Significance
Cadherin–catenin complexes (CCCs) are a central component of adherens junctions. To produce an adhesive cell–cell contact, the CCC forms clusters, “E clusters,” driven by cooperative cis and trans interactions in the cadherin ectodomain and by α-catenin–actin interactions inside cells. We analyze whether E clustering is compatible with CCC-associated proteins (CAPs) and show that space constraints preclude the integration of many CAPs into E clusters. Using natural (scribble and erbin) and “artificial” (cadherin ectodomain antibodies) CAPs, we provide evidence that CAPs form separate “C clusters,” which are unable to intermix with E clusters. Our results thus suggest that CAP-dependent CCC clustering serves as a mechanism for sorting cellular proteins into distinct domains within cell–cell contacts.
Keywords: cadherin, catenins, cadherin-associated proteins, adherens junctions, protein sorting
Abstract
The cytoplasmic tails of classical cadherins form a multiprotein cadherin–catenin complex (CCC) that constitutes the major structural unit of adherens junctions (AJs). The CCC in AJs forms junctional clusters, “E clusters,” driven by cis and trans interactions in the cadherin ectodomain and stabilized by α-catenin–actin interactions. Additional proteins are known to bind to the cytoplasmic region of the CCC. Here, we analyze how these CCC-associated proteins (CAPs) integrate into cadherin clusters and how they affect the clustering process. Using a cross-linking approach coupled with mass spectrometry, we found that the majority of CAPs, including the force-sensing protein vinculin, interact with CCCs outside of AJs. Accordingly, structural modeling shows that there is not enough space for CAPs the size of vinculin to integrate into E clusters. Using two CAPs, scribble and erbin, as examples, we provide evidence that these proteins form separate clusters, which we term “C clusters.” As proof of principle, we show, by using cadherin ectodomain monoclonal antibodies (mAbs), that mAb-bound E-cadherin forms separate clusters that undergo trans interactions. Taken together, our data suggest that, in addition to its role in cell–cell adhesion, CAP-driven CCC clustering serves to organize cytoplasmic proteins into distinct domains that may synchronize signaling networks of neighboring cells within tissues.
The core structural unit of adherens junctions (AJs), the cadherin–catenin complex (CCC), consists of four proteins—a classical cadherin (E-cadherin in epithelia), β-catenin, α-catenin, and p120-catenin (1–4). In the process of cell–cell adhesion, the CCC forms clusters driven by both extracellular and intracellular binding events (5–8). The clustering of cadherin molecules is essential to reinforce weak individual trans adhesive bonds (9–12). In addition, the continuous and fast reassembly of CCC clusters within AJs renders them both highly adhesive and yet flexible (7, 13). While the importance of CCC clustering in cell–cell adhesion was demonstrated more than two decades ago (14), many of the molecular events associated with clustering are still poorly understood. One critical question, which is the focus of this work, is the role of proteins that associate with the CCC, CCC-associated proteins (CAPs), and, in particular, how these proteins change the properties of CCC clusters.
While several mechanisms for CCC clustering have been proposed (12), the best-characterized involves the formation of cis interaction between E-cadherin ectodomains. Cooperative cis and trans interactions arrange cadherin trans dimers into a paracrystalline lattice with a lateral intercadherin (center-to-center) spacing of ∼7 nm (15). The stability of these extracellular clusters is further enhanced by the binding of α-catenin to actin filaments (16–18). Accumulating data suggest that AJs consist of numerous such paracrystalline nanoclusters interspersed with less dense CCC regions (7, 15, 19–21). However, under certain conditions, cadherin clusters can be formed that do not seem to require the formation of ordered ectodomain lattices. For example, clusters are observed in cells expressing a cis interaction–incompetent cadherin mutant although they are less stable than wild-type paracrystalline clusters (20, 22). The underlying clustering mechanism in these cases is unclear.
Here we identified CAPs using a cross-linking agent that only detects proteins up to about 1.5 nm from a target. We provide evidence that most of these CAPs interact with the CCCs outside of cadherin clusters. Our results indicate that CCC clusters that integrate CAPs (C clusters) have fundamentally different structures from the “canonical structures” constrained by cadherin cis interactions. We term the latter “E clusters” to indicate that they are driven by extracellular interactions. We found that two CAPs, scribble and erbin, produced a set of CCC clusters that are spatially distinct from E clusters and from one another. It then appears that C clusters have distinct properties that depend on those of the CAPs themselves. To establish proof of principle, we show that anti-cadherin monoclonal antibodies (mAbs), which, similar to CAPs, are too large to be compatible with an E-cluster lattice, generate distinct adhesive clusters. Taken together, our data show that CAPs are both able to spatially separate C from E clusters and form CAP-dependent C clusters that are separate from one another. In addition to their role in cell–cell adhesion, our results thus suggest that CCC clustering serves as a mechanism for organizing cellular proteins into distinct domains within cell–cell contacts.
Results
Most CCC-Associated Proteins Interact with Adhesion-Competent and Adhesion-Incompetent CCCs.
Our goal in this section is to determine whether clustering affects the list of CAPs that bind to CCCs. To this end, we compared the CAPs that associated with functional monomeric GFP (mGFP)-tagged E-cadherin (EcGFP) with those associating with the nonfunctional WK-EcGFP whose two mutations, W2A and K14E, have been shown to completely abolish E-cadherin trans interactions (15, 23). WK-EcGFP was expressed in A431(EP)-KO (knockout) cells lacking E- and P-cadherins, so as to ensure that P-cadherin–adhesive clusters would not recruit the WK mutant through indirect intracellular interactions. To detect even weak, detergent-sensitive interactions, we used an “in-cell” cross-linking approach where the cells, before anti–GFP-specific precipitation, were cross-linked using the homobifunctional cysteine-specific cross-linker BMPEO3. Due to its short BMPEO3 spacer arm (14.7 Å), this approach most likely identifies proteins that directly interact with the CCC. The BioID-based technology, which was used to identify CAPs in previous studies (24–26), detects proteins located up to 30 nm away from the target (27). Therefore, the interactome identified in those experiments may reflect the protein composition of the cell cortex and may include proteins that do not interact directly with the CCC. Moreover, previous studies (24) have used low-calcium media as a means of generating adhesion-incompetent cadherins. However, this condition produces atypical cis and trans cadherin ectodomain interactions, which cannot realistically represent cadherins that do not undergo clustering (28, 29). The use of the WK cadherin mutant ensures that trans binding cannot occur and thus provides a more realistic proxy for E-cadherin that is not engaged in cell–cell adhesion.
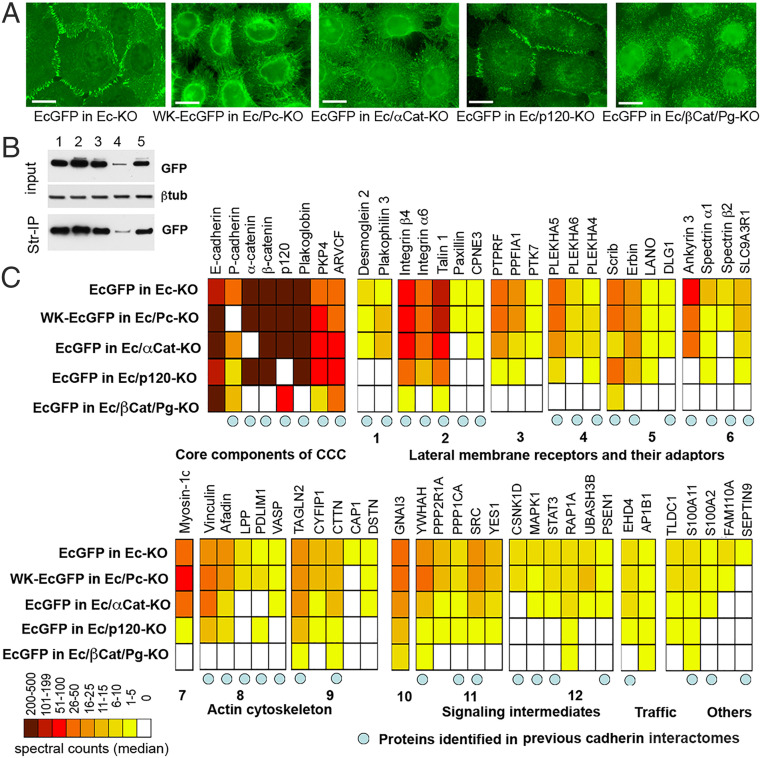
In agreement with previous studies, the GFP tag in EcGFP did not affect localization of E-cadherin in AJs. Also, as expected, WK-EcGFP was randomly located at the cell surface and unable to form AJs (15, 23) (Fig. 1 A and B). After filtering the data through control protein sets obtained from identical pull-down experiments with wild-type A431 cells and with A431(EP)-KO cells expressing a catenin-uncoupled “tailless” E-cadherin mutant, EcΔCytoGFP, a list of 59 proteins specifically cross-linked to the cytosolic portion of the CCC was obtained (Fig. 1B; see also SI Appendix, Table S1). Forty-nine of these proteins had been previously detected in association with CCCs using BioID (24–26). The most abundant proteins were core CCC members themselves—E- and P-cadherins, α-, β-, and p120-catenins, and their orthologs present in the majority of epithelial cells, plakoglobin, PKP4, and ARVCF. The remaining 51 proteins could be roughly divided into four functional groups: various adhesion receptors and their adaptors (21 proteins), components of the actin cytoskeleton (11 proteins), different signaling intermediates (12 proteins), and proteins involved in trafficking (2 proteins). Remarkably, the interactomes obtained for EcGFP and WK-EcGFP were nearly identical with respect to both the protein repertoire and spectral counts for individual proteins. Only P-cadherin, that was knocked out in WK-EcGFP–expressing cells, and two low-abundant proteins, Septin-9 and CAP1, were undetectable in association with WK-EcGFP (Fig. 1).
