Significance
Cholesterol is present within the cell nucleus, where it associates with chromatin, but to date, a direct role for cholesterol in nuclear processes has not been identified. We demonstrate that the transcriptional repressor brain acid soluble protein 1 (BASP1) directly interacts with cholesterol within the cell nucleus through a consensus cholesterol interaction motif. BASP1 recruits cholesterol to the promoter region of target genes, where it is required to mediate chromatin remodeling and transcriptional repression. Our work demonstrates that cholesterol plays a direct role in transcriptional regulation.
Keywords: BASP1, WT1, cholesterol, transcription, chromatin
Abstract
Lipids are present within the cell nucleus, where they engage with factors involved in gene regulation. Cholesterol associates with chromatin in vivo and stimulates nucleosome packing in vitro, but its effects on specific transcriptional responses are not clear. Here, we show that the lipidated Wilms tumor 1 (WT1) transcriptional corepressor, brain acid soluble protein 1 (BASP1), interacts with cholesterol in the cell nucleus through a conserved cholesterol interaction motif. We demonstrate that BASP1 directly recruits cholesterol to the promoter region of WT1 target genes. Mutation of BASP1 to ablate its interaction with cholesterol or the treatment of cells with drugs that block cholesterol biosynthesis inhibits the transcriptional repressor function of BASP1. We find that the BASP1–cholesterol interaction is required for BASP1-dependent chromatin remodeling and the direction of transcription programs that control cell differentiation. Our study uncovers a mechanism for gene-specific targeting of cholesterol where it is required to mediate transcriptional repression.
It has been known for several decades that lipids and the enzymes that regulate lipid metabolism are present in the nucleus (1–3). The pool of nuclear lipids includes phospholipids, sphingolipids, gangliosides, and cholesterol that associate with nuclear proteins, chromatin, and the recently described nuclear lipid droplets (4). Several studies have reported that nuclear lipids associate with components of the transcription machinery, but their function(s) in gene regulation are not clear (1–3).
Brain acid soluble protein 1 (BASP1) is a transcriptional cofactor that associates with Wilms tumor 1 (WT1), converting it from a transcriptional activator to a repressor (5). Although BASP1 was initially identified as a cytoplasmic protein enriched in neuronal cells (6), it has since been found to be widely expressed and also enters the nucleus through a functional nuclear localization sequence (NLS; Fig. 1A; 7). BASP1 is N-terminally myristoylated and uses this motif to interact with membrane phospholipids within the cytoplasm (8). BASP1 also contains a cholesterol recognition amino acid consensus (CRAC) motif adjacent to its N-terminal myristoyl moiety (Fig. 1A) that binds to cholesterol within cell membranes (6, 9, 10). Thus, BASP1 can localize to cell membranes through simultaneous interactions with phospholipids and cholesterol. Previous studies have reported the binding of BASP1 to different cholesterol species, including cholesterol esters and oxysterols (8), which are also present within the nucleus (1). The N-terminal region of BASP1 containing the myristoyl motif, lipid-binding region, and NLS is termed the effector domain (6, 8; sequence shown in Fig. 1A). A similar effector domain is also found in GAP-43 and myristoylated alanine rich protein kinase C substrate (MARCKS), but outside this region, BASP1 shows no similarity to other proteins (6).
Fig. 1.
BASP1 interacts with cholesterol via a CRAC motif. (A) Schematic of BASP1 with N-terminal myristoyl (red), NLS, and PEST motifs. The N-terminal 25 residues—myristoylated G2 (red), PIP2-binding domain, bipartite NLS, and CRAC motif (central Tyr underlined in green)—are below. (B) Myristoylated and nonmyristoylated wtBASP1 or BASP1-Y12L prepared from E. coli (Coomassie gel at Left) were used to probe membranes dot blotted with cholesterol (0.04, 0.2, and 1 nmol). + is a positive control for BASP1 binding (sheep BASP1 antibody). Ctrl is a membrane probed with BSA, while the other panels were probed with the BASP1 derivatives and detected with rabbit BASP1 antibody. ImageJ quantitation of the relative signal in the dot blots is below for three independent experiments. Error bars are SDM of three independent experiments. *** is Student’s t test P < 0.001. (C) Control IgG (αCtrl) or WT1 (αWT1) antibodies were used for immunoprecipitation with nuclear extracts derived from B-K562 or Y-K562 cells and the content probed for BASP1 by immunoblot. Input is 5% of the nuclear extract used in each immunoprecipitation. (D) Control (αCtrl) or HA (αHA) antibodies were used for BASP1 immunoprecipitation with nuclear extracts derived from B-K562 or Y-K562 cells and the content probed for BASP1 or HDAC1 by immunoblot. Input is 5% of the nuclear extract used in each immunoprecipitation.
N-terminal myristoylation of BASP1 is also required for its function as a transcriptional repressor, and it uses this motif to interact with and recruit phosphatidylinositol 4,5-bisphosphate (PIP2) to gene promoters (11, 12). The promoter-bound BASP1–PIP2 complex supports the assembly of chromatin remodeling proteins, including histone deacetylase 1 (HDAC1) and Brahma-related gene 1 (BRG1), to WT1 target genes (11, 12). BASP1 can also repress the function of other transcriptional activators, including c-myc (13, 14) and estrogen receptor α (15), suggesting a broad role for this lipidated cofactor in the regulation of transcription.
WT1 and BASP1 cooperate to drive transcriptional programs that control differentiation in several model cell line systems (7, 11, 12, 16–18). Recent studies in mice found that knockout of BASP1 in taste receptor cells leads to loss of differentiated phenotype and the reactivation of WT1 target genes that are normally associated with the progenitor cells (19). A fundamental role for WT1-BASP1 in promoting the differentiated state is demonstrated by the finding that depletion of BASP1 in adult fibroblasts replaces the requirement of SOX2, c-Myc, or OCT4 in the induction of fibroblasts to pluripotency (20). In these cells, BASP1 normally represses WT1 target genes and blocks pluripotency in favor of cell maintenance in their differentiated state.
As mentioned above, BASP1 contains a functional CRAC motif adjacent to its N-terminal myristoyl moiety and binds to both cholesterol and PIP2 within cell membranes (9, 10; Fig. 1A). In this study, we demonstrate that BASP1 interacts with cholesterol via its CRAC motif within the cell nucleus, recruiting it to the promoter region of WT1 target genes. We find that the BASP1–cholesterol interaction is required for transcriptional repression and that it plays a role in BASP1-dependent differentiation in vitro. Our results demonstrate direct gene-specific control by cholesterol through interaction with the lipidated transcriptional repressor BASP1.
Results
BASP1 Interacts With Cholesterol in the Cell Nucleus through a Conserved CRAC Motif.
BASP1 interacts with cholesterol-rich domains (6, 8–10) and contains a conserved CRAC motif [L/V-X(1-5)-Y-X(1-5)-R/K] located within the N terminus close to the myristoyl motif (21; Fig. 1A). Using Escherichia coli engineered to express N-myristoyl transferase (NMT), we generated recombinant myristoylated wild-type (wt) BASP1 containing a carboxyl-terminal His-tag (myrBASP1) and the myristoylated mutant derivative Y12L (myrBASP1 Y12) that ablates the critical tyrosine residue of the CRAC motif. We also expressed the same proteins in E. coli lacking NMT to produce nonmyristoylated wtBASP1 and BASP1 Y12L. The purified proteins are shown in Fig. 1B. In filter binding assays, recombinant myristoylated wtBASP1 bound directly to cholesterol, but the nonmyristoylated forms of BASP1 did not. Moreover, the interaction of myristoylated BASP1-Y12L with cholesterol was significantly reduced (Fig. 1B; see quantitation below). Thus, both myristoylation of BASP1 and an intact CRAC motif are required for its direct interaction with cholesterol.
