761667 in the CD36 gene is associated to changes in fatty acid metabolism and circulating endocannabinoid levels distinctively in normal weight and obese subjects. Front Physiol. 8:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis M, Mastinu M, Sollai G, Paduano D, Chicco F, Magrì S, Usai P, Crnjar R, Tepper BJ, Barbarossa IT. 2020. Taste changes in patients with inflammatory bowel disease: associations with PROP phenotypes and polymorphisms in the salivary protein, gustin and CD36 receptor genes. Nutrients. 12(2):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis M, Sollai G, Muroni P, Crnjar R, Barbarossa IT. 2015. Associations between orosensory perception of oleic acid, the common single nucleotide polymorphisms (rs1761667 and rs1527483) in the CD36 gene, and 6-n-propylthiouracil (PROP) tasting. Nutrients. 7(3):2068–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrizak I, Šerý O, Plesnik J, Arfa A, Fekih M, Bouslema A, Zaouali M, Tabka Z, Khan NA. 2015. The A allele of cluster of differentiation 36 (CD36) SNP 1761667 associates with decreased lipid taste perception in obese Tunisian women. Br J Nutr. 113(8):1330–1337. [DOI] [PubMed] [Google Scholar]
- Mulligan AA, Luben RN, Bhaniani A, Parry-Smith DJ, O’Connor L, Khawaja AP, Forouhi NG, Khaw KT; EPIC-Norfolk FFQ Study . 2014. A new tool for converting food frequency questionnaire data into nutrient and food group values: FETA research methods and availability. BMJ Open. 4(3):e004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LP, Bolhuis DP, Torres SJ, Keast RS. 2016. Dietary fat restriction increases fat taste sensitivity in people with obesity. Obesity (Silver Spring). 24(2):328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LP, Keast RSJ. 2013. The test-retest reliability of fatty acid taste thresholds. Chemosensory Perception. 6(2):70–77. [Google Scholar]
- O’Brien SA, Feeney EL, Scannell AG, Markey A, Gibney ER. 2013. Bitter taste perception and dietary intake patterns in Irish children. J Nutrigenet Nutrigenomics. 6(1):43–58. [DOI] [PubMed] [Google Scholar]
- Pepino MY, Love-Gregory L, Klein S, Abumrad NA. 2012. The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J Lipid Res. 53(3):561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesník J, Šerý O, Khan AS, Bielik P, Khan NA. 2018. The rs1527483, but not rs3212018, CD36 polymorphism associates with linoleic acid detection and obesity in Czech young adults. Br J Nutr. 119(4):472–478. [DOI] [PubMed] [Google Scholar]
- Pioltine MB, de Melo ME, Santos A, Machado AD, Fernandes AE.Fujiwara CT, Cercato C,Mancini MC,. 2016. Genetic variation in CD36 is associated with decreased fat and sugar intake in obese children and adolescents. J Nutrigenet Nutrigenomics. 9(5-6):300–305. [DOI] [PubMed] [Google Scholar]
- Ramos-Lopez O, Roman S, Martinez-Lopez E, Fierro NA, Gonzalez-Aldaco K, Jose-Abrego A, Panduro A. 2016. CD36 genetic variation, fat intake and liver fibrosis in chronic hepatitis C virus infection. World J Hepatol. 8(25):1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso DS, Mezzavilla M, Pagani L, Robino A, Morini G, Tofanelli S, Carrai M, Campa D, Barale R, Caradonna F, et al. 2016. Global diversity in the TAS2R38 bitter taste receptor: revisiting a classic evolutionary PROPosal. Sci Rep. 6:25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura E, Aldayyani A, Thavaraj P, Prakash S, Greenway D, Thomas WG, Meyerhof W, Roudnitzky N, Foster SR. 2015. Variability in human bitter taste sensitivity to chemically diverse compounds can be accounted for by differential TAS2R activation. Chem Senses. 40(6):427–435. [DOI] [PubMed] [Google Scholar]
- Rozengurt E. 2006. Taste receptors in the gastrointestinal tract. I. Bitter taste receptors and α-gustducin in the mammalian gut. Am J Physiol Gastrointest Liver Physiol. 291(2):171–177. [DOI] [PubMed] [Google Scholar]