Fig. 1.
Proteins associated with EcGFP and with its adhesion-incompetent mutant WK-EcGFP. (A) Immunofluorescence microscopy of A431 cells expressing GFP-tagged cadherin (EcGFP) or its adhesion-defective mutant (WK-EcGFP) in different genetic backgrounds. EcGFP imaged in the cells with the E-cadherin knockout (EcGFP in Ec-KO); WK-EcGFP imaged in the cells with the combined E- and P-cadherin knockout (WK-EcGFP in Ec/Pc-KO); EcGFP imaged in the cells with the combined E-cadherin and α-catenin knockout (EcGFP in Ec/αCat-KO); EcGFP imaged in the cells with the combined E-cadherin and p120 knockout (EcGFP in Ec/p120-KO); EcGFP imaged in the cells with the combined E-cadherin, β-catenin, and plakoglobin knockout (EcGFP in Ec/βCat/Pg-KO). (Scale bars, 10 μm.) (B) Cell lysates (input) obtained from surface-biotinylated cells shown in A were analyzed for GFP and for the loading control, β-tubulin (βtub). The second parts of the lysates were precipitated using streptavidin agarose (Str-IP) and analyzed for GFP. Note that the p120 knockout, in contrast to other manipulations, results in dramatic reduction of both total and cell-surface E-cadherin levels. (C) Diagram showing the median of spectral counts for each protein identified as associating with cadherin CCCs in our cross-linking experiment. Protein names (or their gene symbols) are given above the diagram, while the genetic backgrounds of the cross-linking experiments are given to the left of the diagram. Proteins are grouped according to their function (given below the diagram). The group of “lateral membrane receptors and adaptors” is further split into components of desmosomes (1), focal adhesions (2), transmembrane phosphatases and their adaptors (3), membrane adaptors of the PLEKHA family (4), polarity regulators (5), and spectrin cytoskeleton (6). The “actin cytoskeleton” is split into actin motors (7), actin-binding proteins (8), and actin dynamic regulators (9). The “signaling components” group includes G protein (10), cortical kinases and phosphatases (11), and intermediates of specific signaling pathways (12). Blue circles indicate proteins previously found in BioID-based classical cadherin interactomes (24–26).
Most CAP Binding Is Independent of α-Catenin.
Three proteins from our list, vinculin, afadin, and VASP, have been shown to be recruited into AJs in response to applied mechanical forces (30–33). Experiments with a molecular tension sensor suggest that CCCs outside of adhesive clusters might also be stretched by α-catenin–actin–dependent pooling forces. Such stretching would be relieved upon uncoupling E-cadherin from the actin cytoskeleton by an α-catenin knockout (34). Therefore, we tested the effects of knocking out α-catenin and, surprisingly, while preventing AJ formation (Fig. 1 A and B), the knockout removed only 8 proteins from the EcGFP interactome, with vinculin, afadin, and VASP remaining bound to the CCC. By contrast, knockout of p120 shortened the list by 22 proteins, including VASP, but did not affect vinculin or afadin. The combined knockout of β-catenin and plakoglobin abolished binding to nearly all proteins with the notable exception of those that interact with the juxtamembrane E-cadherin region. Taken together, these data show that the majority of CAPs revealed by our approach interact to a similar extent with both adhesion-competent and adhesion-incompetent CCCs. Furthermore, most of these interactions occur independent of the forces that are generated by the actin cytoskeleton through α-catenin anchorage.
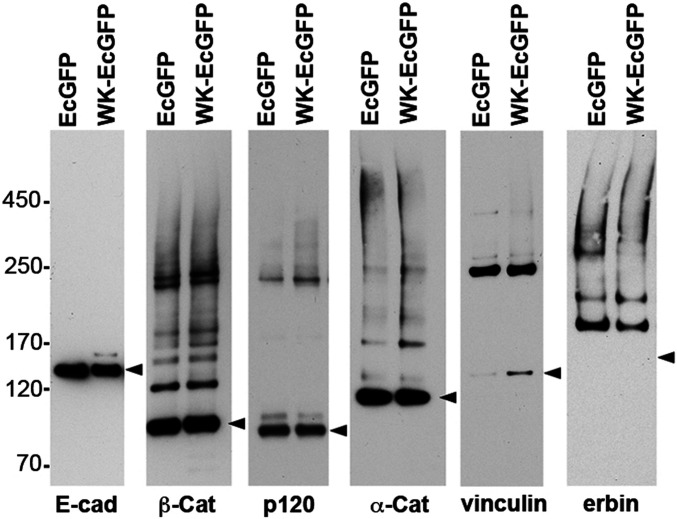
The surprising trans bond–independent presence of vinculin and other force-dependent proteins in our cadherin interactome could in principle be due to the spatial proximity of these proteins in the cell cortex rather than direct physical interactions. To test this scenario, we used Western blotting to analyze BMPEO3-induced adducts containing vinculin or erbin, two CAPs from our list. If these two proteins interact with CCCs that space their thiols about 15 Å apart, BMPEO3 treatment should efficiently generate specific adducts migrating as sharp bands by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). If cross-linking is due just to the coalescence of multiple noninteracting proteins, one would expect a smear of accidental adducts. Western blotting of the GFP precipitates obtained from the BMPEO3-treated cells expressing EcGFP and WK-EcGFP confirmed that BMPEO3 generated high amounts of specific vinculin–catenin and erbin–catenin adducts, which were very similar in both cell types (Fig. 2). We also note that the pattern of major BMPEO3 adducts of p120, β-catenin, and α-catenin also showed no significant differences in cells expressing EcGFP or its WK-EcGFP mutant (Fig. 2). These results strongly suggest that, at least, vinculin and erbin form protein complexes with the CCC and their formation is independent of cadherin trans interactions.
Fig. 2.
Characterization of the BMPEO3 adducts of the major CCC proteins and some CAPs. EcGFP and WK-EcGFP cells shown in Fig. 1 were cross-linked using BMPEO3 and their lysates were precipitated using GFP-trap as in the proteomics experiments. The resulting precipitates were run on SDS-PAGE, transferred onto nitrocellulose, and blotted with antibodies specific to E-cadherin, β-catenin, p120, α-catenin, vinculin, and erbin. Note that in all cases the adducts run as distinct bands. The monomeric form of each protein (including a position for monomeric erbin) is indicated by arrowheads. E-cadherin does not form any adducts since its intracellular region does not have cysteines. Also note that the single vinculin-containing adduct corresponds in size to the one of the major β-catenin adducts. The relative positions of marker proteins, the same for all panels, are indicated (Left).
CAP-Bound Clusters Segregate from CAP-Free CCC Clusters.
The binding of CAPs to extrajunctional CCCs points to the existence of a diverse array of extrajunctional cadherin–catenin supercomplexes (CCSCs), each containing a small number of different CAPs (perhaps only one). Can these CCSCs form distinct C clusters? Supporting this notion are observations that cell–cell contact localization of vinculin, afadin, LPP, and PS1, in contrast to α- and β-catenins, does not strictly match that of E-cadherin but is concentrated in specific subregions either within or around CAP-free E clusters (19, 35–37).
