Significance
The cap structure present at the 5′ end of eukaryotic messenger RNAs (mRNAs) regulates RNA stability, translation, and marks mRNA as self, thereby impeding recognition by the innate immune system. Cellular transcripts beginning with adenosine are additionally modified at the N6 position of the 2’-O methylated cap-proximal residue by the methyltransferase PCIF1 to m7Gpppm6Am. We define a function for this N6-adenosine methylation in attenuating the interferon-β–mediated suppression of viral infection. Cells lacking PCIF1, or defective in its enzymatic activity, augment the cell-intrinsic suppressive effect of interferon-β treatment on vesicular stomatitis virus (VSV) and rabies virus (RABV) gene expression. VSV and RABV mRNAs are efficiently methylated by PCIF1, suggesting this contributes to viral evasion of innate immune suppression.
Keywords: innate immunity, RNA modifications, host–pathogen interactions, nonsegmented negative-sense RNA virus, rhabdovirus
Abstract
Interferons induce cell-intrinsic responses associated with resistance to viral infection. To overcome the suppressive action of interferons and their effectors, viruses have evolved diverse mechanisms. Using vesicular stomatitis virus (VSV), we report that the host cell N6-adenosine messenger RNA (mRNA) cap methylase, phosphorylated C-terminal domain interacting factor 1 (PCIF1), attenuates the antiviral response. We employed cell-based and in vitro biochemical assays to demonstrate that PCIF1 efficiently modifies VSV mRNA cap structures to m7Gpppm6Am and define the substrate requirements for this modification. Functional assays revealed that the PCIF1-dependent modification of VSV mRNA cap structures is inert with regard to mRNA stability, translation, and viral infectivity but attenuates the antiviral effects of the treatment of cells with interferon-β. Cells lacking PCIF1 or expressing a catalytically inactive PCIF1 exhibit an augmented inhibition of viral replication and gene expression following interferon-β treatment. We further demonstrate that the mRNA cap structures of rabies and measles viruses are also modified by PCIF1 to m7Gpppm6Am. This work identifies a function of PCIF1 and cap-proximal m6Am in attenuation of the host response to VSV infection that likely extends to other viruses.
Eukaryotic messenger RNAs (mRNAs) possess a 5′ cap structure that functions in their stability and translation and helps discriminate host from aberrant RNA by the innate immune system (1–4). That mRNA cap structure is formed by the actions of an RNA triphosphatase that converts pppRNA to ppRNA, which serves as substrate for an RNA guanylyltransferase to transfer GMP derived from GTP onto the 5′ end of the RNA to yield GpppRNA (1, 3, 5). Methylation of that 5′ cap structure by a guanine-N-7 methylase yields m7GpppRNA, which is modified by a ribose-2’-O methylase to yield m7GpppNm (1, 3). Known activators of the innate immune system include triphosphate RNA, which is recognized by the host pattern recognition receptor, retinoic acid inducible gene-1 (RIG-I) (4, 6), and cap structures that lack ribose-2’-O methylation, which renders translation of those RNAs susceptible to inhibition by interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) (4, 7).
Internal RNA modifications also have important functional consequences for the fate of mRNA, among which is N6-methyladenosine (m6A). The methyltransferase complex METTL3/METTL14 is responsible for m6A methylation, which regulates diverse functions in mRNA localization, stability, splicing, and translation (8, 9). The RNA modification N6, 2’-O-dimethyladenosine (m6Am) present at the cap-proximal position (m7Gpppm6Am) is regulated separately from m6A (10). Cap-proximal m6Am is present on ∼30% of cellular mRNA (11–16), but its function is enigmatic. The host RNA polymerase II–associated phosphorylated-CTD interacting factor 1 (PCIF1) catalyzes the formation of cap-proximal m6Am (11–14) that is reported to increase (11, 12, 17), decrease (13), or have no consequence (18) for mRNA stability and translation.
