Significance
The tumor suppressor p53 prevents tumorigenesis, while inactivation of p53 promotes cancer development and drug resistance. Here, we identify that a long noncoding RNA, the RNA component of mitochondrial RNA-processing endoribonuclease (RMRP), promotes growth and proliferation of colorectal cancer cells by inhibiting p53 activity. Mechanistically, RMRP retains SNRPA1 in the nucleus, thus preventing its lysosomal degradation. The nuclear SNRPA1 then prompts MDM2-mediated p53 ubiquitination and degradation. Remarkably, RMRP expression is induced by poly (ADP-ribose) polymerase (PARP) inhibitors, a group of targeted anticancer drugs, through the transcription factor C/EBPβ. Targeting RMRP significantly enhances sensitivity of colorectal cancer cells to PARP inhibition by reactivating p53. Our study provides a possible mechanism underling tumor resistance to PARP inhibitors.
Keywords: p53, long noncoding RNA, RMRP, SNRPA1, PARP inhibition
Abstract
p53 inactivation is highly associated with tumorigenesis and drug resistance. Here, we identify a long noncoding RNA, the RNA component of mitochondrial RNA-processing endoribonuclease (RMRP), as an inhibitor of p53. RMRP is overexpressed and associated with an unfavorable prognosis in colorectal cancer. Ectopic RMRP suppresses p53 activity by promoting MDM2-induced p53 ubiquitination and degradation, while depletion of RMRP activates the p53 pathway. RMRP also promotes colorectal cancer growth and proliferation in a p53-dependent fashion in vitro and in vivo. This anti-p53 action of RMRP is executed through an identified partner protein, SNRPA1. RMRP can interact with SNRPA1 and sequester it in the nucleus, consequently blocking its lysosomal proteolysis via chaperone-mediated autophagy. The nuclear SNRPA1 then interacts with p53 and enhances MDM2-induced proteasomal degradation of p53. Remarkably, ablation of SNRPA1 completely abrogates RMRP regulation of p53 and tumor cell growth, indicating that SNRPA1 is indispensable for the anti-p53 function of RMRP. Interestingly and significantly, poly (ADP-ribose) polymerase (PARP) inhibitors induce RMRP expression through the transcription factor C/EBPβ, and RMRP confers tumor resistance to PARP inhibition by preventing p53 activation. Altogether, our study demonstrates that RMRP plays an oncogenic role by inactivating p53 via SNRPA1 in colorectal cancer.
The tumor suppressor p53 plays an essential role in maintaining genomic integrity, preventing malignant transformation, and inhibiting cancer cell growth, proliferation, and motility. Mutation of its encoding gene, TP53, occurs in more than 50% of human cancers. This not only abolishes the wild-type activity of p53 but also endows some of the mutants with the oncogenic function, namely, “gain of function” (1). p53 can be activated in response to a variety of stress signals leading to induction of a whole range of genes involved in tumor suppression (2, 3). For instance, CDKN1A, also known as p21, induces cell growth arrest by inhibiting cyclin-dependent kinases, while PUMA and BAX that belong to the Bcl-2 family are pivotal to mitochondrial outer-membrane permeabilization and consequent apoptosis (2, 3).
Owing to the extreme cytotoxic effect, p53 activity is usually restrained through multiple mechanisms under physiological and pathological (cancerous) conditions. The E3 ubiquitin ligase MDM2, encoded by a p53-responsive gene, is the master antagonist of p53 by promoting its ubiquitination and proteasomal degradation (4–8). MDM2 also insulates p53 from the transcription-related DNA elements by directly associating and concealing its transactivation domain (9) or prompting its cytosolic accumulation through monoubiquitination (10). In addition, MDM2 inhibits p53 messenger RNA (mRNA) translation by perturbing the interaction of the ribosomal protein RPL26 and p53 mRNA (11). Genetic studies showed that depletion of the p53 gene completely rescues embryonic lethality of Mdm2-knockout mice, gracefully validating the central role of MDM2 in the control of p53 activity (12, 13). Cancer cells also utilize diverse oncogenic molecules to modulate the MDM2–p53 axis. For instance, NGFR and PHLDB3 that are highly expressed in multiple human cancers were found to undermine the stability and transcriptional activity of p53 by directly interacting with both MDM2 and p53 (14–16). In our recent attempt to uncover regulators of the MDM2–p53 circuit in colorectal cancer as further described below, we identified a long noncoding RNA (lncRNA) as a player crucial for cancer development and progression.
lncRNAs are a group of regulatory RNAs that are involved in the regulation of almost all aspects of cancer, including genomic instability, cell growth and immortality, angiogenesis, metastasis, and chemoresistance (17). They have also been shown to play various roles in the p53 network (18). While several lncRNAs, such as MALAT1 and MEG3, acted as upstream players of p53, other lncRNAs, including lincRNA-p21, RoR, PANDA, NEAT1, and GUARDIN, functioned as its downstream effectors whose expression is transcriptionally activated by p53 (18–20). Our study herein unveiled a lncRNA called the RNA component of mitochondrial RNA-processing endoribonuclease (RMRP) as a p53 inactivator that plays a protumorigenic role. It was previously shown to be responsible for the cleavage of the RNA primer for mitochondrial DNA replication (21) and the precursor of ribosomal RNA (rRNA) (22). Mutation of the RMRP gene was characterized as a causative event for cartilage–hair hypoplasia (CHH), a recessively inherited developmental disorder characterized by metaphyseal dysplasia, anemia, and immune dysregulation (23). Recently, RMRP was shown to play a role in cancer by competitively sponging microRNAs (24–26). Our further characterization of this lncRNA demonstrated that RMRP can promote cancer cell growth and tumor formation by restricting p53 activity in vitro and in vivo. Interestingly, we also found that poly (ADP-ribose) polymerase (PARP) inhibitors induce the expression of RMRP through the transcription factor (TF) C/EBPβ, and targeting RMRP significantly enhances the cytotoxic effect of PARP inhibitors by activating the p53 pathway. Thus, our study as detailed below unveils RMRP as an inhibitor of p53 via a unique mechanism engaging its binding partner SNRPA1, potentially important for drug resistance of malignant colorectal cancer.
Results
Highly Expressed RMRP Is Associated with Unfavorable Cancer Prognosis.
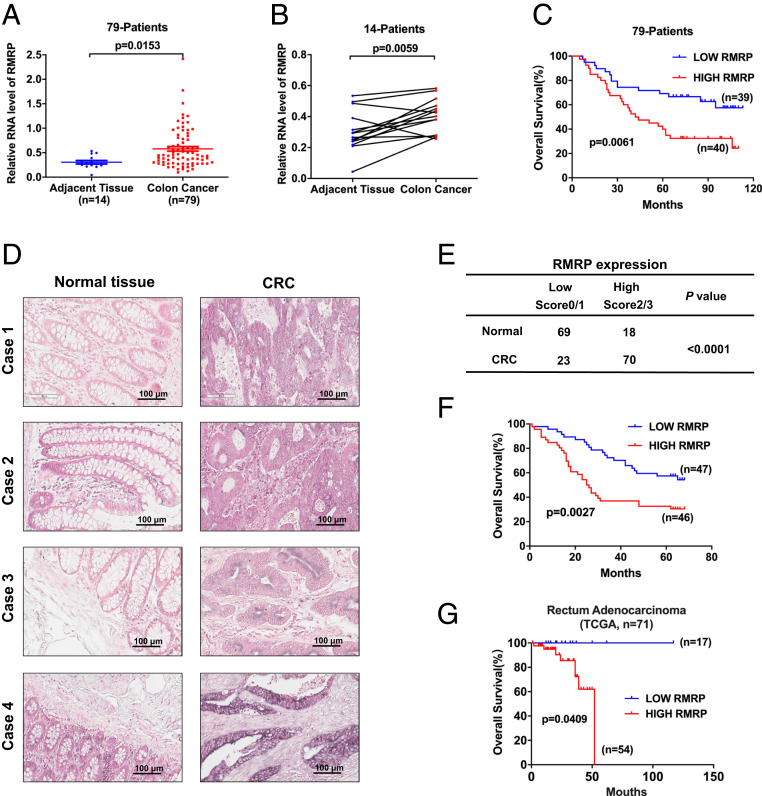
The transcriptomic data of our previous study (14) suggested that the lncRNA RMRP might be involved in the p53 pathway and cancer development. To test this possibility, we first evaluated the expression of RMRP through the complementary DNA (cDNA) array of 79 colorectal cancer samples (SI Appendix, Table S1) with 14 matched adjacent normal tissues. The average level of RMRP determined by qPCR was significantly up-regulated in colorectal cancer compared with the noncancerous tissues (Fig. 1A). This significant difference was more apparently pronounced in the 28 paired samples (Fig. 1B). The univariate and multivariate analyses of the overall survival of the 79 patients indicated RMRP as a negative prognostic factor (SI Appendix, Table S2), although a larger sample size may be needed for more accurate statistical power. Also, the Kaplan–Meier survival analysis revealed that higher expression of RMRP is significantly associated with worse prognosis (Fig. 1C). A set of colorectal cancer tissue array from an independent patient cohort was employed to validate the above observations by RNA in situ hybridization analysis. Again, the RMRP level was elevated in the cancerous tissues over the adjacent normal tissues (Fig. 1 D and E). Consistently, higher expression of RMRP in tumors predicted poorer colorectal cancer prognosis in this patient cohort (Fig. 1F). In line with our results, the TCGA dataset also showed that RMRP expression is negatively correlated with patient survival of rectum adenocarcinoma (Fig. 1G). Thus, these analyses of the lncRNA level and prognostic value in cancer specimens suggest that RMRP might play an oncogenic role.