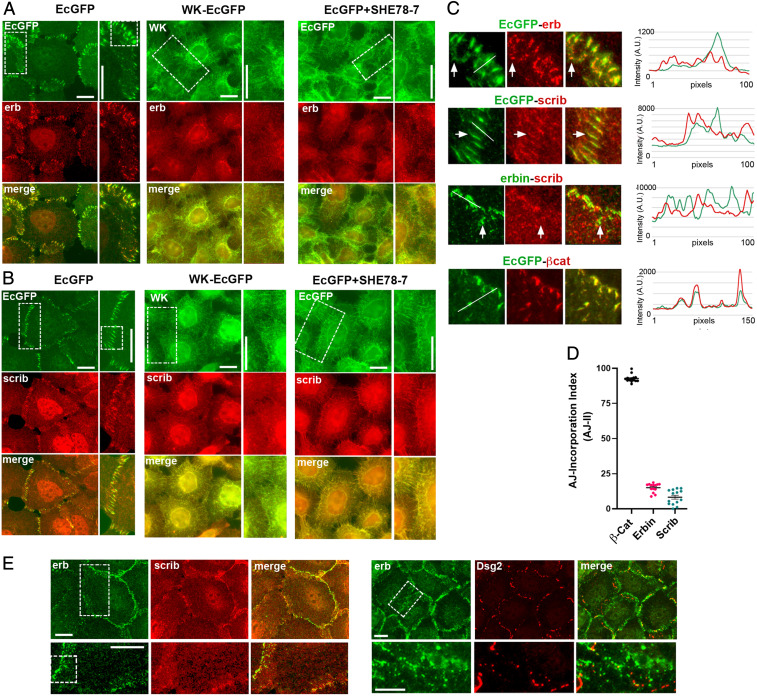
To test whether the sorting of CCSCs into distinct clusters is a common phenomenon, we examined the subcellular localization of two related CAPs from our list, scribble and erbin, both of which had been previously identified in AJs (38–40). No actin-binding activities were noted in these proteins. Immunofluorescence analysis of EcGFP-expressing A431 cells showed that these proteins indeed were localized in AJs but appeared in C clusters, which overlapped only with a small fraction of the CCCs in AJs (Fig. 3 A and B and line scans in Fig. 3C). Inspection of the images and their line scans showed that well-isolated erbin and scribble C clusters (some indicated by arrows in Fig. 3C) exhibited much lower EcGFP fluorescence than neighboring E clusters. To assess the scribble and erbin C-cluster abundance within AJs, we determined a scribble and erbin AJ incorporation index (AJ-II), which we defined as the ratio between the area of erbin- or scribble-specific fluorescence (representing C clusters) of the selected AJ and the total area of this AJ defined by EcGFP fluorescence, which represents the sum of all C and E clusters. The average AJ-II of erbin and scribble was 25 and 10%, respectively, suggesting that the clusters containing either or both proteins represent a relatively small fraction of E-cadherin clusters in AJs (Fig. 3D). Erbin and scribble costaining showed that their clusters were also distinct from one another (Fig. 3E). Finally, since in some cells erbin is located in desmosomes (41), we verified that in A431 cells, erbin clusters and desmosomes exhibited different distributions (Fig. 3E).
Fig. 3.
Subcellular localization of erbin and scribble clusters. (A and B) Immunofluorescence microscopy of A431 cells expressing the GFP-tagged form of cadherin (green) and erbin (erb; red; A) or scribble (scrib; red; B) in various backgrounds (abbreviations are as in Fig. 1A): EcGFP in Ec-KO (EcGFP), WK-EcGFP in Ec/Pc-KO (WK-EcGFP), or EcGFP in Ec-KO, which were cultured for 20 min with the function-blocking E-cadherin mAb SHE78-7 (EcGFP+SHE78-7). The boxed regions indicated in the green images are magnified (Right) (A, B, and E). The boxed regions in the magnified portions are further enlarged in C. (C) Cell–cell contact regions showing relative localization for pairs of proteins: E-cadherin and erbin (EcGFP-erb), E-cadherin and scribble (EcGFP-scrib), erbin and scribble (erb-scrib), and E-cadherin and β-catenin as a control (EcGFP-βCat). In all cases but the control (Bottom), the red clusters only partially correspond to AJs. The arrows show one of the AJ-separated erbin or scribble clusters (in EcGFP-erb or EcGFP-scrib) or scribble-deficient erbin clusters (in erbin-scrib). Line scans performed along the white lines are shown (Right). A.U., arbitrary unit. (D) The AJ incorporation index, which is defined as the ratio between the area of erbin- or scribble-specific fluorescence of the selected AJs and the total area of the same AJs defined by EcGFP fluorescence. Median values are indicated by horizontal bars; n = 15. (E) Immunofluorescence microscopy of A431 cells double-stained for erbin (green) and scribble (red) or for erbin (green) and Desmoglein-2 (Dsg2; red). Erbin clusters do not correspond to scribble clusters or to desmosomes. The boxed regions are enlarged (Right) and further zoomed-in (C). (Scale bars, 10 μm.)
Scribble and Erbin Clustering Depends on E-Cadherin trans Interactions.
To test whether erbin and scribble clustering depends on the E-cadherin trans interactions, we studied A431(EP)-KO cells expressing the adhesion-incompetent WK-EcGFP mutant. In contrast to observations in EcGFP-expressing A431 cells, in WK-EcGFP cells erbin and scribble as well as the mutant itself did not accumulate in cell–cell contact clusters but were, rather, perinuclear or scattered along the entire plasma membrane (Fig. 3 A and B). The loss of erbin and scribble C clusters was also revealed upon acute disruption of AJs in EcGFP cells by a function-blocking E-cadherin mAb, SHE78-7 (Fig. 3 A and B). Altogether, these data strongly suggest that the erbin- and scribble-bound CCC forms C clusters mediated, at least in part, by E-cadherin trans interactions. These two types of C clusters sort away from one another and from the E clusters that apparently comprise the bulk of AJs.
There Is Not Enough Space for Large CAPs within E Clusters.
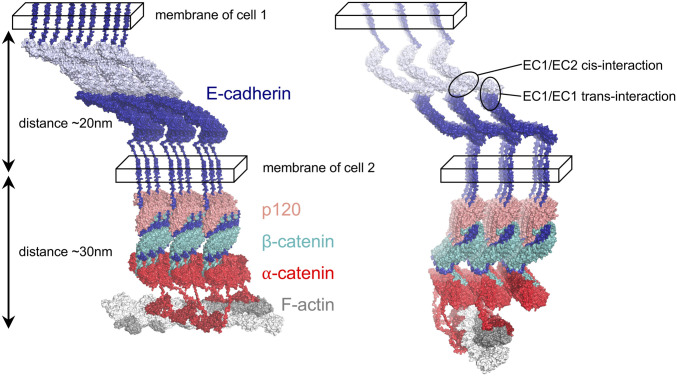
Perhaps the simplest explanation for the sorting of C clusters from E clusters is that steric constraints imposed by the 7-nm intercadherin spacing in the extracellular lattice preclude the integration of CAPs into the cytoplasmic region of E clusters. To evaluate this hypothesis, we built molecular models of E clusters using structural information from X-ray crystallographic and cryoelectron microscopic (cryo-EM) structures wherever possible (Fig. 4 and SI Appendix, Fig. S1). The model was built by stitching together structures of CCC components reported in the literature (15, 42–48) while connecting them with linkers predicted to be unstructured and, as described in detail in Materials and Methods, allowing the maximum possible space between structural regions. We used the 7-nm spacing in the extracellular lattice formed by E-cadherin ectodomains as a constraint for positioning p120, β-catenin, and α-catenin in the model (see details in Materials and Methods). The E cluster was further connected to F-actin via α-catenin ABD domains. The model is almost certainly just a crude representation of an E cluster but the positions of all proteins in the model with respect to the membrane are in agreement with experimentally determined distances (49–51) and the protein–protein interfaces are taken from experimentally determined structures (15, 43–47). Finally, the membrane is represented schematically in Fig. 4 as a box whose width corresponds approximately to the size of E-cadherin’s transmembrane helix (Materials and Methods).
Fig. 4.
Structural model of the E cluster connected to actin. All components of the 3 × 3 lattice, E-cadherin, p120, β-catenin, and α-catenin (color-coded, labeled, and shown in surface representation), satisfy constraints imposed by cis and trans interactions of cadherins (encircled; Right) in the extracellular space (between the two cellular membranes denoted as parallelepipeds). Two views of the lattice are shown. The E cluster is connected to F-actin (shown in gray surface representation) by ABD domains of α-catenin (red) via flexible linkers. F-actin is positioned in such a way that ABD domains from two adjacent rows of CCCs connect to one actin filament by binding the closest available ABD-binding site. The ABD domains (Far Right row of the lattice; dark red) are shown as bound to the rest of α-catenin.
A few details of the model are worth noting. First, only small proteins can fit into the lattice; for example, a single PDZ domain is comparable in size to the maximum spacing between β-catenins (SI Appendix, Fig. S2). Second, we are able to build models that accommodate α-catenin in its folded form, but just barely. Third, since α-catenin is known to partially unfold when binding the E-cadherin–β-catenin complex (52) and vinculin (47), we evaluated whether α-catenin with an unfurled M1 domain (ready to bind vinculin) can form a lattice with vinculin given the distance constraints imposed by cadherins. However, we could not find an arrangement without serious clashes (see Materials and Methods for details).
We also assessed whether erbin and scribble would be able to fit into the cytoplasmic region of the E cluster. These two CAPs are composed of leucine-rich repeat (LRR) and PDZ domains, whose sizes are compared with inter-CCC spacings in SI Appendix, Fig. S2. The figure shows two views of CCCs in a row along two distinct dimensions of the lattice. The view in SI Appendix, Fig. S2, Left has larger spacings between adjacent CCCs, and it appears from the figure that there is room for a PDZ domain of erbin or scribble there, but not LRR. We have tried fitting the domains into the lattice, but we found no orientation for the LRR domain without clashes (see Materials and Methods for details), suggesting that erbin and scribble cannot be accommodated in the E cluster.
The 67A4 mAb Is Recruited into AJs through Junction Reassembly.
