Abstract
Simple Summary
The Ephemeroptera is an ancient lineage of insects, among which the Heptageniidae is one of the most species-rich families, although its phylogenetic relationships have been controversial. The mitogenomes of Heptageniidae were found gene rearrangements of CR-I-M-Q-M-ND2 and a conserved intergenic gap between trnA and trnR. Thus, 15 complete and two nearly complete mitogenomes of Heptageniidae were used to explore mitogenome structures and clarify the disputes of phylogenetic relationships among Heptageniidae. Additionally, the Heptageniidae samples collected from habitats with significant temperature differences were applied to investigate the adaptive evolution of mitochondrial PCGs under low-temperature stress.
Abstract
We determined 15 complete and two nearly complete mitogenomes of Heptageniidae belonging to three subfamilies (Heptageniinae, Rhithrogeninae, and Ecdyonurinae) and six genera (Afronurus, Epeorus, Leucrocuta, Maccaffertium, Stenacron, and Stenonema). Species of Rhithrogeninae and Ecdyonurinae had the same gene rearrangement of CR-I-M-Q-M-ND2, whereas a novel gene rearrangement of CR-I-M-Q-NCR-ND2 was found in Heptageniinae. Non-coding regions (NCRs) of 25–47 bp located between trnA and trnR were observed in all mayflies of Heptageniidae, which may be a synapomorphy for Heptageniidae. Both the BI and ML phylogenetic analyses supported the monophyly of Heptageniidae and its subfamilies (Heptageniinae, Rhithrogeninae, and Ecdyonurinae). The phylogenetic results combined with gene rearrangements and NCR locations confirmed the relationship of the subfamilies as (Heptageniinae + (Rhithrogeninae + Ecdyonurinae)). To assess the effects of low-temperature stress on Heptageniidae species from Ottawa, Canada, we found 27 positive selection sites in eight protein-coding genes (PCGs) using the branch-site model. The selection pressure analyses suggested that mitochondrial PCGs underwent positive selection to meet the energy requirements under low-temperature stress.
Keywords: Heptageniidae, mitochondrial genome, gene rearrangement, phylogenetic relationship, non-coding region (NCR), selective stress analysis
1. Introduction
As a primitive group of winged insects, Ephemeroptera comprises 40 families, 460 genera, and 3700 species [1,2,3]. Ephemeroptera is characterized by multiple ancestral signs including extra appendages (seven pairs of gills on larvae along with the forceps and long tails of adults), a unique prometamorphosis development pattern, and wings that do not fold flat over the abdomen, which have been intensely studied in phylogeny and historical processes [4,5]. As one of the most species-rich families among Ephemeroptera, Heptageniidae consists of three subfamilies (Ecdyonurinae, Heptageniinae, and Rhithrogeninae), 37 genera, and 606 described species [1,3,6]. Much effort has been made to figure out the taxonomy and phylogeny of Ephemeroptera using morphology features, molecular proofs, and combined data [7,8,9,10,11,12,13,14]. Nonetheless, the phylogenetic relationships within Heptageniidae remain controversial [6,13]. The phylogenetic systems of both McCafferty [9] and Kluge [10,11] supported Heptageniidae as belonging to Branchitergalia (Heptagenioidea) with a close relationship to Isonychiidae. The phylogenetic relationship developed using combined data of morphological characters and several nuclear genes by Ogden et al. [12] was different from the former hypotheses, supporting Heptageniidae as a monophyletic group, but its phylogenetic position remained uncertain. According to the newly published study by Ogden et al. [15], the phylogenetic results supported Heptageniidae as a sister group to Isonychiidae using over 440 targeted genomic protein coding regions (exons). In addition, the internal phylogenetic relationships within Heptageniidae were divided into three subfamilies (Heptageniinae, Rhithrogeninae, and Ecdyonurinae) and their relationship was shown as (Ecdyonurinae + (Heptageniinae + Rhithrogeninae)) by Wang and McCafferty [13] and Webb and McCafferty [6]. By contrast, the phylogenetic analysis was presented as (Rhithrogenidae + (Ecdyonurinae + Heptageniinae)) by Ogden et al. [15]. In addition, the genera Stenonema was redefined to include Maccaffertium via two mitochondrial genes (COX1 and 16S rRNA) and two nuclear genes (Wingless (Wg) and histone H3) by Zembrzuski and Anderson [16].
The typical mitochondrial genome (mitogenome) of insects is a 14–20 kb double-stranded circular piece of DNA [17,18,19]. It encodes 37 genes including 13 protein-coding genes (PCGs), two ribosomal RNAs (rRNAs, 16S rRNA and 12S rRNA), 22 transfer RNAs (tRNAs), and the A + T-rich region (control region, CR). Since the mitogenome has features such as rapid evolution rates, small genome sizes, and relatively low recombination and maternal inheritance, it is considered to be an excellent molecular marker for studies in phylogeny, evolution, and comparative genomics [18,20,21,22]. Although most insect mitogenomes are conservative, gene rearrangements and long non-coding regions (NCRs) have been variously reported in Coleoptera, Hemiptera, Lepidoptera, Mantodea, Orthoptera, Thysanoptera, etc. [19,23,24,25,26,27,28,29,30,31]. According to published reports, gene rearrangements of tRNA genes including duplication, translocation, and pseudogenization were mainly concentrated in the regions of CR-I-Q-M-ND2, COX1-K-D-ATP8, and ND3-A-R-N-S-E-F-ND5 [31,32].
Within the order Ephemeroptera, most species retain the same 37 genes as the hypothesized ancestral mitogenome of insects except for Siphluriscidae, Baetidae, Leptophlebiidae, Ephemerellidae, and Heptageniidae [32,33,34,35,36,37,38,39,40,41]. The mitogenome of Siphluriscus chinensis (Siphluriscidae) encoded an extra trnK located between trnS and trnE in the minor coding strand [36]. The trnC and trnY in Alainites yixiani (Baetidae) translocated from a position between trnW and COX1 into the gene cluster of I-Q-M, and the gene order was rearranged as I-C-Q-Y-M. Furthermore, one copy of inversion and translocation of trnI was detected in Ephemerella sp. Yunnan-2018, Ephemerella sp. MT-2014, Serratella zapekinae, Serratella sp. Liaoning-2019, and Serratella sp. Yunnan-2018, along with three duplicate copies of inversion and translocation of trnI in Torleya grandipennis and T. tumiforceps (Ephemerellidae) [32]. The trnA and trnR genes switched positions in Habrophlebiodes zijinensis (Leptophlebiidae) resulting in a gene arrangement R-A-N-S-E-F. Within the family Heptageniidae, an extra trnM was observed in the location between trnQ and ND2; thus, the gene arrangement was arranged as I-M-Q-M in Epeorus herklotsi, Epeorus sp. JZ-2014, Epeorus sp. MT-2014, and Parafronurus youi [33,34,35]. Surprisingly, no gene rearrangements were found in Paegniodes cupulatus (Heptageniidae), showing that its gene order was conservative and different from other Heptageniidae species.
Despite the fact that mitogenomes are generally considered to be under neutral or nearly neutral selection [42], several studies have pointed out that positive selection acted on mitochondrial PCGs linked to environmental adaptations [43,44,45]. In this way, as a potential target associated with energy metabolism under environmental selection pressure, the mitogenome may be suitable for studying positive selection or natural selection. According to the mitochondrial PCGs of flying and flightless grasshoppers, a significant positive selection was found in several genes including ND2, ND4, ND4L, ND5, ND6, ATP8, and COX3 in flying lineages [45]. Hence, this indicated that positive selection stimulated mitochondrial genes to better suit the energy demands of flight in grasshoppers. Likewise, the mitochondrial PCGs were affected by positive selection from the last common ancestor of Pterygota and flying insects, which illustrated that those mitochondrial PCGs related to energy metabolism underwent adaptive evolution during the evolution of flight capacity in insects [45]. As aquatic insects, mayflies spend most of their developmental stages in the water. Among various environmental factors, the water temperature was shown to be crucial to the morphology, behavior, growth, and life cycle of mayflies [46,47]. Therefore, mayflies were proposed as appropriate models to investigate the adaptive evolution of aquatic insects in a low-temperature environment.
To explore the characteristics of gene rearrangements and the phylogenetic relationship of subfamilies in Heptageniidae, we determined the mitogenomes of 17 species from all three subfamilies and six genera of Heptageniidae. The phylogenetic relationship within Ephemeroptera was constructed with gene rearrangements and the location of NCRs to clarify the phylogenetic controversies. Moreover, samples of several Heptageniinae (Maccaffertium, Stenacron, and Stenonema) and Leucrocuta were collected from Ottawa, Canada. The climate there is so cold that the lowest temperature is below 0 °C and water temperature is below 10 °C for eight months of the year [48]. Thus, these mayfly nymphs had to be exposed to low water temperature for a long time. Other samples of Epeorus and Afronurus were from southern provinces (Zhejiang and Yunnan) of China where the mean annual water temperature was about 24–26 °C. Accordingly, Heptageniidae samples collected from habitats with significant temperature differences were suitable materials to assess the adaptive evolution of mitochondrial PCGs under low-temperature stress. In brief, our research not only provides novel insight into the gene rearrangements and phylogenetic relationships within Heptageniidae, but also investigates the evolutionary mechanisms of aquatic insect mitochondrial PCGs under low-temperature stress.
2. Materials and Methods
2.1. Sampling Collection and DNA Extraction
Six specimens were collected from the Rideau River, Ottawa, Canada in July 2017. Eleven specimens were collected from Wu River, Jinhua, Zhejiang province, and Chuan River, Jingdong, Yunnan province, China (Table 1). The specimens were all identified on the basis of a combination of nymph morphology and the alignment of COX1 genes. Because some new species and genera were found in this study, we only identified those species at the genus or family level (Table 1). Samples were stored in 100% ethanol at −40 °C in J.Y.Z.’s lab, College of Life Science and Chemistry, Zhejiang Normal University, China. Our study included 17 specimens representing all three subfamilies, nine specimens from the subfamily Ecdyonurinae (Afronurus and Leucrocuta), five specimens from Heptageniinae (Maccaffertium, Stenacron, and Stenonema), and two specimens from Rhithrogeninae (Epeorus). Total DNA was extracted from legs or half of the whole individual of every species using Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China).