Fig. 1.
LncRNA RMRP is overexpressed in colorectal cancer and associated with unfavorable prognosis. (A) RMRP expression is higher in colon cancer (n = 79) compared with normal tissues (n = 14). (B) RMRP expression is higher in colon cancer tissues (n = 14) compared with the paired adjacent tissues (n = 14). Values are expressed as the median with interquartile range in A and B. (C) A higher level of RMRP predicts poorer prognosis in 79 colon cancer patients. (D) RMRP expression is higher in cancerous tissues compared with the adjacent normal tissues through a colorectal cancer tissue array determined by RNA in situ hybridization. (E) Statistical analysis of RMRP staining in the colorectal cancer tissues (n = 93) and the adjacent tissues (n = 87). Statistical significance was assessed using Fisher’ s exact test. (F) Higher level of RMRP is associated with worse overall survival in 93 colorectal cancer patients. (G) Higher expression of RMRP is associated with worse overall survival in the TCGA rectum adenocarcinoma cohort.
RMRP Hampers p53 Activity by Enhancing MDM2 Function.
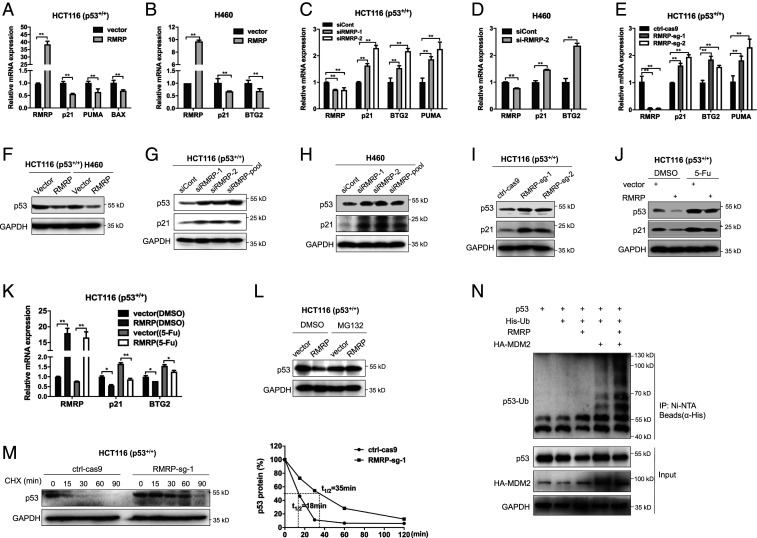
To determine the possible role of RMRP in colorectal cancer, we generated the RMRP-knockout HCT116 cell line by the CRISPR-Cas9 system and performed RNA-sequencing (RNA-seq) analysis of the potential downstream signals of RMRP. A broad panel of genes was dysregulated upon RMRP depletion (SI Appendix, Fig. S1A). The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that the p53 signaling pathway is significantly activated (SI Appendix, Fig. S1B). To validate this observation, we checked whether RMRP regulates p53 target gene expression in colorectal cancer HCT116 and lung cancer H460 cells, both of which sustain wild-type p53, by RT-qPCR analysis. As expected, ectopic expression of RMRP decreased (Fig. 2 A and B), while depletion of RMRP by two independent small interfering RNAs (siRNAs) increased (Fig. 2 C and D), the expression of p53 target genes, such as p21, BTG2, and PUMA. Also, knockout of RMRP in HCT116 cells markedly induced p53 target gene expression (Fig. 2E), which was consistent with the RNA-seq result (SI Appendix, Fig. S1B). However, overexpression or depletion of RMRP did not affect the p53 mRNA level (SI Appendix, Fig. S1C–G), suggesting that RMRP may modulate p53 activity at the posttranslational level. Indeed, overexpression of RMRP decreased (Fig. 2F), whereas knockdown (Fig. 2 G and H) or knockout (Fig. 2I) of RMRP elevated, the protein level of p53 in both HCT116 and H460 cells. Also, RMRP suppressed stress-induced p53 activity, as ectopic RMRP moderately but significantly inhibited p53 and its target gene levels in 5-FU–treated cancer cells (Fig. 2 J and K). Consistent with these results, our analysis of the TCGA database showed a negative correlation between the expression of RMRP and p53 target genes, such as BTG2 and MDM2 (SI Appendix, Fig. S1 H and I). As the TP53 gene is frequently mutated in various cancers, we also wondered whether RMRP is functionally associated with mutant p53. By mining the TCGA database, we found that the expression of RMRP is not associated with the TP53 status (SI Appendix, Fig. S1 J–L). Knockdown of RMRP in p53-R273H–harboring colon cancer HT-29 cells had no impact on the protein level of mutant p53 (SI Appendix, Fig. S1M). Thus, these results suggest that RMRP is a p53 suppressor and may inhibit p53 activity by regulating its protein level.
Fig. 2.
RMRP represses p53 activity by enhancing MDM2-induced p53 proteasomal degradation. (A and B) Overexpression of RMRP reduces p53 target gene expression in HCT116 p53+/+ and H460 cells. (C and D) Knockdown of RMRP increases p53 target gene expression in HCT116 p53+/+ and H460 cells. (E) CRISPR-Cas9–mediated ablation of RMRP induces p53 target gene expression in HCT116 p53+/+ cells. (F) Overexpression of RMRP decreases the protein level of p53 in HCT116 p53+/+ and H460 cells. (G and H) Knockdown of RMRP elevates the protein levels of p53 and p21 in HCT116 p53+/+ and H460 cells. (I) Knockout of RMRP induces p53 and p21 protein levels in HCT116 p53+/+ cell. (J and K) Overexpression of RMRP impairs 5-FU–induced p53 activation determined by IB (J) and RT-qPCR (K). (L) The proteasome inhibitor MG132 blocks RMRP-mediated p53 degradation in HCT116 p53+/+ cells. Cells were treated with MG132 (20 μM) for 6 h before harvested for IB. (M) The p53’s half-life is extended upon RMRP depletion. The ctrl-Cas9 and RMRP-sg-1 cell lines were treated with 100 μg/mL of cycloheximide (CHX) and harvested at the indicated time points for IB (Left). (Right) The ratios of p53/GAPDH. (N) RMRP promotes MDM2-dependent ubiquitination of p53. HCT116 p53−/− cells were transfected with combinations of plasmids encoding p53, RMRP, HA-MDM2, and His-Ub as indicated and treated with MG132 (20 μM) for 6 h before harvested for in vivo ubiquitination assay. *P < 0.05, **P < 0.01 by two-tailed Student’s t test.
Indeed, treatment of cancer cells with the proteasome inhibitor MG132 completely abrogated RMRP-mediated reduction of the p53 protein level (Fig. 2L), indicating that RMRP could be involved in regulation of p53 protein stability. We next tested whether RMRP controls p53 protein half-life by a cycloheximide chase assay. As shown in Fig. 2M, knockout of RMRP drastically prolonged p53 protein half-life. Since MDM2 is a key E3 ubiquitin ligase for p53, we then tested whether RMRP affects MDM2-induced p53 ubiquitination. While ectopic RMRP alone barely affected p53 ubiquitination in the absence of ectopic MDM2 in HCT116 p53−/− cells, it drastically enhanced MDM2-mediated p53 ubiquitination (Fig. 2N). Together, these results demonstrate that RMRP represses p53 activity by prompting its proteasomal degradation mediated by MDM2.
RMRP Promotes Cancer Cell Growth by Inactivating p53.