Although the observations reported above involve cytoplasmic proteins, in order to gain mechanistic insights, and as proof of principle, we studied the effects of mAbs that target the E-cadherin ectodomain but do not block cadherin trans interactions. We anticipated that such mAbs, given their size, could not be incorporated into the ordered extracellular lattice of E clusters and, in parallel to intracellular CAPs, might form separate clusters. We used mAbs 67A4 and 5H9 that recognize E-cadherin epitopes located on the contiguous EC1 and EC2, and EC2 and EC3 domains, respectively, both of which are outside of the E-cadherin trans adhesive interface (53). A standard hanging-drop adhesion assay showed that in contrast to the function-blocking SHE78-7 mAb, which completely abolished aggregation of A431 cells, both 67A4 and 5H9 mAbs did not prevent cell aggregation. The resulting cell aggregates exhibited only minor changes; they were more irregular in shape in the presence of the 67A4 mAb, and were smaller in size in the presence of the 5H9 mAb (SI Appendix, Fig. S3A). Because all results with these mAbs lead to identical conclusions, only experiments with the 67A4 mAb are presented below.
In agreement with the experiments of Petrova et al. (54) with an anti–E-cadherin mAb with similar properties, 30-min-long incubation of A431 cells at 37 °C with the 67A4 mAb resulted in its incorporation into AJs without any obvious effects on cell morphology (SI Appendix, Fig. S3B). Furthermore, complete dissolution of the mAb-labeled AJs after 10 min in low-calcium media (SI Appendix, Fig. S3C) verified that the labeled AJs were calcium-dependent and the mAb did not cross-link E-cadherin between neighboring cells. We incubated live and methanol/acetone–fixed cells with the mAb at 4 and 37 °C. Remarkably, in contrast to the fixed cells at both temperatures or live cells at 37 °C, the mAb was completely unable to label AJs in live culture at 4 °C even after a 1-h-long incubation (SI Appendix, Fig. S3 D and E). This experiment strongly supports the idea that steric constraints impede the direct integration of proteins the size of mAbs, about 15 nm in diameter, into AJs. Continuous remodeling of AJs, including E-cadherin recycling, is apparently needed to deliver mAbs into AJs in living cells.
To fully understand how the mAbs incorporate into the AJs in living cells, we studied the kinetics of this process. The mAb appeared in AJs within 5 min after its addition into the media at 37 °C (SI Appendix, Fig. S4A, 5 min). After this brief incubation, only small clusters within a subset of AJs became labeled (marked by arrowheads in SI Appendix, Fig. S4A; line scan in Fig. S4B). The majority of AJs, as in the cells stained at 4 °C, exhibited only a weak background fluorescence. With time, the number of the mAb-labeled AJs steadily increased (SI Appendix, Fig. S4A, 10, 20, and 40 min). Strikingly, instead of gradually accumulating in AJs, the mAb was delivered into the junctions in the form of separate bright clusters typically located at the periphery of the completely unlabeled AJs (SI Appendix, Fig. S4A, 10 min, and corresponding line scan in SI Appendix, Fig. S4B).
Antibody-Bound and Antibody-Free Cadherin Clusters Are Incompatible.
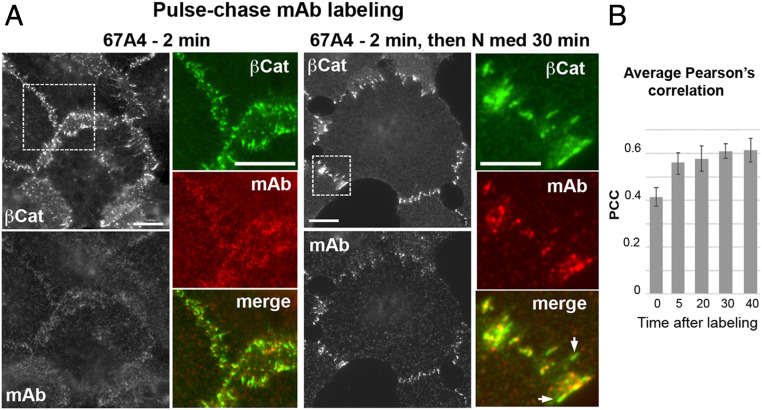
The experiments described in the previous section show that mAb-bound E-cadherin cannot be intermixed with mAb-free E-cadherin and, rather, forms separate clusters. To further investigate this phenomenon, we incubated the cells with the mAb for 2 min at room temperature, resulting, predominantly, in the labeling of the extrajunctional E-cadherin pool (Fig. 5A, 67A4, 2 min). The cells were then chased for an additional 30 min in mAb-free media. If able to intermix, the mAb-labeled extrajunctional pool and unlabeled junctional E-cadherin pool would eventually produce uniformly labeled AJs. However, we found that the labeled E-cadherin, even after 30 min, formed separate clusters or, very often, just small clusters spatially proximal to the completely unlabeled E clusters (Fig. 5A, 30 min). To quantify the intermixing of the mAb-labeled and unlabeled E-cadherin, we monitored the Pearson correlation coefficient (PCC) between the mAb, which marks mAb-bound E-cadherin, and β-catenin, which marks total E-cadherin, in randomly selected individual AJs. Immediately after pulse labeling, the PCC was about 0.4, and increased over the following 5 min to ∼0.5 and then remained constant during the following 25 min of observation (Fig. 5B). These results showed that mAb-bound cadherin cannot be intermixed with unlabeled cadherin in AJs despite the fact that the cadherin half-residence time in AJs of A431 cells, as determined by fluorescence recovery after photobleaching, is on the order of 2 to 3 min (55). Instead, the labeled E-cadherin forms specific clusters, hereafter “mAb-bound clusters.”
Fig. 5.
mAb-free and mAb-bound clusters could not be intermixed. (A) Immunofluorescence microscopy of A431 cells pulse-labeled with the 67A4 mAb. The cells were incubated with the 67A4 mAb for 2 min and were either fixed (67A4 2 min) or further cultured for 30 min in the antibody-free media (67A4 2 min then N med 30 min). Cells were then stained for β-catenin (βCat) and mouse immunoglobulin G antibody (mAb). Note that during the 2-min-long incubation, the mAb predominantly interacted with extrajunctional cadherin and only then integrated into AJ-associated clusters. Boxed regions are magnified (Right). Arrows mark mAb-free AJs. (Scale bars, 10 μm.) (B) Average PCC between β-catenin fluorescence that marks AJs and the mAb-derived fluorescence at different time points after addition of the mAb. The error bars represent SEs (n = 10).
mAb-Bound E-Cadherin Cluster Formation Is Mediated by the Cadherin trans Binding Interface and Binding to F-Actin.
The segregation of mAb-bound and mAb-free E-cadherin clusters suggests that the mAb dramatically changes the cadherin-clustering process. One possible explanation is that mAb-bound clusters cannot be dynamically reassembled and thus cannot be intermixed with highly dynamic E clusters. To determine whether this is the case, EcDn-expressing A431 cells were briefly labeled with the Alexa Fluor 594–labeled 67A4 mAb (0.5 μg/mL) and then imaged in an antibody-free media (Fig. 6A; see also Movie S1). The obtained movies and superimposition of the subsequent frames showed that the patterns of mAb-bound clusters dramatically changed over the 20-s-long time window (Fig. 6B). This result shows that the mAb-bound E-cadherin clusters, similar to E clusters, are continuously and completely reassembled on a subminute timescale.
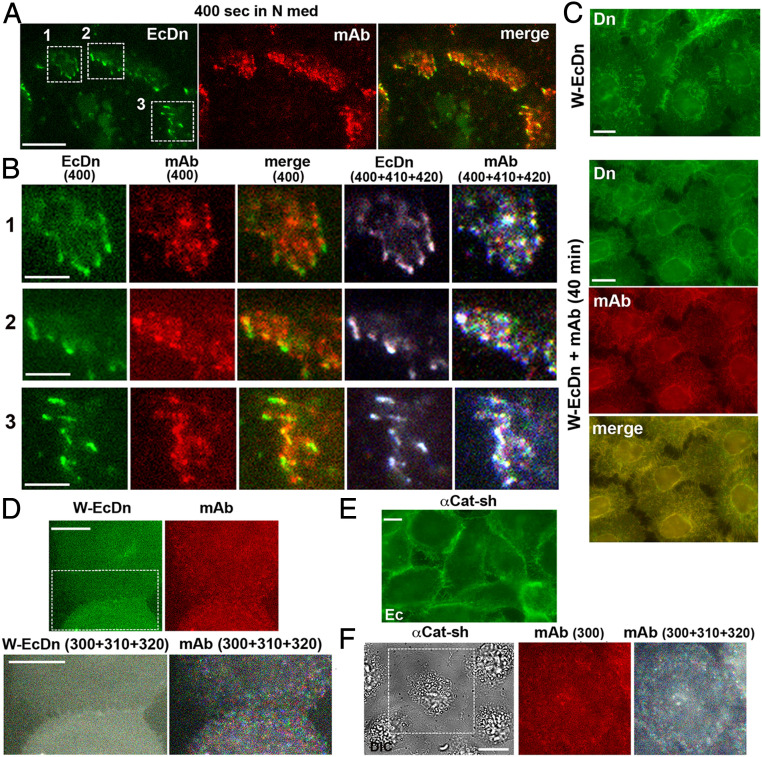
Fig. 6.