Our previous work has used K562 chronic myelogenous leukemia cells, which lack endogenous BASP1 but contain endogenous WT1. Stably introducing wtBASP1 into K562 cells (B-K562) represses WT1-dependent target genes compared to vector control K562 cells (V-K562; 7, 11, 12, 16). We generated a K562 cell line derivative that expresses BASP1-Y12L (Y-K562), which, like wtBASP1, is largely nuclear (SI Appendix, Fig. S1A). Immunoprecipitation of BASP1 from nuclear extracts via a carboxyl-terminal HA-tag followed by dot blotting to detect PIP2 confirmed that both wtBASP1 and BASP1 Y12L bind PIP2 (SI Appendix, S1B; 11, 12). BASP1 Y12L also coimmunoprecipitated with WT1 (Fig. 1C) and HDAC1 (Fig. 1D) from nuclear extracts at a level comparable with wtBASP1. Thus, the interaction of BASP1 with WT1, HDAC1, and PIP2 does not depend on an intact CRAC motif within BASP1, suggesting that these contacts do not require cholesterol.
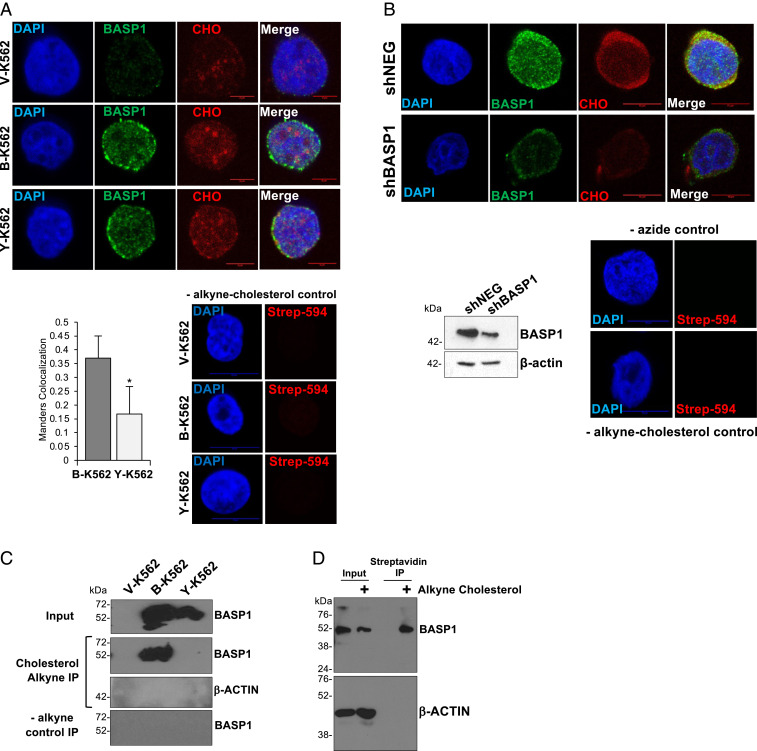
We next sought to determine if BASP1 interacts with cholesterol in the cell nucleus. To resolve nuclear cholesterol from cytoplasmic cholesterol, we isolated nuclei with low cytoplasmic contamination as determined by levels of the Golgi protein TGN46 (SI Appendix, Fig. S1C) and the endoplasmic reticulum (ER) marker calreticulin (SI Appendix, Fig. S1D). The K562 cell line derivatives were incubated in lipid-free media supplemented with a 27-alkyne cholesterol derivative that is utilized by cells as standard cholesterol (22). Nuclei were then prepared and, using click chemistry, the 27-alkyne cholesterol was cross-linked to azide-PEG3-biotin, which was then detected using a streptavidin-linked fluorophore. BASP1 was simultaneously probed using immunocytochemistry. Cholesterol was detected within the purified nuclei of K562 cells (Fig. 2A; cholesterol [CHO]) and showed reduced colocalization with BASP1-Y12L compared to wtBASP1 (Manders quantitation of the proportion of BASP1 signal that overlaps with cholesterol is shown below, Left). Endogenous BASP1 in MCF7 cells also colocalized with 27-alkyne cholesterol within the nucleus (Fig. 2B). Moreover, the nuclear 27-alkyne cholesterol signal in MCF7 cells was reduced when BASP1 expression was knocked down by short hairpin RNA (shRNA). We next used click chemistry combined with immunoprecipitation to test the interaction between BASP1 and nuclear cholesterol. V-K562, B-K562, and Y-K562 cells were cultured with 27-alkyne cholesterol as above for 48 h (as above) and then underwent the click chemistry reaction with azide-PEG3-biotin. Nuclear protein extracts were prepared followed by precipitation with streptavidin-coated beads. wtBASP1, but not BASP1-Y12L, coprecipitated with alkyne cholesterol (Fig. 2C). Similar analysis verified that 27-alkyne cholesterol associates with endogenous BASP1 in the nuclei of MCF7 cells (Fig. 2D). Taken together, the data in Fig. 2 demonstrate that BASP1 interacts with nuclear cholesterol via a functional CRAC motif.
Fig. 2.
BASP1 interacts with nuclear cholesterol. (A) K562 cell derivatives were incubated with 10 mg/mL alkyne cholesterol (CHO) in lipid-free media for 20 h. Nuclei were purified and, after treatment with azide PEG3-biotin and click reaction using 2 mM CuBF4, subjected to immunocytochemistry with streptavidin-linked fluorophore (CHO) or BASP1 antibodies. Scale bar is 5 μm. Below Right, cells treated in the same way, but without CHO-alkyne. Manders analysis to quantitate the colocalization of cholesterol with wtBASP1 and BASP1-Y12L (Below, Left). Error bars are SDM of three independent experiments (n = 35 for each condition). * is Student’s t test P < 0.05. (B) MCF7 cells stably expressing a control shRNA (shNEG) or shRNA targeting BASP1 (shBASP1) were used to prepare nuclear extracts and immunoblotted with BASP1 or b-actin antibodies. Below, shNEG and shBASP1 MCF7 cells were incubated in lipid-free media with 10 μg/mL alkyne cholesterol. Nuclei were prepared followed by click chemistry and immunofluorescence with BASP1 antibodies and streptavidin antibodies to detect nuclear cholesterol (CHO). Scale bars are 10 μm. At Right are control immunofluorescence performed with nuclei purified from cells that were not incubated with alkyne cholesterol. (C) As in A, but nuclear extract was prepared and precipitation performed with streptavidin magnetic beads, then immunoblotted with BASP1 or actin antibodies. −alkyne control IP is the same click chemistry reaction but with cells not incubated with alkyne cholesterol. (D) MCF7 cells were incubated with (+) or without (−) alkyne cholesterol, nuclear extract was prepared followed by click chemistry, and the alkyne cholesterol was captured with streptavidin magnetic beads. The content was probed with BASP1 or β-actin antibodies. Input is 5% of the nuclear extract used in each assay.
BASP1 Recruits Cholesterol to the Gene Promoter Region of WT1 Target Genes.