Table 1.
Information on specimen sources of the samples used in this study and NCBI Genbank accession numbers.
| Subfamily | Species | Specimen No. | Sampling Localities | Accession Number |
|---|---|---|---|---|
| Heptageniinae | Maccaffertium mediopunctatum (McDunnough, 1926) | 03FY33 | Ottawa, Canada | MK642302 |
| Maccaffertium modestum (Banks, 1910) | 03FY62 | Ottawa, Canada | MK642303 | |
| Maccaffertium vicarium (Walker, 1853) | 03FY39 | Ottawa, Canada | MK642304 | |
| Stenacroninterpunctatum (Say, 1839) | 03FY34 | Ottawa, Canada | MK642305 | |
| Stenonema femoratum (Say 1823) | 03FY36 | Ottawa, Canada | MK642306 | |
| Ecdyonurinae | Leucrocuta Aphrodite (McDunnough, 1926) | 03FY51 | Ottawa, Canada | MK642301 |
| Afronurus furcata (Zhou and Zhen, 2003) | 08BF03 | Zhejiang, China | MK642293 | |
| Afronurus rubromaculata (You, Wu, Gui, and Hsu, 1981) | 08BF02 | Zhejiang, China | MK642294 | |
| Afronurus sp1. YW01BF06 | 01BF06 | Zhejiang, China | MK642295 | |
| Afronurus sp2. LS53BF04 | 53BF04 | Zhejiang, China | MK642296 | |
| Afronurus yixingensis (Wu and You, 1986) | 06BF03 | Zhejiang, China | MK642297 | |
| Afronurus sp. 07BF85 | 07BF85 | Yunnan, China | MW450876 | |
| Afronurus sp. 07BF86 | 07BF86 | Yunnan, China | MW450877 | |
| Afronurus sp. 07BF96 | 07BF96 | Yunnan, China | MW450878 | |
| Rhithrogeninae | Epeorus dayongensis (Gui and Zhang, 1992) | 18BF01 | Zhejiang, China | MK642298 |
| Epeorus sp. LA03FY06 | 03FY06 | Zhejiang, China | MK642299 | |
| / | Heptageniidae sp. YW03BF02 | 03BF02 | Zhejiang, China | MK642300 |
2.2. PCR Amplification and Sequencing
This study used both normal polymerase chain reaction (PCR) and long-and-accurate PCR (LA PCR) methods with Takara Taq or Takara LA Taq DNA polymerase (Takara, Dalian, China). Normal PCR (product length < 3000 bp) or LA PCR (product length > 3000 bp) reaction conditions were as in Gao et al. [49]. The mitogenomes were amplified in 700–2000 bp short fragments with universal primers according to the method of Simon et al. [50,51] and Zhang et al. [33]. Afterward, we designed specific primers (Table S1, Supplementary Materials) with Primer Premier 5.0 [52] according to the obtained sequences. All PCR products were obtained in both forward and reverse directions using the primer-walking method and AB13730XL by Sangon Biotech Company (Shanghai, China).
2.3. Mitogenome Annotation and Sequence Analyses
We inspected and assembled mitochondrial sequences using DNASTAR Package v.7.1 [53]. All tRNA genes and their secondary structures were identified by MITOS (http://mitos.bioinf.uni-leipzig.de/index.py, accessed on 11 January 2021) [54]. Two rRNA genes (12S and 16S rRNA) and 13 PCGs were determined by alignments with homologous mtDNA sequences from several species in Heptageniidae using Clustal X [55,56]. The nucleotide composition, codon usage, and relative synonymous codon usage (RSCU) were calculated using Mega 7.0 [56]. The GC skews and AT skews were separately calculated using the following formulae: AT skew = (A − T)/(A + T), and GC skew = (G − C)/(G + C) [57]. Tandem repeats in CRs were detected via Tandem Repeat Finder V 4.09 [58]. The secondary structures of NCRs were found and mapped via RNAstructure Web Servers [59].
2.4. Phylogenetic Analyses
Forty-nine species from Ephemeroptera, including 14 families (Table 2), were used in phylogenetic analyses of Heptageniidae and Ephemeroptera [32,33,34,35,36,37,38,39,40,41]. The taxon of Siphluriscus (Siphluriscidae) was recovered as a sister clade to all other mayflies and, therefore, S. chinensis from the family Siphluriscidae was selected as the outgroup [12,36]. Thirteen PCGs of mayfly mitogenomes were used to construct BI and ML phylogenetic trees. The nucleotide sequences of the 13 PCGs were used for DNA alignment by MAFFT v. 7.475 [60], and the conserved regions were detected by Gblock 0.91b using default settings [61]. The program PartionFinder 1.1.1 was employed on the basis of Bayesian information criterion (BIC) to identify the best partitioning scheme and substitution model, and all 12 partitions were observed (Table S2, Supplementary Materials) [62]. ML analysis was run in RAxML 8.2.0 with a GTRGAMMAI model, and branch support for each node was evaluated with 1000 replicates [63]. BI analysis was performed in MrBayes version 3.2 using a GTR + I + G model [64]. Each of four chains ran for 10 million generations, and sampling every 1000 generations was used for phylogenetic relationship reconstruction [64]. The convergence was evaluated using Tracer version 1.5, and trees from the first 25% of the samples were removed as burn-in during BI analysis.
Table 2.
List of Ephemeroptera mitogenomes used to construct phylogenetic trees.
| Family | Genus | Species | Length (bp) | GenBank No. | References |
|---|---|---|---|---|---|
| Ameletidae | Ameletus | Ameletus sp. MT-2014 | 15,141 | KM244682 | [35] |
| Baetidae | Baetis | Baetis sp. PC-2010 | 14,883 | GU936204 | Unpublished |
| Takobia | Alainites yixiani | 14,589 | GU479735 | Unpublished | |
| Caenidae | Caenis | Caenis sp. JYZ-2018 | 15,254 | MG910499 | [33] |
| Caenis sp. JYZ-2020 | 15,392 | MN356096 | [37] | ||
| Caenis sp. YJ-2009 | 15,351 | GQ502451 | Unpublished | ||
| Ephemerellidae | Ephemerella | Ephemerella sp. MT-2014 | 14,896 | KM244691 | [31] |
| Ephemerella sp. Yunnan-2018 | 15,256 | MT274127 | [32] | ||
| Serratella | Serratella sp. Liaoning-2019 | 15,523 | MT274128 | ||
| Serratella sp. Yunnan-2018 | 15,134 | MT274129 | |||
| Serratella zapekinae | 15,703 | MT274130 | |||
| Torleya | Torleya grandipennis | 15,523 | MT274131 | ||
| Torleya tumiforceps | 15,330 | MT274132 | |||
| Ephemeridae | Ephemera | Ephemera orientalis | 16,463 | EU591678 | Unpublished |
| Ephemera sp. XL-2019 | 15,314 | MK951659 | [35] | ||
| Heptageniidae | Afronurus | Afronurus furcata | 15,420 | MK642293 | This study |
| Afronurus rubromaculata | 15,519 | MK642294 | This study | ||
| Afronurus sp. 07BF85 | 15,473 | MW450876 | This study | ||
| Afronurus sp. 07BF86 | 15,696 | MW450877 | This study | ||
| Afronurus sp. 07BF96 | 15,491 | MW450878 | This study | ||
| Afronurus sp. YW01BF06 | 15,360 | MK642295 | This study | ||
| Afronurus sp. LS53BF04 | 15,866 | MK642296 | This study | ||
| Afronurus yixingensis | 15,883 | MK642297 | This study | ||
| Epeorus | Epeorus dayongensis | 15,337 | MK642298 | This study | |
| Epeorus herklotsi | 15,502 | MG870104 | [30] | ||
| Epeorus sp. JZ-2014 | 15,338 | KJ493406 | Unpublished | ||
| Epeorus sp. MT-2014 | 15,456 | KM244708 | [31] | ||
| Epeorus sp. LA03FY06 | 15,514 | MK642299 | This study | ||
| Leucrocuta | Leucrocuta aphrodite | 15,428 | MK642301 | This study | |
| Maccaffertium | Maccaffertium mediopunctatum | 15,319 | MK642302 | This study | |
| Maccaffertium modestum | 15,324 | MK642303 | This study | ||
| Maccaffertium vicarium | 15,324 | MK642304 | This study | ||
| Paegniodes | Paegniodes cupulatus | 15,715 | HM004123 | Unpublished | |
| Parafronurus | Parafronurus youi | 15,481 | EU349015 | [29] | |
| Stenacron | Stenacron interpunctatum | 15,330 | MK642305 | This study | |
| Stenonema | Stenonema femoratum | 15,332 | MK642306 | This study | |
| Heptageniidae sp. YW03BF02 | 15,663 | MK642300 | This study | ||
| Isonychiidae | Isonychia | Isonychia ignota | 15,105 | HM143892 | Unpublished |
| Isonychia kiangsinensis | 15,456 | MH119135 | [34] | ||
| Leptophlebiidae | Choroterpides | Choroterpides apiculata | 15,199 | MN807287 | [36] |
| Habrophlebiodes | Habrophlebiodes zijinensis | 14,355 | GU936203 | Unpublished | |
| Potamanthidae | Potamanthus | Potamanthus sp. MT-2014 | 14,937 | KM244674 | [35] |
| Siphlonuridae | Siphlonurus | Siphlonurus aestivalis | 15,120 | MT862395 | Unpublished |
| Siphlonurus immanis | 15,529 | FJ606783 | Unpublished | ||
| Siphlonurus sp. MT-2014 | 14,745 | KM244684 | [31] | ||
| Siphluriscidae | Siphluriscus | Siphluriscus chinensis | 16,616 | HQ875717 | [32] |
| Teloganodidae | / | Teloganodidae sp. | 12,435/ | KM244703 | [31] |
| 2817 | KM244670 | ||||
| Vietnamellidae | Vietnamella | Vietnamella dabieshanensis | 15,761 | HM067837 | Unpublished |
| Vietnamella sp. MT-2014 | 15,043 | KM244655 | [31] |
2.5. Positive Selection Analysis
The software EasyCodeML [65] was used to evaluate the selective pressure on the PCGs of Heptageniidae mitogenomes. Due to the significantly lower environment temperatures experienced by the Heptageniidae species from Ottawa, Canada, these were selected as the foreground branch to investigate the molecular evolution trends of mitochondrial PCGs under low-temperature stress. Both the branch model and the branch-site model were employed to explore whether positive selection occurred on specific branches and specific sites at the branch [66,67]. The branch models were performed under the one-ratio model (M0) presuming that ω was fixed over all of the tree or the two-ratio model presuming that ω in specific branches was different from the rest of the tree. Moreover, the branch-site models were combined with heterogeneous ω across sites and branches, which allows positive selection along specified branches (Model A) and can be compared against a null model (Model Anull) that allows neutral evolution and negative selection. Likelihood ratio tests (LRTs) and Bayes empirical Bayes (BEB) were used to assess these models and evaluate the posterior probability of positive selection sites, respectively. Additionally, information on the structure and function of the positively selected genes was acquired using UniProt [68], and 3D structures of the corresponding proteins were built by SWISS-MODEL Workspace [69].
