Significance
Programmed cell death protein-1 (PD-1) inhibitory antibodies, often referred to as checkpoint inhibitors, have revolutionized the treatment of cancer. However, there is an unmet need for PD-1 agonists to treat autoimmune disorders. Herein, we describe the de novo design of a small, stable PD-1 binding protein and development of a synthetic PD-1 agonist.
Keywords: protein design, PD-1, autoimmunity, immunotherapy
Abstract
Programmed cell death protein-1 (PD-1) expressed on activated T cells inhibits T cell function and proliferation to prevent an excessive immune response, and disease can result if this delicate balance is shifted in either direction. Tumor cells often take advantage of this pathway by overexpressing the PD-1 ligand PD-L1 to evade destruction by the immune system. Alternatively, if there is a decrease in function of the PD-1 pathway, unchecked activation of the immune system and autoimmunity can result. Using a combination of computation and experiment, we designed a hyperstable 40-residue miniprotein, PD-MP1, that specifically binds murine and human PD-1 at the PD-L1 interface with a Kd of ∼100 nM. The apo crystal structure shows that the binder folds as designed with a backbone RMSD of 1.3 Å to the design model. Trimerization of PD-MP1 resulted in a PD-1 agonist that strongly inhibits murine T cell activation. This small, hyperstable PD-1 binding protein was computationally designed with an all-beta interface, and the trimeric agonist could contribute to treatments for autoimmune and inflammatory diseases.
Proper immune system function involves a dynamic and delicate balance of stimulatory and inhibitory receptors that generate an appropriate response when a foreign antigen is encountered and then return the system to stasis to prevent chronic overactivation that could damage healthy tissue. Programmed cell death protein-1 (PD-1) is an inhibitory receptor expressed on most immune cells (1), including activated T cells and regulatory T cells. Upon engagement with one of its ligands, programmed cell death-1 ligand 1 (PD-L1) (2) or programmed cell death-1 ligand 2 (PD-L2) (3), on the surface of another cell, PD-1 signals to suppress the T cell, leading to decreased production of proinflammatory cytokines, inhibition of effector function, and apoptosis (4). The PD-1/PD-L1 pathway is also important in the development (5) and maintenance (6) of both central and peripheral tolerance, in part via regulatory T cells. Due to its critical role in maintaining a healthy, balanced immune system, alterations in PD-1 function in either direction can lead to disease. Many tumors use this pathway to evade the immune system by overexpressing PD-L1 and/or PD-L2 to deactivate tumor-infiltrating lymphocytes (TILs). On the other hand, a decrease in function of the PD-1 pathway can lead to autoimmune and inflammatory phenotypes. Mice lacking PD-1 develop various autoimmune symptoms, including autoimmune cardiomyopathy (7) and a lupus-like disease (8), depending on the genetic background (9). In humans, mutations that affect the regulation of PD-1 have been associated with systemic lupus erythematosus (10), rheumatoid arthritis (11), and progression of multiple sclerosis (12).
Antagonistic antibodies targeting PD-1 and PD-L1, often referred to as checkpoint inhibitors, have revolutionized the treatment of cancer (13). Despite the therapeutic potential of this same pathway in autoimmunity, few agonist PD-1 antibodies have been identified, and no PD-1 agonists have yet been approved for clinical use (14). Several groups have demonstrated that treatment with PD-L1-Fc fusions results in varying levels of lymphocyte suppression and reduction in autoimmune phenotypes (15–17). One agonistic antibody has been described that decreased lung inflammation in a mouse model of allergic asthma (18), highlighting the promise of engaging PD-1 signals.
Here, we present a de novo-designed hyperstable miniprotein that specifically binds murine PD-1 on human cells and a trimerized version of the binder that strongly inhibits T cell activation with potential for further development as a synthetic PD-1 agonist for the treatment of autoimmunity.
Results
Computational Design of De Novo PD-1 Binding Proteins.
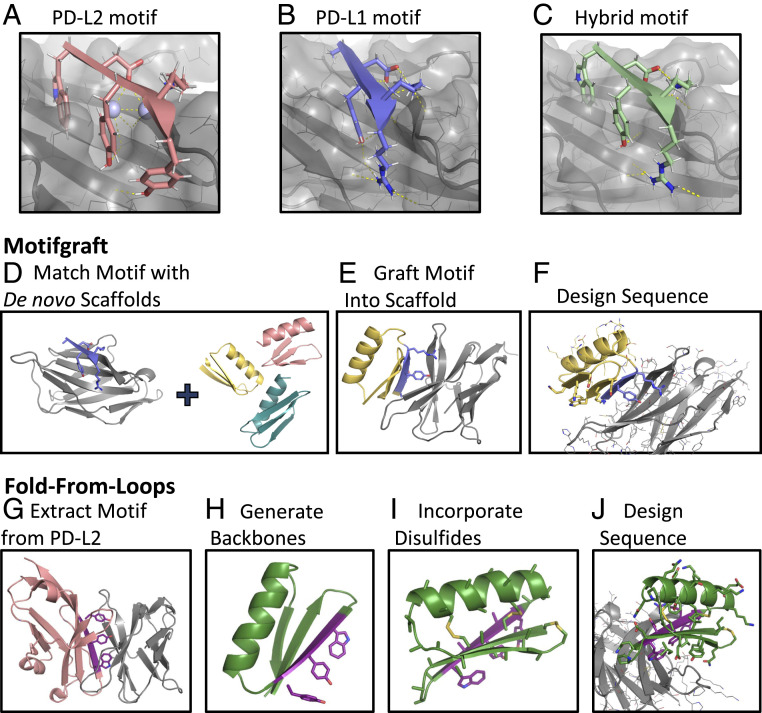
We identified a five-residue edge beta strand in human PD-L1 (19) and murine PD-L2 (20) (positions B110 to B114 in PDB 3BP5 with sequence “WDYKY;” Fig. 1A) that contributes the majority of the binding energy with PD-1. Residues at positions 1, 3, and 5 in this beta strand fit in a pocket on PD-1, and conserved polar residues at positions 2 and 4 make hydrogen bonds across the interface. While the tryptophan at the first position in the PD-L2 motif filled the pocket on PD-1 better than the PD-L1 alanine at this position, the arginine in the fifth position of PD-L1 is involved in an extensive hydrogen bond network with the target; hence, in addition to the “ADYKR” PD-L1 binding motif and the “WDYKY” PD-L2 binding motif, we also scaffolded the hybrid motif of “WDYKR” (Fig. 1C). We built proteins containing these motifs using two approaches as described in the following paragraphs.
Fig. 1.
Computational design of de novo PD-1 binding miniproteins. (A–C) Binding motifs used for design with murine PD-1 shown in gray, binding motif in color, water molecules placed in crystal structure as light blue spheres, and hydrogen bonds as yellow dashed lines. (A) “WDYKY” binding motif from PD-L2 (PDB 3BP5). (B) “ADYKR” binding motif from PD-L1 (PDB 3BIK). (C) “WDYKR” hybrid binding motif. (D–F) Motifgraft computational design protocol. (D) A binding motif (purple) docked to mPD-1 (gray) is aligned to a set of de novo miniprotein scaffolds. (E) A graft is considered successful if both the backbone RMSD between the motif and the scaffold backbone is less than 0.7 Å and the rest of the scaffold does not clash with the target. (F) Motif residues are constrained, and the rest of the interface is designed to optimize interactions with PD-1. (G–J) Fold-From-Loops computational design protocol. (G) A primary binding fragment (purple) is extracted from the crystal structure of mPD-1 (gray) in complex with mPD-L2 (pink) (PDB 3BP5). (H) De novo backbones (green) are generated around the PD-L2 binding fragment. (I) Cysteines are incorporated at geometrically compatible positions for disulfide bond formation. (J) Motif residues and disulfide bonds are first constrained, and then Rosetta sequence design is performed to stabilize the fold and increase interactions with PD-1.