Vesicular stomatitis virus (VSV), a nonsegmented negative-sense (NNS) RNA virus, replicates in the host cell cytoplasm, transcribing five mRNAs from the viral genome (19). The viral large polymerase protein (L) contains the enzymatic activities necessary for transcription of the five mRNAs, including the cotranscriptional addition of a 5′ methylated cap structure (m7GpppAm) and synthesis of the 3′ poly-A tail (20). The polymerase synthesizes the mRNAs by recognizing conserved stop and start sequences within each gene so that each mRNA contains an identical 5′ structure m7GpppAmACAG (21–23). VSV mRNAs isolated from cells are additionally N6-methylated at the cap-proximal Am by a presumed cellular methylase to yield m7Gpppm6AmACAG (24). The efficiency of VSV transcription is such that at least 65% of total cytoplasmic mRNA corresponds to the five VSV mRNAs by 6 h post infection (25). The five VSV mRNAs and their protein products have been extensively characterized biochemically (26) and, as a result, provide unique probes into the function(s) of m6Am.
Here, we demonstrate that VSV mRNAs are efficiently modified at the cap-proximal nucleotide by host PCIF1. In contrast to the substrate requirements for cellular mRNA modification, the PCIF1-dependent N6-methylation of VSV mRNA is independent of prior guanine-N7-methylation of the mRNA cap. Under basal conditions, VSV mRNA stability and translation are unaffected by the presence of m6Am, and viral replication is unaltered. Activation of an antiviral response by the treatment of cells with interferon-β uncovers a function for PCIF1. Cells lacking PCIF1 or expressing a catalytically inactive variant of the protein exhibit an augmented suppression of viral gene expression and infection upon interferon-β treatment. The attenuation of the antiviral activity of interferon-β was dependent upon the catalytic activity of PCIF1, thus defining a functional role of this mRNA cap methylation in evading antiviral suppression of gene expression. This work defines a role of PCIF1-dependent methylation of mRNA cap structures in the attenuation of the antiviral response in VSV-infected cells that likely extends to other viruses.
Results
Analysis of VSV mRNA Cap Structures Isolated From Cells.
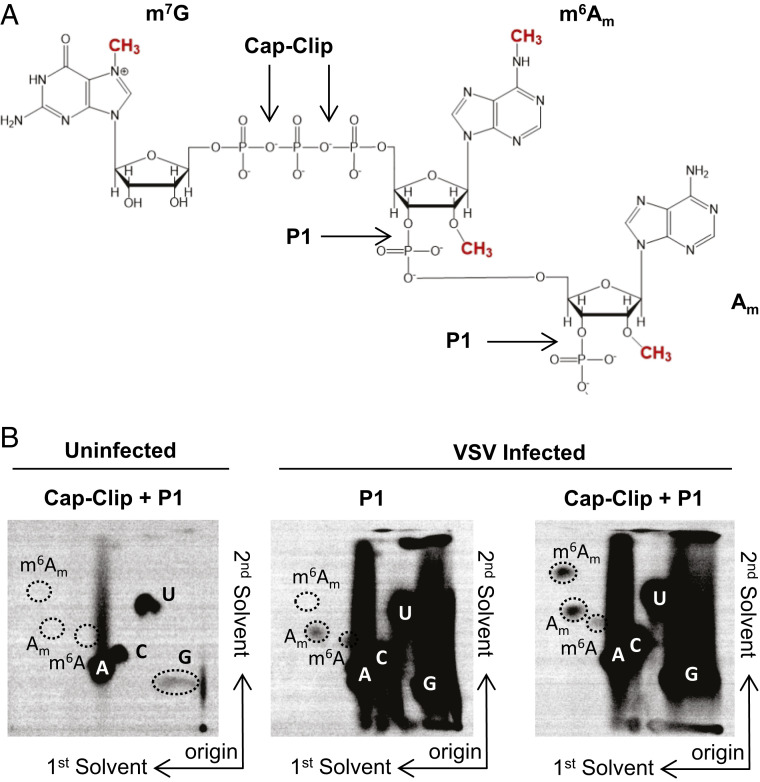
To examine the methylation status of VSV mRNA cap structures, we infected 293T cells at a multiplicity of infection (MOI) of 3, labeled viral RNA by metabolic incorporation of [32P]-phosphoric acid in the presence of actinomycin D from 3 to 7 hours postinfection (hpi), which selectively inhibits host cell transcription, and isolated total poly(A)+ RNA. Following hydrolysis with nuclease P1 to liberate mononucleotides and Cap-Clip acid pyrophosphatase to digest the mRNA cap structure (Fig. 1A), the products were resolved by two-dimensional thin-layer chromatography (2D-TLC) (27) (Fig. 1B). The identity of specific spots was determined by their comigration with co-spotted chemical markers (SI Appendix, Fig. S1A) (17, 27). Analysis of RNA from uninfected cells yields low levels of products that comigrate with pA, pC, pG, and pU, reflecting residual actinomycin D–resistant synthesis of RNA in the cell but no detectable methylated nucleotides (Fig. 1B). Nuclease P1 digestion of RNA from infected cells gave rise to abundant pA, pC, pG, and pU and two additional spots that comigrate with markers for Am and m6A (Fig. 1B). As nuclease P1 leaves the mRNA cap structure intact, the presence of Am and m6A must reflect internal modifications of the viral mRNA, although our analysis cannot discriminate which positions are modified. Prior studies on VSV mRNAs report the presence of Am at the second transcribed nucleotide (24), which may account for some of this internal methylation. Further hydrolysis of the RNA purified from infected cells with Cap-Clip pyrophosphatase leads to the appearance of an additional spot that comigrates with m6Am and an increase in intensity of the Am spot (Fig. 1B). This result demonstrates that VSV mRNA cap structures contain m7Gpppm6Am, as m6Am only appears upon cap-structure hydrolysis. We interpret the increase in Am following Cap-Clip treatment as reflective of the presence of a minority of transcripts containing only Am at the cap-proximal nucleotide (24). Quantitative analysis of each spot reveals over 85% of the cap-proximal nucleotides are m6Am, consistent with previous reports (24). Experiments performed in HeLa cells gave similar results, though no internal m6A was detectable in these cells (SI Appendix, Fig. S1B). We conclude that VSV mRNAs synthesized in 293T and HeLa cells contain primarily m7Gpppm6Am and m7Gpppm6AmAm cap structures.
Fig. 1.
VSV mRNAs contain a 5′ m7Gpppm6Am cap structure. (A) VSV mRNA cap structures present in infected cells. VSV mRNAs contain the conserved 5′ gene start sequence AACAG, including the m7GpppAm cap structures synthesized by the VSV polymerase, N6-methylation at the cap-proximal first nucleotide, and 2’O methylation at the second nucleotide made by the cell. Sites of nuclease P1 and Cap-Clip pyrophosphatase cleavage are marked. (B) VSV contains m6Am at the cap-proximal nucleotide. 293T cells were infected with VSV at a MOI of 3, cellular transcription was halted by adding 10 µg ⋅ ml−1 actinomycin D at 2.5 hpi, and viral RNA was labeled by metabolic incorporation of 100 µCi ⋅ ml−1 [32P] phosphoric acid from 3 to 7 hpi. Total cellular RNA was extracted and, following poly(A) selection, hydrolyzed by the indicated nucleases into monophosphates that were resolved by 2D-TLC and detected by phosphorimaging (representative image; n = 3). Solvents run in the first and second dimensions are marked.
PCIF1 Modifies VSV mRNA.