3. Results
3.1. General Features of the Mitogenomes
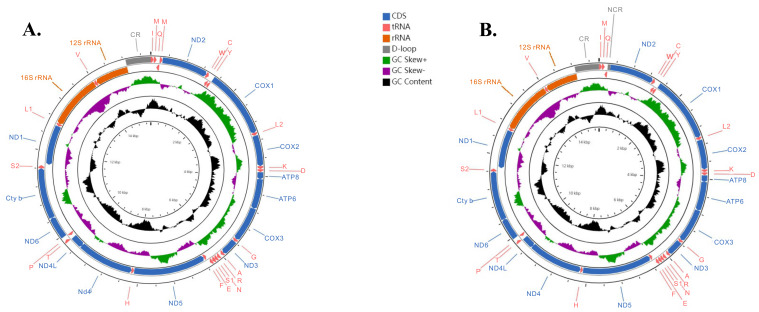
The 17 mitogenomes of Heptageniidae used in this study ranged in length from 15,319 bp in Maccaffertium mediopunctatum (McDunnough, 1926) [70] to 15,883 bp in Afronurus yixingensis (Wu and You, 1986) [71] (Tables S3 and S4, Supplementary Materials) and encoded 13 PCGs, two rRNA genes, 22 or 23 tRNA genes (containing an extra trnM gene in some species), and one CR (Figure 1). Most of the genes were encoded on the major strand, also called the J strand, whereas the minor strand (N strand) carried the remaining genes (four PCGs and eight tRNAs). The A + T content, AT-skew, and GC-skew of corresponding regions (mitogenomes, PCGs, and rRNAs) were separately calculated for each mayfly species and shared conserved characteristics with others (Table 3). The tRNAs of these mayflies all showed the classical cloverleaf secondary structures.
Figure 1.
Circular visualization and organization of the complete mitogenome. External genes on the circle are encoded by the positive strand (5′→3′) and internal genes are encoded by the negative strand (3′→5′). (A) Mitogenomes in the subfamily Ecdyonurinae (Afronurus and Leucrocuta) and Rhithrogeninae (Epeorus); (B) mitogenomes in the subfamily Heptageniinae (Maccaffertium, Stenacron, and Stenonema).
Table 3.
Base composition of 17 mayfly mitochondrial genomes, with a greater separation of each of the three Mitogenome-PGC-rRNA-CR groups and separate underlining to mark the three different groups.
| Species Name | Mitogenome-PGC-rRNA-CR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A + T (%) | AT-Skew | GC-Skew | ||||||||||
| Afronurus furcata | 64.5 | 64.4 | 64.9 | 56.9 | 0.005 | −0.198 | −0.024 | 0.23 | −0.215 | −0.004 | 0.339 | −0.16 |
| Afronurus rubromaculata | 64.5 | 64.6 | 65.0 | 65.9 | 0.009 | −0.193 | −0.025 | 0.23 | −0.218 | −0.010 | 0.327 | −0.11 |
| Afronurus sp. 07BF85 | 63.3 | 62.8 | 65.5 | 60.0 | −0.03 | −0.210 | 0.032 | 0.02 | −0.16 | −0.012 | 0.284 | 0.05 |
| Afronurus sp. 07BF86 | 64.0 | 64.4 | 65.7 | 54.1 | −0.006 | −0.191 | 0.02 | 0.13 | −0.202 | −0.017 | 0.310 | 0.02 |
| Afronurus sp. 07BF96 | 62.9 | 62.4 | 65.3 | / | −0.027 | −0.213 | 0.035 | / | −0.16 | −0.010 | 0.288 | / |
| Afronurus sp. YW01BF06 | 64.6 | 64.7 | 64.4 | / | 0.012 | −0.186 | 0.002 | / | −0.225 | −0.010 | 0.317 | / |
| Afronurus sp. LS53BF04 | 63.3 | 63.3 | 64.7 | 60.1 | 0.012 | −0.198 | −0.022 | 0.16 | −0.227 | −0.010 | 0.329 | −0.30 |
| Afronurus yixingensis | 65.0 | 65.1 | 66.0 | 63.2 | 0.003 | −0.203 | −0.002 | −0.01 | −0.218 | −0.004 | 0.305 | −0.21 |
| Epeorus dayongensis | 64.8 | 64.2 | 66.1 | 73.6 | 0 | −0.191 | 0.012 | 0.04 | −0.212 | −0.024 | 0.269 | 0.01 |
| Epeorus sp. LA03FY06 | 67.1 | 66.3 | 67.6 | 77.8 | −0.014 | −0.188 | 0.02 | 0.00 | −0.24 | −0.003 | 0.307 | −0.19 |
| Leucrocuta aphrodite | 65 | 65.0 | 65.1 | 65.4 | −0.001 | −0.187 | −0.001 | 0.02 | −0.186 | −0.001 | 0.284 | −0.18 |
| Maccaffertium mediopunctatum | 61.7 | 61.5 | 60.7 | 65.9 | −0.002 | −0.190 | 0.015 | 0.02 | −0.178 | −0.045 | 0.234 | −0.35 |
| Maccaffertium modestum | 61.3 | 61.1 | 60.7 | 64.0 | 0.004 | −0.191 | 0.025 | 0.01 | −0.177 | −0.038 | 0.231 | −0.33 |
| Maccaffertium vicarium | 62.3 | 62.3 | 61.8 | 65.4 | 0.005 | −0.169 | 0.013 | 0.00 | −0.172 | −0.046 | 0.253 | −0.25 |
| Stenacron interpunctatum | 59.7 | 59.5 | 58.8 | 61.7 | 0.021 | −0.184 | 0.007 | 0.05 | −0.19 | −0.057 | 0.266 | −0.24 |
| Stenonema femoratum | 62.1 | 61.9 | 61.1 | 66.4 | 0.005 | −0.190 | −0.011 | 0.01 | −0.166 | −0.039 | 0.234 | −0.30 |
| Heptageniidae sp. YW03BF02 | 64.2 | 64.1 | 64.3 | 68.5 | −0.006 | −0.191 | −0.011 | −0.09 | −0.183 | 0.002 | 0.27 | −0.19 |
All these mitochondrial PCGs used conventional invertebrate ATN as start codons, except for COX1 which started with CCG in seven species: Afronurus (A. furcata (Zhou and Zhen, 2003) [72], A. rubromaculata (You, Wu, Gui, and Hsu, 1981) [73], Afronurus sp. LS53BF04, Afronurus sp. YW01BF06, Afronurus sp. 07BF85, Afronurus sp. 07BF86, and Afronurus sp. 07BF96). ATP8 started with GTG in most mayflies except for M. mediopunctatum (McDunnough, 1926) [70], M. modestum (Banks, 1910) [13], M. vicarium (Walker, 1853) [74], Stenacron interpunctatum (Say, 1839) [75], Stenonema femoratum (Say 1823) [76], and Afronurus sp. YW01BF06. ND2 started with GTG except for Epeorus dayongensis (Gui and Zhang, 1992) [77], Epeorus sp. LA03FY06, and Afronurus sp. 07BF96. ND5 started with GTG except for Leucrocuta aphrodite (McDunnough, 1926) [70], and ND6 started with GTT in S. interpunctatum. Typical stop codons TAA and TAG were observed in the majority of PCGs, whereas incomplete stop codons T or TA were assigned in Cyt b (M. mediopunctatum, M. modestum, M. vicarium, and S. femoratum), COX2 (all species), ND4 (all species), and ND5 (all species). The codon number and RSCU in mitochondrial PCGs were conservative among these species (Table S5, Supplementary Materials).
The CRs of Heptageniidae mitogenomes ranged from 487 bp to 1037 bp, with the location between 12S rRNA and trnI. Almost all CRs of these mitogenomes showed the highest A + T content compared to other regions (PCGs, rRNA genes, and tRNA genes) except for A. furcata, A. yixingensis, Afronurus sp. 07BF85, Afronurus sp. 07BF86, and Afronurus sp. LS53BF04. The A + T contents of these CRs ranged from 54.1% in Afronurus sp. 07BF86 to 77.8% in Epeorus sp. LA03FY06. The AT-skew values of the CRs were a little positive except for A. yixingensis, Epeorus sp. LA03FY06, and Heptageniidae sp. YW03BF02, whereas the GC-skew was strongly negative except for Afronurus sp. 07BF85, Afronurus sp. 07BF86, and E. dayongensis. Additionally, tandem repeats were detected in the CRs of Afronurus sp. 07BF85, Afronurus sp. 07BF86, Afronurus sp. LS53BF04, A. furcata, A. rubromaculata, A. yixingensis, and Heptageniidae sp. YW03BF02 (Figure S1, Supplementary Materials).