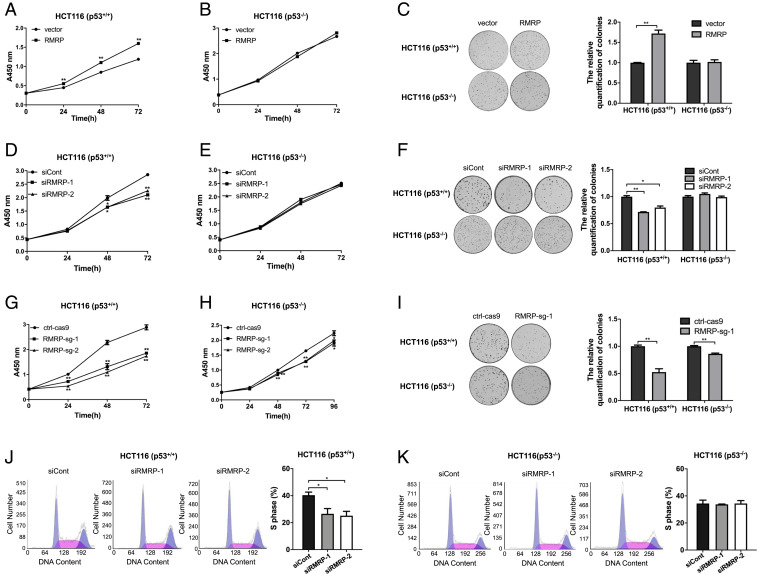
Next, we tested whether RMRP promotes cancer cell growth and proliferation dependently of p53 by employing both p53–wild-type and p53-null HCT116 cells. Ectopic RMRP significantly enhanced the proliferative and colony-forming ability of HCT116 p53+/+ cells but had little effect on proliferation or colony formation of HCT116 p53−/− cells (Fig. 3 A–C). In line with these results, knockdown of RMRP markedly impaired the proliferative and colony-forming ability of HCT116 p53+/+ cells but not HCT116 p53−/− cells (Fig. 3 D–F). Also, knockout of RMRP achieved a more profound inhibitory effect on HCT116 p53+/+ cell proliferation and colony formation than did siRNA-mediated gene silence (Fig. 3 G and I) because of more complete ablation of RMRP expression by CRISPR-Cas9 (Fig. 2E). Of note, RMRP knockout had some marginal effects on the growth and colony formation of HCT116 p53−/− cells (Fig. 3 H and I), suggesting that RMRP might possess some p53-independent functions as shown in the RNA-seq result (SI Appendix, Fig. S1B). Furthermore, depletion of RMRP led to significant reduction of S-phase population in HCT116 p53+/+ cells but not in HCT116 p53−/− cells, as shown by the flow cytometry analysis of the cells (Fig. 3 J and K). Together, these results demonstrate that RMRP can promote the growth and proliferation of colorectal cancer cells by inhibiting p53 activity.
Fig. 3.
RMRP promotes colorectal cancer cell growth and proliferation through inactivation of p53. (A) Overexpression of RMRP accelerates proliferation of HCT116 p53+/+ cells by the cell viability assay. (B) Overexpression of RMRP has no effect on HCT116 p53−/− cell proliferation. (C) Overexpression of RMRP enhances the colony-forming ability of HCT116 p53+/+ cells but not HCT116 p53−/− cells. The quantification of colonies is shown in the right panel. (D) Knockdown of RMRP inhibits proliferation of HCT116 p53+/+ cells. (E) Knockdown of RMRP has no effect on proliferation of HCT116 p53−/− cells. (F) Knockdown of RMRP impedes the colony-forming ability of HCT116 p53+/+ cells but not HCT116 p53−/− cells. (Right) The quantification of colonies. (G) Knockout of RMRP by CRISPR/Cas9 dramatically prompts proliferation of HCT116 p53+/+ cells. (H) RMRP knockout had a marginal inhibitory effect on HCT116 p53−/− cell proliferation. (I) RMRP knockout drastically inhibits the colony-forming ability of HCT116 p53+/+ cells but has a marginal effect on colony formation of HCT116 p53−/− cells. (Right) The quantification of colonies. (J) Knockdown of RMRP leads to reduction of S-phase population in HCT116 p53+/+ cells. The quantification of S-phase is shown in the right panel. (K) Knockdown of RMRP has no effect on HCT116 p53−/− cell cycle progression. The quantification of S-phase is shown in the right panel. *P < 0.05, **P < 0.01 by two-tailed Student’s t test.
RMRP Fosters Tumor Development In Vivo by Inactivating p53.
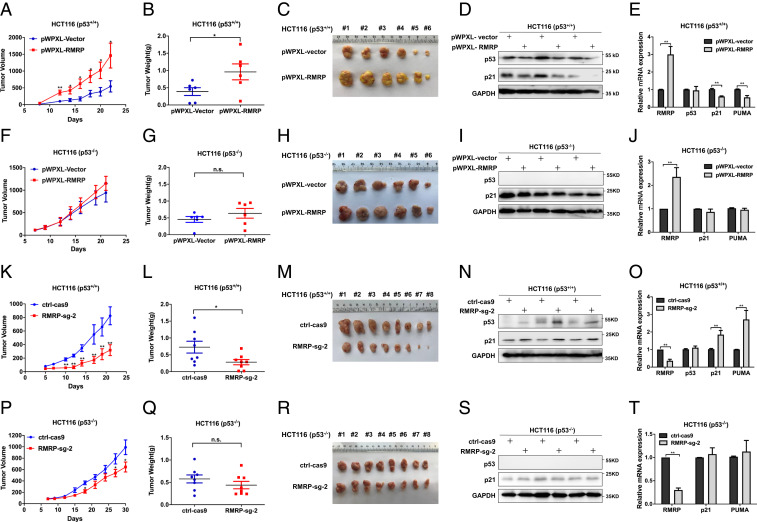
To translate the above results obtained from cultured cancer cells into a more biological setting, we established a set of xenograft models by bilaterally and subcutaneously inoculating HCT116 p53+/+ or HCT116 p53−/− cells with overexpressed or depleted RMRP into nude mice. In agreement with the cell-based results above, ectopic RMRP dramatically accelerated the growth of xenograft tumors derived from HCT116 p53+/+ cells as measured by the tumor volume, weight, and mass (Fig. 4 A–C). This must be due to the inhibition of the p53 pathway, because ectopic RMRP reduced p53 protein level and the expression of its target genes in the tumor tissues examined (Fig. 4 D and E). Indeed, as shown in Fig. 4 F–H, overexpression of RMRP barely affected the growth, weight, and mass of the xenograft tumors derived from HCT116 p53−/− cells. Consistently, none of the p53 target genes tested was influenced in the dissected tumors in the p53-null cells (Fig. 4 I and J). Conversely, knockout of RMRP drastically suppressed growth of the tumors derived from HCT116 p53+/+ cells compared with the control group (Fig. 4 K–M). In accordance, the expression of p53 and its target genes were elevated upon RMRP depletion in these tumors (Fig. 4 N and O). Interestingly, knockout of RMRP could slightly suppress growth of the tumors derived from HCT116 p53−/− cells (Fig. 4 P–R) without affecting the expression of p53 target genes (Fig. 4 S and T), again suggesting its p53-independent activity. Altogether, these results demonstrate that RMRP promotes colorectal cancer development in vivo primarily by inactivating p53.
Fig. 4.
RMRP endorses tumor growth in vivo by inactivating p53. (A) Lentivirus-based overexpression of RMRP in HCT116 p53+/+ cells significantly elevates tumor volume in average compared with the control group. (B and C) The dissected tumors show that RMRP overexpression increases the weight and mass of tumors derived from HCT116 p53+/+ cells. Data are represented as mean ± SD, n = 6. (D) Overexpression of RMRP inhibits p53 and p21 protein expression in vivo. (E) Overexpression of RMRP inhibits the mRNA expression of p21 and PUMA examined in three pairs of xenograft tumors (mean ± SD). (F–H) Overexpression of RMRP has a marginal effect on the growth of tumors derived from HCT116 p53−/− cells. Data are represented as mean ± SD, n = 6. (I and J) Overexpression of RMRP does not affect p53 target gene expression in three pairs of xenograft tumors derived from HCT116 p53−/− cells (mean ± SD). (K) CRISPR/ Cas9–mediated depletion of RMRP in HCT116 p53+/+ cells significantly suppresses tumor volume in average compared with the control group. (L and M) The dissected tumors show that knockout of RMRP diminishes the weight and mass of tumors derived from HCT116 p53+/+ cell. Data are represented as mean ± SD, n = 8. (N) RMRP knockout bolsters p53 and p21 protein expression in vivo. (O) RMRP knockout activates the mRNA expression of p21 and PUMA examined in three pairs of xenograft tumors (mean ± SD). (P–R) Knockout of RMRP has a marginal effect on the growth of tumors derived from HCT116 p53−/− cells. Data are represented as mean ± SD, n = 8. (S and T) Knockout of RMRP does not affect p53 target gene expression in three pairs of xenograft tumors derived from HCT116 p53−/− cells (mean ± SD). *P < 0.05, **P < 0.01 by two-tailed Student’s t test. n.s. indicates no significance.
Small Nuclear Ribonucleoprotein Polypeptide A' Is Required for RMRP-Mediated Inhibition of p53.