Dynamics of the mAb-bound cadherin clusters. (A) A single frame taken from Movie S1. Immunofluorescence microscopy of A431 cells expressing Dendra2-tagged E-cadherin (EcDn) labeled with the Alexa Fluor 594–conjugated 67A4 mAb for 1 min and then imaged in mAb-free media at 10-s resolution for EcDn (green) and for the mAb (red). The presented frame was taken 400 s after labeling. (Scale bar, 10 μm.) (B) Magnification of three areas indicated by the boxed regions in A (numbered) and their time evolution. Each area is presented in five different ways: EcDn (400): a single frame of EcDn fluorescence; mAb(400): a parallel single frame of mAb fluorescence; merge: combined green and red fluorescence of the same frame; EcDn (400+410+420): Three consecutive frames (taken 400, 410, and 420 s after labeling) of EcDn fluorescence were colorized in red, green, and blue (correspondingly) and merged. Note that the combined image is mostly black and white, showing that AJs were structurally stable during the 20-s time frame; mAb (400+410+420): The same three frames as above but taken in the mAb fluorescence channel were similarly colorized and merged. Note that mAb-bound E-cadherin clusters are multicolored, showing their high dynamics. (Scale bars, 5 μm.) (C) A431Ec-KO cells expressing the adhesion-incompetent W-EcDn mutant were fixed and imaged for W-EcDn using Dendra2 fluorescence (Dn) in standard culture (W-EcDn) or after 40 min with the 67A4 mAb (W-EcDn+mAb, 40 min). In the latter case, the cells were stained for mouse IgG (mAb). Note that in both cultures the W-EcDn mutant is unable to form AJs. (Scale bars, 10 μm.) (D) A431Ec-KO cells expressing the W-EcDn mutant were labeled and imaged as indicated in A. (D, Upper) Single frames of the W-EcDn and the mAb fluorescence taken 300 s after labeling are shown. Note that the mutant and the mAb do not show specific enrichment in the cell–cell contact. (D, Lower) Overlay of three consecutive frames of W-EcDn and the mAb fluorescence, W-EcDn (300+310+320), and mAb (300+310+320), correspondingly, were processed as in B. Only the area indicated by a dashed box in W-EcDn is shown. (Scale bars, 10 μm.) (E) Immunofluorescence microscopy of E-cadherin in α-catenin–depleted A431 cells (αCat-sh) using an anti-cadherin antibody. Note that cadherin cannot form well-defined AJs in these cells. (Scale bar, 10 μm.) (F) A single time-lapse differential interference contrast (DIC) image of α-catenin–depleted A431 cells taken 300 s after the cells were labeled by the 67A4 mAb as indicated in A; mAb (300): the mAb fluorescence of the area indicated by a dashed box 300 s after labeling; mAb (300+310+320): three consecutive frames of the mAb fluorescence processed as in B. (Scale bar, 10 μm.)
We then tested the role of the E-cadherin strand-swapped trans interaction in mAb-bound E-cadherin clustering and found that the mAb did not rescue cadherin clustering in A431EP-KO cells expressing the strand swap–incompetent W-EcDn mutant (Fig. 6C). In these cells, the Alexa Fluor 594–labeled mAb produced only faint fluorescence randomly distributed along the entire surface of the cells (Fig. 6D). This fluorescence, undetected in the control EcDn-expressing A431 cells, was apparently caused by the elevated level of surface E-cadherin that resulted from the inability of the mutant to form AJs.
To test whether mAb-bound cadherin clustering is also α-catenin–dependent, we used α-catenin–depleted A431 cells available in our laboratory (6). Cadherin in these cells, while enriched in some cell–cell contacts, was unable to form AJs (Fig. 6E). Notably, similar to the cells expressing W-EcDn, α-catenin–depleted cells failed to form mAb-bound clusters (Fig. 6F). Taken together, these live-imaging experiments showed that mAb-bound clusters are formed through endogenous clustering mechanisms, strand-swapped trans dimerization, and α-catenin–mediated interaction with actin filaments. Nevertheless, despite these commonalities, mAb-bound clusters sort away from E clusters.
Discussion
While many of the key protein components of AJs and their interactions have been characterized in atomic detail, the mechanism that coordinates extracellular and intracellular events in cell–cell adhesion is still poorly understood. Although it is clear that the formation of an extracellular paracrystalline cadherin lattice driven by cis and trans interactions mediates the formation of canonical E clusters, CCC clustering is still observed for cis mutants which ablate the cis interaction (20, 22). It may be that in this case the CCC aggregates into amorphous clusters driven by nonspecific cis interactions on the ectodomain, by trans membrane helical interactions, and/or by other CCC components. Independent of the detailed mechanism, the formation of cadherin clusters under conditions where ordered cis interaction–driven ectodomain lattices cannot be formed suggests that additional clustering mechanisms play a role in AJ structure and dynamics. Here we have shown that CCC-associated proteins can facilitate clustering and that the properties of the resulting clusters are dependent on the nature of individual CAPs.
How are CAPs integrated into CCC clusters? In Fig. 4, we have built a crude model of an E cluster constrained by a 70-Å distance between cadherin tails emerging from the membrane as defined by the ectodomain lattice. It seems clear from the model that the binding of many CAPs to the CCC is incompatible with a cadherin lattice so that direct integration into E clusters is highly unlikely. For example, there is no room for vinculin in E clusters so that its binding to α-catenin could only occur outside the paracrystalline cadherin lattice. Moreover, separate C clusters, which incorporate vinculin, or other CAPs must be organized in a very different way from lattice-driven E clusters.
Our data suggest that CAPs interact with the extrajunctional CCCs forming CCC supercomplexes, which then assemble into C clusters. Supporting this model is our comparison of the interactomes of functional E-cadherin, EcGFP, with that of the adhesion-incompetent E-cadherin mutant WK-EcGFP, which is unable to undergo trans interactions (23, 56). Strikingly, the list of proteins associated with WK-EcGFP is nearly identical to that associated with EcGFP. Taken together, our data clearly indicate that junctional C clusters are formed, not through selective binding of a particular CAP to an E cluster, but through the self-assembly of preformed CCC–CAP supercomplexes.
Of note, the list of CAPs is shortened by only 8 proteins in α-catenin–knockout cells. Interestingly, vinculin, afadin, and VASP, which were shown to interact with the CCC through α-catenin in a tension-dependent manner (31, 46), bind to the CCC even in the knockout. By contrast, the list of CAPs is shortened by 20 proteins in p120-deficient cells, despite the fact that these cells retain AJs. Combined β-catenin/plakoglobin knockout results even in more dramatic reduction in the list of CAPs, suggesting that these two proteins provide binding sites for most of the CAPs. Since, as we argue in Fig. 4, there is no room for most CAPs within E clusters, we are led to the conclusion that most CAPs interact with β-catenin and/or p120 in extrajunctional space and then form distinct C clusters. Consistent with our results, vinculin was shown to interact with the CCC even in the absence of α-catenin (57–59). Taken together, the available data thus suggest that a vinculin–CCC supercomplex delivers vinculin to the sites of cell–cell adhesion, where vinculin binds to α-catenin in a process related in a still undefined way to C-cluster formation. The latter process is apparently force-dependent. Of note, both force-independent and force-dependent steps have recently been shown to participate in the recruitment of vinculin into focal adhesions (60).
To probe the formation of C clusters with CAPs that are not known to interact with actin, we studied the localization of erbin and scribble, that we identified in the CCC interactome. They have both been previously detected in AJs (38–40, 61) and do not possess known actin-binding domains. Immunofluorescence microscopy confirmed previous observations that scribble and erbin are localized in AJs. However, we show here that the distribution of these proteins in AJs is not uniform. In fact, they both are integrated into only a subset of C clusters that are either proximal to or surrounded by other types of C clusters, or by E clusters, which comprise the bulk of AJs. Moreover, the C clusters incorporating erbin and scribble do not overlap with one another. The existence of both erbin- and scribble-specific clusters clearly depends on cadherin-adhesive interactions since they are lost in cells expressing the adhesion-incompetent WK-EcGFP mutant or immediately disassemble after administration of the function-blocking E-cadherin mAb.
Taken together, our results suggest that CAPs associate with extrajunctional CCCs and only then, in the form of CCSCs, assemble C clusters that are spatially and structurally distinct from E clusters. At this stage, we have no direct evidence as to how CCC clusters assemble but we speculate that assembly is facilitated by CAP-specific cis interactions, thus providing a mechanism for the formation of individual clusters. For example, two of the four PDZ domains of scribble interact with β-catenin and another one interacts with p120 orthologs, δ-catenin, PKP4, and ARVCF (62). It is possible that these interactions play a role in the assembly of scribble-specific clusters. Similarly, dimerization of vinculin upon binding to F-actin might also play a “CCC-organizing” role (63).