3.2. Gene Arrangements and NCRs
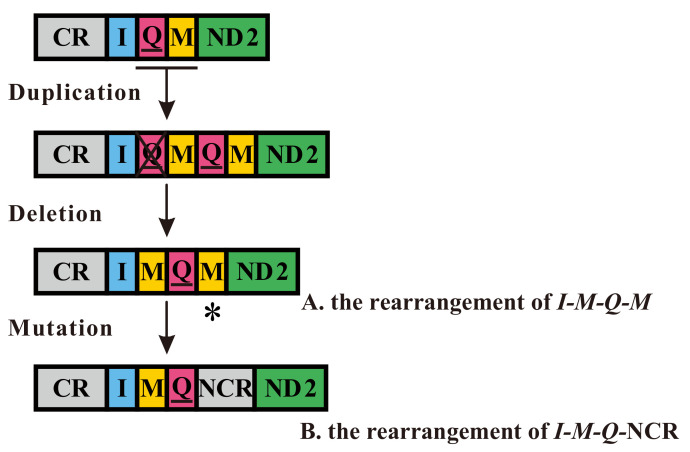
Two types of gene rearrangements occurred in the I-Q-M tRNA cluster and were found in all 17 freshly sequenced mitogenomes of Heptageniidae (Figure 2). The extra trnM was observed in the 11 mitogenomes of the subfamily Ecdyonurinae (Afronurus species and L. aphrodite) and Rhithrogeninae (Epeorus species). As for the location of two trnM copies, one was situated between trnI and trnQ, with another between trnQ and ND2. Thus, their tRNA cluster was shown as I-M-Q-M. The two copies of trnM genes showed high similarity (>70%) and had the same anti-codon (CAU) in nearly all species except for the second trnM (UAU) in L. aphrodite (Figure S2, Supplementary Materials). However, in mitogenomes of the subfamily Heptageniinae (Maccaffertium species, S. interpunctatum, and S. femoratum), a translocation of trnM was found and the trnM gene translocated into the position between trnI and trnQ. Furthermore, the NCR of 55–57 bp located between trnQ and ND2 was detected in these species but showed low similarity to adjacent genes. Hence, the gene order in the species of Heptageniinae was shown as I-M-Q-NCR, and this is the first report of this novel gene rearrangement (I-M-Q-NCR) among mayfly mitogenomes.
Figure 2.
Proposed mechanism of gene rearrangements in the seventeen Heptageniidae mitogenomes. Gene sizes are not drawn to scale. Genes encoded by the L-strand are underlined, whereas those without underline are encoded on the H-strand. Different colored boxes represent different genes. The remaining genes and gene orders that were identical to the ancestral insect are left out. Horizontal lines, asterisk symbols, and crossed-out symbols represent gene duplications, gene mutations, and gene deletions, respectively. (A) Rearrangement of I-M-Q-M; (B) rearrangement of I-M-Q-NCR.
The length, number, and distribution of the NCRs in these mitogenomes of Heptageniidae were relatively conservative. The number of NCRs in every mayfly species ranged from 7 to 12, whereas the length varied from 1 bp to 57 bp. Excluding the NCRs of short length (<15 bp) and the NCR located between trnQ and ND2 (mentioned above), the NCRs located between trnA and trnR were observed in all Heptageniidae mitogenomes with lengths ranging from 25 bp (M. vicarium) to 47 bp (E. herklotsi). Interestingly, the NCRs could be folded as stem-loop secondary structures (Figure S3, Supplementary Materials) and were highly similar (>70%) to CR according to a comparison among the mitochondrial genomic sequences of most species (Figure S4, Supplementary Materials). Notwithstanding, the similarity between the NCR and CR was not exactly high (<70%) or the similarity sequence was short (<20 bp) in A. furcata, A. rubromaculata, Afronurus sp. 07BF86, E. dayongensis, Epeorus sp. LA03FY06, M. mediopunctatum, M. modestum, M. vicarium, S. interpunctatum, and S. femoratum. We also observed NCRs located between trnS2 and ND1 in all mitogenomes of Heptageniidae, which were of 16 bp in length.
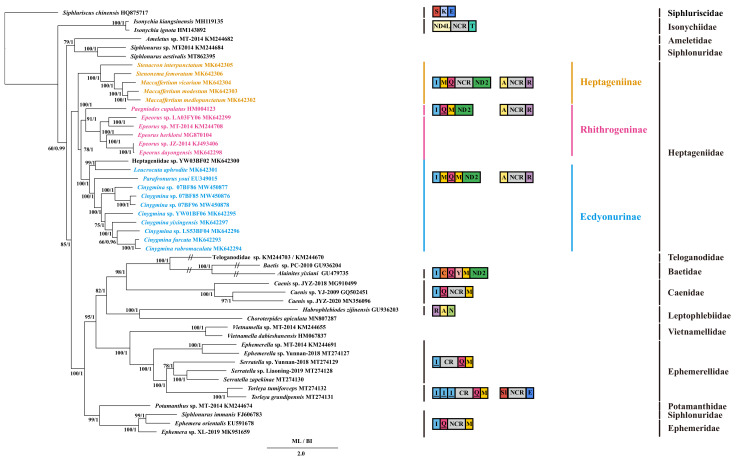
3.3. Phylogenetic Analyses
The BI and ML phylogenetic relationships showed identical topologies (Figure 3). However, long-branch attraction (LBA) has been observed in Baetidae (Baetis sp. PC-2010 and Alainites yixiani) and Teloganodidae sp. MT-2014; thus, their phylogenetic positions still remain uncertain. In general, the monophyly of most families was supported in these phylogenetic trees except for Ephemeridae and Siphlonuridae, but the availability of only one species in Ameletidae, Polymitarcyidae, and Teloganodidae restricted a discussion of their monophyly and phylogenetic relationships.
Figure 3.
Phylogenetic tree of the relationships among 49 species of Ephemeroptera according to the nucleotide dataset of the 13 mitochondrial PCGs. Siphluriscus chinensis was used as the outgroup. The numbers above branches specify bootstrap percentages from ML (left) and posterior probabilities as determined from BI (right). The GenBank accession numbers of all species are shown in the figure. Box images on the right show gene rearrangements and the location of the NCR for the various mayfly species. Genes encoded by the minority strand are underlined, and those without underline are encoded by the majority strand. Different colored boxes represent different genes. The remaining genes and gene orders that were identical to the ancestral insect are left out. Gene sizes are not drawn to scale. The asterisks (*) by Siphlonuridae on the far-right side denote the separation of these sequences.
Within Ephemeroptera, Isonychiidae was a sister group to the other families, according to the phylogenetic topologies. Then, Ameletidae (Ameletus sp. MT-2014) and one branch of Siphlonuridae (Siphlonurus aestivalis and Siphlonurus sp. MT-2014) were found to be a sister group. Heptageniidae was supported as a sister clade to the remaining Ephemeroptera (Baetidae, Caenidae, Ephemerellidae, Ephemeridae, Leptophlebiidae, Potamanthidae, Teloganodidae, and Vietnamellidae). Potamanthidae was the sister clade to (Ephemeridae + Siphlonurus immanis), whereas the remaining families formed another large clade. (Ephemerellidae + Vietnamellidae) was supported as a sister clade to (Leptophlebiidae + (Caenidae + (Baetidae + Teloganodidae))).
Concentrating on the phylogenetic relationship within Heptageniidae, the monophyly of three subfamilies (Ecdyonurinae, Heptageniinae, and Rhithrogeninae) and the genera Afronurus, Epeorus, and Maccaffertium was supported. The branch of Heptageniidae was divided into three clades as follows: (Heptageniinae + (Rhithrogeninae + Ecdyonurinae)). The first branch of Heptageniinae supported (S. interpunctatum + (S. femoratum + Maccaffertium species)). Then, Paegniodes cupulatus and Epeorus species formed the second branch of Rhithrogeninae. The third branch of Ecdyonurinae supported ((Heptageniidae sp. YW03BF02 + L. aphrodite) + (P. youi + Afronurus species)). Significantly, the phylogenetic relationships coincided with the gene order and the location of the NCRs. The lineage of Ephemerellidae was consistent with rearrangements of the trnI gene. The NCRs located between ND4L and trnT were found in the branch of Isonychiidae along with the NCRs located between trnQ and trnM in the branch of Caenidae. Within Heptageniidae, these branches corresponded to different gene arrangements: I-M-Q-NCR in Heptageniinae and I-M-Q-M in the remaining species of Rhithrogeninae and Ecdyonurinae, except for I-Q-M in P. cupulatus.
3.4. Positive Selection Analysis
According to the branch model, 3722 amino-acid sites were used to analyze selective pressure on the basis of the alignment of 13 PCGs in 22 species of Heptageniidae. The results were as follows: p < 0.001, ω0 = 0.02021, ω = 0.02123 < 1, illustrating that the foreground branch (the Heptageniinae species from Ottawa, Canada) was not subject to positive selection (Table S6, Supplementary Materials). On the contrary, we observed that 27 amino-acid sites were under positive selection (p < 0.001, BEB value > 0.95) in the analyses of the branch-site models, of which five amino-acid sites were under highly positive selection (BEB value > 0.99) (Table 4). The 27 positive selection sites corresponding to the mitochondrial PCGs were divided into eight genes, namely, COX1 (two sites), Cyt b (two sites), ND1 (two sites), ND2 (five sites), ND3 (one site), ND4 (two sites), ND5 (six sites), and ND6 (seven sites). Accordingly, the mitochondrial complex I was the main protein complex under selective pressure, including 23 positive selection sites. To determine the functional meaning of these positive selection sites, we explored the feature keys of eight positively selected PCGs from low-temperature branches. The majority of the positive selection sites located within or near to the functional domains of the proteins were encoded by these genes, 14 of which were situated in the protein transmembrane domain of the encoding genes, with an additional six positive selection sites situated in other domains of corresponding genes (Table 5, Figure S5, Supplementary Materials).