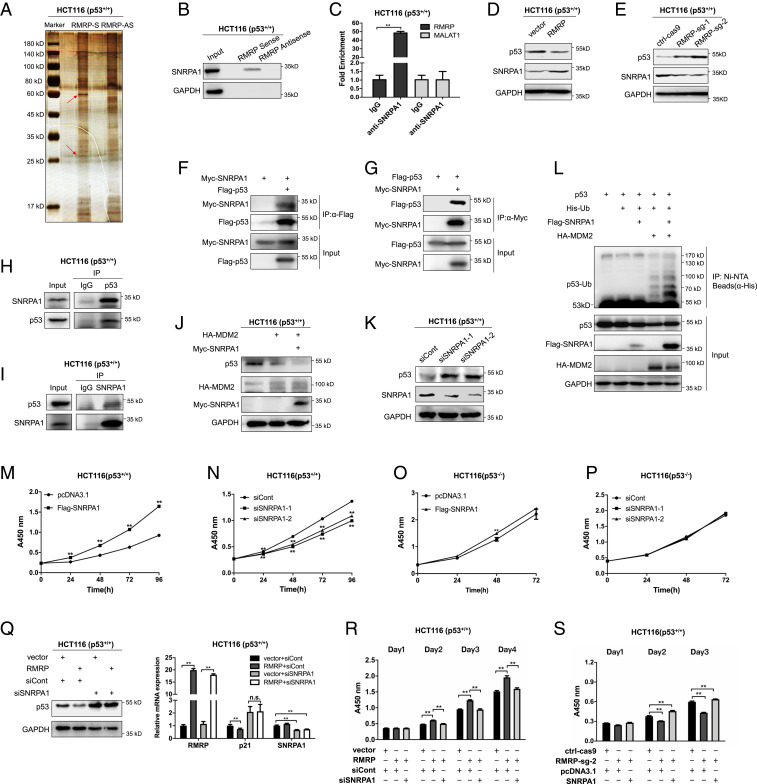
It was puzzling to us how the lncRNA might destabilize p53 by influencing MDM2’s E3 ligase activity. To solve this puzzle, we conducted an RNA pull-down assay coupled with mass spectrometry (MS) in order to identify potential RMRP-interacting proteins that might be involved in this regulation. The silver staining of RMRP-bound proteins showed two distinct bands at the 30 kDa and 60 kDa positions, respectively, on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, which did not appear on the lane for the RMRP antisense sequence control (Fig. 5A). The MS analysis of the two bands revealed some RMRP-interacting candidate proteins. We then performed an RNA pull-down assay and identified SNRPA1 (small nuclear ribonucleoprotein polypeptide A') as a top candidate by verifying its binding to RMRP (Fig. 5B). This result was also confirmed by the RNA immunoprecipitation (RIP) assay using the anti-SNRPA1 antibody (Fig. 5C). Their interaction was specific, because neither the antisense RMRP nor a well-known lncRNA MALAT1 could bind to SNRPA1 (Fig. 5 B and C).
Fig. 5.
RMRP inhibits p53 activity through SNRPA1. (A) Identification of RMRP-interacting proteins. RMRP and the antisense of RMRP were synthesized and biotinylated in vitro followed by the RNA pull-down assay. The silver staining reveals the specific bands (red arrows) that were subjected to MS analysis. (B and C) RMRP interacts with SNRPA1. SNRPA1 is pulled down with RMRP, but not with the antisense of RMRP, by the RNA pull-down assay (B). RMRP, but not another lncRNA MALAT1, is coimmunoprecipitated with SNRPA1 by the RIP assay using an anti-SNRPA1 antibody (C). (D and E) p53 expression is negatively correlated with SNRPA1. Overexpression of RMRP reduces the p53 level, whereas increases the SNRPA1 level in HCT116 p53+/+ cells (D). Knockout of RMRP elevates p53 expression but decreases SNRPA1 expression in HCT116 p53+/+ cells (E). (F and G) Exogenous SNRPA1 interacts with exogenous p53 by reciprocal IP assays. (H and I) Endogenous interactions between SNRPA1 and p53 in HCT116 p53+/+ cells. (J) Overexpression of SNRPA1 promotes p53 protein degradation in the presence of MDM2 in HCT116 p53+/+ cell. (K) Knockdown of SNRPA1 increases p53 protein level in HCT116 p53+/+ cells. (L) SNRPA1 promotes MDM2-induced ubiquitination of p53. (M) Overexpression of SNRPA1 accelerates proliferation of HCT116 p53+/+ cells by the cell viability assay. (N) Knockdown of SNRPA1 inhibits proliferation of HCT116 p53+/+ cells by the cell viability assay. (O) Overexpression of SNRPA1 has no effect on HCT116 p53−/− cell proliferation by the cell viability assay. (P) Knockdown of RMRP has no effect on HCT116 p53−/− cell proliferation by the cell viability assay. (Q) Knockdown of SNRPA1 abolishes RMRP inhibition of p53. (R) Knockdown of SNRPA1 abrogates RMRP-induced cancer cell proliferation. (S) Overexpression of SNRPA1 restores cell proliferation impaired by RMRP depletion. **P < 0.01 by two-tailed Student’s t test. n.s. indicates no significance.
SNRPA1 was previously shown as a component of the spliceosome to play a role in spermatogenesis (27) and reprogramming of pluripotent stem cells (28) by maintaining the integrity of the spliceosome. However, the role of SNRPA1 in cancer remains largely unknown. Interestingly, we observed a negative correlation between the expression of p53 and SNRPA1 upon overexpression or depletion of RMRP (Fig. 5 D and E). These results prompted us to investigate whether SNRPA1 negatively regulates p53 expression via physical interaction with the latter by performing a set of co-IP–IB (immunoblotting) assays. Exogenously expressed Myc-SNRPA1 could be coimmunoprecipitated with Flag-p53 using an anti-Flag antibody (Fig. 5F) and vice versa using an anti-Myc antibody (Fig. 5G). The endogenous SNRPA1-p53 complex was also detected through reciprocal co-IP assays (Fig. 5 H and I). Their interaction was further validated by mapping their binding domains, as the N-terminal amino acids from 1 to 175 of SNRPA1 (SI Appendix, Fig. S2 A and B) interacted with the C-terminal amino acids from 301 to 393 including the oligomerization domain of p53 (SI Appendix, Fig. S2 C and D).
Next, we found that ectopic SNRPA1 enhanced p53 degradation in the presence of MDM2 (Fig. 5J), while knockdown of SNRPA1 drastically induced the p53 protein level (Fig. 5K). Consistently, SNRPA1 promoted MDM2-induced p53 ubiquitination (Fig. 5L). It is likely that SNRPA1 may boost the interaction between MDM2 and p53 or enhance the E3-ubiquitin ligase activity of MDM2 toward p53. Functionally, ectopic SNRPA1 promoted (Fig. 5M), whereas knockdown of SNRPA1 inhibited (Fig. 5N), colorectal cancer cell proliferation. The oncogenic role of SNRPA1 was abated in cancer cells without functional p53, because overexpression or knockdown of SNRPA1 did not impact HCT116 p53−/− cell proliferation (Fig. 5 O and P). Thus, these results evince that the RMRP-binding protein SNRPA1 can promote cancer cell proliferation by interacting with p53 and negating its activity.
To check whether SNRPA1 is required for RMRP regulation of p53 activity, we performed another set of cell-based assays. As shown in Fig. 5Q, while ectopic RMRP significantly suppressed the expression of p53 and p21, knockdown of SNRPA1 completely abolished the ability of RMRP to regulate the p53 pathway, indicating that RMRP suppresses p53 in a SNRPA1-dependent manner. These results may also explain why RMRP did not regulate the level of mutant p53 (SI Appendix, Fig. S1M), as MDM2 usually maintains at a very low expression level in mutant p53-harboring cancer cells. Remarkably, overexpression of SNRPA1 could completely restore cancer cell proliferation that was impaired by RMRP knockout (Fig. 5R), while ablation of SNRPA1 significantly undermined RMRP-induced cancer cell proliferation (Fig. 5S). Together, these results demonstrate that SNRPA1, as a binding partner and an antagonist of p53, is required for RMRP-mediated p53 inactivation.