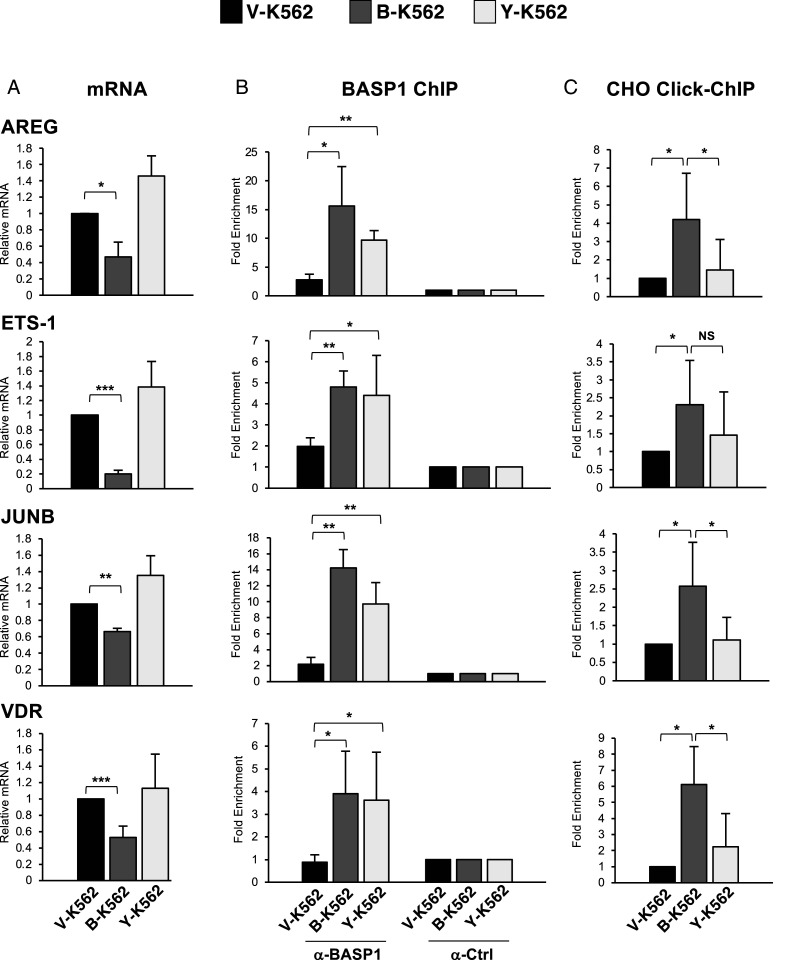
We next investigated whether an intact CRAC motif in BASP1 is required for transcriptional repression. As we have previously shown, wtBASP1 repressed transcription of the WT1 target genes AREG, ETS-1, JUNB, and VDR (7, 11, 12; Fig. 3A). In contrast, BASP1 Y12L was defective in transcriptional repression. Chromatin immunoprecipitation (ChIP) analysis determined that both wtBASP1 and BASP1 Y12L were recruited to the promoter region of all the genes tested (Fig. 3B). Combining click chemistry with ChIP (Click-ChIP), we found that cholesterol associates with the AREG, ETS-1, JUNB, and VDR promoters at a significantly increased level when wtBASP1 is also present at the promoter (Fig. 3C; compare V-K562 with B-K562). In K562 cells that express BASP1-Y12L, cholesterol was recruited at a significantly reduced level at the AREG, JUNB, and VDR promoters compared to B-K562 cells and, although not statistically significant, was also reduced at the ETS-1 promoter. Thus, BASP1 recruits cholesterol to the promoter region of WT1 target genes, and this requires an intact CRAC motif in BASP1.
Fig. 3.
BASP1 Y12L is defective in transcriptional repression and fails to recruit cholesterol to the promoter of WT1 target genes. (A) cDNA was prepared from V-K562, B-K562, or Y-K562 cells. Expression of AREG, ETS-1, JUNB and VDR were compared to control GAPDH. (B) Cells as above were subjected to ChIP analysis with αBASP1 or control (αCtrl) IgG. Presented as fold enrichment over a control genomic region. (C) K562 cell lines were incubated with alkyne cholesterol as in Fig. 1E. After the click chemistry reaction, the cells were subjected to ChIP with streptavidin-linked beads (Click-ChIP). Results are fold enrichment over a control genomic region. Error bars are SDM from at least three independent experiments. Student’s t test; *<0.05, **<0.01, ***<0.001, not significant (NS).
The BASP1–Cholesterol Interaction Is Required for the Control of Differentiation.
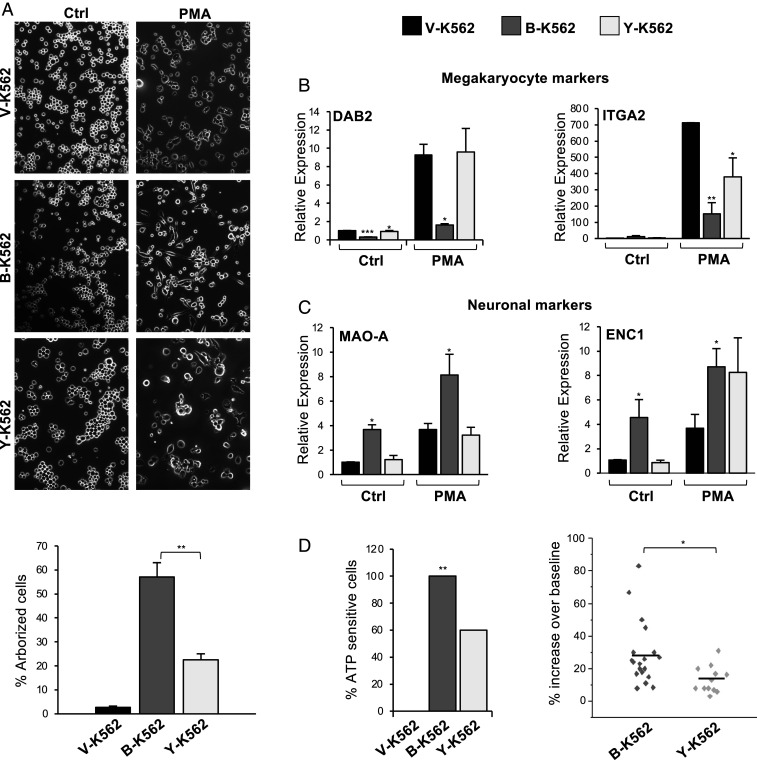
A central function of the WT1–BASP1 complex is to drive cell differentiation programs (5). BASP1 cooperates with WT1 to divert the phorbol ester–induced differentiation program of K562 cells away from megakaryocytes to neuronal-like cells in both morphology and function (7, 11, 12). This was driven by the WT1/BASP1-dependent down-regulation of genes that specify megakaryocyte identity accompanied by the up-regulation of neuronal markers. We tested whether BASP1 Y12L was able to support the neuronal-like differentiation program of K562 cells. The K562 cell derivatives were induced to differentiate with phorbol 12-myristate 13-acetate (PMA) for 72 h, and the extent of cellular arborization was visualized by phase microscopy (Fig. 4A, quantitation shown below). wtBASP1 expression in K562 cells led to induction of an arborized phenotype compared to V-K562 cells, and that arborization was significantly reduced in Y-K562 cells.
Fig. 4.
An intact CRAC motif in BASP1 is required to induce a neuronal-like differentiation program in K562 cells. (A) V-K562, B-K562, or Y-K562 cells were treated with 200 nM PMA or DMSO for 72 h and then observed by phase microscopy. Percentage arborized cells is below. Error bars are SDM of three independent experiments and ** indicates Student’s t test P < 0.01. (B) Cells were treated as in A, but 48 h later, cDNA was prepared and qPCR used to determine expression of the megakaryocyte markers DAB2 and ITGA2 relative to GAPDH. Error bars are SDM of three independent experiments with Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001. (C) As in B, except that neuronal markers MAO-A and ENC1 were analyzed. (D). The K562 derivatives were treated as in A, ATP (10 mM) was applied, and calcium responses were measured by fluorescence imaging. At Left, the percentage of cells that generated a calcium signal to ATP. * indicates χ2 test at P < 0.01. At Right, the responsive B-K562 cells (n = 19 cells) and Y-K562 cells (n = 12 cells) are shown as a scatter plot against the amplitude of response. * indicates P < 0.05 by Student’s t test.
