Significance
Embryonic development relies on a hierarchy of transcription factor expression and chromatin structure changes to establish unique patterns of gene expression in different cell types. The mechanisms regulating these changes are poorly understood. We compared two mechanosensory cell types, inner ear hair cells and epidermal Merkel cells, to demonstrate a feed-forward mechanism involving the transcription factors ATOH1, a “master regulator” of differentiation, and POU4F3 during mechanosensory differentiation. We show that downstream POU4F3 pioneer factor activity allows ATOH1 to access much of its enhancer network in developmentally closed chromatin. Our data suggest this shared feed-forward mechanism preceded the evolutionary divergence of hair cells and Merkel cells from an ancient mechanosensory receptor type and thus may have enabled their divergence.
Keywords: feed-forward epigenetic transcriptional control, ATOH1 and POU4F3, inner ear hair cells, Merkel cells, mechanoreceptor evolution
Abstract
During embryonic development, hierarchical cascades of transcription factors interact with lineage-specific chromatin structures to control the sequential steps in the differentiation of specialized cell types. While examples of transcription factor cascades have been well documented, the mechanisms underlying developmental changes in accessibility of cell type–specific enhancers remain poorly understood. Here, we show that the transcriptional “master regulator” ATOH1—which is necessary for the differentiation of two distinct mechanoreceptor cell types, hair cells in the inner ear and Merkel cells of the epidermis—is unable to access much of its target enhancer network in the progenitor populations of either cell type when it first appears, imposing a block to further differentiation. This block is overcome by a feed-forward mechanism in which ATOH1 first stimulates expression of POU4F3, which subsequently acts as a pioneer factor to provide access to closed ATOH1 enhancers, allowing hair cell and Merkel cell differentiation to proceed. Our analysis also indicates the presence of both shared and divergent ATOH1/POU4F3-dependent enhancer networks in hair cells and Merkel cells. These cells share a deep developmental lineage relationship, deriving from their common epidermal origin, and suggesting that this feed-forward mechanism preceded the evolutionary divergence of these very different mechanoreceptive cell types.
During embryonic development, a hierarchy of transcription factor expression and changes in chromatin structure establish unique patterns of gene expression in different mature cell types as they differentiate from stem or progenitor cells. The establishment of lineage-specific chromatin structures is a powerful mechanism to orchestrate the correct appearance of differentiated cells in response to inductive signals. It first requires the establishment of a lineage-specific epigenetic landscape and, subsequently, the sequential opening and closing of regions of DNA within a constantly changing chromatin structure. This process is regulated by a specialized class of transcription factors known as pioneer factors (1–3). Pioneer factors can engage silent and unmarked nucleosomal DNA and allow “opening” of enhancers that are then bound by transcription factors which initiate differentiation. They are often expressed early in the differentiation of a particular lineage—for example, FOXA transcription factors engage enhancers of liver-specific genes very early in endoderm differentiation (4, 5). However, pioneer factors could also be used to unlock gene regulatory networks in terminally differentiating postmitotic progenitors once they have committed to their final fates. In this paper, we provide evidence for a conserved role for another pioneer factor, POU4F3, in the terminal differentiation of two highly diverged mechanosensory cell types, hair cells of the inner ear and Merkel cells of the skin.
Inner-ear hair cells and the supporting cells that surround them develop from a distinct region of progenitors in the embryonic inner ear (6–8). The first sign of hair cell differentiation in these prosensory regions is the expression of ATOH1, a bHLH transcription factor that is necessary and sufficient for hair cell differentiation (9–13). Lineage tracing with Atoh1-Cre and Atoh1-CreER knock-in mice suggests that many sensory progenitor cells initially express Atoh1, but ultimately differentiate as supporting cells (14, 15) as a result of Atoh1’s repression through Notch-mediated lateral inhibition (16–19). The genetic and epigenetic mechanisms that select some ATOH1+ cells for a hair cell fate are currently unknown. In addition to hair cells in the inner ear, ATOH1 is also necessary for differentiation of mechanosensory Merkel cells from epidermal precursors in the skin (20–22).
We have performed a transcriptional and epigenetic analysis of embryonic cochlear progenitor cells as they begin to express ATOH1 and transition toward differentiated hair cells. We were surprised to find that about one-half of all hair cell enhancers containing ATOH1 binding motifs are epigenetically inaccessible and transcriptionally silent in differentiating hair cells. We show that POU4F3, a known hearing loss gene (DFNA15) (23) and direct transcriptional target of ATOH1 (24, 25), binds to these enhancers and acts as a pioneer factor to make them epigenetically accessible, which allows ATOH1 to bind, initiate transcription, and complete the hair cell differentiation program. Loss of Pou4f3 prevents these hair cell enhancers from becoming accessible and blocks hair cell differentiation. We show that this feed-forward mechanism between ATOH1 and POU4F3 is conserved in Merkel cells of the skin, suggesting that some of the shared gene regulatory networks specifying mechanosensory cells, as well as the feed-forward relationship between ATOH1 and POU4F3, may have an ancient origin at the base of the vertebrate lineage.
Results
ATOH1 Is Unable to Access Much of its Required Targetome at the Onset of Differentiation in Postmitotic Sensory Hair Cell Progenitors.
We purified embryonic day 13.5 (E13.5) Cdkn1b+ (p27Kip1) sensory progenitors and E17.5 Atoh1+ hair cells from the embryonic mouse cochlea using reporter mice expressing Green Fluorescent Protein (GFP) (Fig. 1A) (26, 27) and performed transcriptomic and epigenetic analysis on each population. We adapted the Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATACseq) technique (28) to profile chromatin accessibility with small numbers of Fluorescence-Activated Cell Sorting (FACS)-purified cells (1,000 to 3,000 cells/replicate) from the embryonic cochlea (µATACseq, SI Appendix, Fig. S1 and Materials and Methods). We used μATACseq to investigate the overall changes in chromatin structure during the transition from Cdkn1b-GFP+ cochlear sensory progenitors at E13.5 to nascent ATOH1-GFP+ hair cells at E17.5. We identified 16,006 open promoters and found that open promoter regions were highly correlated between E13.5 sensory progenitors and E17.5 hair cells (R = 0.81, SI Appendix, Fig. S2 A and B), suggesting that changes in promoter accessibility are not sufficient to explain the transcriptional changes seen during the transition from E13.5 progenitors to E17.5 hair cells.
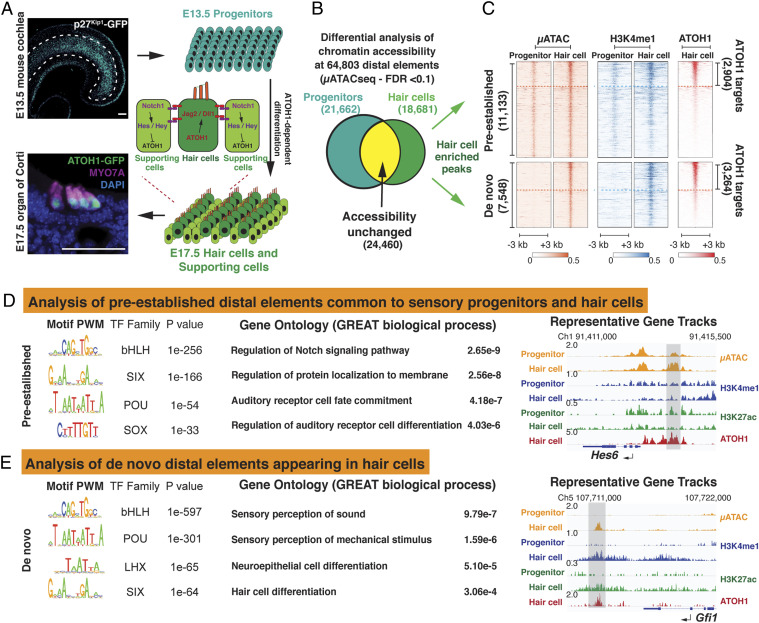
Fig. 1.