In a first design approach, the three binding motifs were grafted onto a set of 34,840 de novo-designed scaffolds (21) containing three beta strands and a single alpha helix using the Motifgraft protocol (22) (Fig. 1D). The miniprotein scaffolds were 35 to 41 residues in length and contained one to four disulfide bonds for stability. All five residue-contiguous segments of each design were superimposed on the motif in complex with PD-1, and such scaffold/PD-1 docks were selected if the backbone RMSD was less than 0.7 Å and the scaffold did not clash with PD-1 (Fig. 1E). Multiple grafts were allowed for the same backbone if there was good alignment at multiple positions along the peptide chain. Sequences were designed for 25,709 selected grafts by constraining the residues in the binding motif and all disulfide bonds and then optimizing interactions with murine PD-1 using Rosetta Monte Carlo sequence design (23) (Fig. 1F). Designs were filtered for interface size (buried solvent accessible surface area) above 1,100 Å, shape complementarity above 0.6, and binding energy below −6 kcal/mol (as computed by Rosetta) to yield a total of 1,058 Motifgraft designs to evaluate experimentally for binding to PD-1.
In the second design approach, the binding motif was used as a seed around which de novo backbones were generated, as in the Fold-From-Loops (24) protocol. In contrast to the Motifgraft design protocol, we picked just the PD-L2 binding motif, as it has the stronger binding affinity of the two native ligands (Fig. 1G). A secondary structure definition (or “blueprint”) file was created to designate the length, identity, and register of each secondary structure element to be constructed on either end of the motif. We chose to focus on topologies consisting of a beta sheet containing two five-residue strands in addition to the motif strand stabilized by a 14-residue alpha helix packing on the side of the sheet not contacting PD-1. Monte Carlo–based fragment assembly was used to generate thousands of backbones with this topology around the motif (Fig. 1H). To increase stability, cysteines were added by scanning for pairs of positions that were geometrically compatible with disulfide bond formation (Fig. 1I). As in the grafting approach, the resulting de novo backbones were superimposed onto the motif strand in the PD-1/PD-L2 crystal structure and those that clashed with PD-1 eliminated (25). The sequences of the nonclashing docked backbones were then designed (Fig. 1J) as described above for the grafting approach. Designs were filtered for interface binding energy below −10 kcal/mol and the disulfide geometry and core packing in the monomer, resulting in 3,999 final designs. The Fold-From-Loops protocol generally yielded larger interfaces (SI Appendix, Fig. S1 A and B).
Genes encoding 1,058 Motifgraft designs and 3,999 Fold-From-Loops designs were ordered as an oligonucleotide array pool and transformed into yeast with a linearized yeast display vector. Following sorting for display on the yeast cell surface using a fluorescein isothiocyanate (FITC)-conjugated antibody against a C-terminal cMyc tag, the library was incubated with 1 µM mPD1-Fc or 1 µM hPD1-Fc, and cells with binding signal above that of a negative control sample incubated with only the secondary fluorophore labels were collected. To distinguish binders to the Fc domain in the PD-1 construct used for screening, we also carried out a binding sort with 1 µM of the Fc isotype control. DNA was extracted from the yeast in the unsorted library as well as all four selected pools. The gene inserts were amplified and a unique barcode for each pool added using qPCR. Next-generation sequencing was used to sequence all five barcoded pools (unsorted, displaying, mPD-1 binding, hPD-1 binding, and Fc binding) simultaneously. Raw sequencing reads were filtered by quality score, and then the count and frequency for each sequence in each pool was calculated. Enrichment ratios were generated by dividing the frequency of a given sequence in one of the binding selections by its frequency in the display reference sort. One Motifgraft design and three Fold-From-Loops designs were enriched in the murine PD-1 selections (enrichment ratio above 2) and depleted in the Fc binding selection (enrichment ratio less than 2), and one Motifgraft design was found to bind hPD-1 specifically. While all the Fold-From-Loops designs contained the “WDYKY” binding fragment extracted from PD-L2, both Motifgraft hits used the “ADYKR” hybrid motif. Of the filters used, there was no single metric that delineated working designs from nonworking designs.
Loop Redesign and Affinity Maturation Yield Higher-Affinity, Cross-Reactive PD-1 Binder.
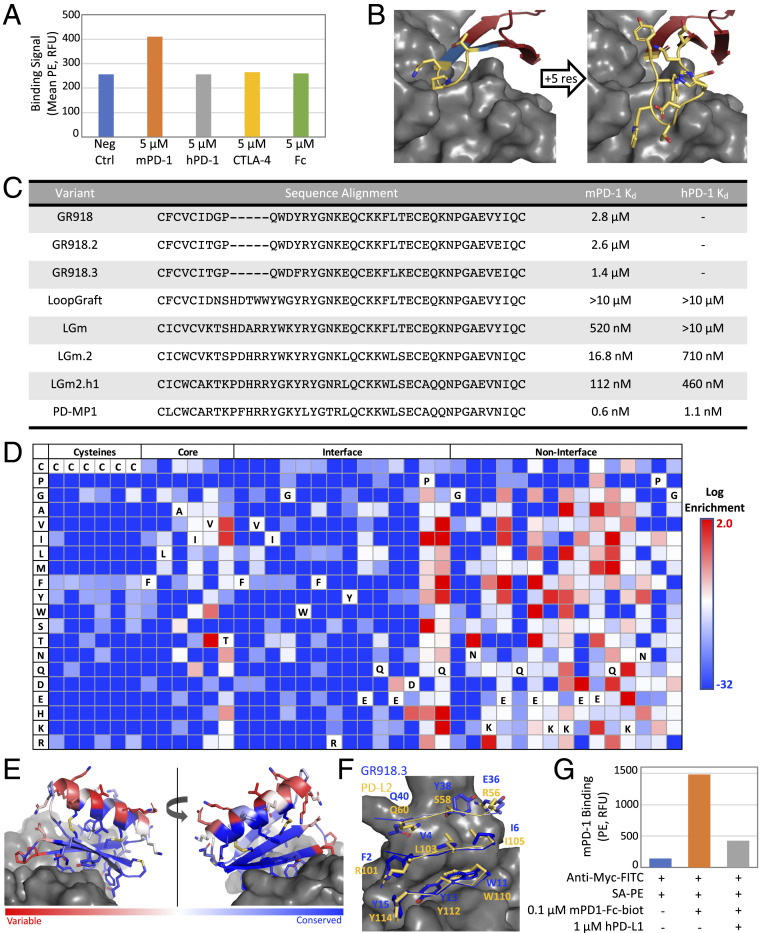
One of the Fold-From-Loops designs, GR918, was chosen for further optimization as this design had the highest mPD-1 enrichment. The original design was found to bind mPD-1 on yeast with a Kd of 2.8 µM (Fig. 2 A, orange and C) but showed no affinity for hPD-1 (Fig. 2A, gray). GR918 did not bind to the Fc isotype control (Fig. 2A, yellow) or a protein with a similar immunoglobulin fold, CTLA-4 (Fig. 2A, green). Due to the high error rate of the gene synthesis technology employed, the initial high-throughput screen also included several variants of each design ordered. We identified a variant, GR918.2, containing two substitutions (D7T and Y38E) screened in the original design pool that slightly increased binding to mPD-1 (Fig. 2C).