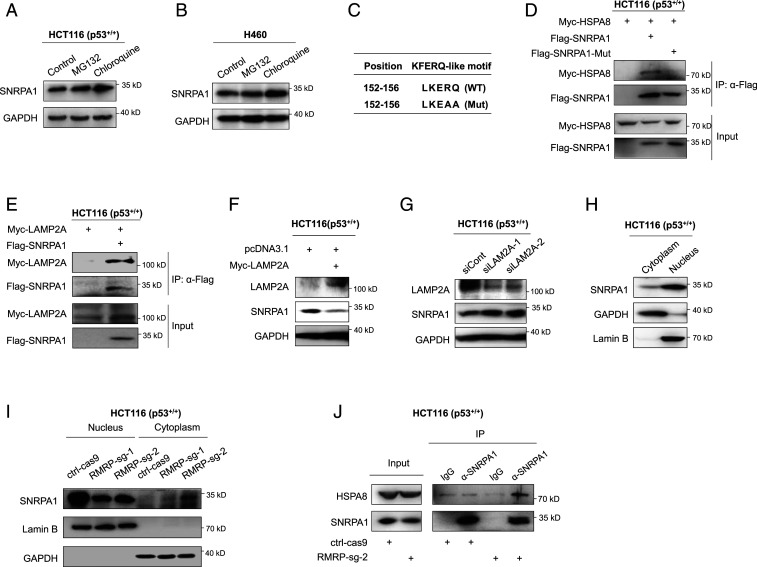
RMRP Prevents SNRPA1 Degradation via Chaperone-Mediated Autophagy.
Since RMRP appeared to increase the protein, but not RNA, level of SNRPA1 (Fig. 5 D, E, and Q), we sought to elucidate the molecular basis. First, we tested whether SNRPA1 is degraded via the proteasome or the lysosome by treating HCT116 p53+/+ and H460 cells with the proteasome inhibitor MG132 or the autophagy inhibitor chloroquine (CQ). Interestingly, the SNRPA1 protein level was increased in response to CQ, but not MG132, treatment of both of the cell lines (Fig. 6 A and B), indicating that SNRPA1 might undergo autophagy-related lysosomal degradation. Accordingly, we identified a KFERQ-like motif, LKERQ, in SNRPA1 (Fig. 6C), which is a requisite for chaperone-mediated autophagy (CMA), a selective pathway for lysosomal proteolysis (29, 30). The KFERQ-like motif can mediate the interaction of a substrate protein with the chaperone protein HSPA8, also known as HSC70. This complex is recruited to and pulled across the lysosome membrane with the assistance of the lysosome membrane protein LAMP2A and consequently subjected to degradation by the lysosome (29). We therefore tested whether SNRPA1 binds to HSPA8 via its LKERQ motif. As shown in Fig. 6D, Myc-HSPA8 could be coimmunoprecipitated with Flag-SNRPA1 using the anti-Flag antibody, while mutation of the LKERQ motif to LKEAA (Fig. 6C) (31) completely disrupted the SNRPA1–HSPA8 interaction (Fig. 6D). Also, SNRPA1 was shown to interact with LAMP2A (Fig. 6E). Furthermore, overexpression of LAMP2A markedly reduced (Fig. 6F), while knockdown of LAMP2A dramatically elevated, the expression of SNRPA1 (Fig. 6G). Thus, these results ascertain that SNRPA1 is a substrate of the CMA pathway for lysosomal proteolysis.
Fig. 6.
RMRP prevents lysosomal proteolysis of SNRPA1 by perturbing CMA. (A and B) SNRPA1 protein is stabilized by the autophagy inhibitor CQ but not by the proteasome inhibitor MG132. HCT116 p53+/+ (A) and H460 (B) cells were treated with MG132 (20 μM, 6 h) and CQ (50 μM, 8 h) before harvested for IB. (C) The presence of a KFERQ-like motif, LKERQ, in SNRPA1 and mutation of this motif to LKEAA. (D) Exogenous SNRPA1 interacts with exogenous HSPA8 via the LKERQ motif. HCT116 p53−/− cells were transfected with combinations of plasmids encoding Myc-HSPA8, Flag-SNRPA1, and Flag-SNRPA1-Mut followed by co-IP–IB assays. (E) Exogenous SNRPA1 interacts with exogenous LAMP2A. HCT116 p53−/− cells were transfected with plasmids encoding Flag-SNRPA1 and Myc-LAMP2A followed by co-IP–IB assays. (F) Overexpression of LAMP2A reduces the protein level of SNRPA1 in HCT116 p53+/+ cells. (G) Knockdown of LAMP2A increases the expression of SNRPA1 in HCT116 p53+/+. (H) The cellular distribution of SNRPA1 protein in HCT116 p53+/+ cells. GAPDH and Lamin B indicate the cytosolic and nuclear fractions, respectively. (I) CRISPR-Cas9–mediated ablation of RMRP reduces the SNRPA1 level in the nucleus, while increases the cytosolic accumulation of SNRPA1, in HCT116 p53+/+ cells. (J) Knockout of RMRP enhances the endogenous interaction of SNRPA1 and HSPA8.
Next, we tested whether RMRP controls SNRPA1 protein turnover via the CMA pathway. Given that both RMRP and SNRPA1 are mainly located in the nucleus (SI Appendix, Fig. S3A and Fig. 6H), we postulated that RMRP may sequester SNRPA1 in the nucleus, thus preventing the latter from being captured by the chaperone protein HSPA8 in the cytoplasm. To test this conjecture, we conducted subcellular fractionation and showed that knockout of RMRP markedly diminishes nuclear enrichment of SNRPA1 but increases the cytosolic SNRPA1 level (Fig. 6I and SI Appendix, Fig. S3B). Drastically, knockout of RMRP enhanced the interaction between endogenous SNRPA1 and HSPA8 (Fig. 6J). Collectively, these findings demonstrate that RMRP binds to and detains SNRPA1 in the nucleus, consequently stabilizing SNRPA1 by blocking the CMA pathway–mediated proteolysis.
The C/EBPβ-RMRP-p53 Pathway Determines Tumor Sensitivity to PARP Inhibition.
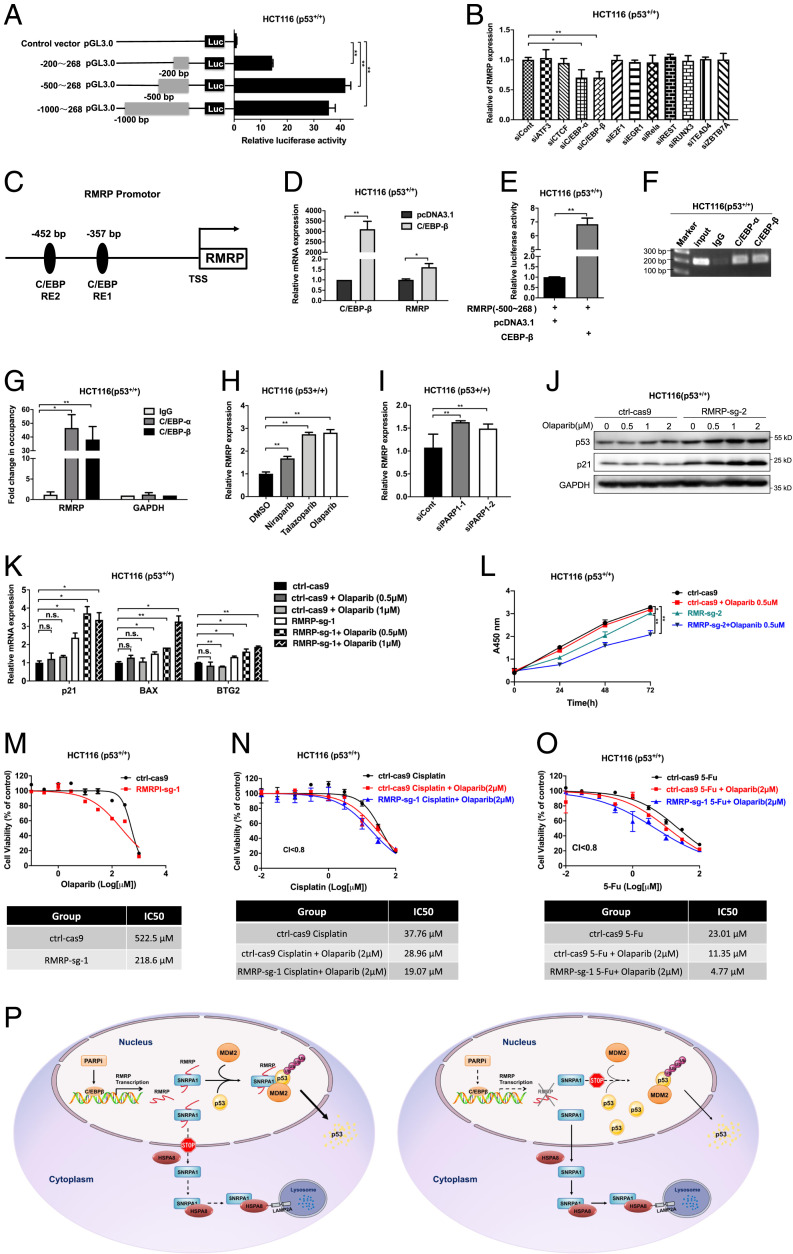
To figure out how RMRP is activated to negate p53 activity in cancer cells, we first evaluate the RMRP promoter activity by conducting a set of luciferase reporter assays driven by different promoter regions. While the 200-bp promoter upstream of the transcription initiation site exhibited some luciferase activity, a considerable higher activity was observed when the 500-bp promoter was used (Fig. 7A). The result suggested that the −200- to −500-bp region of the RMRP promoter is crucial for its expression. Analysis of this region revealed potential DNA elements for 11 different TFs (32). By knocking down each of these TFs, we found that C/EBPα and β is crucial for RMRP transcription, as ablation of C/EBPα or β significantly reduced the RMRP level (Fig. 7 B and C). Indeed, C/EBPα and β induced RMRP expression when individually overexpressed in HCT116 p53+/+ cells (Fig. 7D and SI Appendix, Fig. S4A). Also, overexpression of C/EBPα or β significantly boosted luciferase activity driven by this RMRP promoter region (Fig. 7E and SI Appendix, Fig. S4B). Furthermore, both C/EBPα and β associated with the RMRP promoter as revealed by a chromatin immunoprecipitation (ChIP) assay (Fig. 7 F and G). In line with these results, analysis of the CCLE and TCGA databases showed that the RMRP level is positively correlated with the expression of C/EBPs (SI Appendix, Fig. S4 C and D) and that C/EBPβ is associated with unfavorable prognosis of colorectal cancer (SI Appendix, Fig. S4 E and F). These results indicate that RMRP is a bona fide target gene of C/EBPs.