ATOH1 binds to pre-established and de novo distal regulatory elements in differentiating hair cells. (A) Schematic of early mouse organ of Corti development. Photomicrograph of E13.5 cochlear whole mount expressing Cdkn1b-GFP (p27Kip1-GFP) within the prosensory domain. Hair cell differentiation as seen in an E17.5 cross-section of organ of Corti expressing ATOH1-GFP, stained with antibody against the hair cell marker MYO7A, and counter stained by DAPI. The cartoon depicts Atoh1-dependent, Notch-mediated lateral inhibition. Atoh1 is up-regulated in the undifferentiated progenitors (E13.5), which then differentiate into a mosaic of hair cells and supporting cells. (Scale bar, 50 μm.) (B) Venn diagram illustrating the quantitative comparison of chromatin accessibility at the 64,803 total distal elements present in sensory progenitors and nascent hair cells (FDR < 0.1). A total of 18,681 distal elements are significantly more accessible in the nascent hair cells (hair cell–enriched distal elements) relative to 21,662 enriched sites in sensory progenitors and 24,460 sites remaining unchanged between the two populations. (C) Hair cell–enriched distal elements (18,681), shown as heatmaps, are subclustered into “pre-established” and “de novo” sites (see Results) (μATACseq, IDR < 0.1). Heatmaps also indicate the presence of H3K4me1 and ATOH1 binding at the 18,681 hair cell–enriched distal elements. The dashed line separates the ATOH1 targets from non-ATOH1 targets in each group (IDR < 0.1). (D) Motif enrichment analysis of the 2,904 pre-established ATOH1 targets (Left). GO results associated with the pre-established ATOH1 targets suggest that enhancers for Notch signaling components are poised to be activated in the sensory progenitors (Middle). Hes6 locus serves as an example of the pre-established ATOH1 targets, which are already open, and H3K4me1-labeled, in the sensory progenitors (Right, gray bar). (E) Motif enrichment analysis of the 3,264 de novo ATOH1 targets (Left) indicating that POU-domain transcription factors are more enriched in the de novo ATOH1 targets compared to the pre-established ATOH1 targets. GO results indicate that ATOH1 targets within the de novo group are associated with hair cell differentiation and mechanosensation (Middle). The Gfi1 locus is representative of distal elements within the group of de novo ATOH1 targets, which are not open or labeled by active histone modifications in the sensory progenitor cells but become open and active (H3K27ac) following hair cell differentiation (Right, gray bar).
We next analyzed the changes in accessibility of distal regulatory elements (enhancers) to further explore the mechanisms that control the transition from E13.5 progenitors to E17.5 hair cells. We detected 64,803 distal elements from both progenitor and hair cell populations and observed that, in contrast to our promoter analysis, the accessibility of distal elements was highly dynamic (R = 0.41, Fig. 1B and SI Appendix, Fig. S2 C and D). We found that 21,662 distal elements were less accessible in E17.5 hair cells relative to E13.5 progenitors, while 18,681 distal elements were significantly more accessible (False Discovery Rate (FDR) < 0.1; Fig. 1B), and an additional 24,460 distal elements were not significantly changed during the transition from E13.5 progenitors to E17.5 hair cells.
We focused our analysis on the hair cell–enriched distal elements, as their gain in accessibility suggests a potential role in regulating changes in gene expression during the differentiation of hair cells (29–31). Of the 18,681 distal elements that were significantly enriched in E17.5 hair cells, around 60% (11,133) were also discernible (peak Irreproducible Discovery Rate (IDR) < 0.1) in undifferentiated E13.5 sensory progenitors (Fig. 1C), suggesting that these “pre-established” elements are potential enhancers, which are already primed for activation. To test this, we analyzed the undifferentiated E13.5 progenitors with CUT&RUN (C&R; ∼5,000 cells/replicate) (32) and found that these pre-established elements contain nucleosomes marked by H3K4me1 (Fig. 1C), a marker for primed enhancers (33, 34). In addition, these 11,133 pre-established chromatin regions include 2,904 distal elements that become targets of ATOH1, shortly after ATOH1 is first expressed in differentiating hair cells (Fig. 1C; ATOH1 C&R; IDR < 0.1). Several of these elements are associated with genes of the Notch signaling pathway in hair cells, such as Jag2, Dll1, Mfng, and Hes6 (Fig. 1D).
While around 60% of distal elements enriched in hair cells were pre-established in the progenitor population, the remaining ∼40% (7,548/18,681) of hair cell–enriched distal elements were not accessible in E13.5 sensory progenitors and appeared de novo in differentiating hair cells (Fig. 1 C, Bottom). Moreover, these elements were not labeled by H3K4me1 or other histone modifications (H3K27ac, H3K4me3, and H3K27me3; Fig. 1C and SI Appendix, Fig. S3 A and B) in E13.5 progenitors. This suggests these “de novo” distal elements are silent and inaccessible in sensory progenitors but become accessible and primed as the progenitors are induced to differentiate into hair cells. Of these 7,548 distal elements, we identified 3,264 elements as direct targets of ATOH1 by C&R (IDR < 0.1, Fig. 1C). This group of de novo ATOH1 targets, which are activated in differentiating hair cells, contain many putative enhancers near genes responsible for hair cell maturation and the development of mechanosensory machinery, such as Gfi1 (Fig. 1E), Lhx3, Tmc1, Insc, Lhfpl5, and Myo6 (SI Appendix, Fig. S4).
POU4F3 Is Necessary to Enable ATOH1 Access to its Targetome Early in Differentiating Hair Cells.
We next asked how ATOH1 is able to recognize its targets in inaccessible chromatin regions during the transition from sensory progenitors to differentiating hair cells. Structural analysis suggests that most bHLH transcription factors, including ATOH1, contain an extended recognition α-helix in the basic domain that limits their ability to bind nucleosome-occupied DNA (3, 35). Therefore, we hypothesized that other hair cell–specific transcription factors may be required to act as pioneer factors to overcome nucleosomal barriers and allow ATOH1 to fully engage its transcriptional targets. Indeed, we observed that the vast majority of the de novo distal elements lacked any of the active or repressive histone modifications we examined in sensory progenitors (SI Appendix, Fig. S3). This is similar to the requirement for pioneer factor activity during reprogramming of somatic cells to pluripotency (3). We performed motif enrichment analysis on all identified ATOH1 target elements and found that POU-domain transcription factor family motifs were more highly enriched in the de novo ATOH1 targets that become accessible in hair cells compared to pre-established ATOH1 targets that are already accessible in sensory progenitors (Fig. 1 E compared to D, 16.67% de novo sites contain POU motifs, while only 3.82% pre-established sites contain POU motifs).
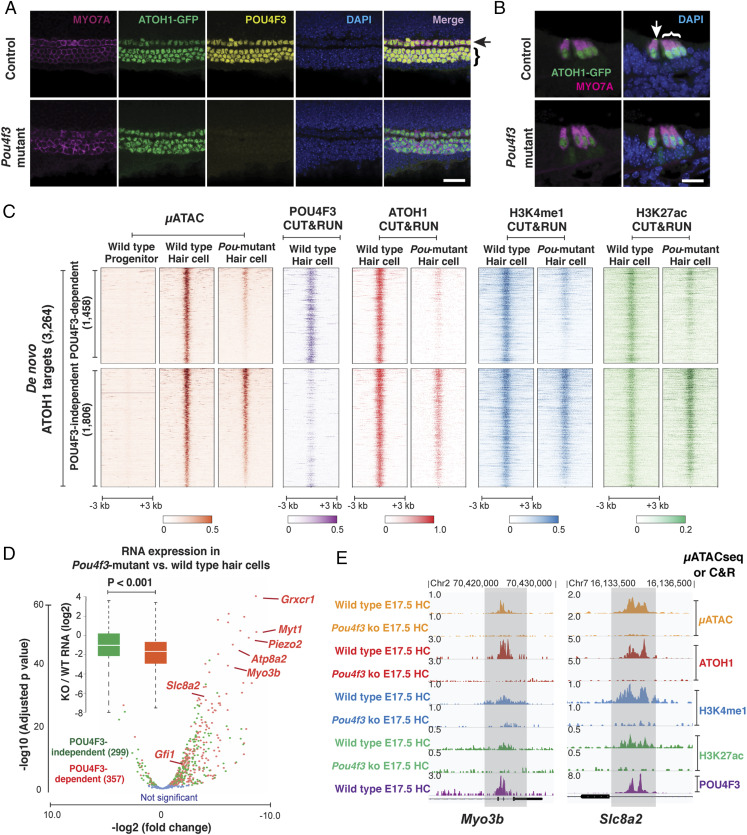
POU4F3 is an excellent candidate to act as a pioneer factor in hair cell differentiation, as it is the only POU-domain transcription factor highly expressed in hair cells (Fragments Per Kilobase of transcript per Million mapped reads (FPKM) > 400, SI Appendix, Fig. S5A). We therefore tested whether POU4F3 is necessary for the acquisition of chromatin accessibility at de novo ATOH1 targets in differentiating hair cells (Fig. 2). Previous studies have shown that cochlear hair cells begin to develop normally in the absence of Pou4f3 but then die rapidly after birth (36, 37). We used Atoh1-Gfp reporter mice (27) to show that the ATOH1-GFP fusion protein is still strongly expressed in four rows of MYO7A-positive hair cells in Pou4f3 mutant cochleae at E17.5 (Fig. 2 A and B), confirming that ATOH1 expression is not dependent on POU4F3 and enabling us to FACS-purify and analyze GFP+ hair cells from wild-type and Pou4f3 mutant cochleae at E17.5 (SI Appendix, Fig. S5B). Quantitative analysis of μATACseq profiles revealed that around 45% (1,458/3,264) of the de novo ATOH1 targets were significantly less accessible in Pou4f3 mutant hair cells (POU4F3-dependent ATOH1 targets; FDR < 0.1, fold change > 2; Fig. 2C). C&R profiling confirmed that POU4F3 preferentially bound to these sites (Fig. 2C) and is thus likely to directly regulate their chromatin accessibility. In addition to the reduced chromatin accessibility, ATOH1 binding at these 1,458 POU4F3-dependent ATOH1 targets was dramatically reduced in Pou4f3 mutant hair cells (Fig. 2C). In contrast, the binding of ATOH1 to the other 1,806 POU4F3-independent de novo ATOH1 targets was far less reduced in Pou4f3 mutant hair cells (Fig. 2C). These results indicate that POU4F3 binds nucleosomal DNA at inaccessible sites in progenitors (Fig. 1C) and is necessary for the chromatin rearrangement that occurs in many ATOH1 targets as progenitors differentiate into hair cells.