Fig. 2.
Loop redesign and affinity maturation. (A) The original GR918 design binds mPD-1 (orange) above background (blue) in a yeast display assay but not hPD-1 (gray), CTLA-4 (yellow), or the Fc isotype control (green). A total of 50,000 cells were measured for each sample. (B) The Direct Segment Lookup protocol was used to extend a four-residue beta hairpin loop (Left, yellow) by matching neighboring beta strand residues (Left, blue) in the miniprotein binder with large, conformationally constrained loops from the PDB. A nine-residue loop (Right) was found in a yeast display screen of loop extension designs that showed binding to both mPD-1 and hPD-1. (C) Sequence alignment of all variants derived from the original GR918 design along with murine and human PD-1 binding affinities on yeast. (D) Heat map representing the log enrichments for the GR918.3 SSM library selected with 250 nM mPD-1-Fc-biot. Enriched mutations are shown in red and depleted in blue. (E) The Shannon Entropy of each position derived from the SSM data are mapped onto the model of GR918.3 ranging from conserved positions in blue with a minimum value of 0.01 to variable positions in red with a maximum value of 3.11. (F) Interface residues of GR918.3 colored by Shannon Entropy as in (E) (blue = conserved) overlaid with PD-L2 interface residues in yellow (mPD-1 in gray). (G) Flow cytometry data of the hPD-L1 yeast competition assay. Background fluorescence of yeast displaying GR918.3 incubated with SA-PE alone was 143 RFU (relative fluorescence units) (blue). The same cells labeled with 0.1 µM mPD-1-Fc-biot and SA-PE gave 1,482 RFU (orange), and this was reduced to 426 RFU after coincubation with 1 µM hPD-L1 (gray). A total of 50,000 cells were measured for each sample.
A site saturation mutagenesis (SSM) library containing every single-point substitution of GR918.2 was screened for increased binding to mPD-1 on yeast (SI Appendix, Fig. S2A). Combinations of four mutations identified in the SSM to be beneficial to binding were tested and a triple mutant, referred to here as GR918.3, consisting of Y13F, K22E, and T26K was found to have a Kd of 1.4 µM for mPD-1 (Fig. 2C), which is approximately threefold higher than that of mPD-L2, the higher affinity of the two native ligands, on yeast (SI Appendix, Fig. S2B). GR918.3 did not have any detectable binding to hPD-1.
To obtain higher-affinity binders, we sought to extend a four-residue beta hairpin loop (Fig. 2 B, Left, yellow) that is in close proximity to PD-1, such that sidechains in the loop could interact with pockets in PD-1. This loop was quite tolerant to mutation in the GR918.2 and GR918.3 SSM screens. The Direct Segment Lookup protocol in Rosetta was used to identify six- to nine-residue conformationally constrained loops observed in the PDB matching the two-residue terminal segments of the GR918 loop with backbone atom RMSD less than 1.25 Å (Fig. 2 B, Left, blue). Matching loops were grafted onto GR918, and the sequences were designed to increase contacts with PD-1 while preserving graft structure via a structure-based position-specific scoring matrix (PSSM) with explicit preservation of proline or glycine residues. We screened 689 loop designs via yeast display for both mPD-1 and hPD-1 binding. A design with a nine-residue loop (Fig. 2 B, Right) containing five mutations arising from the high gene synthesis error rate (SI Appendix, Fig. S2C), called LoopGraft, had binding signal to hPD-1 while maintaining binding to mPD-1. After two rounds of affinity maturation for mPD-1 binding (Fig. 2C, LGm and LGm.2) followed by two rounds of hPD-1 affinity maturation (Fig. 2C, LGm2.h1), the final protein binds mPD-1 with a Kd of 0.6 nM and hPD-1 with a Kd of 1.1 nM on yeast (Fig. 2C). We refer to this final variant as PD-MP1 for Programmed Cell Death Miniprotein 1.
Design Binds at PD-L1/2 Interface on mPD-1.
The beta sheet of GR918 was designed to bind the ligand interface on PD-1 to block PD-L1/2 binding. GR918.3 SSM data (Fig. 2D) was consistent with GR918 binding to PD-1 as anticipated. The Shannon entropy (26) of each position along the peptide is shown in Fig. 2E; interface beta sheet residues are highly conserved while noninterface, solvent-exposed residues on the helix are highly variable, suggesting that the beta strands of GR918.3 form the interface with PD-1 as designed. All cysteine residues involved in the three designed disulfide bonds and the rest of the core residues are highly conserved, suggesting that the protein folds as designed. We constructed and screened an SSM for LGm.2 and observed a similar pattern of cysteine, core, and interface conservation with noninterface residues being much more variable (SI Appendix, Fig. S2D), suggesting that the binding mode is not altered by the loop extension.
To assess whether GR918 binds the intended site on PD-1, we performed a competition assay on yeast using soluble PD-L1. Yeast cells displaying GR918.3 were incubated with 0.1 µM biotinylated mPD1-Fc in the absence or presence of 1 µM unbiotinylated hPD-L1 competitor, then the secondary label streptavidin-phycoerythrin (SA-PE) was added and binding assayed by flow cytometry (Fig. 2G). A large decrease in signal was observed in the presence of a 10-fold excess of hPD-L1 (Fig. 2G, gray), suggesting that GR918.3 binds the intended region of PD-1.
X-ray Crystal Structure and Stability of GR918.2.
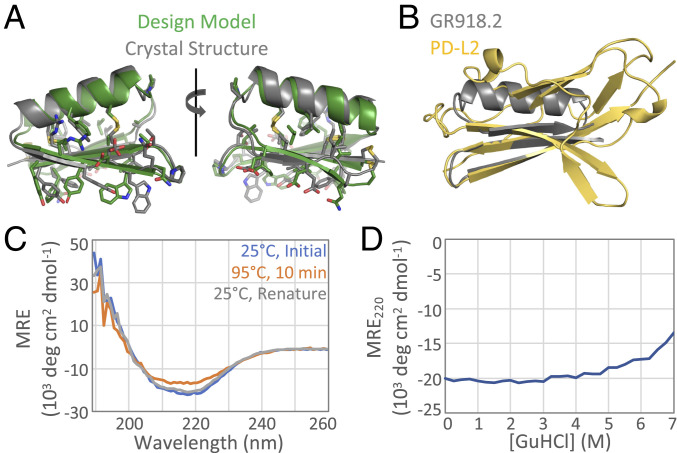
After the first round of affinity maturation, soluble GR918.2 was expressed using the Daedalus mammalian expression system (27) to confirm the fold, stability, and monomeric state of the protein in vitro. The apo crystal structure at 1.07-Å resolution confirms the protein folds as designed with a backbone RMSD of 1.33 Å between the crystal structure and the computational model (Fig. 3A). All three disulfides are clearly formed between the correct pairs of cysteines, and the rest of the core residues are packed as designed. GR918.2 forms a dimer in the asymmetric unit, but the protein elutes as a monomer in size exclusion chromatography (SI Appendix, Fig. S3A), suggesting that the dimer interface is only formed at the very high concentrations used in crystallography. Quite notably, GR918.2 captures the functional interface of PD-L2 using a much smaller, idealized protein backbone (Fig. 3B). Despite considerable effort, we were not able to obtain a crystal structure of any variant postloopgraft or a cocrystal structure with murine or human PD-1. However, as noted above, the SSM of LGm.2 shows conservation of core and cysteine residues, indicating that the protein folds as design after the loop redesign (SI Appendix, Fig. S2D).