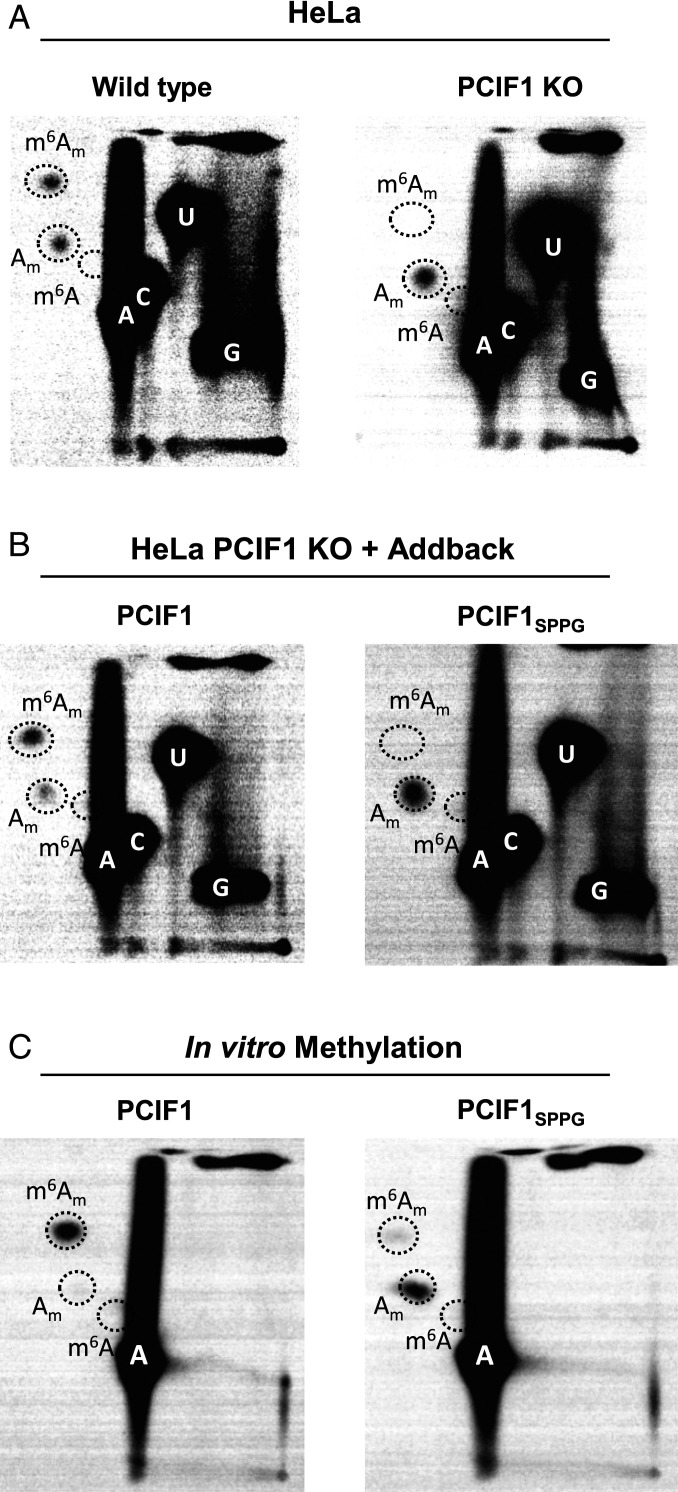
To determine whether VSV mRNA are modified by PCIF1, the cellular N6-methyltransferase for mRNA (SI Appendix, Fig. S2) (11–14), we infected PCIF1 knockout (KO) cells and performed RNA analysis as above. Viral mRNA isolated from PCIF1 KO cells (HeLa or 293T) lacked detectable levels of m6Am (Fig. 2A and SI Appendix, Fig. S1C). Cap-proximal m6Am was restored upon addback of PCIF1 but not a catalytically inactive mutant PCIF1SPPG (Fig. 2B). Methylation reactions performed in vitro demonstrate that purified PCIF1 but not PCIF1SPPG are responsible for m6Am on VSV mRNA (Fig. 2C and SI Appendix, Fig. S1D). Collectively, these results demonstrate that PCIF1 is necessary and sufficient for formation of cap-proximal m6Am on VSV mRNA.
Fig. 2.
PCIF1 is the cap-proximal N6-methyltransferase. (A) CRISPR-mediated PCIF1 KO HeLa cells, or WT parental cells, were infected with VSV at a MOI of 3. Viral RNA was radiolabeled and analyzed as in Fig. 1 (representative images; n = 3). (B) Addback of PCIF1, but not a catalytically inactive mutant PCIF1SPPG restores m6Am on VSV mRNA. HeLa PCIF1 KO cells stably expressing 3X-FLAG-PCIF1, or 3X-FLAG-PCIF1SPPG were infected with VSV, and RNA was radiolabeled and digested, and 2D-TLC was performed as in A (representative images; n = 3). (C) PCIF1 N6-methylates VSV mRNA in vitro. Purified VSV mRNA, transcribed in vitro from viral particles in the presence of [32P]-α-ATP, was used as a template for in vitro methylation with 50 nM purified PCIF1, or PCIF1SPPG. Following hydrolysis, the products were visualized by 2D-TLC and phosphorimaging as in Fig. 1 (representative images; n = 3).