As proof of principle, we studied two E-cadherin mAbs, 67A4 and 5H9, which abolish cadherin cis interactions but leave the trans binding interface unaltered (53). Our results show that the mAbs are unable to interact with E-cadherin in AJs but rather bind to extrajunctional E-cadherin. Furthermore, instead of being intermixed with the mAb-free E-cadherin in AJs, the mAb-bound E-cadherin generates separate adhesion clusters. Under conditions of mAb excess, the mAb-bound clusters dominate and eventually replace the mAb-free AJs. When mAb-bound and mAb-free E-cadherin pools coexist, the cells continue to exhibit two pools of clusters. The data show that both the mAb-bound and mAb-free E-cadherin still use trans interactions and α-catenin binding to F-actin for their clustering. Importantly, the mAbs, which are applied from outside, preclude any possibility that the observed segregation of mAb-bound and mAb-free E-cadherin clusters occurs as a result of antibody-mediated binding of the CCC to any specific intracellular structures. While the mAb experiments involve extracellular phenomena and our focus in this work is on CAPs that associate with the cytosolic portion of CCCs, the experiments with mAbs demonstrate the possibility of coexistence of different types of CCC clusters, all of which are based on the same cadherin trans interactions.
In conclusion, we present strong evidence that at least some CAPs cannot be integrated into E clusters but form CAP-specific C clusters. We speculate that formation of these C clusters is driven by cis interactions provided by the CAPs themselves, that play a similar role as cadherin cis interactions in E clusters. An important consequence of this model is that the structural organization of CAP-specific clusters must be matched on two sides of an adhesion interface. Such complementarity in composition would ultimately equalize the number of C clusters containing particular CAPs in neighboring cells. The structural synergism between trans and cis interactions that has been shown to play a role in adhesion (8, 64) could function, therefore, as a sorting mechanism that arranges CAPs—various receptors, signaling intermediates, and their adaptors—into specialized clusters that are equivalent in structure and number to those in neighboring cells. This feature of cadherin-based adhesion might potentially synchronize signaling events in individual cells in a given tissue.
Materials and Methods
Plasmids.
The plasmids encoding Ec-mGFP (denoted EcGFP), EcΔCytoGFP, and Ec-Dn have been reported (7, 17). Mutations W2A inserted into Ec-Dn (W-EcDn) and W2A/K14E inserted into Ec-mGFP (WK-EcGFP) were described by Hong et al. (23). mCherry-tagged E-cadherin, EcCH, was constructed from pRc-EcDn by replacing the tags. The plasmid pRcCMV-P1EcDn was constructed by the replacement of the N-terminal region of E-cadherin in EcDn with the homological region of P1Ec-Myc (65). All plasmid inserts were verified by sequencing.
Cell Culture, Transfection, and Cell Labeling.
The cells, A431D, A431, and A431-EcDn, and α-catenin–deficient A431 cells (A431aCat-sh) have been previously described (6, 19, 66). Ec-mGFP– (EcGFP) and W-EcDn–expressing A431E-KO cells were obtained using stable transfection with the corresponding plasmids of the A431E-KO cells in which endogenous E-cadherin was knocked out using the Alt-R CRISPR-Cas9 System (IDT) (7). The catenin-knockout cells were obtained using the same protocol. In brief, the EcGFP-A431E-KO cells were transfected with an RNA complex consisting of a gene-specific CRISPR RNA (crRNA; designed by software of the Broad Institute of Harvard and the Massachusetts Institute of Technology) and transactivating RNA. The following crRNAs were used: p120–5′-GTGAAGCTCGCCGGAAACTT-3′; β-catenin–5′-GAAACAGCTCGTTGTACCGC-3′; plakoglobin–5′-CATGGCCTCCCGCACCCGTT-3′; and α-catenin–5′-GAAGGGGGATAAAATTGCGA-3′. Similarly, WK-EcGFP cells were obtained after transfection of the A431(EP)-KO cells in which both E- and P-cadherin were silenced using the same approach. The hanging-drop assay was performed as described by Kim et al. (67). In brief, about 1.5 × 105 cells in 30 mL were seeded onto the inner surface of 35-mm culture dishes as hanging drops and allowed to aggregate overnight. To assay for tightness of cell–cell adhesion, the drops were passed 10 times through a standard 200-μL pipet tip. The resulting suspension was imaged through a 10× phase-contrast objective.
For mAb-binding experiments, the 67A4 (Millipore; MAB-3199Z), 5H9 (Santa Cruz; sc-52327), or SHE78-7 (Zymed Laboratories) mAbs (azide-free) were added into the culture media at a final concentration of 2 μg/mL and incubated as indicated. For live-cell imaging, the cells were labeled by adding into the cultures the Alexa Fluor 594–conjugated 67A4 mAb (azide-free; BioLegend) at a final concentration of 1.2 μg/mL for 2 min. The cells were incubated for 1 to 2 min in the labeling media and, after a brief washing, the labeled cells were imaged in the label-free imaging media.
Immunofluorescence Microscopy.
For immunofluorescence, cells were fixed and permeabilized with 3% formaldehyde, 1% Triton X-100. See a study by Indra et al. for details (19). Wide-field images were taken using an Eclipse 80i Nikon microscope (Plan Apo 100×/1.40 objective lens) and a digital camera (CoolSNAP EZ; Photometrics). The images were then processed using Nikon NIS-Elements software. For immunostaining the following antibodies were used: mouse mAb anti–E-cadherin clone HECD1 (Zymed Laboratories) and anti-p120 (BD Transduction Laboratories); sheep anti-erbin (R&D Systems; AF7866); rabbit anti-Dendra2 (Evrogen, Moscow, Russia); anti–β-catenin and anti-Dsg2 (Invitrogen; MA5-14461 and 21880-1-AP); anti-scribble and anti–α-catenin (Abcam; ab36708 and ab51032). All secondary antibodies were produced in donkey (Jackson ImmunoResearch Laboratories).
Live-Cell Imaging and Data Processing.
The experiments were performed essentially as described earlier (17, 55) using halogen light source (Fig. 6A and SI Appendix, Movie S1). In brief, cells were imaged (in L-15 media with 10% fetal bovine serum) with an Eclipse Ti-E microscope (Nikon) at 37 °C controlled with Nikon NIS-Elements software. The microscope was equipped with an incubator chamber, CoolSNAP HQ2 camera (Photometrics), and Plan Apo VC 100×/1.40 lens. The 2 × 2 binning mode was used in all live-imaging experiments. At this microscope setting the pixel size was 128 nm. All images were saved as Tiff files and processed using ImageJ software (NIH).
For standard line-scan analysis, the Fiji (NIH) plot profile tool was used. Briefly, one pixel-width line was drawn along the selected contact on the merged images. The line was restored accurately on the green and red images separately using the “restore previous selection” command. Individual plots were then created using the “plot profile” command for red and green images. The final graphs were created with Microsoft Excel with the calculated plot values. AJ-II determination was performed using Fiji (NIH). For each protein staining, 14 individual AJs of 25 to 50 pixels in length were cropped using a manual cropping tool. The fraction of these AJs occupied by red fluorescence was calculated using the color threshold tool by measuring the total green area and red area within the green area. The SE was calculated and plotted accordingly.
Proteomics.
The confluent cultures of the indicated cells grown on 10-cm plates were cross-linked using the BMPEO3 cross-linker as we described previously (66), and then lysed with lysis buffer (LB; 20 mM Tris⋅HCl, 150 mM NaCl, 2 mM ethylenediaminetetraacetate, 1% Triton X-100), cleared by centrifugation, and incubated for 1 h with 30 μL GFP-trap beads (Chromotek). After incubation, the beads were washed four times in LB, boiled in 30 mL of SDS-sample buffer, and loaded for SDS-PAGE. The samples were run through 4 to 12% SDS-PAGE and the samples were submitted to the proteomics facility of Northwestern University, where they were subjected to in-gel reduction, alkylation, tryptic digestion, and subsequent quantitative MS analyses.
Raw quantitative MS/MS data were obtained via Scaffold Viewer’s (version Scaffold_4.9.0) complete export function. Total spectral counts were automatically normalized through Scaffold’s algorithm, where the UniProt database (the uniprot-SP-human_20180326_20190417 database; unknown version, 20,303 entries) was applied for the purpose of identification. The protein identification threshold was set to 1% false discovery rate.
A total of seven samples independently obtained from EcGFP-A431E-KO cells were quantified. Using RStudio (version 3.6.0, 2019-04-26), each sample column was joined by their respective unique ID (gene name) to reproduce a merged data frame. Proteins with fewer than or equal to four identifications across all seven samples were excluded from further data processing. Mean spectral counts were then calculated for the remaining proteins. A similar technique as described above was applied to obtain maximum spectral count values for the samples obtained from A431 cells (11 samples) and mean spectral counts for the samples obtained from A431E-KO cells expressing EcΔCytoGFP (6 samples). These values were then applied against mean values of E-cadherin to identify contaminants and nonspecific proteins. In both combinations, proteins with spectral counts greater than 20% of the E-cadherin mean were subjected to removal, with the exception of CDH1 and CDH3. A total of 59 proteins remained. Using the spectral counts of the E-cadherin sample set as a source, two-tailed t test was applied to the remaining 61 proteins to observe individual P values (at α = 0.05). The same procedure was used for analyses from three to five samples of EcGFP-A431Ec-KO cells with additional knocked-out α-catenin, p120, and a combination of plakoglobin and β-catenin. For Western blot analysis, the GFP-trap precipitates together with a sample of protein markers (10 to 450 kDa; Invitrogen) were separated on 3 to 8% Tris-acetate gels (Invitrogen) and transferred to nitrocellulose membrane (Millipore). Cell-surface proteins were biotinylated exactly as described (36). In brief, the confluent cultures (grown on 6-cm dishes) were washed and incubated at 4 °C with 2 mL of 0.5 mg/mL sulfo-NHS-LC-biotin (Pierce Chemical) in PBS-Ca for 5 min. The cell lysates were obtained using LB (see above) and biotinylated proteins were then precipitated by streptavidin agarose and analyzed by immunoblotting.