Fig. 3.
Crystal structure and stability of GR918.2. (A) GR918.2 crystal structure (gray) aligned to the design model (green) with a backbone RMSD of 1.33 Å. (B) At only 5.6 kDa, GR918.2 (gray) captures the functional beta sheet interface of the 11.6-kDa binding domain of PD-L2 (yellow). (C) CD spectra at an initial temperature of 25 °C (blue), after incubating at 95 °C for 10 min (orange), and then after cooling back down to 25 °C (gray). (D) Chemical denaturation with GuHCl, monitoring CD signal at 220 nm. MRE = molar residue ellipticity.
GR918.2 was found to be hyperstable to both thermal and chemical denaturation. Thermal denaturation was performed by heating the protein from 25 to 95 °C, and the secondary structure was monitored using circular dichroism spectroscopy (CD) at 222 nm. The protein gave a CD spectra characteristic of an alpha-beta fold, and only ∼20% of the signal was lost at 95 °C, which was fully regained upon cooling back down to 25 °C (Fig. 3C). Chemical denaturation was performed in a similar manner, and the protein only lost ∼35% of the CD signal at 220 nm in 7 M GuHCl (Fig. 3D); however, longer incubation times may be necessary to fully reach equilibrium. This high stability is due to the three disulfide bonds and is not retained upon reduction of these bonds.
Monomeric and Trimeric PD-MP1 Binds PD-1 In Vitro and on Mammalian Cells.
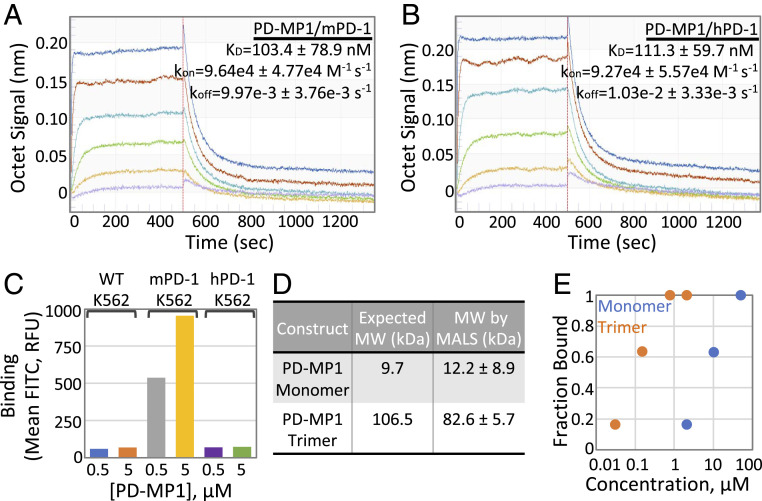
In vitro binding of soluble PD-MP1 to murine and human PD-1 was measured using Octet Bio-Layer Interferometry (BLI). Soluble PD-MP1 bound to biotinylated mPD-1-Fc loaded onto Streptavidin Octet sensors with a Kd of 103.4 ± 78.9 nM (Fig. 4A) and to hPD-1-Fc-biot loaded onto sensors with a Kd of 111.3 ± 59.7 nM (Fig. 4B) in this assay. This is 100- to 200-fold higher than the Kd’s on yeast, likely because of avidity effects in the flow cytometry experiments.
Fig. 4.
Monomeric and Trimeric PD-MP1 Bind PD-1 in vitro and on mammalian cells. Three independent BLI binding titrations were performed to determine on and off rates for PD-MP1 binding to mPD-1 (A) and hPD-1 (B). The on rates (kon), off rates (koff), and dissociation constants (Kd) listed are the average and SD from three independent titrations. Purple = 10 nM PD-MP1, yellow = 30 nM PD-MP1, green = 90 nM PD-MP1, light blue = 270 nM PD-MP1, red = 810 nM PD-MP1, and dark blue = 2.43 µM PD-MP1. (C) Monomeric PD-MP1 binds K562 cells expressing mPD-1 in a concentration-dependent manner (gray and yellow) but not wild-type K562 cells (blue and orange) or cells expressing hPD-1 (light blue and green). A total of 50,000 cells were measured for each sample. (D) Monomeric and trimeric PD-MP1 constructs were confirmed to form the correct oligomeric species by gel filtration followed by multiangle light scattering (MALS). (E) Monomeric and trimeric LGm.2 proteins bind mPD-1/K562 cells with increasing affinities correlating with increasing avidity. LGm.2 monomer has a Kd for mPD-1 on K562 cells of 6 µM, and LGm.2 trimer has a Kd of 90 nM. A total of 50,000 cells were measured for each sample.
We tested whether soluble PD-MP1 could bind PD-1 on K562 human stable cell lines expressing murine or human PD-1. Anti–mPD-1-FITC and anti–hPD-1-FITC antibodies were used to confirm expression of PD-1 for both transduced cell lines by flow cytometry (SI Appendix, Fig. S3B), and an anti–myc-FITC antibody was used to detect binding of the myc-tagged monomeric PD-MP1 to the cells. Concentration-dependent binding was observed to the K562 cells expressing murine PD-1 (Fig. 4C, gray and yellow) that was not observed to the wild-type K562 cells (Fig. 4C, blue and orange), indicating that this interaction is mPD-1 dependent. However, no binding was detected above background levels to the K562 cells expressing human PD-1 (Fig. 4C, purple and green). Given that the design binds hPD-1 in the biolayer interferometry experiments (Fig. 4B), the reasons for this lack of binding on cells are not clear, and for the experiments described in the remainder of this paper, we focus on binding to murine PD-1.
To increase the apparent affinity of PD-MP1 and to investigate possible agonism by receptor clustering, we generated a PD-1–binding trimer. The Spycatcher/Spytag system (28) was used to covalently attach PD-MP1 or LGm.2 to the designed homotrimer 2L6HC3_13 (29). 2L6HC3_13 was expressed and purified from bacteria as a genetic fusion with a Spycatcher domain, and LGm.2 and PD-MP1 were expressed and purified with a Spytag and Myc-tag from mammalian cells. Spycatcher_2L6HC3_13 was mixed with a slight excess of Spytag_PD-MP1 or Spytag_LGm.2, and the conjugated product was purified by gel filtration (SI Appendix, Fig. S3 C–E). Monomeric PD-MP1 was stable after isolation of the monomeric fractions (∼12.7 mL elution volume) on a Superdex75 column (SI Appendix, Fig. S3D), and no aggregation was observed after conjugating to stable, purified trimer (SI Appendix, Fig. S3E). The purified PD-MP1_2L6HC3_13 was confirmed to form a trimer by multiangle light scattering (MALS) (Fig. 4D). As assessed by flow cytometry, monomeric LGm.2 had a Kd of 6 µM to mPD-1 on K562 cells (Fig. 4E, blue), while the trimeric LGm.2 protein had a ∼65-fold boost in apparent affinity with a Kd of 90 nM (Fig. 4E, orange).