Fig. 2.
POU4F3 is necessary to provide access to ∼45% of de novo ATOH1 targets in nascent hair cells. (A and B) Confocal images of the E17.5 cochlea show ATOH1-GFP is highly expressed in one row of inner hair cells and three rows of outer hair cells in both wild-type and Pou4f3 mutant cochlea. Whole mount (A) and cryo-section (B) stained with antibody to hair cell marker MYO7A, confirming that the early specification of hair cells is not dependent on the expression of Pou4f3. (Scale bar, 20 μm.) (C) Heatmap representations of chromatin accessibility at the 3,264 de novo ATOH1 targets in sensory progenitors, wild-type hair cells, and Pou4f3 mutant hair cells (µATACseq). Distal elements are clustered into two groups, indicating a large group (1,458) of POU4F3-dependent loci (FDR < 0.1); Heatmap representation of POU4F3 C&R in wild-type hair cells, indicating preferential binding of POU4F3 to the POU4F3-dependent ATOH1 targets; Heatmap representations of C&R for ATOH1 in wild-type and Pou4f3 mutant hair cells, indicating that the loss of ATOH1 binding at these sites correlates with loss of Pou4f3 expression and loss of chromatin accessibility (μATACseq); Heatmap representations of C&R for H3K4me1 and H3K27ac in wild-type and Pou4f3 mutant hair cells, indicating that acquisition of active enhancer marks at POU4F3-dependent sites is correlated with Pou4f3 expression and chromatin accessibility (μATACseq). (D) Scatter plot and box plot representations comparing transcriptomes of wild-type and Pou4f3 mutant hair cells (RNA-seq). The red dots represent hair cell–specific genes within 50 kb of POU4F3-dependent ATOH1 targets. The green dots represent the hair cell–specific genes within 50 kb of POU4F3-independent ATOH1 targets. The blue dots represent the genes that are not significantly changed. Box plot Insert with median and quartile statistics, indicating that genes associated with POU4F3-dependent ATOH1 targets are specifically affected by the loss of Pou4f3 expression (P < 0.001). (E) Representative gene tracks at the putative enhancers for two deafness-related genes, Myo3b and Slc8a2. μATACseq and C&R data illustrate enhancer changes at genes whose expression is reduced in response to loss of POU4F3 binding at ATOH1 targets.
To investigate whether changes in gene expression occurred preferentially at POU4F3-dependent ATOH1 targets in Pou4f3 mutant hair cells, we compared the transcriptome of E17.5 hair cells from wild-type and Pou4f3 mutant cochleae by RNA sequencing (RNA-seq) (Fig. 2D). We focused our analysis on the 656 genes that were highly up-regulated during the transition from progenitors to nascent hair cells (E13.5 progenitors versus E17.5 hair cells, fold change > 4, FDR <0.1, FPKM >5; Dataset S1) and which lay within 50,000 base pairs (bp) of the 3,264 de novo ATOH1 targets (Dataset S2). The expression of most of these hair cell–specific genes was significantly reduced in Pou4f3 mutant hair cells (Fig. 2D). Notably, genes associated with POU4F3-dependent ATOH1 targets were predominately reduced in Pou4f3 mutants (P < 0.001, Fig. 2D, boxplot), including many genes related to mechanotransduction and human deafness (e.g., Grxcr1, Atp8a2, Adcy1, Ush2a, Myo3b, and Piezo2) (Fig. 2E).
The reduced expression of ATOH1 target genes in Pou4f3 mutant hair cells could be a consequence of the loss of enhancer activity at these POU4F3-dependent targets. We tested this by profiling active histone modifications at these elements (H3K4me1 and H3K27ac). We found the levels of H3K4me1 and H3K27ac at the POU4F3-dependent ATOH1 targets were significantly reduced in Pou4f3 mutants, whereas the levels of these marks of active enhancers remained largely unchanged at POU4F3-independent ATOH1 target elements (Fig. 2 C and E and SI Appendix, Fig. S6). Taken together, our results indicate that POU4F3 is necessary for accessibility of about one-half of de novo ATOH1 targets in nascent hair cells and is required for ATOH1 binding to these sites. In the absence of POU4F3 to open these distal elements, expression of these ATOH1 target genes is significantly reduced.
POU4F3 Acts as a Pioneer Factor to Bind to Closed Chromatin in a Heterologous Cell Type and Synergizes with ATOH1 to Open Inaccessible Chromatin.
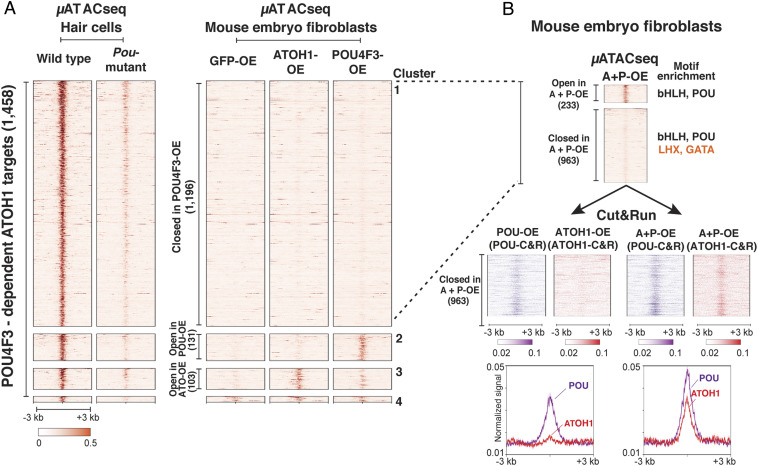
Previous unbiased screening for nucleosome binding ability of transcription factors predicts that POU-domain transcription factors have strong nucleosome binding potential (35). Therefore, we sought to determine if POU4F3 is associated with pioneer factor activity at ATOH1 target elements that require POU4F3 for their accessibility in hair cells, as shown (Fig. 2C). We overexpressed ATOH1 and POU4F3, alone and together in mouse embryonic fibroblasts (MEFs), followed by μATACseq to assess chromatin structure changes at the 1,458 POU4F3-dependent ATOH1 targets that we identified in hair cells (Figs. 2C and 3A).
Fig. 3.
POU4F3 binds closed chromatin and facilitates ATOH1 binding at many ATOH1-targets in a heterologous cell type. Viral overexpression of POU4F3 and ATOH1, alone and together, in MEFs was analyzed by μATACseq and C&R to determine the extent of transcription factor binding to inaccessible chromatin regions (B), and the requirement for cofactor activity to induce chromatin accessibility (A). (A) Heatmap representations of the μATACseq landscape at the 1,458 POU4F3-dependent ATOH1 targets identified in Fig. 2C. Data from viral overexpression of GFP (GFP-OE, control), ATOH1 (ATOH1-OE), and POU4F3 (POU4F3-OE) in MEFs was compared to μATACseq from wild-type hair cells and Pou4f3 mutant hair cells collected at E17.5 (provided for comparison; from Fig. 2C). The 1,458 distal elements that were dependent on Pou4f3 expression for chromatin accessibility in hair cells were reclustered on the basis of their chromatin accessibility in the infected MEFs. Cluster 1 (1,196), regions remaining closed in ATOH1-, POU4F3-, or GFP-overexpressing MEFs; Cluster 2 (131), regions opened by overexpression of POU4F3 alone; Cluster 3 (103), regions induced by overexpression of ATOH1 alone; and Cluster 4 (28), regions already accessible in control. (B) Heatmap representations of μATACseq results (Upper) from MEFs overexpressing ATOH1 and POU4F3 together at the 1,196 regions that remain closed following overexpression of individual transcription factors (Group 1 in A). Results show that a small number of these regions (233) were made accessible by the synergy between POU4F3 and ATOH1; however, the majority remained closed. C&R results for ATOH1 and POU4F3, shown as heatmaps and signal profiles (Lower Left arrow in B) at the 963 distal elements that fail to open when POU4F3 and ATOH1 are expressed independently, indicate that POU4F3 binds closed chromatin and ATOH1 does not (Left arrow). ATOH1 and POU4F3 synergize to provide access for ATOH1 when coexpressed with POU4F3 (Lower Right arrow in B) without affecting chromatin accessibility (B, Upper).