Structural Modeling of E Clusters Bound to Actin.
The extracellular E-cadherin lattice was built using the “generate symmetry mates” option in the PyMOL Molecular Graphics System (version 2.2.0; Schrödinger) as applied to a crystal structure of E-cadherin featuring both trans and cis interactions among ectodomains [Protein Data Bank (PDB) ID code 3Q2V (15)]. Cα-atoms at the C terminus of EC5 domains of E-cadherins in a 3 × 3 lattice were used as guides for constraints when placing cytoplasmic components into the lattice, with every protein structure translated according to the distances and angles shown in SI Appendix, Fig. S1. The following crystal structures were used (PDB ID codes are given in parentheses) when combining fragments of cytoplasmic components together: p120 bound to E-cadherin [3L6X (44)], β-catenin bound to E-cadherin [1I7W, 1I7X (43)], head domain of α-catenin bound to β-catenin [4ONS, chains C and D (46)], full-length α-catenin [4IGG (48)], full-length vinculin [1TR2 (42)], and human vinculin head domain in complex with an unfurled M-domain fragment of α-catenin [4EHP (47)].
All structures were stitched together either by structural superposition (whenever structures had overlapping regions) or by building linkers to connect the fragments in unstructured regions. The following C-terminal sequence stretches were modeled as linkers in PyMOL: 691 to 705, 732 to 755, 774 to 781, and 839 to 851. The transmembrane (TM) portion of E-cadherin, 707 to 731, predicted based on its helical secondary structure and high positive TMpred scores (https://embnet.vital-it.ch/software/TMPRED_form.html), was modeled as a single-pass helix in Coot [version 0.8.9.1 (68)]. The helix is about ∼33 Å in length, flanked by unstructured polar (Gln) or charged (Arg) residues, and oriented perpendicular to the plane of the membrane in Fig. 4. The residue numbers correspond to the human E-cadherin sequence (UniProt ID code P12830). The geometry of all modeled regions was further regularized in Coot.
Arrangement of p120 with Respect to β-Catenin.
A short E-cadherin linker of eight amino acids connects the two complexes. The linker ends are located near the N terminus of p120 and the middle of the β-catenin armadillo repeat domain, thus placing p120 and β-catenin in close proximity (it is also evident from the presented cross-linking experiment; Fig. 2), but whether an interface is formed between the two is unknown. We built a p120–β-catenin complex by docking the p120–E-cadherin heterodimer against the β-catenin–E-cadherin complex using ClusPro (69) and picking a top-scoring model that would satisfy two conditions: 1) Orientation between p120 and β-catenin would be compatible with an eight-amino-acid-long linker between E-cadherin tails that were structurally unresolved; and 2) the orientation should be as extended as possible to provide maximum space between the lattice components.
Incorporating α-Catenin into the Lattice.
It was difficult to find orientations of CCC components that would allow folded α-catenin to exist in the lattice without steric clashes. Small-angle X-ray scattering (SAXS) data (52) suggest that when α-catenin binds E-cadherin–β-catenin, structural rearrangement occurs leading to separation of the head and M domain. However, we did not consider this possibility in our modeling due to the lack of available coordinates of the E-cadherin–β-catenin–α-catenin complex. Thus, the folded form of α-catenin was used in our modeling.
To find orientations that might allow α-catenin to fit into the lattice without clashes, we created 1,728 2 × 2 lattices of full-length α-catenin–β-catenin heterodimers that are compatible with extracellular E-cadherin spacing. Rigid heterodimers were rotated by different combinations of roll, pitch, and yaw Euler angles (from α, β, γ = 0° to α, β, γ = 330° with a step of 30°) and then translated into the 2 × 2 lattice (SI Appendix, Fig. S5A). Each orientation was then evaluated for clashes by counting the number of atoms that are in close proximity to one other between distinct heterodimers in the lattice. All lattice configurations had clashes defined by a 4-Å-distance cutoff between atoms, approximately the sum of the effective van der Waals radius of CH2 groups (SI Appendix, Fig. S5B). A model with a minimal number of four such clashing atoms (SI Appendix, Fig. S5C) was chosen to represent the orientation of α-catenin–β-catenin in the lattice.
Incorporating Vinculin into the Lattice.
We first built a model of an α-catenin–vinculin complex (SI Appendix, Fig. S6A). In this model, the M1 domain of α-catenin is unfolded, in agreement with the crystal structure of the complex-containing protein fragments (47) (SI Appendix, Fig. S6 A, Top Left), while the head and M domain of α-catenin no longer share an interface [in agreement with SAXS data (52)]. Using the same approach as discussed in the paragraph above (SI Appendix, Fig. S5A), we generated 1,728 2 × 2 lattices using the α-catenin–vinculin heterodimer shown in SI Appendix, Fig. S5B. All lattice configurations resulted in severe clashes (SI Appendix, Fig. S6C). We then built a more compact model of α-catenin–vinculin (SI Appendix, Fig. S6D) where the M domain of α-catenin was reoriented with respect to the original model (SI Appendix, Fig. S6B) so as to allow the fewest possible clashes as determined by visual inspection. However, extended sampling still failed to produce a clash-free structure (SI Appendix, Fig. S6E). We note that all α-catenin–vinculin heterodimers we considered lacked ABD domains. These domains have previously been shown (44, 52, 70–73) to be easily detachable from the rest of the protein so as to bind actin (SI Appendix, Fig. S7).
Incorporating Erbin and Scribble into the Lattice.
Erbin has an LRR domain followed by a PDZ domain. Scribble has an LRR domain followed by four PDZ domains comparable in size to that of erbin. SI Appendix, Fig. S2 compares the size of the PDZ and LRR domains with spacings in the lattice. We show a representative PDZ domain of scribble [PDB ID code 5VWC; orange (74)] and a top structural template [E value <10−28 by HHpred search (75)] found for LRR domains of both scribble and erbin sequences [PDB ID code 4U09; yellow (76)]; structures of LRR domains of scribble and erbin have not been solved yet. The view in SI Appendix, Fig. S2 was chosen to show the maximum distance available between CCCs along two distinct dimensions in the lattice, and by visual observation PDZ domains can barely fit into the maximum spacing while LRR domains cannot. We also tried fitting these two domains manually into the lattice using PyMOL and were unable to find orientations with no clashes for the LRR domain. Moreover, if p120 and β-catenin were not to form an interface (as modeled in the lattice) and instead assumed a more distant orientation with respect to one another, there would be even no space for a PDZ domain of erbin and scribble in the E cluster, as it barely fits between CCCs of the current model.
Attachment to Actin.
An actin filament decorated by ABDs (Fig. 4) was built by combining three identical cryo-EM structures of the F-actin–ABD complex [PDB ID code 6UPV (45)] via superimposition of G-actins in PyMOL. F-actin–ABD was oriented manually in PyMOL to be parallel and equidistant from two CCC rows of the 3 × 3 lattice. α-Catenin ABD domains in these two CCC rows were then removed and the M domains of folded α-catenins were attached to ABDs on F-actin via flexible linkers modeled in Coot (these linkers, longer than 60 amino acids, have no crystal structure available).