Analysis of the level of messenger RNA (mRNA) encoding the megakaryocyte-specific markers DAB2 and ITGA2 revealed that, whereas wtBASP1 blocked their induction (as we previously reported in ref. 7), BASP1 Y12L failed to repress transcription of DAB2 and was defective in repression of ITGA2 (Fig. 4B). Analysis of neuronal-specific markers (MAO-A and ENC1) showed that BASP1 induced their expression as we previously reported (Fig. 4C; 7). BASP1 Y12L was defective in inducing MAO-A but still able to support induction of ENC1. These data suggest that BASP1 Y12L is selective in its effects in diverting the PMA-induced K562 cell differentiation program to neuronal-like cells. We next measured the ability of the PMA-treated cells to respond to the neurotransmitter adenosine triphosphate (ATP). In agreement with our earlier work (7), none of the control V-K562 cells generated a Ca2+ signal in response to ATP, while 100% of the B-K562 cells responded (19 of 19 cells tested; Fig. 4D, Left). Y-K562 cells showed an intermediate response rate (60%; 12 of 20 cells tested). We then analyzed the Ca2+ signals of the cells that responded to ATP. Comparisons of the peak amplitude of the measured responses in the B-K562 and Y-K562 cells found the size of the Ca2+signals in BASP1 Y12L cells were significantly reduced compared to wtBASP1 cells, suggesting the Y-K562 cells are less responsive to ATP (Fig. 4D, Right). Thus, the CRAC motif in BASP1 plays a role in the diversion of the differentiation program of K562 cells to a neuronal-like cell.
Inhibition of Cholesterol Synthesis Abolishes Transcriptional Repression by BASP1.
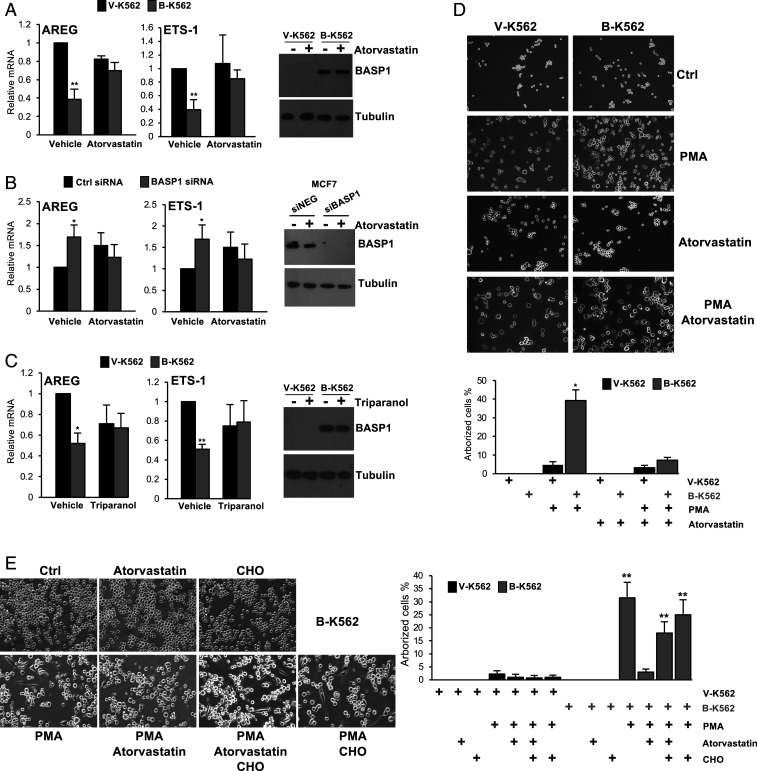
Our results so far have shown that mutation of the critical tyrosine residue of the BASP1 CRAC motif disrupts the interaction with cholesterol and its recruitment to the promoter and causes a defect in transcriptional repression and BASP1-induced differentiation. To confirm and extend these findings, we determined the effect of cholesterol-depleting drugs on BASP1 function. We first confirmed that the drugs were used at an effective concentration to significantly reduce nuclear cholesterol (SI Appendix, Fig. S2A). We treated V-K562 cells or B-K562 cells with atorvastatin, an inhibitor of HMG-CoA reductase (the rate-limiting enzyme in cholesterol biosynthesis) and determined the effect on BASP1-mediated transcriptional repression. Atorvastatin treatment abolished transcriptional repression of AREG and ETS-1 by BASP1 in K562 cells (Fig. 5A). Similar experiments with lovastatin (which also inhibits HMG-CoA reductase) produced comparable results (SI Appendix, Fig. S2B). We also tested the effect of atorvastatin on BASP1 transcriptional repressor function in MCF7 cells transiently transfected with either a control small interfering RNA (siRNA) or siRNA targeting BASP1. Knockdown of endogenous BASP1 in MCF7 cells led to relief of repression of WT1 target genes as previously reported (Fig. 5B; 12). When the cells were treated with atorvastatin, the relief of transcriptional repression upon BASP1 knockdown did not occur. We observed similar effects when MCF7 cell line derivatives that stably express either a control shRNA (shNEG) or BASP1 shRNA (shBASP1), targeting a different sequence to the siRNA used in Fig. 5B), were treated with lovastatin (SI Appendix, Fig. S2C). Statins inhibit the biosynthesis of both cholesterol and isoprenoids (23). We therefore next tested the effect of triparanol, an inhibitor of 24-dehydrocholesterol reductase, which catalyzes the final step in cholesterol biosynthesis pathway and does not inhibit isoprenoid production. As we had observed with the statins, triparanol blocked the transcriptional repressor function of BASP1 (Fig. 5C). We next determined whether interference with cholesterol biosynthesis in K562 cells affects the BASP1-dependent differentiation to neuronal-like cells. As before, treatment of B-K562 cells, but not V-K562 cells, with PMA induced an arborized phenotype (Fig. 5D). This was blocked if the cells were simultaneously treated with atorvastatin (see quantitation below). To confirm that the effect of atorvastatin on the differentiation of B-K562 cells was due to reduced cholesterol availability, we determined whether its effects could be reversed by the coaddition of cholesterol to the cell culture media. We used an established protocol that solubilizes cholesterol with methyl-β-cyclodextrin (MβCD) at levels that lead to cellular accumulation of cholesterol (24). Addition of cholesterol through this method led to the PMA-induced arborization of the B-K562 cells even when atorvastatin was present in the media (Fig. 5E; quantitation shown at Right. Images of the V-K562 cells are shown in SI Appendix, Fig. S2D). Taken together, the data in Fig. 5 demonstrate that inhibition of cholesterol biosynthesis blocks the transcriptional repressor function of BASP1 and impedes BASP1-induced differentiation.
Fig. 5.
Statins specifically inhibit the transcriptional repressor function of BASP1. (A) V-K562 or B-K562 cells were treated with 20 μM atorvastatin or drug vehicle (DMSO) for 48 h. cDNA was prepared, and expression of AREG and ETS-1 was determined relative to GAPDH. At Right, a Western blot of BASP1 is shown. (B) MCF7 cells were transfected with either a control siRNA (siNEG) or siRNA targeting BASP1 (siBASP1). 24 h later, the cells were treated with 20 μM atorvastatin or drug vehicle (DMSO) for 48 h. cDNA was prepared, and expression of AREG and ETS-1 was determined relative to GAPDH. At Right, a Western blot of BASP1 is shown. (C) As in A, except that cells were treated with 20 μM triparanol. (D) V-K562 cells or B-K562 cells were treated with PMA (200 nM) and/or atorvastatin (20μM) as indicated. 72 h later, the cells were analyzed as in Fig. 4A. Percentage arborized cells is below (error bars are SDM of three independent experiments, and * indicates Student’s t test P < 0.05.) (E) As in D, except that cholesterol was also added to the media (100 μM) where indicated. Percentage arborized cells is at Right (error bars are SDM of four independent experiments, and ** indicates Student’s t test P < 0.01). Graph also includes quantitation from similarly treated V-K562 cells (images shown in SI Appendix, Fig. S2D).