Fig. 7.
The C/EBPβ-RMRP axis confers colorectal cancer resistance to PARP inhibition. (A) Evaluation of the RMRP promoter activity by the luciferase reporter assay. (B) RMRP expression was determined by individually knocking down 11 TFs in HCT116 p53+/+ cells. (C) Schematic of the ∼500-bp region of the RMRP promoter. Two potential consensus binding sites of C/EBPs are indicated. (D) Overexpression of C/EBPβ up-regulates the RMRP level in HCT116 p53+/+ cells. (E) Overexpression of C/EBPβ triggers RMRP promoter (∼500 bp) activity as determined by the luciferase reporter assay. (F and G) C/EBPα and β associate with the RMRP promoter by the ChIP assay. The bound DNA elements were analyzed by PCR. (H) The expression of RMRP was induced by PARP inhibitors, including Olaparib, Niraparib, and Talazoparib. HCT116 p53+/+ cells were treated with the PARP inhibitors for 16 h before harvested for RT-qPCR. (I) Knockdown of PARP-1 up-regulates the RMRP level in HCT116 p53+/+ determined by RT-qPCR. (J and K) Knockout of RMRP sensitizes cancer cells to Olaparib-induced p53 activation. (L) A low dose of Olaparib significantly inhibits proliferation of RMRP-knockout, but not RMRP-proficient, colorectal cancer cells. (M) Knockout of RMRP sensitizes colorectal cancer cells to Olaparib as determined by the IC50. (N and O) Knockout of RMRP sensitizes colorectal cancer cells to the combination use of Olaparib with the genotoxic agents, Cisplatin (N) and 5-FU (O), as determined by the IC50. *P < 0.05, **P < 0.01 by two-tailed Student’s t test. (P) A schematic for RMRP-induced colorectal cancer resistance to PARP inhibitors by preventing p53 activation. Treatment of cancer cells with PARP inhibitors induces RMRP expression through the TF C/EBPβ. RMRP interacts with and sequesters SNRPA1 in the nucleus, thus blocking CMA-mediated lysosomal proteolysis of SNRPA1. The nuclear SNRPA1 then binds to p53 and promotes MDM2-induced p53 ubiquitination and proteasomal degradation (Left). Knockout of RMRP prompts cytosolic enrichment and lysosomal degradation of SNRPA1, leading to reactivation of p53 and tumor cell sensitization to PARP inhibitors (Right).
Recently, PARP-1 was shown to inactivate C/EBPβ by ADP-ribosylating (or PARylating) it (33, 34). We thus tested whether PARP inhibitors could elicit RMRP expression by treating HCT116 p53+/+ cells with or without three PARP inhibitors: Olaparib, Niraparib, and Talazoparib. As shown in Figs. 7H and SI Appendix, Fig. S4 G and H, the expression of RMRP was significantly induced by these PARP inhibitors. The effect was specific to PARP inhibition, because other genotoxic agents, glucose deprivation, or serum starvation did not influence RMRP expression (SI Appendix, Fig. S4 I–K). Consistently, knockdown of PARP-1 also increased the level of RMRP (Fig. 7I). Several studies reported that PARP inhibitors could induce p53 activation through replication stress (19, 35), while others suggested that they may impair p53 activity by triggering its nuclear export (36, 37). Thus, we tested whether RMRP is involved in PARP inhibitor regulation of p53 in colorectal cancer. Indeed, a low dose of Olaparib markedly activated the p53 pathway in RMRP-knockout HCT116 p53+/+ cells, whereas having a marginal effect on RMRP-proficient colorectal cancer cells (Fig. 7J), which was also verified by the expression of p53 target genes (Fig. 7K). These results strongly suggested that RMRP could confer resistance to PARP inhibitors. To test this assumption, we performed cell viability assays and showed that a low dose of Olaparib significantly suppresses proliferation of RMRP-knockout colorectal cancer cells but has no influence on RMRP-proficient colorectal cancer cells (Fig. 7L and SI Appendix, Fig. S4L). Consistent with these results, depletion of RMRP dramatically sensitized colorectal cancer cells to Olaparib, as evidenced by the decrease in half-maximal inhibitory concentration (IC50) from 522.5 μM to 218.6 μM in HCT116 p53+/+ cells (Fig. 7M) and from 63.34 μM to 15.97 μM in LOVO cells (SI Appendix, Fig. S4M). In contrast, knockout of RMRP barely influenced HCT116 p53−/− cell sensitivity to Olaparib (SI Appendix, Fig. S4N), suggesting that the effect of RMRP on Olaparib sensitivity is largely dependent on the regulation of p53. Moreover, targeting RMRP also significantly improved the cytotoxic effect of combination use of Olaparib with genotoxic agents, including Cisplatin and 5-FU (Fig. 7 N and O). Taken together, these results demonstrate that RMRP activation as a result of PARP1 inhibition accounts for drug resistance by inactivating p53.
Discussion
p53 inactivation is pivotal to cancer onset and progression and also highly associated with the resistance of several anticancer therapies. In this study, we uncovered RMRP that is highly expressed in colorectal cancer as an oncogenic lncRNA by inactivating p53 (Fig. 1). RMRP promoted colorectal cancer cell survival and proliferation in vitro and in vivo (Figs. 3 and 4) by opposing p53 activity (Fig. 2), whereas it had only a trivial effect on growth of cancer cells depleted of p53. We further identified SNRPA1 as an RMRP-interacting protein that bridges the interplay between RMRP and the p53 pathway (Fig. 5 A–E). Biochemically, RMRP prevented CMA-mediated lysosomal proteolysis of SNRPA1 by sequestering the latter in the nucleus (Fig. 6), where SNRPA1 could physically interact with p53 and bolster MDM2-induced proteasomal degradation of p53 (Fig. 5 F–L). Moreover, inhibition of PARP-1 by siRNAs or PARP inhibitors elevated RMRP expression by derepressing the C/EBPβ TF (Fig. 7 A–I). Finally, targeting RMRP enhanced sensitivity of colorectal cancer cells to PARP inhibitor–triggered p53 activation and cytotoxicity (Fig. 7 J–O). Taken together, our study as presented here unravels a hitherto unappreciated role of RMRP in provoking resistance to PARP inhibitors by attenuating p53 activation (Fig. 7P).
RMRP is vital to embryonic development due to its essential role in the processing of mitochondria RNA and rRNA (23, 38, 39). Our finding herein unveils an important role of RMRP in orchestrating the MDM2–p53 circuit. First, we showed that ectopic expression of RMRP significantly dampens, while depletion of RMRP boosts, the p53 protein level and transcriptional activity via multiple approaches in different cancer cell lines (Fig. 2 A–K). Second, RMRP enhanced MDM2-mediated ubiquitination and proteasomal degradation of p53 (Fig. 2 L–N). Finally, RMRP displayed a tumorigenic effect on growth and proliferation of colorectal cancer in vitro and in vivo (Figs. 3 and 4). In agreement with these results, RMRP was highly expressed in colorectal cancer tissues compared with the adjacent normal tissues (Fig. 1 A, B, D, and E), and a high level of RMRP in tumors is significantly associated with unfavorable prognosis (Fig. 1 C, F, and G). This finding is also consistent with a recent study showing that a recurrent mutation in the RMRP promoter leads to higher expression of RMRP in breast cancer (40). Our results underscored a p53-dependent function of RMRP, as ectopic expression or RNA interference (RNAi)-mediated knockdown of RMRP selectively regulates growth and proliferation of colorectal cancer cells harboring wild-type p53 (Figs. 3 A–F, J, and K and 4 A–J). Interestingly, knockout of RMRP by CRISPR-Cas9 had a moderate growth-inhibitory effect on p53-null HCT116 cells (Figs. 3 H and I and 4P). Since one of the RMRP functions is implicated in ribosome biogenesis, it is possible but not yet determined that complete loss of RMRP might evoke nucleolar stress leading to both p53-dependent and -independent pathways (41, 42). Alternatively, knockout of RMRP may also trigger other cancer-associated signaling pathways as shown in the RNA-seq results (SI Appendix, Fig. S1 A and B). These findings convincingly demonstrate that RMRP promotes colorectal cancer development mainly by inactivating p53.