We clustered the 1,458 hair cell elements into four groups based on their µATACseq accessibility in MEFs (Fig. 3A). Although expressing POU4F3 or ATOH1 alone was able to “open” a small number of these sites (103 sites by ATOH1; 131 sites by POU4F3; and 28 sites in Control) (Fig. 3A), presumably through collaboration with unknown cofactors present in MEFs, while a large majority of POU4F3-dependent ATOH1 targets (1,196/1,458) remained closed in MEFs (Fig. 3A). Among these 1,196 sites that failed to open in response to ATOH1 or POU4F3 alone, 233 sites became accessible when ATOH1 and POU4f3 were overexpressed together in MEFs (Fig. 3B, Group 1), suggesting a synergy between these two factors. However, 963 remained closed, suggesting that additional factors not present in MEFs are needed to induce chromatin rearrangement at these sites. Motif enrichment analysis indicated that in addition to POU4F3 and ATOH1 cognate motifs, these 963 regions were also enriched for LHX and GATA transcription factor motifs (Fig. 3 B, Upper). Consistent with this, studies from our laboratory and others have shown that Lhx3 and Gata3 (38–41) are expressed in nascent hair cells but that these factors are not expressed in MEFs (41).
Despite the fact that ATOH1 and POU4F3 are unable to induce accessibility at these 963 sites, we tested whether either POU4F3 or ATOH1 are capable of nucleosomal binding to these sites using C&R (Fig. 3 B, Upper, group 2). POU4F3, when exogenously expressed in MEFs, was able to bind closed chromatin at these sites (Fig. 3 B, Lower), demonstrating a key characteristic of pioneer factor activity (1, 2). In contrast, ATOH1 was unable to bind to these sites in MEFs by itself (Fig. 3 B, Lower). Interestingly, when expressed together, ATOH1 and POU4F3 were able to synergize and allow ATOH1 binding, although the pair remained unable to increase chromatin accessibility based on the μATACseq profile (Fig. 3 B, Upper). Together, these data show that POU4F3 is able to bind to closed chromatin, while ATOH1 is not. However, POU4F3 can synergize with ATOH1 to allow ATOH1 engagement with closed chromatin.
ATOH1 and POU4F3 Synergize to Form a Feed-Forward Regulatory Circuit in Differentiating Hair Cells.
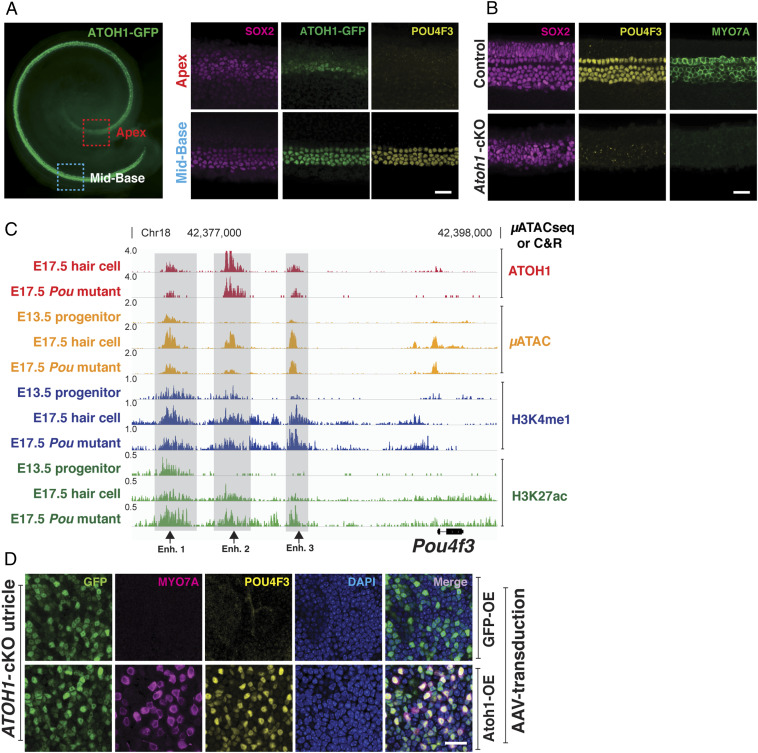
At the time that Atoh1 expression is initially up-regulated in progenitors, more than 50% of its putative enhancer targetome resides in closed chromatin (Fig. 1C) and thus cannot be bound by ATOH1 (Fig. 2C). Our data also show that POU4F3 is required to open chromatin associated with many of these distal elements that are bound by ATOH1 (Fig. 2); however, Pou4f3 messenger RNA has been reported to be expressed in the cochlea at a later time than ATOH1 (42, 43). To help explain this conundrum, we first confirmed that Atoh1 expression precedes Pou4f3 in the developing cochlea. Hair cell differentiation in the cochlea is known to propagate in a basal-to-apical gradient (9, 11, 44). At E15.5, we found that ATOH1 and POU4F3 proteins colocalized in four rows of hair cells at the base of the cochlea (Fig. 4A). However, in the less-differentiated apex, only ATOH1 could be detected in a group of progenitors/nascent hair cells (Fig. 4A). Moreover, when we used a Pax2-Cre transgenic mouse (45) to conditionally delete Atoh1 starting at E8.5, we were unable to detect POU4F3 protein in the E15.5 cochlear sensory epithelia (Fig. 4B). Together, these data indicate that ATOH1 expression precedes and is necessary for the expression of Pou4f3 in differentiating hair cells.
Fig. 4.
ATOH1 and POU4F3 synergize to form a feed-forward regulatory circuit in differentiating hair cells. (A) Whole mount of developing cochlea expressing ATOH1-GFP (green) at E15.5 (Leftl; red dashed rectangle, apical region; blue dashed rectangle, midbasal region). (Scale bar, 100 μm.) Higher-magnification images indicate that in the midbase, where hair cell differentiation first initiates, ATOH1 and POU4F3 are coexpressed; while in the apex, ATOH1 is expressed, but POU4F3 expression is not detected at this time within the basal-apical gradient characteristic of cochlear development. (Scale bar, 20 μm.) (B) Midbasal region of the cochlea at E15.5 in both wild-type and Atoh1 conditional knockout (Pax2-Cre) is shown. No hair cells are formed in the Atoh1 conditional knockout cochlea (MYO7A, green), and POU4F3 (yellow) is not expressed. (Scale bar, 20 μm.) (C) Genome browser (IGV) representation of the μATACseq and C&R results at the Pou4f3 locus. Putative ATOH1-bound enhancers for Pou4f3 up-regulation in the hair cells are indicated (gray bars). ATOH1-bound enhancers 1 and 3 are accessible in progenitors and labeled by H3K4me1 (black arrows). Binding of ATOH1 to all three indicated sites is not affected in Pou4f3 mutant. These data suggest that Pou4f3 expression in the hair cells is directly controlled by ATOH1. (D) Confocal images of utricular explant from Atoh1 conditional knockout mice. The utricles are overexpressed with GFP (control) or ATOH1-GFP, encoded in the AAV. Overexpression of ATOH1 but not GFP (control) induced the expression of POU4F3, suggesting that ATOH1 is sufficient for Pou4f3 expression in the inner ear. (Scale bar, 20 μm.)
Our data confirm that Atoh1 is epistatic to Pou4f3 (Fig. 4 A and B) and show that POU4F3 is required for the accessibility of ∼45% of the ATOH1 binding sites that appear de novo during hair cell differentiation (Fig. 2C). Moreover, we show that POU4F3 can bind to closed chromatin and enable the sequential binding of ATOH1 (Fig. 3 A and B). These observations invite the question of how these “closed” sites become accessible to ATOH1. To answer this, we hypothesized a model in which an epigenetic feed-forward circuit is initiated by the direct binding of ATOH1 to a preexisting open enhancer in sensory progenitors near the Pou4f3 locus. This binding of ATOH1 is sufficient to stimulate POU4F3 expression, which in turn makes more ATOH1 targets accessible through its pioneer activity. Consistent with this model, µATACseq and C&R results showed that ATOH1 directly bound to several accessible distal elements upstream of the Pou4f3 promoter in a POU4F3-independent manner (Fig. 4C), including one site (Fig. 4C; enhancer 3) which has previously been shown to be a bona fide hair cell–specific Pou4f3 enhancer (24). Analysis of µATACseq and H3K4me1 peaks showed that two of these distal elements (Fig. 4C; enhancer 1 and 3) were pre-established (µATACseq) and primed in E13.5 progenitors (H3K4me1-positive) (33, 34) prior to Pou4f3 expression. These data suggest that the initial up-regulation of Pou4f3 in the developing cochlea is directly controlled by ATOH1.