Trimeric PD-MP1 Exhibits Agonistic Activity in Mouse T Cell Activation Assay.
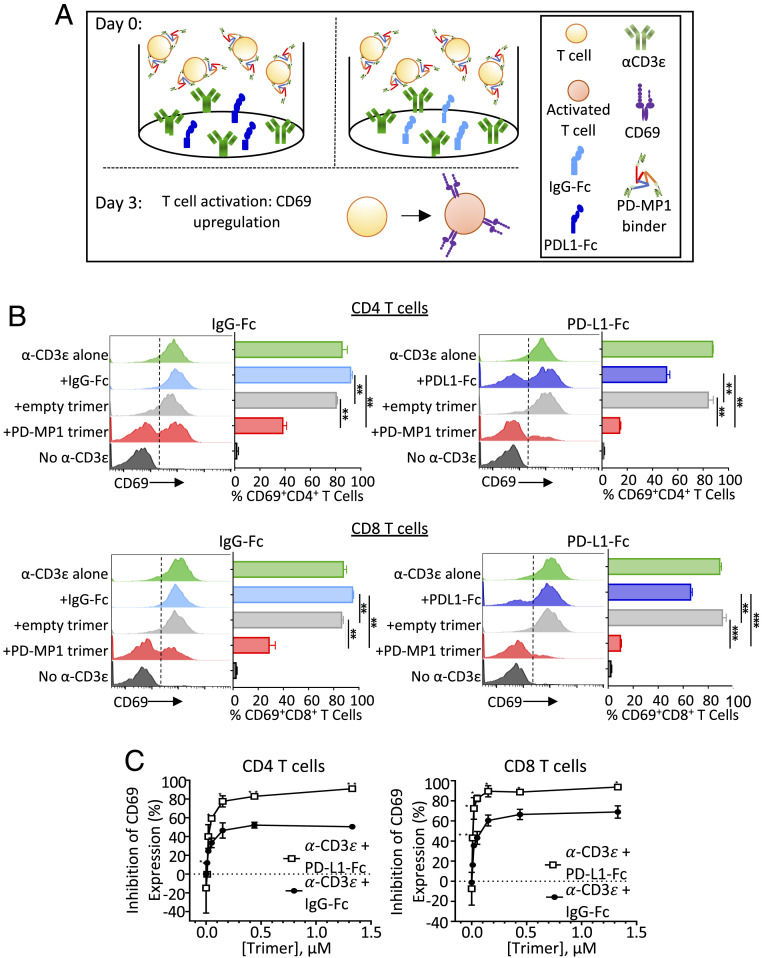
PD-MP1 trimer was cultured with T cells during activation to test for PD-1 agonist or antagonist activity (Fig. 5A). To test potential PD-1 antagonism, PD-MP1 trimers were added to mouse T cells stimulated with anti-CD3 antibody in the presence of plate-bound mouse PD-L1-Fc to see if they would block the PD-L1–driven inhibition of T cell activation. Potential PD-1 agonism was tested by adding trimers to stimulated T cells without PD-L1-Fc. After 3 d, activation of both CD4+ and CD8+ T cells was measured via CD69 expression (Fig. 5B). As expected, T cells stimulated with an anti-CD3ɛ antibody alone or with addition of control IgG-Fc showed high levels of T cell activation as indicated by up-regulation of CD69, and stimulation of T cells in the presence of PD-L1-Fc elicited lower CD69 expression.
Fig. 5.
Agonistic effect of PD-MP1 trimers on mouse T cells. (A) Experiment schematic. Mouse T cells were incubated in plates coated with anti-CD3ɛ and PD-L1-Fc (Left) or IgG-Fc (Right). PD-MP1 trimer or empty trimer control were added for 3 d, then T cell activation status was measured via up-regulation of CD69 protein. (B) Histograms and bar graphs showing T cell activation expressed as the percentage of CD69+ T cells. Mouse T cells were added to tissue culture wells coated with either α-CD3ɛ (green), α-CD3ɛ + IgG-Fc (light blue), α-CD3ɛ + PD-L1-Fc (dark blue), or no α-CD3ɛ (black). Titrations of PD-MP1 trimer (red) or empty trimer (gray) were added to the wells immediately after. CD69 expression levels were analyzed via flow cytometry in CD4+ T cells (Top) and CD8+ T cells (Bottom). Results shown are from the 0.44-µM concentration of each trimer formulation, n = 2. The error bars indicate the mean ± SD. Comparison between groups was calculated using the two-tailed unpaired Student’s t test. ***P ≤ 0.0001, **P ≤ 0.001, *P ≤ 0.05. (C) Percent of inhibition of T cell activation (compared to control trimers) induced by PD-MP1 trimer in CD4+ T cells (Left) or CD8+ T cells (Right). Setting the average %CD69+ T cells treated with the empty trimer as baseline activity, the percent inhibition was calculated by subtracting the %CD69+ T cells treated with PD-MP1 trimer from the baseline, then dividing the difference by the baseline activity. n = 2, error bars indicate the mean ± SD. Comparison between groups was calculated using the two-tailed unpaired Student’s t test. ***P ≤ 0.0001, **P ≤ 0.001, *P ≤ 0.05.
Addition of the PD-MP1 binders considerably reduced the CD69 induction elicited by anti-CD3ɛ in both CD4 and CD8 T cells compared to empty control trimers. This inhibition of T cell activation was dose dependent (Fig. 5C) and stronger than observed with plate-bound PD-L1-Fc. For T cells treated with anti-CD3ɛ and IgG-Fc, addition of the PD-MP1 trimers inhibited activation of CD4+ T cells up to 50% and inhibited activation of CD8+ T cells up to 70% more than empty trimers. In comparison, the PD-L1-Fc led to 40% inhibition in CD4 T cells and 28% in CD8 T cells. The strongest inhibition was in the presence of both the PD-MP1 trimers and PD-L1-Fc, which inhibited activation of both CD4+ T cells and CD8+ T cells up to 90%. Taken together, this indicates that the PD-MP1 trimer is an agonist for PD-1 that inhibits T cell activation in both CD4 and CD8 T cells.
Discussion
PD-MP1 is a de novo-designed binder with an all-beta protein interface. A challenge in designing interfaces containing beta sheets is that, if the target surface is nonpolar, the beta strands will have hydrophobic residues both in the interior hydrophobic core of the monomer and on the exterior to interact with the target and hence there is no hydrophobic/hydrophilic patterning to guide folding (30). This problem is exacerbated by the propensity of beta strands to aggregation (31). In accordance with these observations, we found core stabilization through the use of disulfide bonds to be very important to obtaining a soluble PD-1 binder that folds as designed. While the original GR918 design had weak affinity to mPD-1, following affinity maturation and loop extension, we obtained a higher-affinity, cross-reactive PD-1 binder. Further methods development is needed to improve design protocols so that high affinity binders to challenging targets like PD-1 can be obtained directly from design calculations without further optimization.