To elucidate the mechanism underlying RMRP inhibition of p53, we first conducted an RNA pull-down assay followed by the MS analysis. Although a distinct band of ∼60 kDa was observed (Fig. 5A), p53 was ruled out by the MS result. Interestingly, we identified SNRPA1 from the band of ∼30 kDa and validated it as an interacting protein of RMRP. Although SNRPA1 was previously reported as a component of the spliceosome important for cell fate determination (27, 28), the role of SNRPA1 in cancer remained unknown. We uncovered SNRPA1 as a direct target of the lysosome for its proteolysis via the CMA pathway. First, we showed that CQ but not MG132 led to stabilization of SNRPA1 (Fig. 6 A and B). Also, SNRPA1 harbored a typical KFERQ-like motif that was critical for binding to the chaperone protein HSPA8 (Fig. 6C). Indeed, SNRPA1 bound with HSPA8 and LAMP2A, whereas mutation of the KFERQ-like motif completely abrogated the SNRPA1–HSPA8 interaction (Fig. 6 D and E). Remarkably, knockdown of LAMP2A, which specifically blocks CMA, elevated the protein level of SNRPA1 (Fig. 6G). Intriguingly, RMRP sequestered SNRPA1 in the nucleus, as RMRP depletion prompted translocation of SNRPA1 to the cytoplasm in which it underwent lysosomal degradation through the interaction with HSPA8 (Fig. 6 I and J). The nuclear SNRPA1 was able to bind to p53 and promote MDM2-induced p53 proteasomal degradation (Fig. 5 F–L), consequently leading to augmented growth of cancer cells (Fig. 5 M–P). Importantly, ablation of SNRPA1 completely annihilated the effect of RMRP on the regulation of p53 pathway and cancer cell proliferation (Fig. 5 Q–S), strongly suggesting the interplay between RMRP and SNRPA1 in the regulation of p53 stability and activity. Thus, our results demonstrate that RMRP suppresses p53 activity by recruiting SNRPA1 as its partner protein and preventing it from CMA-mediated degradation by the lysosome, consequently boosting MDM2-dependent degradation and inactivation of p53 and promoting cancer cell growth.
Through a screening for regulators of RMRP, we identified C/EBPα and β as TFs for RMRP expression (Fig. 7 A–G). While C/EBPα and β may form heterodimers to activate gene transcription in cooperation, they were also reported to have distinct functions. Recently, it has been reported that PARP-1 mediates ADP ribosylation of C/EBPβ and thus impairs its DNA binding and transcriptional activity during adipogenesis and in cancer cells (33, 34). Interestingly and consistently with the two studies, we found that inhibition of PARP-1 by siRNAs or PARP inhibitors induces RMRP expression (Fig. 7 H and I). These PARP inhibitors are widely used for treatment of various tumors harboring BRCA1/2 mutations or with homologous recombination deficiency (43). Recently, several studies showed that PAPR inhibitors can activate the p53 pathway probably by inducing replication stress (19, 35), and functional p53 is crucial to the antitumor effect of PARP inhibitors in some types of cancer (44, 45). Our study not only further advances this knowledge but also offers molecular insights into how malignant cancers might be resistant to the treatment with these inhibitors (i.e., RMRP at a higher level in cancer prevents p53 activation [Fig. 7 J and K] and consequently confers tumor resistance to PARP inhibition [Fig. 7L]) by partnering with SNRPA1. Remarkably, targeting RMRP significantly improved the cytotoxic efficacy of Olaparib or combination use of Olaparib with genotoxic agents in colorectal cancer cells (Fig. 7 M–O). Hence, these findings unravel a mechanism of cancer cell resistance to PARP inhibitors and strongly suggest that targeting RMRP may bolster PARP inhibition-related cancer therapies by reactivating the p53 pathway.
In summary, our study presented here identifies RMRP as an oncogenic lncRNA that can suppress p53 activity by stabilizing SNRPA1 that in turn assists MDM2 in degrading p53. Since PARP inhibitors induced RMRP expression through the TF C/EBPβ, consequently impairing p53 activation and diminishing cytotoxic effect, our study also offers a possible molecular mechanism underling the resistance of some malignant cancers to PARP inhibitors. Our study suggests the RMRP–SNRPA1 pathway as a potential target for future development of a therapeutic approach for cancers that are resistant to these inhibitors.
Materials and Methods
Plasmids and Antibodies.
The plasmid expressing RMRP was purchased from Shanghai Genechem Co., LTD). The Flag-tagged plasmids expressing SNRPA1, C/EBP-α, and C/EBP-β were purchased from Vigene Biosciences, Inc.. The Myc-tagged SNRPA1, HSPA8, and LAMP2A plasmids were generated by inserting the full-length cDNA amplified by PCR into the pcDNA3.1/Myc-His vector. The Myc-tagged plasmids expressing SNRPA1 fragments, aa 1 to 175 and aa 176 to 225, were generated by the same approach using the corresponding primers. The plasmids expressing HA-MDM2, Flag-p53, and His-Ub were described previously (14, 46). The Flag-tagged SNRPA1 plasmid was a gift from Laixin Xia (27). The anti-Flag (Catalog F1804, Sigma-Aldrich), anti-Myc (Catalog 60003–1, Proteintech), anti-SNRPA1 (Catalog 17368–1-AP, Proteintech), anti-LAMP2A (Catalog ab18528, Abcam), anti-HSPA8 (Catalog ab19136, Abcam), anti-p53 (DO-1, Catalog sc-126, Santa Cruz Biotechnology), anti-p21 (Catalog #2947, Cell Signaling Technology), anti-Lamin B (Catalog 66095–1-Ig, Proteintech), anti-MDM2 (Catalog ab16895, Abcam), anti-GAPDH (Catalog 60004–1, Proteintech), and the secondary antibodies for rabbit (Catalog ARG65351, Arigo) and mouse (Catalog ARG65350, Arigo), and the light chain–specific secondary mouse antibody (Catalog 115–035-174, Jackson) were commercially purchased.
IB.
Cells were harvested and lysed in lysis buffer consisting of 50 mM Tris/HCl (pH7.5), 0.5% Nonidet P-40 (Nonidet P-40), 1 mM ethylene diamine tetraacetic acid (EDTA), 150 mM NaCl, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 10 mM pepstatin A, and 1 mM leupeptin. Equal amounts of clear cell lysate (20 to 80 mg) were used for IB analysis as described previously (47).
Immunoprecipitation.
The immunoprecipitation (IP) assay was conducted using antibodies as indicated in the Figs. 5 and 6 and SI Appendix, Fig. S2. Briefly, proteins of ∼500 to 1,000 mg were incubated with the indicated antibodies at 4 °C for 4 h or overnight. Protein A or G beads (Santa Cruz Biotechnology) were then added, and the mixture was incubated at 4 °C for additional 1 to 2 h. Beads were washed at least three times with lysis buffer. Bound proteins were detected by IB with antibodies as indicated.
RNA Pull-Down Assay.
Sense or antisense RMRP was transcribed in vitro using T7 RNA Polymerase (New England Biolabs, Ipswich) and labeled by the Biotin RNA Labeling Mix (Roche). Biotinylated RNA was pretreated with RNA structure buffer (Beyotime) to obtain an appropriate secondary structure. Protein extracts of 1 mg were incubated with the pretreated biotinylated RNAs at 4 °C for 1 h. Then, 40-μL prewashed streptavidin magnetic beads (Invitrogen) were gently added, and the mixture was incubated on a rotator at 4 °C overnight. The beads were washed three times with NT2 buffer. Bound proteins were eluted in 60 μL protein lysis buffer, separated by the SDS-PAGE, and visualized by silver staining using the Fast Silver Stain Kit (Beyotime). Specific bands were collected for MS analysis or for IB analysis. The primers of sense or antisense RMRP for in vitro transcription are provided in SI Appendix, Table S6.
RNA IP Assay.