- Sayed A, Khan NA. 2015. Is the single nucleotide polymorphism (SNP) of CD36, a lipid-receptor, a predictor of obesity in adulthood? Med Sci. 31(12):1072–1074. [DOI] [PubMed] [Google Scholar]
- Sayed A, Šerý O, Plesnik J, Daoudi H, Rouabah A, Rouabah L, Khan NA. 2015. CD36 AA genotype is associated with decreased lipid taste perception in young obese, but not lean, children. Int J Obes (Lond). 39(6):920–924. [DOI] [PubMed] [Google Scholar]
- Shen Y, Kennedy OB, Methven L. 2017. The effect of genotypical and phenotypical variation in taste sensitivity on liking of ice cream and dietary fat intake. Food Quality and Preference. 55:79–90. [Google Scholar]
- Shim JS, Oh K, Kim HC. 2014. Dietary assessment methods in epidemiologic studies. Epidemiol Health. 36:e2014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollai G, Melis M, Mastinu M, Pani D, Cosseddu P, Bonfiglio A, Crnjar R, Tepper BJ, Barbarossa IT. 2019. Human tongue electrophysiological response to oleic acid and its associations with PROP taster status and the CD36 polymorphism (rs1761667). Nutrients. 11(2):315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JE, Newman LP, Keast RS. 2011. Oral sensitivity to oleic acid is associated with fat intake and body mass index. Clin Nutr. 30(6):838–844. [DOI] [PubMed] [Google Scholar]
- Tepper BJ. 2008. Nutritional implications of genetic taste variation: the role of PROP sensitivity and other taste phenotypes. Annu Rev Nutr. 28:367–388. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Koelliker Y, Zhao L, Ullrich NV, Lanzara C, d’Adamo P, Ferrara A, Ulivi S, Esposito L, Gasparini P. 2008. Variation in the bitter-taste receptor gene TAS2R38, and adiposity in a genetically isolated population in Southern Italy. Obesity (Silver Spring). 16(10):2289–2295. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Nurse RJ. 1997. Fat perception is related to PROP taster status. Physiol Behav. 61(6):949–954. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Nurse RJ. 1998. PROP taster status is related to fat perception and preference. Ann N Y Acad Sci. 855:802–804. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Nurse NJ. 2006. PROP taster status is related to fat perception and preference. Ann N Y Acad Sci. 5:802–804. [DOI] [PubMed] [Google Scholar]
- Timpson NJ, Christensen M, Lawlor DA, Gaunt TR, Day IN, Ebrahim S, Davey Smith G. 2005. TAS2R38 (phenylthiocarbamide) haplotypes, coronary heart disease traits, and eating behavior in the British Women’s Heart and Health Study. Am J Clin Nutr. 81(5):1005–1011. [DOI] [PubMed] [Google Scholar]
- Tucker RM, Kaiser KA, Parman MA, George BJ, Allison DB, Mattes RD. 2017. Comparisons of fatty acid taste detection thresholds in people who are lean vs. overweight or obese: a systematic review and meta-analysis. PLoS One. 12(1):e0169583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RM, Mattes RD. 2013. Influences of repeated testing on nonesterified fatty acid taste. Chem Senses. 38(4):325–332. [DOI] [PubMed] [Google Scholar]
- Williams JA, Bartoshuk LM, Fillingim RB, Dotson CD. 2016. Exploring ethnic differences in taste perception. Chem Senses. 41(5):449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2018. WHO | Obesity and overweight. http://www.who.int/mediacentre/factsheets/fs311/en/
- World Health Organisation . 2020. Healthy diet. Fact Sheet. https://www.who.int/news-room/fact-sheets/detail/healthy-diet
- Yang Q, Williamson AM, Hasted A, Hort J. 2020. Exploring the relationships between taste phenotypes, genotypes, ethnicity, gender and taste perception using Chi-square and regression tree analysis. Food Quality and Preference. 83:103928. [Google Scholar]
- Zhou X, Yeomans M, Thomas A, Wilde P, Linter B, Methven L. 2020. Individual differences in oral tactile sensitivity and gustatory fatty acid sensitivity and their relationship with fungiform papillae density, mouth behaviour and texture perception of a food model varying in fat. Food Quality and Preference. 90:104116.. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.