We next tested if ATOH1 overexpression is sufficient to drive Pou4f3 expression in the inner ear. We generated AAV2/Anc80L65 viruses (46) expressing ATOH1-T2A-GFP or GFP alone and used these to infect inner ear tissue from Atoh1 mutant mice. Since cochlear sensory progenitors undergo apoptosis in the absence of ATOH1 (SI Appendix, Fig. S7A) (11, 44), we overexpressed ATOH1 in the Atoh1 mutant utricle, a vestibular sensory organ in the inner ear containing hair cells. Unlike the cochlea, the utricular sensory epithelium in Atoh1 mutant mice does not degenerate embryonically but remains as a monolayer of SOX2-positive progenitor cells with a normal morphology (SI Appendix, Fig. S7B) (9). Reintroducing ATOH1 into Atoh1 mutant utricles induced expression of POU4F3 and other hair cell–specific proteins including MYO7A and LHX3 (Fig. 4D and SI Appendix, Fig. S7C), indicating that sensory epithelia from Atoh1 mutant utricles retain their potential to express Pou4f3 and differentiate into hair cells. Taken together, these data demonstrate that Pou4f3 is a direct downstream target of ATOH1 in differentiating hair cells and is part of a feed-forward mechanism that is deployed by ATOH1 to gain access to its 1,458 de novo targets.
The ATOH1-POU4F3 Feed-Forward Circuit Is Conserved in Mechanosensory Merkel Cells.
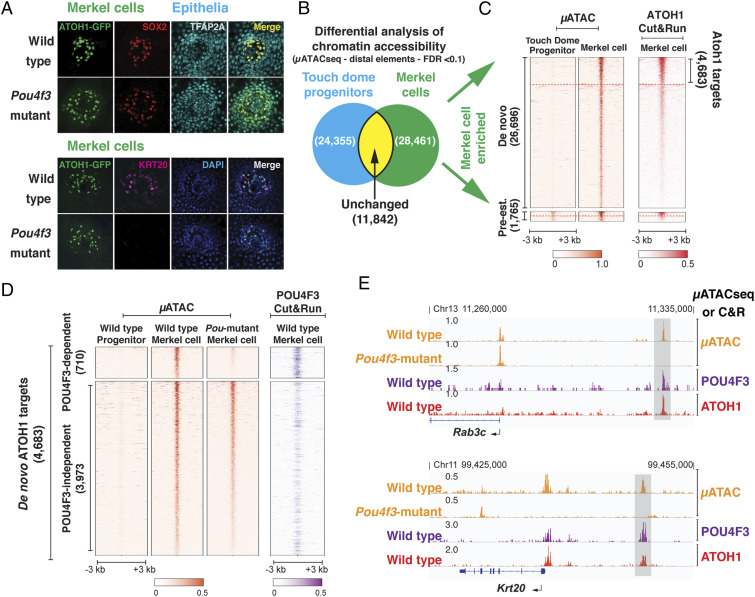
Merkel cells are a population of epidermally derived secondary receptor cells specialized for tactile discrimination and the sensation of light touch (47–49). Merkel cells and sensory hair cells share an ectodermal origin, but their lineages diverge in early embryogenesis, with Merkel cells deriving from nonneural ectoderm, while sensory hair cells derive entirely from the otic placode (7, 20, 21). Despite their different embryonic origins and their division of labor in perceiving different external mechanical stimuli, hair cells and Merkel cells share cellular and molecular features (50) and are suggested to be “sister” cell types, derived from an ur-mechanoreceptor (51). Notably, Merkel cells also express ATOH1 and POU4F3 and require ATOH1 for their differentiation (22). Immunostaining of developing touch domes in the epidermis of Pou4f3 mutant mice also containing an Atoh1-Gfp reporter show that Merkel cells lacking POU4F3 still express the early Merkel cell markers ATOH1 and SOX2 (Fig. 5A) (52) but not KRT20, a more-mature Merkel cell keratin (Fig. 5A) (53). This suggests that, like hair cells, Merkel cells are specified in the absence of Pou4f3 but cannot complete their program of differentiation. Thus, the feed-forward role of POU4F3 seen in hair cells may also be important in the maturation of Merkel cells.
Fig. 5.
The ATOH1-POU4F3 feed-forward circuit is conserved in Merkel cells. (A). Confocal images of the touch domes from wild-type and Pou4f3-mutant hairy skin. The expression of early Merkel cell markers (ATOH1 and SOX2) in the touch dome is not dependent on Pou4f3 expression. However, the mature Merkel cell marker KRT20 is not expressed in Pou4f3 mutant Merkel cells. (Scale bar, 40 μm.) (B) Venn diagram representing a quantitative comparison of chromatin accessibility at the distal elements between the TDEPs and Merkel cells. A total of 28,461 distal elements are significantly more open in the differentiating Merkel cells (Merkel cell–enriched distal elements), compared to 24,355 distal elements that are more enriched in progenitors. A total of 11,842 distal elements remain unchanged in the two populations (E17.5; FDR < 0.1). (C) Heatmap representation of chromatin accessibility (µATACseq) and ATOH1 binding (C&R) at the 28,461 Merkel cell–enriched distal elements. These elements were separated into pre-established (1,765) and de novo (26,696) clusters. C&R results show ATOH1 targets (4,683) in the de novo cluster (IDR < 0.1). As shown in the hair cells (Fig. 1C), ATOH1 binds to both pre-established and de novo distal elements in Merkel cells. (D) Heatmap representation of the μATACseq results at the 4,683 de novo ATOH1 targets (Left), from wild-type TDEPs, wild-type Merkel cells, and Pou4f3 mutant Merkel cells. Distal elements are clustered into POU4F3 dependent (710) and independent (3,973) based on whether the chromatin accessibility is significantly reduced in Pou4f3 mutant Merkel cells compared to wild-type Merkel cells (FDR < 0.1). POU4F3 C&R from wild-type Merkel cells (Right) indicates direct binding at POU4F3-dependent ATOH1 targets in Merkel cells. (E) Representative genome browser tracks (IGV) of μATACseq and C&R data at two POU4F3-dependent ATOH1 targets in Merkel cells, Rab3c, and Krt20 (Dataset S3).
These findings prompted us to investigate whether POU4F3 also regulates the accessibility of ATOH1 targets in a feed-forward manner during the maturation of Merkel cells. Merkel cell progenitors (touch dome epithelial progenitors, TDEP) reside among the epithelial precursors of touch domes in E17.5 epidermis and can be isolated by FACS purification using a unique combination of expressed antigens (Integrin-α-6, Cd34, Sca-1, and CD200) (54), whereas differentiating Merkel cells can be isolated by their expression of ATOH1-GFP. We used Atoh1-Gfp mice and Atoh1-Gfp; Pou4f3 mutant mice to FACS purify TDEPs and Merkel cells from wild-type and Pou4f3 mutant epidermis. We analyzed accessible chromatin with μATACseq and identified 28,461 out of a combined 64,658 open distal elements that were enriched in E17.5 Merkel cells compared to TDEPs (DESeq2, FDR < 0.1; Fig. 5B), while 24,355 elements were more accessible in touch dome epithelial precursors, and 11,842 distal elements remained unchanged during the differentiation process.
We focused our analysis on the 28,461 Merkel cell–enriched distal elements and found that a small portion of these were already accessible (pre-established) in the TDEPs (1,756/28,461), while the majority arose de novo during Merkel cell differentiation (26,696/28,461) (Fig. 5C), of which 4,683 (IDR < 0.1) were highly enriched for ATOH1 binding (Fig. 5C). To determine the number of these de novo ATOH1 targets that are dependent on POU4F3 for accessibility, we compared their µATACseq profile between wild-type and Pou4f3 mutant Merkel cells. We found that the accessibility of 710 of these 4,683 de novo ATOH1 target elements was dependent on the expression of POU4F3 and that these 710 elements bound POU4F3 (Fig. 5D). Many of these POU4F3-dependent de novo ATOH1 target elements are near genes critical for Merkel cell maturation and mechanotransduction, such as Krt20 and Rab3c (Fig. 5E) (53, 55) and the cardinal light-touch mechanotransduction channel Piezo2 (Fig. 6D) (47–49, 56). RNA-seq confirmed that expression of Piezo2, Rab3c, and Krt20 was significantly reduced in Pou4f3 mutant Merkel cells (FDR < 0.1; Dataset S3 and SI Appendix, Fig. S8). Together, these results suggest that, just as in hair cells, the pioneer factor activity of POU4F3 is required for ATOH1 to correctly coordinate mechanosensory differentiation in Merkel cells.