Table 4.
Positive selection analysis of mitochondrial protein-coding genes based on the branch-site model.
| Tree | Model | Ln L | Estimates of Parameters | Model Compared | 2ΔL | LRT p-Value | Positive Sites | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ML | Model A | −121,931.169602 | Site class | 0 | 1 | 2a | 2b | Model A vs. Model A null |
17.1487 | 0.00004434 | 682 Q 0.988 *, 749 S 0.979 *, 1340 L 0.977 *, 1636 V 0.954 *, 1827 E 0.989 *, 1843 A 0.983 *, 2118 P 0.995 **, 2123 F 0.983 *, 2167 S 0.979 *, 2288 T 0.983 *, 2311 T 0.991 **, 2398 A 0.970 *, 2613 S 0.962 *, 2619 M 0.986 *, 3001 L 0.971 *, 3005 S 0.964 *, 3155 S 0.969 *, 3313 S 0.984 *, 3444 S 0.989 *, 3466 H 0.978 *, 3557 L 0.976 *, 3566 I 0.996 **, 3582 C 0.985 *, 3664 E 0.996 **, 3665 Q 0.984 *, 3679 I 0.993 **, 3712 Q 0.981 * |
| Proportion | 0.9141 | 0.0377 | 0.0463 | 0.0019 | |||||||
| Background ω | 0.0137 | 1.0000 | 0.0137 | 1.0000 | |||||||
| Foreground ω | 0.0137 | 1.0000 | 2.7648 | 2.7648 | |||||||
| Model A Null | −121,939.505555 | / | / | ||||||||
Note: * and ** indicate BEB values > 0.95 and > 0.99, respectively.
Table 5.
The features and description of the positive selection sites detected in the mitochondrial PCGs of Heptageniidae species.
| Genes | Positive Selection Sites | Amino Acids | BEB Value | Feature Key * | Description | |
|---|---|---|---|---|---|---|
| Foreground | Background | |||||
| COX1 | 408 | N | Q/K | 0.988 * | Domain | COX1 |
| 475 | A | S | 0.979 * | Domain | COX1 | |
| Cyt b | 62 | T | L | 0.977 * | Domain | CYTB_NTER |
| 358 | T/I | V/T/I | 0.954 * | Transmembrane | Helical | |
| ND1 | 173 | Y | E/K/Q | 0.989 * | / | / |
| 189 | S | A/T | 0.983 * | Transmembrane | Helical | |
| ND2 | 148 | S/G | P/S | 0.995 ** | Transmembrane | Helical |
| 153 | T/S | F | 0.983 * | Transmembrane | Helical | |
| 197 | N | S/P/N | 0.979 * | Domain | Proton_antipo_M | |
| 318 | G/N | S/L/T/A | 0.983 * | / | / | |
| 341 | S | S/P/T/L | 0.991 ** | / | / | |
| ND3 | 85 | Q | A/N/S/T | 0.970 * | / | / |
| ND4 | 183 | S | G/S/K | 0.962 * | Transmembrane | Helical |
| 189 | G | M/L | 0.986 * | Transmembrane | Helical | |
| ND5 | 25 | T | L | 0.971 * | Transmembrane | Helical |
| 29 | L/A | S | 0.964 * | Transmembrane | Helical | |
| 179 | E/H | S/G/T | 0.969 * | Transmembrane | Helical | |
| 337 | S | S/N/I/T | 0.984 * | Domain | Proton_antipo_M | |
| 468 | L | S/V/I/A | 0.989 * | Transmembrane | Helical | |
| 490 | F | N/H/Q/G/S | 0.978 * | Domain | NADH5_C | |
| ND6 | 3 | T | L/F/M | 0.976 * | / | / |
| 12 | L | T/I | 0.996 ** | Transmembrane | Helical | |
| 28 | I/V | C/S/I/V | 0.985 * | Transmembrane | Helical | |
| 110 | S | E/D | 0.996 ** | / | / | |
| 111 | D | Q | 0.984 * | / | / | |
| 125 | P | N/T/I/G | 0.993 ** | Transmembrane | Helical | |
| 158 | N | Q/N | 0.981 * | Transmembrane | Helical | |
Note: * and ** indicate BEB values > 0.95 and > 0.99, respectively.
4. Discussion
4.1. Gene Arrangements and NCRs
The typical gene arrangement occurred in most mayfly mitogenomes, except for Siphluriscidae, Baetidae, Leptophlebiidae, Ephemerellidae, and Heptageniidae [32,33,34,35,36,37,38,39,40,41]. Gene rearrangements in these groups were mainly concentrated in the regions of CR-I-Q-M-ND2 and A-R-N-S-E-F. In our study, the gene rearrangements in the mitogenomes of Heptageniidae were divided into two types: a gene arrangement of I-M-Q-M in the subfamily Ecdyonurinae (Afronurus, Parafronurus, and Leucrocuta) and Rhithrogeninae (Epeorus), and a novel gene arrangement of I-M-Q-NCR in the subfamily Heptageniinae (Maccaffertium, Stenacron, and Stenonema). Moreover, two copies of trnM genes had the same anti-codon (CAU) in almost all species excluding the second trnM (UAU) in L. aphrodite. The codon AUA is translated as Met in the invertebrate mitochondrial genetic code, like the normal codon AUG, as reported in the fruit fly Drosophila melanogaster [78,79]. Therefore, the second trnM with anti-codon (UAU) was considered to be functional in L. aphrodite. Furthermore, similar gene rearrangements occurring in the region of CR-I-Q-M-ND2 were also reported in other orders of insects, e.g., M-I-Q tRNA cluster in Lepidoptera (Manduca sexta) [80], Q-I-M in Hemiptera (Neuroctenus parus) [81], and I-I-I-I-I-Q-M in Mantodea (Schizocephala bicornis) [31]. Consequently, the region of CR-I-Q-M-ND2 is regarded as a hotspot for gene rearrangements in insects.
The tandem duplication–random loss (TDRL) model [82,83] was proposed and used to explain similar gene rearrangements in other insects [81,84]. Therefore, the TDRL model can be reasonably used to explain the gene rearrangements of Heptageniidae (Figure 2). The region of CR-I-Q-M-ND2 was presumed to be the original gene arrangement. The mechanisms of gene rearrangement of I-M-Q-M were assumed to be as follows: a tandem duplication of Q-M happened, followed by random loss of the first trnQ, resulting in the gene order I-M-Q-M, as reported in Zhang et al. [33]. As for the gene rearrangement of I-M-Q-NCR, we propose that the tandem duplication of Q-M happened, followed by the random loss of the first trnQ and the mutation of the second trnM. Consequently, the translocation of trnM and the NCR located between trnQ and ND2 was observed in the subfamily Heptageniinae (Maccaffertium, Stenacron, and Stenonema). Notwithstanding, the mitogenome of P. cupulatus showed the typical insect gene order and was different from other mitogenomes of Heptageniidae. The conservative gene order of P. cupulatus was proposed to occur as follows: the extra trnM between trnI and trnQ was lost from the ancestral I-M-Q-M type and, thus, formed the I-Q-M gene arrangement. Therefore, the gene arrangements among genera of Heptageniidae need further study.
Generally, insect mitogenomes are of high compaction with rare and short NCRs except for the CR [17]. The great majority of mayfly mitogenomes featured short NCR lengths. However, NCRs of 25–47 bp were located between trnA and trnR and observed in all mitogenomes of Heptageniidae [33,34,35]. This feature is rarely observed in other mayfly mitogenomes. Thus, this NCR located between trnA and trnR may be a synapomorphy for Heptageniidae. The NCRs can form stem-loop secondary structures (Figure S3, Supplementary Materials), which may contribute to the progress of replication slippage and then an increase in duplicate copies [85]. Furthermore, the NCR was proposed as an alternative replication origin for mtDNA [86,87]. As for the occurrence of the NCR, it was inferred to derive mainly from the corresponding CR because of the high similarity between the two (>70%), such as the complete sequence (37 bp) in L. aphrodite (similarity 94.59%) and the partial sequence (23 bp) in P. youi (similarity 100%) (Figure S4, Supplementary Materials). Considering the long distance between the NCR and CR, the recombination model may be more suitable to explain NCR production [88,89]. The creation of the NCR was presumed to occur as follows: a fragment containing the CR was cleaved out and then inserted into a location between trnA and trnR. Although there is a low similarity between the NCR and CR (<70%) or a short similar sequence (<20 bp) in several species (as mentioned in the results), the NCR was proposed to have evolved independently under relaxed selective pressure instead of evolving in concert with the CR [89]. In addition, the short NCR located between trnS2 and ND1 was detected in all Heptageniidae mitogenomes, which has also been reported in Ephemeroptera and other insects [23,32,33,34,49]. According to the alignments of these NCRs of all Heptageniidae species (Figure S6), a highly conserved motif of 16 bp (TACTTAAAAARKTCAR) may be the binding site of the transcription termination factor (DmTTF) [90].
4.2. Phylogenetic Analyses
Higher-level phylogenetic relationships within Ephemeroptera have not been generally accepted [8,9,10,11,12]. In our results, the BI and ML phylogenetic analyses shared congruent topologies (Figure 3). S. chinensis, the only species of Siphluriscidae, was deemed as the basal group of Ephemeroptera from the study of Ogden et al. [12] and Zhang et al. [33]. The next were Isonychiidae, Ameletidae, and one species of Siphlonuridae, as indicated by our results. The phylogenetic position of Isonychiidae was convincingly supported by Ogden et al. [12] as the primitive clade except for Siphluriscidae and Baetidae from topologies. Then, Heptageniidae was supported as a sister clade to the remaining Ephemeroptera by our results, contrary to the topologies constructed by Kluge [10,11] and McCafferty [8,9], as well as Ogden et al. [15], which suggested that Heptageniidae was sister to Isonychidae based on morphological characteristics and nuclear data, respectively. According to a comparison of our results and other studies, the phylogenetic position of Heptageniidae is still challenging to determine.