The BASP1–Cholesterol Interaction Is Required for Chromatin Remodeling.
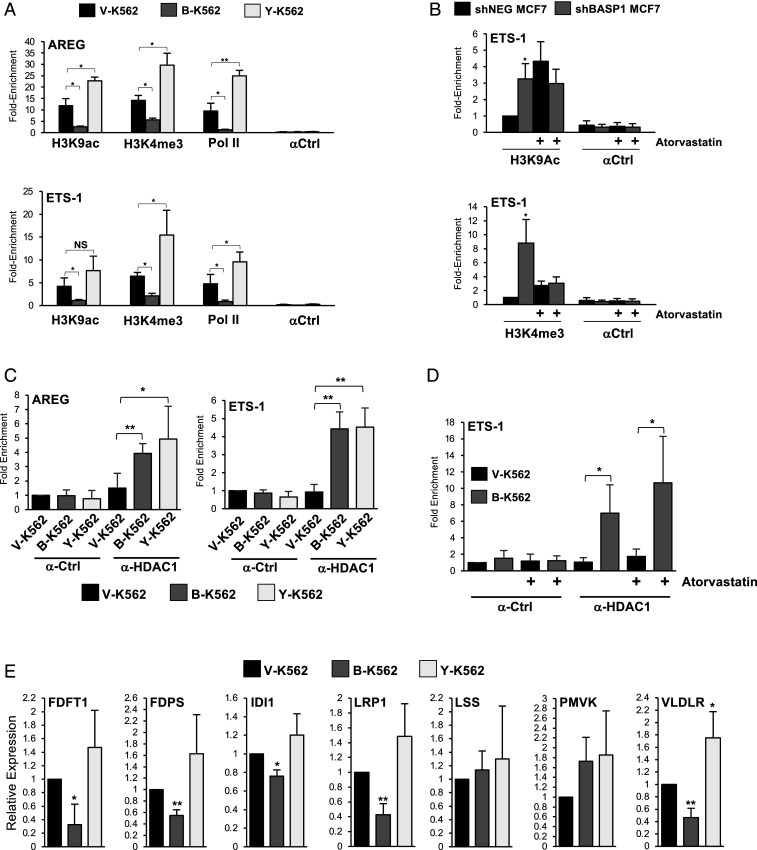
BASP1 acts as a transcriptional repressor by remodeling chromatin and blocking the recruitment of RNA polymerase II (Pol II) at the promoter region of WT1 target genes (11, 12, 17). We next performed ChIP analysis with the K562 cell line derivatives to determine the effect of wtBASP1 and BASP1-Y12L on Pol II recruitment as well as two histone modifications indicative of transcriptional activity (H3K9Ac and H3K4me3) at the AREG and ETS-1 promoters (Fig. 6A). Compared to V-K562 cells, B-K562 cells show significantly reduced H3K9Ac, reduced H3K4me3, and reduced Pol II binding at the AREG and ETS-1 promoters. BASP1 Y12L did not support the reduction in either the active chromatin marks or in Pol II recruitment. Indeed, there was a significant increase over the vector control cells, suggesting the possibility that BASP1 Y12L has transcriptional activator properties. We next performed ChIP analysis using MCF7 cell line derivatives that express either a control shRNA (shNEG MCF7) or an shRNA that targets BASP1 (shBASP1) to determine the effect of atorvastatin on H3K9Ac and H3K4me3 marks at the ETS-1 promoter. Consistent with the observations in K562 cells, shRNA-mediated knockdown of BASP1 in MCF7 cells caused an increase in H3K9Ac and H3K4me3 marks at the ETS-1 promoter (Fig. 6B). However, treatment of the MCF7 cell line derivatives with atorvastatin abolished the BASP1-dependent changes in these marks. We note that atorvastatin treatment caused a general increase in H3K9 acetylation and to a lesser extent H3K4 trimethylation, suggesting that it also elicits BASP1-independent functions on these histone modifications. Taken together, the data in Fig. 6 A and B suggest that the BASP1–cholesterol interaction is required for BASP1-dependent histone modification.
Fig. 6.
A role for cholesterol in BASP1-dependent chromatin remodeling and cholesterol homeostasis. (A) ChIP was performed with V-K562, B-K562, and Y-K562 cells with control IgG (αCtrl), H3K9Ac, H3K4me3, and RNA Pol II antibodies. Data are presented as fold enrichment at the AREG (Top) and ETS-1 (Bottom) promoters over a control genomic region. (B) MCF7 cells stably expressing either control shRNA (shNEG) or BASP1 shRNA (shBASP1) were treated with 20 μM atorvastatin or vehicle (DMSO). 48 h later, the cells were cross-linked, and ChIP was performed with antibodies against H3K9ac (Top graph) or H3K4me3 (Bottom graph) alongside IgG control (αCtrl). Data are presented as fold enrichment at the ETS-1 promoter against a control genomic region. Error bars are SDM of at least three independent experiments (*P < 0.05 and **P < 0.01, and NS is not significant by Student’s t test). (C) ChIP was performed with V-K562, B-K562, and Y-K562 cells with control IgG (α-Ctrl) or HDAC1 (α-HDAC1) antibodies. Data are presented as fold enrichment at the AREG (Top) and ETS-1 (Bottom) promoters compared to a control genomic region. (D) V-K562 or B-K562 cells were treated with 20 μM atorvastatin or vehicle (DMSO). 48 h later, the cells were cross-linked, and ChIP was performed with antibodies against HDAC1 (α-HDAC1) or control (α-Ctrl) antibodies. Data are presented as fold enrichment at the ETS-1 promoter against a control genomic region. Error bars denote SDM of at least three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test). (E) cDNA was prepared from V-K562, B-K562, or Y-K562 cells. Expression of the indicated genes was compared to control GAPDH. Error bars denote SDM of four independent experiments (*P < 0.05, **P < 0.01 by Student’s t test).
Our previous work has shown that BASP1 interacts with HDAC1 to deacetylate H3K9 (11). BASP1 Y12L coimmunoprecipitated with HDAC1 from nuclear extracts at a level similar to wtBASP1 (Fig. 1D). Consistent with this, we also found that BASP1 Y12L recruited HDAC1 to the promoter region of the AREG and ETS-1 genes to a level similar to wtBASP1 (Fig. 6C). Furthermore, treatment of B-K562 cells with atorvastatin did not significantly affect HDAC1 recruitment at the ETS-1 promoter (Fig. 6D). Thus, the BASP1–cholesterol interaction is not required for HDAC1 recruitment but is required for both the deacetylation of H3K9 and demethylation of H3K4.
Previous studies have identified several genes in the cholesterol biosynthesis and lipid transport pathways as direct targets of WT1 (25, 26). Since BASP1 is a general repressor of WT1 transcription function, we sought to determine whether an intact BASP1 CRAC motif is required for transcriptional repression of a cohort of WT1 target genes identified from these previous studies connecting WT1 with cholesterol homeostasis. In K562 cells, wtBASP1, but not BASP1 Y12L, repressed transcription of key components of the mevalonate pathway including FDFT1, FDPS, and IDI1 and the lipoprotein receptors LRP1 and VLDLR (five of the seven WT1 target genes tested; Fig. 6E).
Discussion
In this study, we provide evidence for the directed recruitment of cholesterol to gene promoters by BASP1 and show that cholesterol is required for BASP1-dependent histone modification and transcriptional repression. Previous studies found that the BASP1–PIP2 interaction is required to mediate contact between BASP1 and HDAC1 (11). We determined that the BASP1–PIP2 interaction is not dependent on the BASP1 CRAC motif and, consistent with this, BASP1 Y12L also interacts with HDAC1. Thus, the BASP1–HDAC1 interaction is not dependent on cholesterol, identifying distinct roles for PIP2 and cholesterol on BASP1 function as a transcriptional corepressor.