The designed PD-1 binding trimer presented here has potential as a PD-1 agonist for the treatment of autoimmune and inflammatory diseases. In contrast, most specific anti–PD-1 antibodies are antagonists (14). PD-MP1 decreased T cell activation up to 70%; in future studies, it will be important to assess immunogenicity and developability compared to other PD-1 affinity reagents. Little is known about the optimal valency and geometry for synthetic PD-1 agonists (18), and it is likely that still-better agonists can be created from our designed PD-1 binding domain. Taking advantage of the small size and modularity of the design, additional molecules could easily be generated by direct or chemical fusion of the modular PD-MP1 binding domain to oligomers with various valencies and geometries in order to study the mechanism of signaling and obtain an even more potent agonist.
Targeting of a PD1 agonist to sites of inflammation would avoid systemic immunosuppression and be advantageous for organ-specific autoimmune diseases, such as diabetes. A targeted therapeutic could be generated by fusing trimeric PD-MP1 with other small binding domains against relevant tissue-specific markers. Due to its small size, PD-MP1 would likely have a short serum half-life, as it would likely be cleared from the blood through the kidneys very quickly (32). If desired, increased half-life in circulation could likely be achieved by fusion to an Fc domain or polyethylene glycol (PEG) conjugation. However, the short lifetime could be an advantage for a targeted therapeutic programmed to act locally but be cleared quickly if it diffused away from the target site. A genetically encodable PD-1 agonist could contribute to CAR-Treg therapies currently being developed for the treatment of several autoimmune diseases, including asthma and multiple sclerosis (33–35): trimeric PD-MP1 can be genetically encoded as a fusion protein whose gene could be introduced into the engineered cells with an inducible promoter (36), allowing specific expression only upon target activation to locally suppress the immune response and reduce inflammation. More generally, the small size and hyperstability of PD-MP1, and the agonist activity of the trimer, could be useful in future research and therapeutic development efforts.
Materials and Methods
Computational Design and Gene Synthesis.
The Rosetta protein structure prediction and design software suite (23) was used for all design calculations using the Talaris2013 energy function (37). The Motifgraft (38) (SI Appendix, Fig. S4) and Fold-From-Loops (24) (SI Appendix, Fig. S5) computational protocols were performed as previously described using the MotifGraft and BluePrintBDR movers, respectively. The Remodel mover was used to add disulfide bonds to de novo backbones. Two rounds of design and all-atom minimization using the PackRotamersMover and TaskAwareMinMover were performed to generate final sequences for the Motifgraft designs. The Fold-From-Loops backbones were sequence designed using three rounds of the FastDesign mover followed by 15,000 rounds of the GenericSimulatedAnnealer to optimize the SSprediction, CavityVolume, SSShapeComplementarity, PackStat, hbond_lr_bb/residue, dslf_fa13/residue, TotalHydrophobic/TotalSasa_Hydrophobic (buried nonpolars), Holes, Ddg, and Rmsd Rosetta filter scores.
The Direct Segment Lookup protocol was used for loop redesign (SI Appendix, Fig. S6). First, the DirectSegmentLookupMover was used to graft up to 10-residue loops into GR918 between positions 6 and 11 that matched the backbone of neighboring residues 5 and 6 and 11 and 12 with an RMSD of less than 0.75 Å. The grafted loops were Cartesian-space minimized using the TaskAwareMinMover, and a structure-based PSSM was obtained using the SegmentSequenceProfileMover. Four cycles of FastDesign in conjunction with the FavorSequenceProfile mover were used to design the grafted loop sequence using the PSSM. Redesigned loop sequences were filtered for ShapeComplementarity above 0.65, Ddg below −30, and Sasa above 1,500.
Genes encoding all 5,057 initial designs and 689 loop redesigns to be tested were codon optimized for Saccharomyces cerevisiae (39) and ordered as a single oligo pool from CustomArray. SSM genes were amplified from Agilent oligo array pools. Combination Libraries were generated from Integrated DNA Technologies (IDT) ultramers containing degenerate codons. Genes for all individual mutants to be tested were constructed through oligo assembly using IDT oligos.
Yeast Surface Display.
Oligo pools were amplified and transformed into yeast as previously described (25). Initial testing of all designs and affinity maturation of GR918 was performed using yeast surface display (40). For the high-throughput screen, yeast-displaying designs were labeled with either 1 µM mPD-1-Fc, 1 µM hPD-1-Fc, or 1 µM IgG2a Fc (expressed from HEK 293F cells) as well as 5 µM biotinylated protein ZZ (41) (expressed from Escherichia coli BL21 cells) which specifically binds Fc and acts as an intermediate label for 1 h at 23 °C. Cells were then washed and labeled with 0.01 mg/mL SA-PE (ThermoFisher) and 0.01 mg/mL anti–cmyc-FITC (Immunology Consultants Laboratory) as a control for display of the proteins on ice for 10 min under aluminum foil. Two consecutive rounds of fluorescence-activated cell sorting (FACS) were performed under these labeling conditions and all FITC+/PE+ cells collected using a Sony SH800 cell sorter. A single reference sort of all FITC+-displaying cells was also done on the naïve library. Sequencing of the FITC+ pool and both binding selections was performed on an Illumina MiSeq.
The loop redesigns were similarly screened in high throughput as a single yeast-display pool. One reference sort was done to collect all FITC+-displaying cells. Two consecutive sorts were done for mPD-1 binding, labeling with 250 nM for the first sort and 100 nM mPD-1-Fc-biot for the second sort. Two consecutive sorts were also performed with 10 µM hPD-1-Fc-biot. All five sorted pools were then deep sequenced.
All SSM libraries went through two subsequent rounds of FACS. First, a titration was done to determine a Kd on yeast for the parent sequence. The first sort was labeled with mPD-1-Fc-biot or hPD-1-Fc-biot at approximately one-half of the parent Kd, and the top 5% FITC+PE+ were collected. The second sort was labeled with a fourfold lower concentration than the first, and the top 1% FITC+PE+ were collected. Both selections were deep sequenced along with a FITC+ reference sort. Enrichment ratios were calculated for each single mutant by dividing the counts in the selected pool by the counts in the reference pool. The Shannon entropy (26) was calculated for each position as previously described.
Combination Libraries containing all possible combinations of beneficial mutations from the SSM screens were sorted to convergence by doing 4 to 5 consecutive sorts at decreasing target concentration and collecting only the top 0.2 to 1% FITC+PE+ cells. Library convergence was monitored by plating the sorted cultures on C-Trp-Ura and Sanger sequencing 24 clones.
Yeast display titrations were conducted by incubating yeast cells displaying the GR918 variant with a range of concentrations of biotinylated mPD-1-Fc or hPD-1-Fc for 1 h at 23 °C. Cells were washed and then incubated with 0.01 mg/mL SA-PE and 0.01 mg/mL anti–cmyc-FITC on ice for 10 min under aluminum foil. Two final washes were done prior to measuring FITC and PE fluorescence on a BD Accuri C6 flow cytometer.
For the competition assay, yeast displaying GR918.3 were incubated with 0.1 µM mPD-1-Fc-biot alone or coincubated with 1 µM hPD-L1 (expressed in E. coli) for 1 h at 23 °C. After secondary labeling with SA-PE and anti–cmyc-FITC, the cells were analyzed using a BD Accuri C6 flow cytometer.
Protein Expression and Oligomer Conjugation.