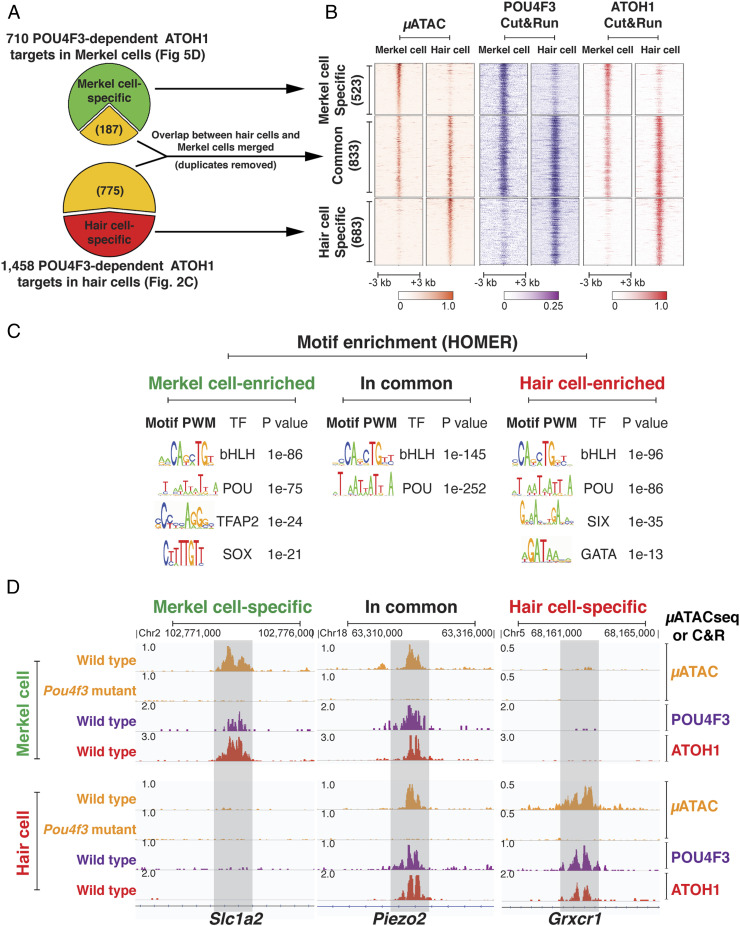
Fig. 6.
The ATOH1-POU4F3 synergy represents a conserved gene regulatory mechanism directing shared and divergent enhancer networks in different mechanosensory cell types. (A) Diagrams show the differential comparison of chromatin accessibility at the POU4F3-dependent ATOH1 targets specific to Merkel cells and hair cells. Of the 710 (Fig. 5D) POU4F3-dependent ATOH1 binding sites in Merkel cells, 523 were specific to Merkel cells compared to hair cells, while 187 were also accessible in hair cells. Of the 1,458 (Fig. 2C) POU4F3-dependent ATOH1 binding sites in hair cells, 683 were specific to hair cells, while 775 were also accessible in Merkel cells. Of the 187 + 775 common sites present in the two cell types, 833 were nonoverlapping (common). (B) Heatmap shows the chromatin accessibility and the binding profile of POU4F3 and ATOH1 at Merkel cell–specific, hair cell–specific, and common sites of POU4F3-dependent ATOH1 targets revealed in A. POU4F3 is able to weakly bind to Merkel cell–specific sites which are closed in hair cells and hair cell–specific sites which are closed in Merkel cells, consistent with its pioneer factor activity. (C) Motif enrichment analysis of POU4F3-dependent ATOH1 targets sites among Merkel cell–specific, Hair cell–specific, and common sites revealed in A. Different sets of transcription factor motifs are enriched in Merkel cell–specific and hair cell–specific POU4F3-dependent ATOH1 targets, suggesting a context-dependent specificity for the opening and activation of POU4F3-dependent ATOH1 targets in different cell types. (D) Genome browser representation of the μATACseq and C&R data at examples of the POU4F3-dependent ATOH1-targets that are Merkel cell specific (Slc1a2), common (Piezo2), and hair cell specific (Grxcr1).
The ATOH1-POU4F3 Synergy Represents a Conserved Gene Regulatory Mechanism Directing Shared and Divergent Enhancer Networks in Different Mechanosensory Cell Types.
It has been suggested that sensory hair cells and Merkel cells arose evolutionarily from a common mechanoreceptor cell type (51). One mechanism for such a divergence would be the appearance of unique enhancer targets in each sister cell that depend on ATOH1/POU4F3 synergy to potentiate a pool of regulatory elements that contain the motifs of both transcription factors but ultimately diverge in their utilization.
Of the 710 POU4F3-dependent ATOH1 targets identified in Merkel cells (Fig. 5D), only 523 distal elements are specific to Merkel cells compared to hair cells (Fig. 6A), while the remaining 187 are shared with hair cells. Of the 1,458 POU4F3-dependent ATOH1 targets identified in hair cells (Fig. 2C), only 683 distal elements are specific to hair cells compared to Merkel cells (Fig. 6A), while the remaining 775 are shared with Merkel cells. C&R analysis showed that POU4F3 and ATOH1 binding also diverged in a cell type–specific manner (Fig. 6 B and D). Among the 683 hair cell–specific POU4F3-dependent ATOH1 targets were genes encoding unique components of the hair cell mechanotransduction machinery, for instance Grxcr1 (Fig. 6D), Lhfpl5, Ush2a, Clrn1, Cdh23, and Myo3b (57, 58). Gene ontology (GO) terms associated with these distal elements included “stereocilium” and “cluster of actin-based cell projections” (SI Appendix, Fig. S9 A and B), while the 523 putative regulatory elements enriched in Merkel cells generated GO terms such as glutamatergic synapses, integrin complex, and inward rectifier potassium channel complex (SI Appendix, Fig. S9A). Motif enrichment analysis revealed that the binding sites for SIX and GATA transcription factor family members were differentially represented among the 683 hair cell–specific POU4F3-dependent ATOH1 targets, compared to those in Merkel cells (Fig. 6C). Immunostaining confirmed that SIX1 was broadly expressed in the cochlea at E13.5 and E17.5 (SI Appendix, Fig. S10) (42, 59), as is GATA3 (39, 40). In contrast, TFAP2 and SOX-factor motifs were differentially enriched in the Merkel cell–specific sites (Fig. 6C), and we found that TFAP2A is expressed in Merkel cells as well as the epidermal cells within and surrounding the touch domes (Fig. 5A) (60).
In contrast to the POU4F3-dependent ATOH1 targets that are differentially open between hair cells and Merkel cells, we found that 833 distal elements were commonly accessible and appear POU4F3 dependent in at least one cell type (μATACseq; Fig. 6 A and B). C&R analysis showed that POU4F3 and ATOH1 bound to these common regions in both hair cells and Merkel cells (Fig. 6B). GO analysis showed that these shared regulatory elements are enriched near genes responsible for synaptogenesis and ion channels (SI Appendix, Fig. S9A). For example, the PIEZO2 channel protein is present in both Merkel cells and hair cells. While PIEZO2 is crucial for Merkel cell mechanosensitivity, it nonetheless contributes to the function of both cell types (47–49, 56, 61). We identified a potential intronic enhancer in the Piezo2 locus (H3K27ac+ and H3K4me1+ in hair cells) that was bound by ATOH1 and POU4F3 in both hair cells and Merkel cells (Fig. 6D) and whose chromatin was not accessible in either cell type in Pou4f3 mutant mice (Fig. 6D). This suggests that the ATOH1/POU4F3 feed-forward circuit may affect Piezo2 expression in both mechanosensory cell types by regulating accessibility and activity at the same regulatory element.
Interestingly, POU4F3 seems to bind weakly to many of the Merkel cell–specific targets that lie in closed chromatin in hair cells and vice versa (Fig. 6B), consistent with the ability of POU4F3 to bind to closed chromatin. Taken together, we propose that the ATOH1/POU4F3 feed-forward circuit may constitute part of the core gene regulatory mechanism for mechanosensation, which is complemented by distinct enhancer networks that are regulated by lineage-specific transcription factors to allow the differentiation of specific mechanosensory cell types.
Discussion
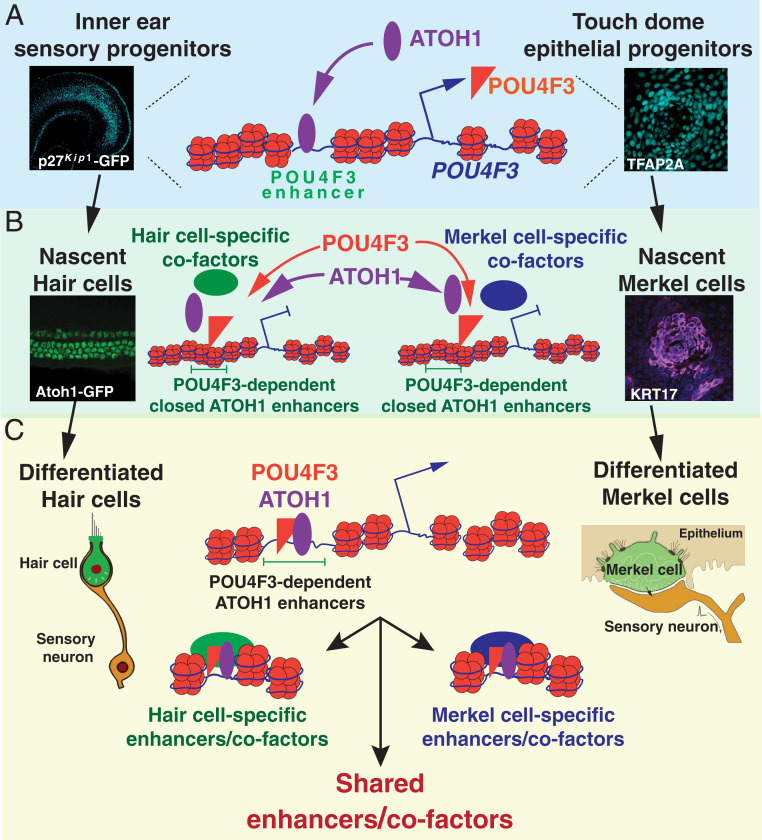
During development and regeneration, the correct coordination of progenitor cell differentiation relies on changing patterns of gene expression, which in turn relies on specific patterns of chromatin accessibility and epigenetic modifications (31, 62). We used the differentiation of sensory hair cells in the developing inner ear to show that a significant number of chromatin targets of the bHLH transcription factor ATOH1 are located in inaccessible chromatin when Atoh1 is first up-regulated in undifferentiated cochlear sensory progenitors (Fig. 1C). We found a feed-forward synergy between ATOH1 and a downstream transcription factor POU4F3, in which ATOH1 first stimulates the expression of Pou4f3 in differentiating hair cells, with POU4F3 then making additional ATOH1 targets accessible to ATOH1 binding by acting as a pioneer factor (Fig. 7). Remarkably, this feed-forward mechanism is conserved in another mechanosensory cell type, Merkel cells. We hypothesize that this feed-forward mechanism arose in an ancestral neurosensory mechanoreceptor cell type and may have provided a basis for sister-cell evolution through enhancer network divergence between modern mechanoreceptors (51).