It is possible that the BASP1–cholesterol interaction facilitates the recruitment of other chromatin remodeling complexes that are a prerequisite for the action of HDAC1 at the promoter. For example, the enzymatic activities of HDAC1 and the histone demethylase LSD1 within the CoREST complex are interdependent (27). Moreover, the activity of class I HDACs within different complexes exhibit distinct specificities of action (28). It is therefore possible that the lack of HDAC1-mediated H3K9 deacetylation in Y-K562 cells is due to the failure of BASP1 Y12L to recruit other chromatin remodeling factors that have yet to be identified.
Alternatively, cholesterol might be required to facilitate nucleosome modification through direct effects on chromatin. Previous work has reported that cholesterol binds directly to histones and drives the compaction of nucleosomes in vitro (2, 29). Cholesterol interacts with at least six sites on the nucleosome and is proposed to facilitate dewetting of hydrophobic surfaces on the histones, leading to enhanced histone–histone contacts that drive chromatin condensation. Thus, BASP1-mediated recruitment of cholesterol might elicit direct effects on the nucleosome that contribute to establishing transcriptionally repressive condensed chromatin at specific genes. Further studies are needed to address this question.
A role for lipids in regulating the affinity and/or specificity of protein–protein interactions in transcription regulation is reminiscent of the action of lipid-derived hormones on steroid receptor transcription factors (30). However, the widely reported roles of nuclear lipids in transcription suggest a more general function in transcription control. The regulation of transcription by lipids might also have additional physiological importance. Knockout of WT1 in adult mice leads to rapid fat loss through direct effects on adipocyte homeostasis and lipid metabolism (31). Indeed, several genes in the cholesterol biosynthesis pathway and lipid transport are target genes of WT1 (25, 26). We find that BASP1 represses transcription of several of these WT1 target genes and that this is dependent on an intact CRAC motif in BASP1. These results suggest that the control of WT1-BASP1 by lipids provides a feedback mechanism to regulate lipid metabolism.
Our data suggest that either BASP1 associates with cholesterol in the cytoplasm and then moves to the nucleus or that it first associates with cholesterol that is already present within the nucleus. How lipids are sequestered within the nucleus and the regulation of their subnuclear localization is not yet clear, but lipidated transcription factors such as BASP1 can provide at least part of the mechanism. Nuclear lipid droplets (NLDs) have recently been described that may play a role in the regulation of transcription factor accessibility (4, 32, 33). BASP1 is a component of cytoplasmic cholesterol-rich lipid droplets (34), which raises the prospect that BASP1 might also be incorporated into cholesterol-rich NLDs. NLDs associate with promyelocytic leukemia bodies (35), which can also contain BASP1 (18). It is therefore possible that BASP1–lipid interactions may control transcription processes through the formation of phase-separated nuclear bodies (36). Lipidated transcription factors like BASP1 have the potential to play important roles in liquid-liquid phase separation in chromatin remodeling and transcription control (37, 38).
Materials and Methods
DNA Constructs.
pcDNA3 BASP1-HA is described in Goodfellow et al. (7). pcDNA3-BASP1 Y12L-HA was generated using the Quickchange site-directed mutagenesis kit (Agilent, 200518) and verified by DNA sequencing. The coding sequence of wtBASP1 and BASP1-Y12L lacking the stop codon was amplified and cloned in frame into the NcoI/XbaI sites of pET23d bacterial expression vector to generate BASP1 and BASP1-Y12L with a carboxyl-terminal His-tag. pRSF-1-NMT for expression of N-myristoyl transferase in bacteria was purchased from Addgene (42578).
Cell Lines, Transfection, and Antibodies.
K562 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 (Thermo Fisher, 11875101) supplemented with 10% (vol/vol) fetal calf serum (Sigma-Aldrich, F9665), 1% (vol/vol) penicillin–streptomycin (Pen-Strep) (Sigma-Aldrich, P4333), and 1% (vol/vol) L-glutamine (Sigma-Aldrich, G7513). MCF7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Thermo Fisher, 11960044) supplemented with 10% (vol/vol) fetal calf serum and 1% (vol/vol) Pen-Strep. Stably transfected K562 and MCF7 cell lines were additionally supplemented with 1 mg/mL G-418 (Sigma-Aldrich, A1720). All cells were kept at 37 °C in humidified 95% air and 5% CO2 atmosphere. Cells were transfected with plasmids using Effectene (Qiagen, 301425) and with siRNA using HiPerFect (Qiagen, 301704). Antibodies used are listed in SI Appendix, Table S1. For cell treatments, atorvastatin (Sigma-Aldrich, PZ0001), lovastatin (Sigma-Aldrich, M2147), triparanol (Sigma-Aldrich, T5200), and phorbol 12-myristyl 3-acetate (PMA; Sigma-Aldrich, 524400) were dissolved in dimethyl sulfoxide (DMSO) and added to the cell culture media at the concentrations indicated in the figure legends. Addition of cholesterol to cell culture media was as described by Weidenmaier et al. (24). Cholesterol (Sigma-Aldrich, C8667; dissolved in ethanol) was added to methyl-β-cyclodextrin (MβCD, Sigma-Aldrich, C4555) dissolved in water to a final concentration of 5 mM cholesterol/42 mg/mL MβCD. This was added to cell culture media at a final concentration of 100 μM cholesterol.
Chromatin Immunoprecipitation.
K562 cells were collected and resuspended to a density of 1 × 106 cells/mL in phosphate buffered saline (PBS). Cross-linking was achieved by adding formaldehyde to a final concentration of 1.42% (vol/vol) and incubating at room temperature for 15 min. Formaldehyde was quenched with ice-cold 125 mM glycine for 5 min at room temperature. Cells were collected by centrifugation at 2,000 × g for 5 min at 4 °C and washed with ice-cold PBS. Cells were resuspended to a concentration of 1 × 107 cells/mL and lysed in 1 mL immunoprecipitation (IP) buffer [150 mM NaCl, 50 mM Tris⋅HCl (pH 7.5), 5 mM ethylenediamine tetraacetic acid (EDTA), 0.5% (vol/vol) Nonidet P-40, and 1% (vol/vol) Triton X-100] plus protease inhibitor mixture for 15 min on ice. Lysed samples were centrifuged at 2,000 × g for 5 min at 4 °C, and the pellet resuspended in 1 mL IP buffer plus protease inhibitors for sonication. Chromatin was sheared via sonication using a QSonica Q500 at 60% amplitude. Successful sonication giving fragments 200 to 500 bp in length was confirmed by resolving a small (de-cross-linked) sample on a 1.5% (wt/vol) agarose gel. Following sonication, the lysate was cleared by centrifugation at 12,000 × g for 10 min at 4 °C. Samples were precleared by incubating with 10 μL Protein G magnetic beads (Thermo Fisher, 88847) for 1 h, rotating at 4 °C. Five μL Protein G magnetic beads, 600 μL IP buffer, 1 μL 10 mg/mL acetylated bovine serum albumin (BSA), and appropriate antibody were also incubated together in one microtube per desired chromatin immunoprecipitation for a minimum of 4 h, rotating at 4 °C. 200 μL of precleared chromatin was then added to the incubated microtube of antibody and beads and rotated at 4 °C overnight. A 2% input sample was also stored for later de-cross-linking and processing.