Supplementary Material
Acknowledgments
We thank Dr. B. Mitchell for valuable discussions and suggestions. Proteomics, sequencing, and flow cytometry were performed at the Northwestern University Proteomics Center of Excellence, Genetic and Flow Cytometry Facilities. The work was supported by NIH Grants AR44016 and AR057992 (to S.M.T.), NSF Grant MCB-1914542 (to B.H.), and NIH Grants R01GM118584 and R01MH114817 (to L.S.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105550118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Green K. J., Getsios S., Troyanovsky S., Godsel L. M., Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb. Perspect. Biol. 2, a000125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gumbiner B. M., Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6, 622–634 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Harris T. J., Tepass U., Adherens junctions: From molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11, 502–514 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Niessen C. M., Leckband D., Yap A. S., Tissue organization by cadherin adhesion molecules: Dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91, 691–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley C. D., et al., Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 346, 1254211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C. S., et al., α-Catenin-mediated cadherin clustering couples cadherin and actin dynamics. J. Cell Biol. 210, 647–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Indra I., et al., Spatial and temporal organization of cadherin in punctate adherens junctions. Proc. Natl. Acad. Sci. U.S.A. 115, E4406–E4415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y., Vendome J., Shapiro L., Ben-Shaul A., Honig B., Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature 475, 510–513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brasch J., Harrison O. J., Honig B., Shapiro L., Thinking outside the cell: How cadherins drive adhesion. Trends Cell Biol. 22, 299–310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mège R. M., Ishiyama N., Integration of cadherin adhesion and cytoskeleton at adherens junctions. Cold Spring Harb. Perspect. Biol. 9, a028738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troyanovsky S., Adherens junction assembly. Subcell. Biochem. 60, 89–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap A. S., Gomez G. A., Parton R. G., Adherens junctions revisualized: Organizing cadherins as nanoassemblies. Dev. Cell 35, 12–20 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Indra I., Troyanovsky R. B., Shapiro L., Honig B., Troyanovsky S. M., Sensing actin dynamics through adherens junctions. Cell Rep. 30, 2820–2833.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yap A. S., Brieher W. M., Pruschy M., Gumbiner B. M., Lateral clustering of the adhesive ectodomain: A fundamental determinant of cadherin function. Curr. Biol. 7, 308–315 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Harrison O. J., et al., The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 19, 244–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen S. D., et al., αE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol. Biol. Cell 24, 3710–3720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong S., Troyanovsky R. B., Troyanovsky S. M., Binding to F-actin guides cadherin cluster assembly, stability, and movement. J. Cell Biol. 201, 131–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X. P., et al., Structural basis of αE-catenin-F-actin catch bond behavior. eLife 9, e60878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Indra I., Hong S., Troyanovsky R., Kormos B., Troyanovsky S., The adherens junction: A mosaic of cadherin and nectin clusters bundled by actin filaments. J. Invest. Dermatol. 133, 2546–2554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strale P. O., et al., The formation of ordered nanoclusters controls cadherin anchoring to actin and cell-cell contact fluidity. J. Cell Biol. 210, 1033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truong Quang B. A., Mani M., Markova O., Lecuit T., Lenne P. F., Principles of E-cadherin supramolecular organization in vivo. Curr. Biol. 23, 2197–2207 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Troyanovsky R. B., Indra I., Chen C. S., Hong S., Troyanovsky S. M., Cadherin controls nectin recruitment into adherens junctions by remodeling the actin cytoskeleton. J. Cell Sci. 128, 140–149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong S., Troyanovsky R. B., Troyanovsky S. M., Cadherin exits the junction by switching its adhesive bond. J. Cell Biol. 192, 1073–1083 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z., et al., E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci. Signal. 7, rs7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., et al., The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics. J. Cell Sci. 132, jcs221606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Itallie C. M., et al., Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J. Cell Sci. 127, 885–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troyanovsky R. B., Klingelhöfer J., Troyanovsky S., Removal of calcium ions triggers a novel type of intercadherin interaction. J. Cell Sci. 112, 4379–4387 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Troyanovsky R. B., Sokolov E., Troyanovsky S. M., Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol. Cell. Biol. 23, 7965–7972 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.le Duc Q., et al., Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J. Cell Biol. 189, 1107–1115 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leerberg J. M., et al., Tension-sensitive actin assembly supports contractility at the epithelial zonula adherens. Curr. Biol. 24, 1689–1699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuzawa K., Himoto T., Mochizuki Y., Ikenouchi J., α-Catenin controls the anisotropy of force distribution at cell-cell junctions during collective cell migration. Cell Rep. 23, 3447–3456 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M., Alpha-catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Borghi N., et al., E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U.S.A. 109, 12568–12573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Indra I., Troyanovsky R., Troyanovsky S. M., Afadin controls cadherin cluster stability using clathrin-independent mechanism. Tissue Barriers 2, e28687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiss A., Troyanovsky R. B., Troyanovsky S. M., p120-catenin is a key component of the cadherin-gamma-secretase supercomplex. Mol. Biol. Cell 19, 4042–4050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oldenburg J., et al., VASP, zyxin and TES are tension-dependent members of focal adherens junctions independent of the α-catenin-vinculin module. Sci. Rep. 5, 17225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi J., Troyanovsky R. B., Indra I., Mitchell B. J., Troyanovsky S. M., Scribble, Erbin, and Lano redundantly regulate epithelial polarity and apical adhesion complex. J. Cell Biol. 218, 2277–2293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laura R. P., et al., The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 277, 12906–12914 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Navarro C., et al., Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene 24, 4330–4339 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Harmon R. M., et al., Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. J. Clin. Invest. 123, 1556–1570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borgon R. A., Vonrhein C., Bricogne G., Bois P. R., Izard T., Crystal structure of human vinculin. Structure 12, 1189–1197 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Huber A. H., Weis W. I., The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105, 391–402 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Ishiyama N., et al., Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 141, 117–128 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Mei L., et al., Molecular mechanism for direct actin force-sensing by α-catenin. eLife 9, e62514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pokutta S., Drees F., Takai Y., Nelson W. J., Weis W. I., Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J. Biol. Chem. 277, 18868–18874 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rangarajan E. S., Izard T., The cytoskeletal protein α-catenin unfurls upon binding to vinculin. J. Biol. Chem. 287, 18492–18499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rangarajan E. S., Izard T., Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 20, 188–193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertocchi C., et al., Nanoscale architecture of cadherin-based cell adhesions. Nat. Cell Biol. 19, 28–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farquhar M. G., Palade G. E., Junctional complexes in various epithelia. J. Cell Biol. 17, 375–412 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNutt N. S., Weinstein R. S., Membrane ultrastructure at mammalian intercellular junctions. Prog. Biophys. Mol. Biol. 26, 45–101 (1973). [DOI] [PubMed] [Google Scholar]
- 52.Bush M., et al., An ensemble of flexible conformations underlies mechanotransduction by the cadherin-catenin adhesion complex. Proc. Natl. Acad. Sci. U.S.A. 116, 21545–21555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laur O. Y., Klingelhöfer J., Troyanovsky R. B., Troyanovsky S. M., Both the dimerization and immunochemical properties of E-cadherin EC1 domain depend on Trp(156) residue. Arch. Biochem. Biophys. 400, 141–147 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Petrova Y. I., Spano M. M., Gumbiner B. M., Conformational epitopes at cadherin calcium-binding sites and p120-catenin phosphorylation regulate cell adhesion. Mol. Biol. Cell 23, 2092–2108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong S., Troyanovsky R. B., Troyanovsky S. M., Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc. Natl. Acad. Sci. U.S.A. 107, 3528–3533 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrison O. J., et al., Two-step adhesive binding by classical cadherins. Nat. Struct. Mol. Biol. 17, 348–357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazan R. B., Kang L., Roe S., Borgen P. I., Rimm D. L., Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 272, 32448–32453 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Peng X., Maiers J. L., Choudhury D., Craig S. W., DeMali K. A., α-Catenin uses a novel mechanism to activate vinculin. J. Biol. Chem. 287, 7728–7737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ray S., Foote H. P., Lechler T., Beta-catenin protects the epidermis from mechanical stresses. J. Cell Biol. 202, 45–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han S. J., et al., Pre-complexation of talin and vinculin without tension is required for efficient nascent adhesion maturation. eLife 10, e66151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y., Aiga M., Yoshida E., Humbert P. O., Bamji S. X., Scribble interacts with beta-catenin to localize synaptic vesicles to synapses. Mol. Biol. Cell 20, 3390–3400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonello T. T., Peifer M., Scribble: A master scaffold in polarity, adhesion, synaptogenesis, and proliferation. J. Cell Biol. 218, 742–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tolbert C. E., Thompson P. M., Superfine R., Burridge K., Campbell S. L., Phosphorylation at Y1065 in vinculin mediates actin bundling, cell spreading, and mechanical responses to force. Biochemistry 53, 5526–5536 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Y., et al., Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc. Natl. Acad. Sci. U.S.A. 107, 17592–17597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klingelhöfer J., Troyanovsky R. B., Laur O. Y., Troyanovsky S., Amino-terminal domain of classic cadherins determines the specificity of the adhesive interactions. J. Cell Sci. 113, 2829–2836 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Troyanovsky R. B., Klingelhöfer J., Troyanovsky S. M., α-Catenin contributes to the strength of E-cadherin-p120 interactions. Mol. Biol. Cell 22, 4247–4255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J. B., et al., N-cadherin extracellular repeat 4 mediates epithelial to mesenchymal transition and increased motility. J. Cell Biol. 151, 1193–1206 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kozakov D., et al., The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ishiyama N., et al., An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288, 15913–15925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janssen M. E., et al., Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell 21, 271–281 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Jockusch B. M., Rüdiger M., Crosstalk between cell adhesion molecules: Vinculin as a paradigm for regulation by conformation. Trends Cell Biol. 6, 311–315 (1996). [DOI] [PubMed] [Google Scholar]
- 73.Nicholl I. D., et al., α-Catenin structure and nanoscale dynamics in solution and in complex with F-actin. Biophys. J. 115, 642–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lim K. Y. B., Gödde N. J., Humbert P. O., Kvansakul M., Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor β-PIX. J. Biol. Chem. 292, 20425–20436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zimmermann L., et al., A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Collins P. J., et al., Recent evolution of equine influenza and the origin of canine influenza. Proc. Natl. Acad. Sci. U.S.A. 111, 11175–11180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.