As for Heptageniidae, the monophyly of three subfamilies Ecdyonurinae, Heptageniinae, and Rhithrogeninae was supported, consistent with the research of Wang and McCafferty [13] and Webb and McCafferty [6]. Nevertheless, the internal phylogenetic classification within Heptageniidae in our study differed from Wang and McCafferty [13]. In our study, the phylogenetic relationship within Heptageniidae was shown as (Heptageniinae + (Ecdyonurinae + Rhithrogeninae)), opposite to the (Ecdyonurinae + (Heptageniinae + Rhithrogeninae)) presented in Wang and McCafferty [13] and (Rhithrogenidae + (Ecdyonurinae + Heptageniinae)) in Ogden et al. [15]. When the phylogenetic classification was combined with gene rearrangements and NCRs, there was a specific correlation. The gene rearrangement of I-M-Q-NCR was concentrated in Heptageniinae, and the gene arrangement of I-M-Q-M was observed in the remaining species of Rhithrogeninae and Ecdyonurinae except for I-Q-M in P. cupulatus. These results illustrated that Ecdyonurinae and Rhithrogeninae were closely related, and the two formed a sister group to Heptageniinae. Phylogenetic relationships highly congruent with gene rearrangements and NCR locations were also reported in other insects [91,92], which suggests that synapomorphic gene rearrangements and NCRs have formed continuously during evolution and could provide effective phylogenetic information. However, the phylogenetic relationship of the three subfamilies within Heptageniidae is also controversial due to the lack of other evidence in our study. In addition, concerning the taxonomy of Stenonema, Stenacron and Maccaffertium, compared to the research of Zembrzuski and Anderson [16], it is a pity that we could not draw a valid conclusion on the basis of our results because of the lack of sequences for these genera. Further morphological and molecular data are required to demonstrate a more exact phylogenetic relationship among Ephemeroptera.
In fact, the gene arrangement of I-Q-M was found only in P. cupulatus of Heptageniidae. This was confusing as to whether such a gene arrangement was specific to the genus Paegniodes or formed during the random mutation progress. More mayfly mitogenomes are expected to be sequenced, which will help to explore the types of gene arrangements and clear phylogenetic classifications within Heptageniidae.
4.3. Positive Selection Analyses
Adaptive evolution of mitochondrial genes under environmental pressure is supported by several studies [43,44,45]. Environmental temperature significantly influences energy requirements and metabolic adaptation, which is essential to mayflies as aquatic insects [46,47]. Multiple subunits of the mitochondrial complexes associated with oxidative phosphorylation are encoded by mitochondrial genes, with the exception of complex II [93]. In this way, positive selection of mitochondrial genes was proposed to be related to temperature and adaptation to the energy demands of mayflies.
Analysis of the branch model showed that there was no positive selection on the foreground branch. It was proposed that information indicating positive selection was possibly covered by continuous neutral evolution or negative selection at most sites in the gene sequence [94]. According to the branch-site model, 27 positive selection sites were found. It is worth noting that 23 of the positive selection sites were concentrated on the coding sequence of mitochondrial complex I. As the first large protein complex of the respiratory chain, complex I provides the proton power for ATP synthesis during electron transfer from NADH to ubiquinone via the transmembrane proton pump [95,96,97]. Therefore, complex I is essential for the energy metabolism of cells and drives more than one-third of the total energy production in the mitochondrion [87]. ND1–ND6 subunits are regarded as the minimal assembly of complex I and form the core of the transmembrane region [98], with the ND2, ND4, and ND5 genes proposed to be main candidates to harbor the proton pump [99]. The importance of complex I and its subunits can explain the reason for more positive selection sites in complex I than in other complexes. In addition, several positive selection sites were also observed in the subunits (Cyt b and COX1) of complex III and complex IV. Cyt b is the main transmembrane subunit of complex III and exerts a crucial function in ATP production [100]. Furthermore, complex IV has regulatory effects in the electron transport chain, and its subunit COX1 starts the assembly process of complex IV [101]. Moreover, the positive selection sites in the eight PCGs were located in or close to the functional domains according to the structural analysis (Table 5). Consequently, the adaptive changes in amino acids at these positive selection sites, especially in the functional regions, were proposed to affect protein stability or even function [102,103]. On the whole, mitochondrial PCGs are related to energy metabolism and can experience positive selection to cope with energy needs under low-temperature stress.
5. Conclusions
Fifteen complete mitogenomes and two nearly complete mitogenomes of Heptageniidae were successfully determined. Gene rearrangements in these mitogenomes of Heptageniidae were divided into two types: one with the commonly reported gene order of I-M-Q-M in the subfamily Ecdyonurinae (Afronurus, Parafronurus, and Leucrocuta) and Rhithrogeninae (Epeorus), and the other with a novel gene order of I-M-Q-NCR in Heptageniinae (Maccaffertium, Stenacron, and Stenonema). These gene rearrangements were explained by the tandem duplication–random loss (TDRL) model. In addition, the NCRs located between trnA and trnR were found in all Heptageniidae species and inferred to be a synapomorphy for Heptageniidae. The phylogenetic relationships within Ephemeroptera were highly congruent with the gene rearrangements and the location of NCRs, supporting the monophyly of Heptageniidae and its internal phylogenetic relationship (Heptageniinae + (Ecdyonurinae + Rhithrogeninae)). The selection pressure analyses indicated that mitochondrial PCGs of mayflies underwent positive selection to cope with potential energy requirements under low-temperature stress.
Acknowledgments
We would like to thank Huan Zhang for her help in the experiments.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/insects12070656/s1: Figure S1. Organization of the repeat regions in the CRs of Heptageniidae species; Figure S2. Alignment between two copies of trnM sequences in Heptageniidae species; Figure S3. The possible secondary structure of the NCRs located between trnA and trnR; Figure S4. Alignment between CRs and the NCRs located between trnA and trnR in Heptageniidae species; Figure S5. Distribution of mutations in the three-dimensional structure of COX1, Cyt b, ND1, ND2, ND3, ND4, ND5, and ND6 genes for the Heptageniidae species from Ottawa, Canada; Figure S6. Alignment of the NCRs located between trnS2 and ND1 in Heptageniidae species; Table S1. Specific primers used to amplify the Heptageniidae mitogenomes; Table S2. The partition schemes and best-fit models selected; Table S3. Location of features in the mitogenomes of the subfamily Ecdyonurinae (Afronurus, Parafronurus, and Leucrocuta) and Rhithrogeninae (Epeorus); Table S4. Location of features in the mitogenomes of the subfamily Heptageniinae (Maccaffertium, Stenacron, and Stenonema); Table S5. The codon number and relative synonymous codon usage in mitochondrial PCGs; Table S6. Positive selection analysis of mitochondrial PCGs based on the branch model.