Fig. 7.
Working model: ATOH1/POU4F3 feed-forward circuit controls the differentiation of hair cells and Merkel cells. (A) POU4F3 is a direct target of ATOH1. ATOH1 is up-regulated in the inner ear sensory progenitors and the TDEPs during their differentiation. ATOH1 binds to pre-established enhancers upstream of the Pou4f3 gene to stimulate the expression of POU4F3. (B) POU4F3 has pioneer factor activity. POU4F3 can recognize its cognate motifs in nucleosome-occupied DNA and is required for ATOH1 binding in closed chromatin. The binding of POU4F3 and ATOH1 does not result in the opening or the activation of the regulatory elements in the closed chromatin. (C) Context-dependent opening and activation of POU4F3-dependent ATOH1 targets. POU4F3, ATOH1, and other hair cell–specific transcription factors/cofactors can open and activate an array of hair cell–specific enhancers for mechanosensory differentiation in hair cells (e.g., stereocilia) tip link mechanotransduction apparatus formation. On the other hand, POU4F3, ATOH1, and other Merkel cell–specific transcription factors/cofactors can open and activate an array of Merkel cell–specific enhancers for mechanosensory differentiation in Merkel cells.
We present several lines of evidence for the feed-forward synergy between ATOH1 and POU4F3. First, our μATACseq analysis of hair cell progenitors shows that over 50% of ATOH1 binding sites in hair cells are in an epigenetically inaccessible state in progenitors. This implies that additional mechanisms must, by necessity, be deployed to make these binding sites accessible before further steps in hair cell development can proceed. Second, we show the distal elements that are inaccessible to ATOH1 are enriched for POU factor binding sites (Fig. 1E) and that POU4F3 is able to bind these elements when overexpressed in MEFs, where ATOH1 is unable to bind them (Fig. 3B). Third, our analysis of Pou4f3-null mice shows that POU4F3 binding is necessary for these elements to become accessible (Fig. 2C) and for the hair cell genes associated with them to be transcribed (Fig. 2D). Finally, we show that Atoh1 is required for Pou4f3 expression and that Pou4f3 is a direct transcriptional target of ATOH1 (Fig. 4). After ATOH1 levels begin to rise in differentiating hair cells, POU4F3 levels rise sufficiently to stimulate the pioneer factor activity needed to make additional hair cell loci accessible, allowing ATOH1 to continue driving the differentiation of hair cells. We suggest that this feed-forward mechanism allows POU4F3-expressing cells to fully commit to a hair cell fate by opening the final sets of enhancers needed for ATOH1 to complete the process of hair cell differentiation. One prediction of this mechanism would be that Cre-Lox lineage labeling of Pou4f3-expressing cells should label significantly fewer supporting cells than Atoh1-Cre mice (14, 15), as cells expressing ATOH1 and POU4F3 would be more committed to a hair cell fate than cells expressing ATOH1 alone.
Our data clearly demonstrate a need for synergy between ATOH1 and POU4F3 in positively promoting hair cell differentiation. However, our RNA-seq analysis of Pou4f3-null mice also supports a role for these factors in repressing alternative neurosensory fates. We show that many genes normally expressed by spiral ganglion neurons that innervate hair cells are ectopically expressed in the Pou4f3 mutant hair cells, including NeuroD1, Pou4f1, and Lhx2 (SI Appendix, Fig. S5A), suggesting that the derepression of a neuronal differentiation program in Pou4f3 mutants is due to the loss of an active inhibitory mechanism. Evidence suggests that this active inhibitory mechanism in wild-type hair cells may be mediated by the transcriptional repressor GFI1 (63–65), which we show is also one of the POU4F3-dependent ATOH1 targets in hair cells (Fig. 2D and Dataset S1). Together, these data suggest that the ATOH1/POU4F3 feed-forward synergy may indirectly repress neuronal programs such as axon formation in order to generate the axonless mechanosensory hair cells.
Atoh1 is expressed in developing hair cells and Merkel cells (9, 22, 66), intestinal secretory cell lineages (67), cerebellar granule precursors (68), and subpopulations of excitatory neurons in the hindbrain (27, 69). Pou4f3 is expressed in a variety of cell types, such as hair cells (36, 70), Merkel cells (24), dorsal root ganglion neurons (71), and some retinal ganglion neurons (72). However, combinatorial expression of Atoh1 and Pou4f3 has so far only been observed in hair cells and Merkel cells, suggesting that the ATOH1/POU4F3 feed-forward synergy that we describe may have a general role of promoting mechanosensory differentiation (71).
In the mechanosensory module of the nematode Caenorhabditis elegans, the single bHLH and POU-IV homologs Lin-32 and Unc-86 are both indispensable for the formation of their six touch receptor neurons (73). In contrast, in the mouse inner ear, distinct combinations of bHLH and POU-IV homologs are required for the differentiation of hair cells (Atoh1 and Pou4f3) and the sensory neurons that innervate them (NeuroD1 and Pou4f1). It is likely that successive duplication of bHLH and POU family members during chordate evolution permitted the division of labor of a mechanosensitive sensory neuron into two cell types—a mechanosensitive secondary receptor cell (hair cells or Merkel cells) and a primary sensory neuron (spiral ganglion neurons of the inner ear, or SA1 afferent neurons of the skin). It is interesting to speculate on the relationship of epidermally derived Merkel cells and placodally derived hair cells to the primary and secondary sensory receptor cells seen in nonvertebrate chordates (74–77) and whether the ATOH1/POU4F3 feed-forward mechanism we describe here also plays a role in the development of mechanosensory cells throughout the chordate lineage. It will be interesting to investigate whether other POU-bHLH family interactions may specify additional terminal developmental outcomes, further suggesting the evolutionarily conserved nature of this feed-forward interaction and its utilization in the evolution of new cell types.
Materials and Methods
A detailed description of materials and methods is included in the SI Appendix accompanying this paper. Animals: Experimental procedures involving animals were approved by the Animal Care and Use Committee of the University of Southern California. Histochemistry: As previously described in Doetzlhofer et al. (78). Cell purification by FACS: Cochlear cells were purified as described (78). Merkel cells were purified as described (54). Chromatin structure and Epigenetic analysis: ATACseq and CUT&RUN were described by Buenrostro et al. (28) and Skene and Henikoff (32), respectively, with modifications described in SI Appendix, Materials and Methods. Bioinformatics: ENCODE pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline) was adapted for quality control and data analysis for next generation sequencing data (see SI Appendix, Materials and Methods for details of analysis). Overexpression of transcription factors: Adeno-associated vrius was prepared as described in Suzuki et al. (46), and lentivirus was produced as described in Menendez et al. (41).
Supplementary Material
Acknowledgments
We thank Dr. Andrew Groves (Baylor College of Medicine), Dr. Ksenia Gnedeva (University of Southern California), and members of the N.S. laboratory for helpful discussion and careful reading/editing of the manuscript. We thank Welly Makmura for technical assistance throughout this project and Francis James for his contributions to constructing the pipelines for the RNA-seq, μATAC-seq, and C&R data and quality control analysis. We thank Dr. Steven Henikoff (Fred Hutchinson Cancer Research Center) for providing the Protein A/MNase fusion construct used for C&R, Dr. Andrew Groves for Pax2-Cre mice, and Drs. Edwin Rubel and Jennifer Stone for Pou4f3-Dtr mice. This work was supported by grants to N.S. from the National Institute on Deafness and Other Communication Disorders (NIDCD) (Grant No. R01DC015829; Grant No. R01DC015530) and from the Hearing Restoration Project of the Hearing Health Foundation. T.T. was supported by Grant Nos. T32DC009975 and F31DC017376 from the NIDCD; J.D.N. was supported by Grant No. T32HD060549 from National Institute of Child Health and Human Development (NICHD) and Grant No. F31DC018703 from the NIDCD.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105137118/-/DCSupplemental.