The Magna RIP RNA-binding protein IP kit was purchased from Millipore. The RIP assays were performed following the manufacturer’s protocol. Cells growing in 10-cm dishes were lysed in 0.5 mL of lysis buffer containing the protease inhibitor and RNase inhibitor. The supernatant was collected by centrifugation for 20 min at 10,000 × g at 4 °C and then incubated with Protein A/G magnetic beads and antibodies at 4 °C overnight with gentle rotation. The beads were washed six times with RIP wash buffer containing the RNase inhibitor. For IB analysis, 1/5 of the beads were lysed in SDS loading buffer, and 4/5 of the beads were lysed in RNAiso Plus. The bound RNA was extracted and analyzed by RT-qPCR.
Chromatin IP Assay.
The Magna ChIP A/G Chromatin IP Kit were purchased form Merck. The chromatin IP (ChIP) assay was performed following the manufacturer’s protocol. Briefly, cells were crosslinked with 1% formaldehyde and then terminated by adding 10× glycine. Cells were scraped off the dish and resuspended in ChIP lysis buffer containing the protease inhibitor. The cell pellet was collected and lysed with nuclear lysis buffer. Chromatin was sheared into ∼200-∼1,000-bp fragments by sonication. The anti-C/EBPα antibody, anti-C/EBPβ antibody, or IgG with magnetic beads was added to the chromatin mixture rotating at 4 °C overnight. The beads were washed four times and then eluted using the elution buffer. The bound DNA was purified and analyzed by PCR. The primers used in the ChIP assay are listed in SI Appendix, Table S5.
CRISPR/Cas9-Mediated Gene Editing.
The CRISPR/Cas9 targeting vector lentiCRISPR version 2 was purchased from Addgene. The small-guide RNA (sgRNA) for RMRP was designed at http://crispr.mit.edu/, and the sequences of sgRNAs are presented in SI Appendix, Table S6. The sgRNA was cloned into the lentiCRISPR version 2 vector at the BsmBI site. The combination of sgRNAs was used to achieve the best efficiency, and two different clones, RMRP-sg-1 and RMRP-sg-2, were selected for future experiments. The cells were infected the lentiviruses encoding the sgRNA and selected by 1 μg/mL puromycin for a week.
Mouse Xenograft Experiment.
Male BALB/c nude mice of 4 to 5 wk old were purchased from the Department of Laboratory Animal Science in Shanghai Medical College of Fudan University. To evaluate the effect of RMRP overexpression on tumor growth, mice were bilaterally and subcutaneously inoculated with 5 × 105 HCT116 p53+/+ or HCT116 p53−/− cells stably expressing pWPXL-vector or pWPXL-RMRP. To determine the effect of RMRP depletion on tumor growth, mice were bilaterally and subcutaneously inoculated with 5 × 105 HCT116 p53+/+ or HCT116 p53−/− cells stably expressing ctrl-cas9 or RMRP-sg-2. Tumor growth was monitored every other day with electronic digital calipers in two dimensions. Tumor volume was calculated according to the formula: volume = length × width2 × 0.5. Tumors were then harvested, weighed, and subjected to IB and RT-qPCR analyses. The animal protocols were in compliance with ethical regulations and approved by the Animal Welfare Committee of Shanghai Medical College at Fudan University.
Cancer Patients.
Informed consent was obtained from all patients, and the study was approved by the ethics committee of Fudan University Shanghai Cancer Center.
Statistics.
All in vitro experiments were performed in biological triplicate and data were presented as mean ± SD. The Student’s t test or one-way ANOVA was performed to evaluate the differences between two groups or more than two groups. The Cox univariate proportional hazards regression model was used to determine the independent clinical factors based on the investigated variables. The Kaplan–Meier statistics were used to analyze the significant difference of patient survival. Pearson’s correlation was performed to analyze the correlation of the gene expression. P < 0.05 was considered statistically significant.
Additional detailed materials and methods are provided in the online supporting information.
Supplementary Material
Acknowledgments
We thank the laboratory members for active and helpful discussion, Yazhen Gui for laboratory assistance, Ping Zhang for flow cytometry, and the innovative research team of the high-level local university in Shanghai. X.Z. was supported by the National Natural Science Foundation of China (Grants 81874053 and 82072879), Q.H. was supported by the National Natural Science Foundation of China (Grant 81702352), and H.L. was supported by the Reynolds and Ryan Families Chair Fund of Translational Cancer.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026813118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Freed-Pastor W. A., Prives C., Mutant p53: One name, many proteins. Genes Dev. 26, 1268–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine A. J., The many faces of p53: Something for everyone. J. Mol. Cell Biol. 11, 524–530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine A. J., p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 20, 471–480 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J., The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Oliner J. D., Kinzler K. W., Meltzer P. S., George D. L., Vogelstein B., Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358, 80–83 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Wu X., Bayle J. H., Olson D., Levine A. J., The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7 (7A), 1126–1132 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Haupt Y., Maya R., Kazaz A., Oren M., Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Kubbutat M. H., Jones S. N., Vousden K. H., Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Oliner J. D., et al., Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362, 857–860 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Li M., et al., Mono- versus polyubiquitination: Differential control of p53 fate by Mdm2. Science 302, 1972–1975 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M. B., Oren M., Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 32, 180–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montes de Oca Luna R., Wagner D. S., Lozano G., Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Jones S. N., Roe A. E., Donehower L. A., Bradley A., Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378, 206–208 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Zhou X., et al., Nerve growth factor receptor negates the tumor suppressor p53 as a feedback regulator. eLife 5, e15099 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang L., et al., Inactivating p53 is essential for nerve growth factor receptor to promote melanoma-initiating cell-stemmed tumorigenesis. Cell Death Dis. 11, 550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao T., et al., Pleckstrin homology domain-containing protein PHLDB3 supports cancer growth via a negative feedback loop involving p53. Nat. Commun. 7, 13755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt A. M., Chang H. Y., Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang A., Xu M., Mo Y. Y., Role of the lncRNA-p53 regulatory network in cancer. J. Mol. Cell Biol. 6, 181–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adriaens C., et al., p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 22, 861–868 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Hu W. L., et al., GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol. 20, 492–502 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Chang D. D., Clayton D. A., A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 6, 409–417 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldfarb K. C., Cech T. R., Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev. 31, 59–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridanpää M., et al., Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104, 195–203 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Shao Y., et al., LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget 7, 37812–37824 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao X., Gu Y., Wang G., Chen S., c-Myc, RMRP, and miR-34a-5p form a positive-feedback loop to regulate cell proliferation and apoptosis in multiple myeloma. Int. J. Biol. Macromol. 122, 526–537 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Tang L., et al., Long noncoding-RNA component of mitochondrial RNA processing endoribonuclease is involved in the progression of cholangiocarcinoma by regulating microRNA-217. Cancer Sci. 110, 2166–2179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H., et al., Major spliceosome defects cause male infertility and are associated with nonobstructive azoospermia in humans. Proc. Natl. Acad. Sci. U.S.A. 113, 4134–4139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y. D., et al., The unique spliceosome signature of human pluripotent stem cells is mediated by SNRPA1, SNRPD1, and PNN. Stem Cell Res. (Amst.) 22, 43–53 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Klionsky D. J., et al., Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dice J. F., Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 15, 305–309 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Kaushik S., Cuervo A. M., The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messeguer X., et al., PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Ryu K. W., et al., Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 360, eaan5780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo X., et al., PARP-1 controls the adipogenic transcriptional program by PARylating C/EBPβ and modulating its transcriptional activity. Mol. Cell 65, 260–271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai X. T., Moles R., Chaib-Mezrag H., Nicot C., Small PARP inhibitor PJ-34 induces cell cycle arrest and apoptosis of adult T-cell leukemia cells. J. Hematol. Oncol. 8, 117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanai M., et al., Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 9, 1175–1183 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Fischbach A., et al., The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 46, 804–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiel C. T., et al., Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am. J. Hum. Genet. 77, 795–806 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermanns P., et al., Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum. Mol. Genet. 14, 3723–3740 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Rheinbay E., et al., Recurrent and functional regulatory mutations in breast cancer. Nature 547, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Lu H., Signaling to p53: Ribosomal proteins find their way. Cancer Cell 16, 369–377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X., Liao W. J., Liao J. M., Liao P., Lu H., Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mateo J., et al., A decade of clinical development of PARP inhibitors in perspective. Ann. Oncol. 30, 1437–1447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sizemore S. T., et al., Synthetic lethality of PARP inhibition and ionizing radiation is p53-dependent. Mol. Cancer Res. 16, 1092–1102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jelinic P., Levine D. A., New insights into PARP inhibitors’ effect on cell cycle and homology-directed DNA damage repair. Mol. Cancer Ther. 13, 1645–1654 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Dai M. S., et al., Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24, 7654–7668 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y., et al., Ubiquitin ligase TRIM71 suppresses ovarian tumorigenesis by degrading mutant p53. Cell Death Dis. 10, 737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.