Author Contributions
Conceptualization, J.-Y.Z., D.-N.Y., X.-D.X. and J.-Y.G.; methodology, J.-Y.Z., D.-N.Y., X.-D.X. and Y.-Y.C.; statistical analysis, J.-Y.Z., J.-Y.G., Z.-Y.Z. and Y.-Y.C.; investigation, D.-N.Y. and Y.-R.C.; data curation, J.-Y.G., X.-D.X., Y.-Y.C., and K.B.S.; writing—original draft preparation, Z.-Y.Z., J.-Y.G., and K.B.S.; writing—review and editing, J.-Y.Z., D.-N.Y., X.-D.X., J.-Y.G., Y.-Y.C. and K.B.S.; maps and graphics, Y.-R.C.; project administration, J.-Y.Z., D.-N.Y. and K.B.S.; funding acquisition, J.-Y.Z. and D.-N.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province (LY18C040004), the College Students’ Innovation and Entrepreneurship Project of China (201910345030 and 202010345026), and the College Students’ Innovation and Entrepreneurship Project of Zhejiang Province (202010345R128). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data supporting the findings of this study are openly available in National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov), accession numbers were MK642293-MK642306 and MW450876-MW450878.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barber-James H.M., Gattolliat J.-L., Sartori M., Hubbard M.D. Global diversity of mayflies (Ephemeroptera, Insecta) in freshwater. Hydrobiologia. 2008;595:339–350. doi: 10.1007/s10750-007-9028-y. [DOI] [Google Scholar]
- 2.Bauernfeind E., Soldan T. The Mayflies of Europe (Ephemeroptera) Apollo Books; Ollerup, Denmark: 2012. [Google Scholar]
- 3.Jacobus L.M., Macadam C.R., Sartori M. Mayflies (Ephemeroptera) and Their Contributions to Ecosystem Services. Insects. 2019;10:170. doi: 10.3390/insects10060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas J.A., Trueman J.W.H., Rambaut A., Welch J.J. Relaxed phylogenetics and the Palaeoptera problem: Resolving deep ancestral splits in the insect phylogeny. Syst. Biol. 2013;62:285–297. doi: 10.1093/sysbio/sys093. [DOI] [PubMed] [Google Scholar]
- 5.Kukalová-Peck J. Fossil history and the evolution of Hexapod structures. In: Naumann I.D.C., Came P.B., Lawrence J.F., Nielsen E.S., Spradberry J.P., Taylor R.W., Whitten M.J., Littlejohn M.J., editors. The Insects of Australia: A Textbook for Students and Research Workers. CSIRO and Melbourne University; Melbourne, Australia: 1991. pp. 141–179. [Google Scholar]
- 6.Webb J.M., McCafferty W.P. Heptageniidae of the world. Part II: Key to the genera. Can. J. Arthropod. Identif. 2008;7:1–55. [Google Scholar]
- 7.Edmunds G.F. Biogeography and evolution of Ephemeroptera. Annu. Rev. Entomol. 1972;17:21–42. doi: 10.1146/annurev.en.17.010172.000321. [DOI] [Google Scholar]
- 8.McCafferty W.P. Toward a phylogenetic classification of the Ephemeroptera (Insecta): A commentary on systematics. Ann. Entomol. Soc. Am. 1991;84:343–360. doi: 10.1093/aesa/84.4.343. [DOI] [Google Scholar]
- 9.McCafferty W.P. Ephemeroptera. In: Poole R.W., Gentili P., editors. Nomina Insecta Nearctica, a Checklist of the Insects of North America. Volume 4. Entomological Information Services; Rockville, MD, USA: 1997. pp. 89–117. [Google Scholar]
- 10.Kluge N. The Phylogenetic System of Ephemeroptera. Kluwer Academic; Dordrecht, The Netherlands: 2004. [Google Scholar]
- 11.Kluge N.J. Phylogeny and higher classification of Ephemeroptera. Zoosyst. Ross. 1998;72:255–269. [Google Scholar]
- 12.Ogden T.H., Gattolliat J.L., Sartori M., Staniczek A.H., Soldán T., Whiting M.F. Towards a new paradigm in mayfly phylogeny (Ephemeroptera): Combined analysis of morphological and molecular data. Syst. Entomol. 2009;34:616–634. doi: 10.1111/j.1365-3113.2009.00488.x. [DOI] [Google Scholar]
- 13.Wang T.Q., McCafferty W.P. Heptageniidae (Ephemeroptera) of the world. Part I: Phylogenetic higher classification. Trans. Am. Entomol. Soc. 2004;130:11–45. [Google Scholar]
- 14.Edmunds G.F. The principles applied in determining the hierarchic level of the higher categories of Ephemeroptera. Syst. Zool. 1962;11:22–31. doi: 10.2307/2411446. [DOI] [Google Scholar]
- 15.Ogden T.H., Breinholt J.W., Bybee S.M., Miller D.B., Sartori M., Shiozawa D., Whiting M.F. Mayfly Phylogenomics: Initial Evaluation of Anchored Hybrid Enrichment Data for the order Ephemeroptera. Zoosymposia. 2019;16:167–181. [Google Scholar]
- 16.Zembrzuski D.C., Anderson F.E. Clarifying the phylogenetic relationships and taxonomy of Stenonema, Stenacron and Maccaffertium, three common eastern North American mayfly genera. Mol. Phylogenet. Evol. 2018;128:212–220. doi: 10.1016/j.ympev.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L.P., Cai Y.Y., Yu D.N., Storey K.B., Zhang J.Y. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae) PeerJ. 2018;6:e4595. doi: 10.7717/peerj.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boore J.L. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 2006;21:439–446. doi: 10.1016/j.tree.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Cheng X.F., Zhang L.P., Yu D.N., Storey K.B., Zhang J.Y. The complete mitochondrial genomes of four cockroaches (Insecta: Blattodea) and phylogenetic analyses within cockroaches. Gene. 2016;586:115–122. doi: 10.1016/j.gene.2016.03.057. [DOI] [PubMed] [Google Scholar]
- 22.Ma Y., He K., Yu P., Yu D., Cheng X., Zhang J. The complete mitochondrial genomes of three bristletails (Insecta: Archaeognatha): The paraphyly of Machilidae and insights into archaeognathan phylogeny. PLoS ONE. 2015;10:e0117669. doi: 10.1371/journal.pone.0117669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Dai X.Y., Xu X.D., Zhang Z.Y., Yu D.N., Storey K.B., Zhang J.Y. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ. 2019;7:e7633. doi: 10.7717/peerj.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron S.L., Dowton M., Castro L.R., Ruberu K., Whiting M.F., Austin A.D., Diement K., Stevens J. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome. 2008;51:800–808. doi: 10.1139/G08-066. [DOI] [PubMed] [Google Scholar]
- 25.Jiang P., Li H., Song F., Cai Y., Wang J.Y., Liu J.P., Cai W.Z. Duplication and remolding of tRNA genes in the mitochondrial genome of Reduvius tenebrosus (Hemiptera: Reduviidae) Int. J. Mol. Sci. 2016;17:951. doi: 10.3390/ijms17060951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L.P., Cai Y.Y., Yu D.N., Storey K.B., Zhang J.Y. The complete mitochondrial genome of Psychomantis borneensis (Mantodea: Hymenopodidae) Mitochondrial DNA B. 2018;3:42–43. doi: 10.1080/23802359.2017.1419094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L.P., Ma Y., Yu D.N., Storey K.B., Zhang J.Y. The mitochondrial genomes of Statilia maculata and S. nemoralis (Mantidae: Mantinae) with different duplications of trnR genes. Int. J. Biol. Macromol. 2019;121:839–845. doi: 10.1016/j.ijbiomac.2018.10.038. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L.P., Yu D.N., Storey K.B., Cheng H.Y., Zhang J.Y. Data for praying mantis mitochondrial genomes and phylogenetic constructions within Mantodea. Data Brief. 2018;21:1277–1285. doi: 10.1016/j.dib.2018.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin Z.Z., Liu Y., Zhang D.Z., Wang Z.F., Tang B.P., Zhang H.B., Zhou C.L., Chai X.Y., Liu Q.N. Comparative mitochondrial genome analysis of Spilarctia subcarnea and other noctuid insects. Int. J. Biol. Macromol. 2018;107:121–128. doi: 10.1016/j.ijbiomac.2017.08.153. [DOI] [PubMed] [Google Scholar]
- 30.Dickey A.M., Kumar V., Morgan J.K., Jara-Cavieres A., Shatters R.G., Jr., McKenzie C.L., Osborne L.S. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): Extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genom. 2015;16:439. doi: 10.1186/s12864-015-1672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L.P., Yu D.N., Storey K.B., Cheng H.Y., Zhang J.Y. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int. J. Biol. Macromol. 2018;111:787–795. doi: 10.1016/j.ijbiomac.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Xu X.D., Jia Y.Y., Cao S.S., Zhang Z.Y., Storey K.B., Yu D.N., Zhang J.Y. Six complete mitochondrial genomes of mayflies from three genera of Ephemerellidae (Insecta: Ephemeroptera) with inversion and translocation of trnI rearrangement and their phylogenetic relationships. PeerJ. 2020;8:e9740. doi: 10.7717/peerj.9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J.Y., Zhou C.F., Gai Y.H., Song D.X., Zhou K.Y. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene. 2008;424:18–24. doi: 10.1016/j.gene.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 34.Gao X.Y., Zhang S.S., Zhang L.P., Yu D.N., Zhang J.Y., Cheng H.Y. The complete mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) and its phylogeny. Mitochondrial DNA B. 2018;3:303–304. doi: 10.1080/23802359.2018.1445482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang M., Tan M.H., Meng G.L., Yang S.Z., Su X., Liu S.L., Song W.H., Li Y.Y., Wu Q., Zhang A.B. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014;42:e166. doi: 10.1093/nar/gku917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D., Qin J.C., Zhou C.F. The phylogeny of Ephemeroptera in Pterygota revealed by the mitochondrial genome of Siphluriscus chinensis (Hexapoda: Insecta) Gene. 2014;545:132–140. doi: 10.1016/j.gene.2014.04.059. [DOI] [PubMed] [Google Scholar]
- 37.Cai Y.Y., Gao Y.J., Zhang L.P., Yu D.N., Storey K.B., Zhang J.Y. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) and the phylogeny of Ephemeroptera in Pterygota. Mitochondrial DNA B. 2018;3:577–579. doi: 10.1080/23802359.2018.1467239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye Q.M., Zhang S.S., Cai Y.Y., Storey K.B., Yu D.N., Zhang J.Y. The complete mitochondrial genome of Isonychia kiangsinensis (Ephemeroptera: Isonychiidae) Mitochondrial DNA B. 2018;3:541–542. doi: 10.1080/23802359.2018.1467233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song N., Li X.X., Yin X.M., Li X.H., Yin J., Pan P.L. The mitochondrial genomes of palaeopteran insects and insights into the early insect relationships. Sci. Rep. 2019;9:17765. doi: 10.1038/s41598-019-54391-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao S.S., Xu X.D., Jia Y.Y., Guan J.Y., Storey K.B., Yu D.N., Zhang J.Y. The complete mitochondrial genome of Choroterpides apiculata (Ephemeroptera: Leptophlebiidae) and its phylogenetic relationships. Mitochondrial DNA B. 2020;5:1159–1160. doi: 10.1080/23802359.2020.1730270. [DOI] [Google Scholar]
- 41.Xu X.D., Jia Y.Y., Dai X.Y., Ma J.L., Storey K.B., Zhang J.Y., Yu D.N. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) from Fujian and the phylogeny of Caenidae within Ephemeroptera. Mitochondrial DNA B. 2020;5:192–193. doi: 10.1080/23802359.2019.1698986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballard J.W.O., Kreitman M. Is mitochondrial DNA a strictly neutral marker? Trends Ecol. Evol. 1995;10:485–488. doi: 10.1016/S0169-5347(00)89195-8. [DOI] [PubMed] [Google Scholar]