Amino acid sequences for all protein constructs tested herein are listed in SI Appendix, Fig. S7. Soluble GR918.2 was produced using the Daedalus mammalian expression system (27) by the Molecular Design and Therapeutics core at the Fred Hutchinson Cancer Research Center. All other GR918 monomeric constructs, including the loop redesigned variants, were cloned with a His-tag and Myc-tag as a Siderocalin (Scn) fusion after an IgK secretion signal sequence into the mammalian expression vector CMVR. Polyethylenimine (PEI) transient transfection was used to introduce the vector into Expi293F HEK cells (ThermoFisher). The spent media was harvested 3 d posttransfection, and the soluble protein was purified by immobilized metal affinity chromatography (IMAC) using Ni Sepharose Excel Resin (GE Life Sciences). Spytagged monomers were cleaved with Tev protease (expressed in E. coli) followed by another IMAC step to remove the Scn fusion and His-tag, and then gel filtration was used as a final polishing step for the cleaved protein on a Superdex 75 Increase 10/300 GL column (GE Healthcare Life Sciences) (SI Appendix, Fig. S3 C, Lane 1 and D).
The 2L6HC3_13 trimer was cloned with an N-terminal His-tag and Spycatcher domain into pET29b(+). The protein was expressed in the E. coli expression strain Lemo21(DE3) (New England BioLabs) using IPTG induction and purified via Ni-NTA IMAC followed by gel filtration using a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences) (SI Appendix, Fig. S3 C, Lane 2 and D, orange).
Trimeric PD-MP1 was generated via Spycatcher/Spytag conjugation by incubating the Spycatcher_2L6HC3_13 construct with a 10% molar excess of Spytag_Myc_PD-MP1 at 23 °C for >1 h. Conjugated product was separated from unconjugated components via gel filtration using a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences) (SI Appendix, Fig. S3 C and E).
Crystallography.
Crystal screens were set up using the sitting drop vapor diffusion method by mixing 2.25 mg/mL GR918.2 2:1, 1:1, or 1:2 with reservoir solution from the 96-well Morpheus Crystallization Screen. Crystals grew in several drops (Morpheus D1, E1, F1, and H1) within 3 wk at 18 °C. Optimization screens were set up with each of these four conditions at 1:4, 1:2, 1:1, 2:1, and 4:1 mixtures with 9.85 mg/mL protein solution. Crystals were seen in most of the drops after ∼24 h at 18 °C. Because the Morpheus screen conditions already contain cryoprotectant, the crystals were looped and immediately flash frozen in liquid nitrogen. Diffraction data were collected from a crystal grown in 0.1 M MES/Imidazole pH 6.5, 0.03 M Diethylene Glycol, 0.03 M Triethylene Glycol, 0.03 M Tetraethylene Glycol, 0.03 M Pentaethylene Glycol, 10% PEG 20,000, and 20% PEG MME 500.
X-ray diffraction data were collected at the Advanced Photon Source, Northeastern Collaborative Access Team, Beamline ID-C. Images were processed to 1.07 Å in space group C2 using the program XDS (42). The structure was solved by molecular replacement using PHASER (43) in the PHENIX software suite (44) and the helix (residues 18 to 31) from the GR918 design model as a search model, with SHELXE (45) used for model completion. Two copies of GR918 were placed in the asymmetric unit. Coot (46) and PHENIX.refine (47) were used for several rounds of manual building and refinement. Data collection and refinement statistics are listed in SI Appendix, Fig. S8A.
CD.
All CD experiments were conducted using an AVIV Model 420 CD Spectrometer. Thermal denaturation was conducted by first performing a wavelength scan from 190 to 260 nm, sampling every 1 nm, at 23 °C using 0.23 mg/mL GR918.2 in phosphate buffered saline (PBS) in a 1-mm quartz cuvette. Then, the CD signal at 220 nm was monitored while heating the sample to 95 °C, taking a data point every 2 °C after incubating the sample for 30 s at the new temperature (SI Appendix, Fig. S8B). Another wavelength scan was taken at 95 °C and again after cooling the sample back down to 23 °C. All measurements were also taken with PBS for buffer subtraction.
Chemical denaturation was performed in a similar manner. Initially, a wavelength scan from 190 to 260 nm was taken on 0.05 mg/mL GR918.2 in PBS in the absence of denaturant in a 1-cm quartz cuvette at 23 °C. A solution of 8 M GuHCl with 0.05 mg/mL GR918.2 was then titrated into the cuvette under constant volume and with stirring, and the CD signal at 220 nm was recorded every 0.25 M up to 7 M. A final wavelength scan was performed of the protein solution with 7 M GuHCl.
Octet BLI.
Streptavidin Biosensors (Forte Bio) were loaded with either 50 nM mPD-1-Fc-biot or hPD-1-Fc-biot for 60 s at 25 °C. Loaded sensors were washed in Octet buffer (HBS-EP+, 0.5% dry milk) for 240 s and then a baseline was established in fresh Octet Buffer for 300 s. Loaded sensors were moved to a solution containing PD-MP1 at a range of concentrations between 5 nM and 6 µM and allowed to associate for 500 s. Dissociation was done by moving the sensor back to Octet Buffer for 1,000 s. Association rates, dissociation rates, and dissociation constants are listed in Fig. 4 A and B.
Cell Binding Assays.
Stable K562 cell lines constitutively expressing human or murine PD-1 were generated via lentiviral transduction of wild-type K562 cells with a DNA construct containing the extracellular and transmembrane domains of either murine or human PD-1 followed by an internal ribosome entry site (IRES) sequence and an iRFP gene. Transduced cells were sorted for iRFP expression to establish stable clonal cell lines expressing high levels of PD-1. The K562 cell lines were incubated with varying concentrations of soluble myc-tagged PD-MP1 monomer or trimer at 23 °C for >1 h. Cells were washed and then labeled with 0.01 mg/mL anti–cmyc-FITC (Immunology Consultants Laboratory) for 10 min on ice under aluminum foil. Cells were washed twice before measuring the FITC signal on a BD LSR II flow cytometer.
T Cell Activation Assay.
Mouse T cells from C57BL/6 female mice were isolated using a Pan T cell Isolation kit II (Milyteni Biotec). T cells were then resuspended to 1 × 106 cells/mL and transferred to a 96-well plate precoated with 2.5 µg/mL of anti-mouse CD3-ε antibody (BD Pharmingen, clone 145-2C11) plus 10 µg/mL of either recombinant mouse PD-L1-Fc (R&D Systems) or recombinant human IgG1-Fc (R&D Systems). PD1-MPI trimers and control trimers were thawed to room temperature and diluted to 5.32 µM in warm media. Threefold serial dilutions were performed in warm media to achieve further titrations, then trimers were added to the T cells and incubated at 37 °C, 5% CO2 for 3 d. After incubation, cells were washed in PBS and stained with a fixable live/dead stain (APC-ef780, Invitrogen) for 15 min on ice. Cells were washed in staining buffer (PBS + 0.5% bovine serum albumin [BSA]) and resuspended in a T cell activation antibody mixture (CD3-PerCP-Cy5.5, BD Pharmingen; CD8-FITC, Biolegend; CD4-APC, Invitrogen; CD69-BV421, BD Horizon) on ice for 30 min. Cells were then washed in staining buffer and analyzed using BD FACSymphony A3 Flow Cytometer, FACSDiva software, FlowJo version 10.6.1, and Graphpad Prism version 7.