Immunoprecipitated samples were then magnetized, supernatant was discarded, and beads were sequentially washed once in IP buffer, high-salt IP buffer [500 mM NaCl, 50 mM Tris⋅HCl (pH 8.0), 5 mM EDTA, 0.5% (vol/vol) Nonidet P-40, 1% (vol/vol) Triton × 100], LiCl buffer [10 mM Tris⋅HCl (pH 8.0), 250 mM LiCl, 1 mM EDTA, 1% (vol/vol) Nonidet P-40, 1% (wt/vol) sodium deoxycholate], and Tris-EDTA (TE) buffer [10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA]. After the final wash, beads and 2% input samples were resuspended in 100 μL of proteinase K (PK) buffer [125 mM Tris⋅HCl (pH 8.0), 10 mM EDTA, 150 mM NaCl, 1% (wt/vol) sodium dodecyl sulfate (SDS)] and placed at 65 °C overnight. Samples were then incubated with 1 μL of 20 mg/mL Proteinase K for 3 to 4 h at 55 °C. Finally, the immunoprecipitated DNA was purified using the Qiaquick PCR purification kit (Qiagen, 28104) according to manufacturer’s instructions. Eluted DNA was placed at 95 °C for 10 min and prepared for qPCR using the primers in SI Appendix, Table S2.
Immunoprecipitation.
All protein immunoprecipitation was carried out using nuclear extracts prepared from K562 cells. Cells were collected by centrifugation for 3 min at 1,400 × g. The packed cell volume (PCV) was estimated, and the pellet was incubated with 2/3 PCV of nuclear extraction 1 (NE1) buffer [10 mM Hepes (pH 8.0), 1.5 mM MgCl2, 10 mM KCL, 1 mM dithiothreitol (DTT)] for 15 min on ice, with protease inhibitors. Cells were then passed through a 23-gauge needle 4 to 5 times and microfuged at full speed for 30 s to isolate nuclei. The supernatant was discarded and remaining pellet resuspended in one PCV of NE2 buffer [20 mM Hepes (pH 8.0), 1.5 mM MgCl2, 25% (vol/vol) glycerol, 420 mM NaCl, 0.2 mM EDTA, 1 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] on ice for with regular stirring for 30 min, with protease inhibitor mixture. Nuclear debris was pelleted by 5 min microfuge at full speed. The supernatant (nuclear extract) was dialyzed against buffer D [20 mM Hepes (pH 8.0), 100 mM KCl, 0.2 mM EDTA, 20% (vol/vol) glycerol, and 0.5 mM DTT] for 2 h at 4 °C. Dialyzed samples were cleared by centrifugation at 13,000 × g for 5 min at 4 °C and precleared by incubation with 10 μL Protein G magnetic beads and appropriate IgG antibody for 1 to 2 h at 4 °C. Following the preclear, samples were divided as necessary onto fresh Protein G magnetic beads, IP buffer [20 mM Hepes (pH 8.0), 100 mM KCl, 0.2 mM EDTA, 20% (vol/vol) glycerol, 0.5 mM DTT, and 0.05% (vol/vol) Nonidet P-40] and the appropriate antibody. Samples were rotated at 4 °C overnight. The magnetic beads were collected and washed three times in 1 mL IP buffer. For Western blot, beads were resuspended in 10 μL 1× SDS loading buffer per gel and incubated at 95 °C for 3 min before SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Quantitative RT-PCR Analysis.
Total RNA was prepared using the RNeasy kit (Qiagen, 74104) and complementary DNA (cDNA) prepared using the Iscript cDNA synthesis kit (Bio-Rad, 1708890). qPCR was performed using iQ SYBR green master mix (Bio-Rad, 1725121) with samples run in triplicate on a Bio-Rad Miniopticon system. Melt curve analysis was performed at the end of each run. Data were collected using the BioRad-CFX Manager software (Version 3). The relative abundance of each transcript was compared with control GAPDH. Primers used to amplify cDNA are listed in SI Appendix, Table S2.
Immunofluorescence.
K562 cells grown in suspension culture were collected via centrifugation at 1,400 × g for 3 min. Nuclei were isolated using the Nuclei EZ Prep nuclei isolation kit (Sigma-Aldrich, NUC101-1KT). Nuclei were fixed in 4% (vol/vol) paraformaldehyde and rotated at room temperature for 15 min, then incubated with 50 mM NH4Cl for 15 min, rotated, then washed three times in PBS. The nuclei were then incubated in blocking buffer [2% (wt/vol) BSA, 0.25% (wt/vol) gelatin, 0.2% (wt/vol) glycine, and 0.2% (vol/vol) Triton X-100 in PBS] for 1 h, rotating at room temperature. Primary antibody was diluted in PBS with 1% (wt/vol) BSA, 0.25% (wt/vol) gelatin, and 0.2% (vol/vol) Triton X-100 and incubated for 1 h, rotating at room temperature. Nuclei were washed three times with washing buffer [0.2% (wt/vol) gelatin in PBS]. Fluorescent secondary antibody was then applied for 45 min in the same buffer as the primary antibody, and samples were rotated in the dark. Secondary antibodies were AlexaFluor anti-mouse (594) antibody (Thermo Fisher, 35510) and AlexaFluor anti-rabbit (488) antibody (a21206). Nuclei were washed three times in washing buffer before 10 min incubation with DAPI solution. Nuclei were resuspended in a minimum volume of 1,4-diazabicyclo[2.2.2]-octane (DABCO) mounting media (Sigma-Aldrich) to mount onto poly-lysine–coated slides. Samples were viewed using a Leica SP5-II AOBS confocal laser scanning microscope attached to a Leica DM I6000 inverted epifluorescence microscope with oil 63× lens. Images were processed using ImageJ or Volocity 6.3 software.
Click Chemistry.
K-562 and MCF7 cells were incubated in lipid-free media (RPMI and DMEM, respectively) containing 10 μg/mL alkyne cholesterol (Avanti, 700143) for 16 h. Samples were washed once in PBS and nuclei obtained using the EZ Prep nuclei kit, then incubated with 50 μM Azide-PEG3-biotin conjugate (Sigma-Aldrich, 762024) was dissolved in prewarmed buffer A and added to samples. The click reaction was initiated via addition of 2 mM CuBF4 in 2% (vol/vol) acetonitrile, and the reaction was left to proceed at 43 °C for 30 min with gentle agitation. The nuclei were then washed in buffer A and immediately used for preparation of nuclear extract for immunoprecipitation (using streptavidin magnetic beads, Thermo Fisher, 88816), chromatin immunoprecipitation (Click-ChIP with streptavidin magnetic beads), or immunofluorescence (using Streptavidin Alexa Fluor 594 conjugate, Thermo Fisher, S32356).
Protein Production, Cholesterol Assay, and Cholesterol-Binding Assay.
For production of BASP1 proteins, BL21 DE3 cells were initially transformed with pRSF-1-NMT (Addgene, 42578) for the expression of NMT, then with the pET23d BASP1 expression vectors. 20 mL of culture in Luria Broth (LB) medium was seeded overnight at 37 °C, then added to 200 mL of media and incubated for 1 h in a shaker at 37 °C. When required, myristic acid was added to a final concentration of 0.2 mg/mL The optical density (at 600 nm [OD600]) was monitored, and when OD600 = 0.6, isopropyl ß-D-1-thiogalactopyranoside (IPTG) (1 mM) was added. After incubation at 37 °C for 3 h, the cells were harvested, and the pellet was frozen at −80 °C. The His-tagged BASP1 derivatives were prepared using Ni-nitriloacetic acid (NTA) beads following the manufacturer’s instructions (Qiagen, 30210). Purified proteins were dialyzed into buffer D and stored at −80 °C.