- 43.Cai Y.T., Li Q., Zhang J.Y., Storey K.B., Yu D.N. Characterization of the mitochondrial genomes of two toads, Anaxyrus americanus (Anura: Bufonidae) and Bufotes pewzowi (Anura: Bufonidae), with phylogenetic and selection pressure analyses. PeerJ. 2020;8:e8901. doi: 10.7717/peerj.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y.X., Xu S.X., Xu J.X., Guo Y., Yang G. Adaptive evolution of mitochondrial energy metabolism genes associated with increased energy demand in flying insects. PLoS ONE. 2014;9:e99120. doi: 10.1371/journal.pone.0099120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X.D., Jiang G.F., Yan L.Y., Li R., Mu Y., Deng W.A. Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front. Genet. 2018;9:605. doi: 10.3389/fgene.2018.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bottová K., Derka T., Svitok M. Population dynamics of mayflies in a constant temperature spring stream in West Carpathians. Limnologica. 2013;43:469–474. doi: 10.1016/j.limno.2013.03.002. [DOI] [Google Scholar]
- 47.Scherr M.A., Wooster D.E., Rao S. Effects of temperature on growth rate and behavior of Epeorus albertae (Ephemeroptera: Heptageniidae) nymphs. Environ. Entomol. 2010;39:2017–2024. doi: 10.1603/EN09247. [DOI] [PubMed] [Google Scholar]
- 48.Zhai Y.Y., Huang G., Wang X.Q., Zhou X., Lu C., Li Z. Future projections of temperature changes in Ottawa, Canada through stepwise clustered downscaling of multiple GCMs under RCPs. Clim. Dyn. 2019;52:3455–3470. doi: 10.1007/s00382-018-4340-y. [DOI] [Google Scholar]
- 49.Gao X.Y., Cai Y.Y., Yu D.N., Storey K.B., Zhang J.Y. Characteristics of the complete mitochondrial genome of Suhpalacsa longialata (Neuroptera, Ascalaphidae) and its phylogenetic implications. PeerJ. 2018;6:e5914. doi: 10.7717/peerj.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon C., Buckley T.R., Frati F., Stewart J.B., Beckenbach A.T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006;37:545–579. doi: 10.1146/annurev.ecolsys.37.091305.110018. [DOI] [Google Scholar]
- 51.Simon C., Frati F., Beckenbach A., Crespi B., Liu H., Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- 52.Lalitha S. Primer premier 5. Biotech. Softw. Internet Rep. 2000;1:270–272. doi: 10.1089/152791600459894. [DOI] [Google Scholar]
- 53.Burland T.G. DNASTAR’s Lasergene sequence analysis software. In: Misener S., Krawetz S.A., editors. Bioinformatics Methods and Protocols. Humana Press; Totowa, NJ, USA: 1999. pp. 71–91. [DOI] [PubMed] [Google Scholar]
- 54.Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 55.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perna N.T., Kocher T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 58.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reuter J.S., Mathews D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinf. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 62.Lanfear R., Calcott B., Ho S.Y., Guindon S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 63.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ronquist F., Teslenko M., Mark P.V.D., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao F.L., Chen C.J., Arab D.A., Du Z.G., He Y.H., Ho S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019;9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 67.Zhang J.Z., Nielsen R., Yang Z.H. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 68.The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDunnough J. Notes on North American Ephemeroptera with descriptions of a new species. Can. Entomol. 1926;58:184–196. doi: 10.4039/Ent58184-8. [DOI] [Google Scholar]
- 71.Wu T., You D.S. A new species of the genus Cinygmina from China(Ephemeroptera: Ecdyoneuridae) Acta Zootaxon. Sin. 1986;11:280–282. [Google Scholar]
- 72.Zhou C.F., Zheng L.Y. The genus Cinygmina(Ephemeroptera:Heptageniidae) in China, with a description of a new species. Acta Entomol. Sin. 2003;46:755–760. [Google Scholar]
- 73.You D., Wu T., Gui H., Hsu Y. Two new species and diagnostic characters of genus Cinygmina (Ephemeroptera: Ecdyonuridae) J. Nanjing Norm. Univ. (Nat. Sci.) 1981;3:26–30. [Google Scholar]
- 74.Walker F. Catalogue of the Specimens of Neuropterous Insects in the Collection of the British Museum. British Museum; London, UK: 1853. Part III.—(Termitidae-Ephemeridae) pp. 477–585. [Google Scholar]
- 75.Say T. Descriptions of new North American neuropterous insects and observation on some already described. J. Acad. Nat. Sci. Phila. 1839;8:9–46. [Google Scholar]
- 76.Say T. Descriptions of insects belonging to the order Neuroptera Linne and Latreille, collected by the expedition under the command of Major Long. West. Q. Rep. 1823;2:160–165. [Google Scholar]
- 77.Gui H., Zhang J. A new species of genus Epeorus Eaton (Ephemeroptera: Heptageniidae) Acta Zootaxon. Sin. 1992;17:61–63. [Google Scholar]
- 78.Tomita K., Ueda T., Ishiwa S., Crain P.F., McCloskey J.A., Watanabe K. Codon reading patterns in Drosophila melanogaster mitochondria based on their tRNA sequences: A unique wobble rule in animal mitochondria. Nucleic Acids Res. 1999;27:4291–4297. doi: 10.1093/nar/27.21.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe K., Yokobori S.-I. tRNA modification and genetic code variations in animal mitochondria. J. Nucleic Acids. 2011;2011:1–12. doi: 10.4061/2011/623095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cameron S.L., Whiting M.F. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 2008;408:112–123. doi: 10.1016/j.gene.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 81.Li H., Liu H., Shi A., Stys P., Zhou X., Cai W. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae) PLoS ONE. 2012;7:e29419. doi: 10.1371/journal.pone.0029419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arndt A., Smith M.J. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol. Biol. Evol. 1998;15:1009–1016. doi: 10.1093/oxfordjournals.molbev.a025999. [DOI] [PubMed] [Google Scholar]
- 83.Excoffier L. Evolution of human mitochondrial DNA: Evidence for departure from a pure neutral model of populations at equilibrium. J. Mol. Evol. 1990;30:125–139. doi: 10.1007/BF02099939. [DOI] [PubMed] [Google Scholar]
- 84.Tan M.H., Zhang R., Hardman C., Zhou X. Mitochondrial genome of Hylaeus dilatatus (Hymenoptera: Colletidae) Mitochondrial DNA A. 2016;27:3975–3976. doi: 10.3109/19401736.2014.989511. [DOI] [PubMed] [Google Scholar]
- 85.Zhang B., Ma C., Edwards O., Fuller S., Kang L. The mitochondrial genome of the Russian wheat aphid Diuraphis noxia: Large repetitive sequences between trnE and trnF in aphids. Gene. 2014;533:253–260. doi: 10.1016/j.gene.2013.09.064. [DOI] [PubMed] [Google Scholar]
- 86.Crozier R.H., Crozier Y.C. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dotson E.M., Beard C.B. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata. Insect Mol. Biol. 2001;10:205–215. doi: 10.1046/j.1365-2583.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- 88.Lunt D.H., Hyman B.C. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- 89.Zhang H.L., Ye F. Comparative mitogenomic analyses of praying mantises (Dictyoptera, Mantodea): Origin and evolution of unusual intergenic gaps. Int. J. Biol. Sci. 2017;13:367–382. doi: 10.7150/ijbs.17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roberti M., Polosa P.L., Bruni F., Musicco C., Gadaleta M.N., Cantatore P. DmTTF, a novel mitochondrial transcription termination factor that recognises two sequences of Drosophila melanogaster mitochondrial DNA. Nucleic Acids Res. 2003;31:1597–1604. doi: 10.1093/nar/gkg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng B.Y., Cao L.J., Tang P., van Achterberg K., Hoffmann A.A., Chen H.Y., Chen X.X., Wei S.J. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea) Mol. Phylogenet. Evol. 2018;124:1–9. doi: 10.1016/j.ympev.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 92.Tyagi K., Chakraborty R., Cameron S.L., Sweet A.D., Chandra K., Kumar V. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta) Sci. Rep. 2020;10:695. doi: 10.1038/s41598-020-57705-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scheffler I.E. Molecular genetics of succinate:quinone oxidoreductase in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 1998;60:267–315. doi: 10.1016/s0079-6603(08)60895-8. [DOI] [PubMed] [Google Scholar]
- 94.Shen Y.J., Kou Q., Zhong Z.X., Li X.Z., He L.S., He S.P., Gan X.N. The first complete mitogenome of the South China deep-sea giant isopod Bathynomus sp. (Crustacea: Isopoda: Cirolanidae) allows insights into the early mitogenomic evolution of isopods. Ecol. Evol. 2017;7:1869–1881. doi: 10.1002/ece3.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu. Rev. Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 96.Kühlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015;13:89. doi: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wirth C., Brandt U., Hunte C., Zickermann V. Structure and function of mitochondrial complex I. Biochim. Biophys. Acta. 2016;1857:902–914. doi: 10.1016/j.bbabio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 98.Bridges H.R., Birrell J.A., Hirst J. The mitochondrial-encoded subunits of respiratory complex I (NADH:ubiquinone oxidoreductase): Identifying residues important in mechanism and disease. Biochem. Soc. Trans. 2011;39:799–806. doi: 10.1042/BST0390799. [DOI] [PubMed] [Google Scholar]
- 99.Mathiesen C., Hägerhäll C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta Bioenerg. 2002;1556:121–132. doi: 10.1016/S0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 100.Andreu A.L., Hanna M.G., Reichmann H., Bruno C., Penn A.S., Tanji K., Pallotti F., Iwata S., Bonilla E., Lach B., et al. Exercise intolerance due to mutations in the cytochrome b gene of mitochondrial DNA. N. Engl. J. Med. 1999;341:1037–1044. doi: 10.1056/NEJM199909303411404. [DOI] [PubMed] [Google Scholar]
- 101.Bourens M., Barrientos A. A CMC1-knockout reveals translation-independent control of human mitochondrial complex IV biogenesis. EMBO Rep. 2017;18:477–494. doi: 10.15252/embr.201643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DePristo M.A., Weinreich D.M., Hartl D.L. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat. Rev. Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 103.Bromberg Y., Rost B. Correlating protein function and stability through the analysis of single amino acid substitutions. BMC Bioinf. 2009;10:S8. doi: 10.1186/1471-2105-10-S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are openly available in National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov), accession numbers were MK642293-MK642306 and MW450876-MW450878.