Supplementary Material
Acknowledgments
We want to thank Steve Almo at the Albert Einstein College of Medicine for his gift of mPD-1 protein at the start of this project. Christy Tinberg and Songyu Wang from Amgen tested stability of molecules prior to the functional assays. This project was supported by a grant from the NIH (Grant 5U01GM094665) and Amgen Research. C.M.B. received support from a generous gift from Rocky and Genie Higgins. G.J.R. was funded as a Merck Fellow of the Life Sciences Research Foundation. M.J.B. was supported by the Institute for Protein Design (IPD) Launch Matching Fund. A.F. was funded by the NIH (Grant R01GM092802). L.C. was funded through a grant from the Bill and Melinda Gates Foundation (Grant OPP1156262 AM03). I.G. was supported by the Open Philanthropy Project Improving Protein Design Fund and the Nordstrom Barrier Institute for Protein Design Directors Fund. A.K. was funded by a gift from Silicon Valley Community. C.J.M. was supported by Bill and Melinda Gates Foundation Grant OPP1156262 and Amgen. F.D. was funded by the NIH (Grant R01GM123089).
Footnotes
Competing interest statement: A.V.K. and K.V.T. are current employees, and S.M.-M. is a former employee, of Amgen Inc. M.J.B. is a current employee of Neoleukin Therapeutics.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102164118/-/DCSupplemental.
Data Availability
The atomic coordinates and structure factors for GR918.2 have been deposited in Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) with PDB ID: 6V67 (48). All other study data are included in the article and/or SI Appendix.
References
- 1.Dai S., Jia R., Zhang X., Fang Q., Huang L., The PD-1/PD-Ls pathway and autoimmune diseases. Cell. Immunol. 290, 72–79 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Zak K. M., et al., Structure of the complex of human programmed death 1, PD-1, and its ligand PD-L1. Structure 23, 2341–2348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang S., Kim P. S., A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery. Proc. Natl. Acad. Sci. U.S.A. 116, 24500–24506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokosuka T., et al., Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 209, 1201–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura H., Honjo T., Minato N., Facilitation of β selection and modification of positive selection in the thymus of PD-1-deficient mice. J. Exp. Med. 191, 891–898 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blank C., et al., Absence of programmed death receptor 1 alters thymic development and enhances generation of CD4/CD8 double-negative TCR-transgenic T cells. J. Immunol. 171, 4574–4581 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Nishimura H., et al., Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science 291, 319–322 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Nishimura H., Nose M., Hiai H., Minato N., Honjo T., Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 11, 141–151 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Keir M. E., et al., Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med. 203, 883–895 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prokunina L., et al., A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat. Genet. 32, 666–669 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Lee Y. H., Bae S.-C., Kim J.-H., Song G. G., Meta-analysis of genetic polymorphisms in programmed cell death 1. Associations with rheumatoid arthritis, ankylosing spondylitis, and type 1 diabetes susceptibility. Z. Rheumatol. 74, 230–239 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Kroner A., et al., A PD-1 polymorphism is associated with disease progression in multiple sclerosis. Ann. Neurol. 58, 50–57 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Abril-Rodriguez G., Ribas A., SnapShot: Immune checkpoint inhibitors. Cancer Cell 31, 848–848.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Paluch C., Santos A. M., Anzilotti C., Cornall R. J., Davis S. J., Immune checkpoints as therapeutic targets in autoimmunity. Front. Immunol. 9, 2306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao W., Demirci G., Strom T. B., Li X. C., Stimulating PD-1-negative signals concurrent with blocking CD154 co-stimulation induces long-term islet allograft survival. Transplantation 76, 994–999 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Song M.-Y., et al., Protective effects of Fc-fused PD-L1 on two different animal models of colitis. Gut 64, 260–271 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Wang G., Hu P., Yang J., Shen G., Wu X., The effects of PDL-Ig on collagen-induced arthritis. Rheumatol. Int. 31, 513–519 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Helou D. G., et al., PD-1 pathway regulates ILC2 metabolism and PD-1 agonist treatment ameliorates airway hyperreactivity. Nat. Commun. 11, 3998 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin D. Y., et al., The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. U.S.A. 105, 3011–3016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lázár-Molnár E., et al., Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc. Natl. Acad. Sci. U.S.A. 105, 10483–10488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhardwaj G., et al., Accurate de novo design of hyperstable constrained peptides. Nature 538, 329–335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva D.-A., Correia B. E., Procko E., Motif-driven design of protein-protein interfaces. Methods Mol. Biol. 1414, 285–304 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Leaver-Fay A., et al., ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Correia B. E., et al., Proof of principle for epitope-focused vaccine design. Nature 507, 201–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chevalier A., et al., Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bywater R. P., Prediction of protein structural features from sequence data based on Shannon entropy and Kolmogorov complexity. PLoS One 10, e0119306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandaranayake A. D., et al., Daedalus: A robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res. 39, e143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zakeri B., et al., Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyken S. E., et al., De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science 352, 680–687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udgaonkar J. B., Polypeptide chain collapse and protein folding. Arch. Biochem. Biophys. 531, 24–33 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Bellesia G., Shea J.-E., Effect of beta-sheet propensity on peptide aggregation. J. Chem. Phys. 130, 145103 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Tibbitts J., Canter D., Graff R., Smith A., Khawli L. A., Key factors influencing ADME properties of therapeutic proteins: A need for ADME characterization in drug discovery and development. MAbs 8, 229–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skuljec J., et al., Chimeric antigen receptor-redirected regulatory T cells suppress experimental allergic airway inflammation, a model of asthma. Front. Immunol. 8, 1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esmaeilzadeh A., Tahmasebi S., Athari S. S., Chimeric antigen receptor -T cell therapy: Applications and challenges in treatment of allergy and asthma. Biomed. Pharmacother. 123, 109685 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Duffy S. S., Keating B. A., Moalem-Taylor G., Adoptive transfer of regulatory T cells as a promising immunotherapy for the treatment of multiple sclerosis. Front. Neurosci. 13, 1107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacsovics-Bankowski M., et al., Detailed characterization of tumor infiltrating lymphocytes in two distinct human solid malignancies show phenotypic similarities. J. Immunother. Cancer 2, 38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bazzoli A., Kelow S. P., Karanicolas J., Enhancements to the Rosetta energy function enable improved identification of small molecules that inhibit protein-protein interactions. PLoS One 10, e0140359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Correia B. E., et al., Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18, 1116–1126 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Hoover D. M., Lubkowski J., DNAWorks: An automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 30, e43 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao G., et al., Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Chen C., Huang Q. L., Jiang S. H., Pan X., Hua Z. C., Immobilized protein ZZ, an affinity tool for immunoglobulin isolation and immunological experimentation. Biotechnol. Appl. Biochem. 45, 87–92 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy A. J., et al., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider T. R., Sheldrick G. M., Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine P. V., et al., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bick M. J., et al., Apo structure of the de novo PD-1 binding miniprotein GR918.2. Protein Data Bank. https://www.rcsb.org/structure/6V67. Deposited 4 December 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for GR918.2 have been deposited in Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) with PDB ID: 6V67 (48). All other study data are included in the article and/or SI Appendix.