Cholesterol was solubilized in methanol/chloroform/water (2:1:0.8 vol/vol/v) at a final concentration of 1 mM, diluted as required, and dotted onto polyvinylidene difluoride (PVDF) membrane. The membrane was incubated in binding buffer [20 mM Tris (pH 8), 300 mM NaCl, 0.5% (vol/vol) Tween, and 2.5% (wt/vol) dried milk] for 1 h rocking at room temperature. Fresh binding buffer was added containing the BASP1 derivative at a concentration of 50 ng/μL and incubation continued for 1 h. The membrane was washed three times for 10 min each with binding buffer. The membrane was then incubated with binding buffer containing rabbit anti-BASP1 antibody for 30 min then washed three times for 5 min each. The membrane was then incubated with binding buffer containing anti-rabbit horse radish peroxidase (HRP) for 30 min, then washed three times for 5 min each and subjected to chemiluminescence. Cholesterol content of nuclear extracts was measured using an Amplex Red cholesterol assay kit (Invitrogen, A12216).
Calcium Imaging.
K562 cell line derivatives were grown on coverslips for imaging. Cells were loaded with 2 μM fura2/AM with Pluronic F-127 for 40 min in Tyrode’s solution [140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 3 mM CaCl2, 10 mM Hepes, 10 mM glucose, and 1 mM pyruvic acid (pH 7.4)] and then imaged with an Olympus IX71 microscope. Excitation wavelengths of 340 nm and 380 nm were used with an emission wavelength of 510 nm. Images were collected every 4 s using Imaging Workbench 5.2 (Indec Biosystems). Cells were perfused with Tyrode’s solution and stimulated with 10 mM ATP. Imaging data were collected as a ratio of fluorescence intensities. An evoked response was defined as an increase in fluorescence that was greater than 2 SD values above baseline.
Quantification and Statistical Analysis.
Data are expressed as mean with SD (SDM). Data distribution and significance between different groups was analyzed in Excel or OriginPro 7.5 using Student’s t test. Calcium imaging data were graphed without any filtering using OriginPro 7.5 software, and statistical comparisons were made using a Fisher’s exact test. Autoradiographs were quantitated using Image J (39).
Supplementary Material
Acknowledgments
We are grateful to Adrian Bunzel for help with fluorimetry and to Khamal Ampah for help with phase imaging. This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) to S.G.E.R. (BB/T001925/1), Medical Research Council (MRC) to S.G.E.R. (MR/K001027/1), and NIH to K.F.M. and S.G.E.R. (1R01GM098609). A.E.L. was supported by a Wellcome Trust PhD Studentship for the Dynamic Cell Biology program (083474).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101671118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Garcia-Gil M., Albi E., Nuclear lipids in the nervous system: What they do in health and disease. Neurochem. Res. 42, 321–336 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Fernandes V., Teles K., Ribeiro C., Treptow W., Santos G., Fat nucleosome: Role of lipids on chromatin. Prog. Lipid Res. 70, 29–34 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Fiume R., et al., Nuclear phosphoinositides: Their regulation and roles in nuclear functions. Int. J. Mol. Sci. 20, E2991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbosa A. D., Siniossoglou S., New kid on the block: Lipid droplets in the nucleus. FEBS J. 287, 4838–4843 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Toska E., Roberts S. G., Mechanisms of transcriptional regulation by WT1 (Wilms’ tumour 1). Biochem. J. 461, 15–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosevitsky M. I., Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 245, 245–325 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Goodfellow S. J., et al., WT1 and its transcriptional cofactor BASP1 redirect the differentiation pathway of an established blood cell line. Biochem. J. 435, 113–125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epand R. M., Proteins and cholesterol-rich domains. Biochim. Biophys. Acta 1778, 1576–1582 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Epand R. M., Vuong P., Yip C. M., Maekawa S., Epand R. F., Cholesterol-dependent partitioning of PtdIns(4,5)P2 into membrane domains by the N-terminal fragment of NAP-22 (neuronal axonal myristoylated membrane protein of 22 kDa). Biochem. J. 379, 527–532 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epand R. F., Sayer B. G., Epand R. M., Induction of raft-like domains by a myristoylated NAP-22 peptide and its Tyr mutant. FEBS J. 272, 1792–1803 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Toska E., et al., Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep. 2, 462–469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toska E., Shandilya J., Goodfellow S. J., Medler K. F., Roberts S. G., Prohibitin is required for transcriptional repression by the WT1-BASP1 complex. Oncogene 33, 5100–5108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartl M., Nist A., Khan M. I., Valovka T., Bister K., Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1). Proc. Natl. Acad. Sci. U.S.A. 106, 5604–5609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl M., Puglisi K., Nist A., Raffeiner P., Bister K., The brain acid-soluble protein 1 (BASP1) interferes with the oncogenic capacity of MYC and its binding to calmodulin. Mol. Oncol. 14, 625–644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh L. A., et al., BASP1 interacts with oestrogen receptor α and modifies the tamoxifen response. Cell Death Dis. 8, e2771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belali T., et al., WT1 activates transcription of the splice factor kinase SRPK1 gene in PC3 and K562 cancer cells in the absence of corepressor BASP1. Biochim. Biophys. Acta. Gene Regul. Mech. 1863, 194642 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Essafi A., et al., A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev. Cell 21, 559–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green L. M., Wagner K. J., Campbell H. A., Addison K., Roberts S. G., Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res. 37, 431–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y., Dutta Banik D., Muna M. M., Roberts S. G., Medler K. F., The WT1-BASP1 complex is required to maintain the differentiated state of taste receptor cells. Life Sci. Alliance 2, e201800287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanchard J. W., et al., Replacing reprogramming factors with antibodies selected from combinatorial antibody libraries. Nat. Biotechnol. 35, 960–968 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Fantini J., Epand R. M., Barrantes F. J., Cholesterol-recognition motifs in membrane proteins. Adv. Exp. Med. Biol. 1135, 3–25 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Hofmann K., et al., A novel alkyne cholesterol to trace cellular cholesterol metabolism and localization. J. Lipid Res. 55, 583–591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mullen P. J., Yu R., Longo J., Archer M. C., Penn L. Z., The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer 16, 718–731 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Widenmaier S. B., et al., NRF1 is an ER membrane sensor that is central to cholesterol homeostasis. Cell 171, 1094–1109.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Rae F. K., et al., Anlaysis of complementary expression profiles following WT1 induction versus repression reveals the cholesterol/fatty acid synthetic pathways as a possible major target of WT1. Oncogene 23, 3067–3079 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Tamura I., et al., Wilms tumor 1 regulates lipid accumulation in human endometrial stromal cells during decidualization. J. Biol. Chem. 295, 4673–4683 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Y., et al., Mechanism of crosstalk between the LSD1 demethylase and HDAC1 deacetylase in the CoREST complex. Cell Rep. 30, 2699–2711.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z. A., et al., Diverse nucleosome site-selectivity among histone deacetylase complexes. eLife 9, e57663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva I. T. G., Fernandes V., Souza C., Treptow W., Santos G. M., Biophysical studies of cholesterol effects on chromatin. J. Lipid Res. 58, 934–940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sever R., Glass C. K., Signaling by nuclear receptors. Cold Spring Harb. Perspect. Biol. 5, a016709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chau Y. Y., Hastie N., Wt1, the mesothelium and the origins and heterogeneity of visceral fat progenitors. Adipocyte 4, 217–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welte M. A., Expanding roles for lipid droplets. Curr. Biol. 25, R470–R481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farese R. V. Jr, Walther T. C., Lipid droplets go nuclear. J. Cell Biol. 212, 7–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khor V. K., et al., The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS One 9, e105047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohsaki Y., et al., PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 212, 29–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sztacho M., Sobol M., Balaban C., Escudeiro Lopes S. E., Hozák P., Nuclear phosphoinositides and phase separation: Important players in nuclear compartmentalization. Adv. Biol. Regul. 71, 111–117 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Erdel F., Rippe K., Formation of chromatin subcompartments by phase separation. Biophys. J. 114, 2262–2270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., et al., Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 76, 646–659.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.