Data Availability
NexGen sequence data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GSE150391).
References
- 1.Iwafuchi-Doi M., Zaret K. S., Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwafuchi-Doi M., Zaret K. S., Cell fate control by pioneer transcription factors. Development 143, 1833–1837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soufi A., et al., Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirillo L. A., et al., Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Lee C. S., Friedman J. R., Fulmer J. T., Kaestner K. H., The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Kelley M. W., Regulation of cell fate in the sensory epithelia of the inner ear. Nat. Rev. Neurosci. 7, 837–849 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Groves A. K., Zhang K. D., Fekete D. M., The genetics of hair cell development and regeneration. Annu. Rev. Neurosci. 36, 361–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson P. J., Huarcaya Najarro E., Sayyid Z. N., Cheng A. G., Sensory hair cell development and regeneration: Similarities and differences. Development 142, 1561–1571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bermingham N. A., et al., Math1: An essential gene for the generation of inner ear hair cells. Science 284, 1837–1841 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Zheng J. L., Gao W.-Q., Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3, 580–586 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Chen P., Johnson J. E., Zoghbi H. Y., Segil N., The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495–2505 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Woods C., Montcouquiol M., Kelley M. W., Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci. 7, 1310–1318 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Cai T., et al., Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 35, 5870–5883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Xie X., Deng M., Chen X., Gan L., Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 48, 407–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driver E. C., Sillers L., Coate T. M., Rose M. F., Kelley M. W., The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev. Biol. 376, 86–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanford P. J., et al., Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat. Genet. 21, 289–292 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Kiernan A. E., Cordes R., Kopan R., Gossler A., Gridley T., The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development 132, 4353–4362 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Tateya T., Imayoshi I., Tateya I., Ito J., Kageyama R., Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev. Biol. 352, 329–340 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Abdolazimi Y., Stojanova Z., Segil N., Selection of cell fate in the organ of Corti involves the integration of Hes/Hey signaling at the Atoh1 promoter. Development 143, 841–850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison K. M., Miesegaes G. R., Lumpkin E. A., Maricich S. M., Mammalian Merkel cells are descended from the epidermal lineage. Dev. Biol. 336, 76–83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Keymeulen A., et al., Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J. Cell Biol. 187, 91–100 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maricich S. M., et al., Merkel cells are essential for light-touch responses. Science 324, 1580–1582 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vahava O., et al., Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science 279, 1950–1954 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Masuda M., et al., Regulation of POU4F3 gene expression in hair cells by 5′ DNA in mice. Neuroscience 197, 48–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda R., Pak K., Chavez E., Ryan A. F., Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Mol. Neurobiol. 51, 672–684 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y.-S., Liu F., Segil N., A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 133, 2817–2826 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Rose M. F., et al., Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron 64, 341–354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank C. L., et al., Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat. Neurosci. 18, 647–656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller B., et al., Widespread changes in nucleosome accessibility without changes in nucleosome occupancy during a rapid transcriptional induction. Genes Dev. 31, 451–462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klemm S. L., Shipony Z., Greenleaf W. J., Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Skene P. J., Henikoff S., An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Creyghton M. P., et al., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calo E., Wysocka J., Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez Garcia M., et al., Structural features of transcription factors associating with nucleosome binding. Mol. Cell 75, 921–932.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang M., Gao W. Q., Hasson T., Shin J. J., Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development 125, 3935–3946 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Hertzano R., et al., Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum. Mol. Genet. 13, 2143–2153 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Hertzano R., et al., Lhx3, a LIM domain transcription factor, is regulated by Pou4f3 in the auditory but not in the vestibular system. Eur. J. Neurosci. 25, 999–1005 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Luo X. J., et al., GATA3 controls the specification of prosensory domain and neuronal survival in the mouse cochlea. Hum. Mol. Genet. 22, 3609–3623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bardhan T., et al., Gata3 is required for the functional maturation of inner hair cells and their innervation in the mouse cochlea. J. Physiol. 597, 3389–3406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menendez L., et al., Generation of inner ear hair cells by direct lineage conversion of primary somatic cells. eLife 9, e55249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed M., et al., Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 22, 377–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan N., et al., A novel Atoh1 “self-terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS One 7, e30358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai T., Seymour M. L., Zhang H., Pereira F. A., Groves A. K., Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J. Neurosci. 33, 10110–10122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohyama T., Groves A. K., Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis 38, 195–199 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Suzuki J., Hashimoto K., Xiao R., Vandenberghe L. H., Liberman M. C., Cochlear gene therapy with ancestral AAV in adult mice: Complete transduction of inner hair cells without cochlear dysfunction. Sci. Rep. 7, 45524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda R., et al., Merkel cells transduce and encode tactile stimuli to drive Aβ-afferent impulses. Cell 157, 664–675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maksimovic S., et al., Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature 509, 617–621 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo S.-H., et al., Piezo2 is required for Merkel-cell mechanotransduction. Nature 509, 622–626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lumpkin E. A., Caterina M. J., Mechanisms of sensory transduction in the skin. Nature 445, 858–865 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Arendt D., et al., The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Laga A. C., et al., Expression of the embryonic stem cell transcription factor SOX2 in human skin: Relevance to melanocyte and Merkel cell biology. Am. J. Pathol. 176, 903–913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perdigoto C. N., Bardot E. S., Valdes V. J., Santoriello F. J., Ezhkova E., Embryonic maturation of epidermal Merkel cells is controlled by a redundant transcription factor network. Development 141, 4690–4696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doucet Y. S., Woo S.-H., Ruiz M. E., Owens D. M., The touch dome defines an epidermal niche specialized for mechanosensory signaling. Cell Rep. 3, 1759–1765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haeberle H., et al., Molecular profiling reveals synaptic release machinery in Merkel cells. Proc. Natl. Acad. Sci. U.S.A. 101, 14503–14508 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ranade S. S., et al., Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516, 121–125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pepermans E., Petit C., The tip-link molecular complex of the auditory mechano-electrical transduction machinery. Hear. Res. 330 (Pt A), 10–17 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Petit C., Richardson G. P., Linking genes underlying deafness to hair-bundle development and function. Nat. Neurosci. 12, 703–710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang T., Xu J., Maire P., Xu P.-X., Six1 is essential for differentiation and patterning of the mammalian auditory sensory epithelium. PLoS Genet. 13, e1006967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L., et al., TFAP2C- and p63-dependent networks sequentially rearrange chromatin landscapes to drive human epidermal lineage commitment. Cell Stem Cell 24, 271–284.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Z., et al., Mechanosensory hair cells express two molecularly distinct mechanotransduction channels. Nat. Neurosci. 20, 24–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cantone I., Fisher A. G., Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 20, 282–289 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Wallis D., et al., The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130, 221–232 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Costa A., Powell L. M., Soufi A., Lowell S., Jarman A. P., Atoh1 is repurposed from neuronal to hair cell determinant by Gfi1 acting as a coactivator without redistribution of its genomic binding sites. bioRxiv [Preprint] (2019). 10.1101/767574 (Accessed 6 June 2020). [DOI]
- 65.Matern M. S., et al., GFI1 functions to repress neuronal gene expression in the developing inner ear hair cells. Dev. Camb. Engl. 147, dev186015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lumpkin E. A., et al., Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389–395 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Yang Q., Bermingham N. A., Finegold M. J., Zoghbi H. Y., Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294, 2155–2158 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Ben-Arie N., et al., Math1 is essential for genesis of cerebellar granule neurons. Nature 390, 169–172 (1997). [DOI] [PubMed] [Google Scholar]
- 69.Helms A. W., Johnson J. E., Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development 125, 919–928 (1998).9449674 [Google Scholar]
- 70.Xiang M., et al., Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc. Natl. Acad. Sci. U.S.A. 94, 9445–9450 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Badea T. C., et al., Combinatorial expression of Brn3 transcription factors in somatosensory neurons: Genetic and morphologic analysis. J. Neurosci. 32, 995–1007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sajgo S., et al., Molecular codes for cell type specification in Brn3 retinal ganglion cells. Proc. Natl. Acad. Sci. U.S.A. 114, E3974–E3983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chalfie M., Au M., Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science 243, 1027–1033 (1989). [DOI] [PubMed] [Google Scholar]
- 74.Burighel P., Caicci F., Manni L., Hair cells in non-vertebrate models: Lower chordates and molluscs. Hear. Res. 273, 14–24 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Burighel P., et al., Does hair cell differentiation predate the vertebrate appearance? Brain Res. Bull. 75, 331–334 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Gasparini F., et al., Cytodifferentiation of hair cells during the development of a basal chordate. Hear. Res. 304, 188–199 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Tournière O., et al., NvPOU4/Brain3 functions as a terminal selector gene in the nervous system of the cnidarian Nematostella vectensis. Cell Rep. 30, 4473–4489.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Doetzlhofer A., et al., Hey2 regulation by FGF provides a notch-independent mechanism for maintaining pillar cell fate in the organ of corti. Dev. Cell 16, 58–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NexGen sequence data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GSE150391).