N7-Guanosine Methylation Is Dispensable for PCIF1 Modification of VSV mRNA.
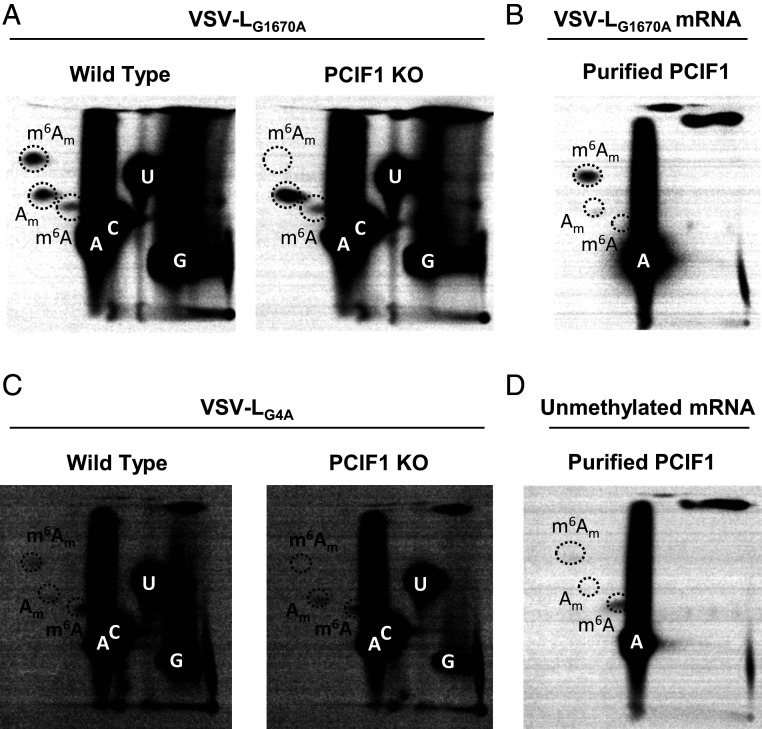
Substrates for modification by PCIF1 require the presence of a methylated m7G mRNA cap structure (11, 12), and capped RNA lacking m7G serve as poor substrates for PCIF1 in vitro (14). The obligatory sequential methylation of cellular mRNAs at m7G and subsequent ribose-2’-O positions (1) precludes tests of the importance of the 2’-O methylation alone in PCIF1 modification of mRNA. By contrast, the mRNA cap methylation reactions of VSV, and by inference, other NNS RNA viruses, proceed in the opposite order. Experiments conducted with viral mutants and by reconstitution of cap methylation in vitro demonstrate that cap ribose-2’-O methylation precedes and facilitates the subsequent guanine-N-7 methylation (28), allowing us to explore the impact of 2’-O methylation alone on PCIF1 modification of VSV mRNAs. mRNA synthesized in cells by the viral mutant VSV-LG1670A contains primarily GpppAm mRNA caps (29), which serve as effective substrates for PCIF1 (Fig. 3A). In agreement with an analysis of viral mRNA made in cells, mRNA transcribed from purified VSV-LG1670A virions are fully methylated by PCIF1 in vitro (Fig. 3B and SI Appendix, Fig. S3). mRNA synthesized by a second viral mutant, VSV-LG4A, that produces unmethylated GpppA cap structures (29) was not modified by PCIF1 (Fig. 3C). A low level of m6A observed following hydrolysis of RNA produced by VSV-LG4A is consistent with modification at internal positions of the mRNA (Fig. 1B). In agreement with this finding, viral mRNA synthesized by VSV in the presence of the methylation inhibitor S-adenosyl-homocysteine is poorly methylated by PCIF1 (Fig. 3D and SI Appendix, Fig. S3). Taken together, this set of results demonstrate in contrast to cellular mRNA modification that VSV mRNA requires ribose-2’-O methylation and not guanine-N-7 methylation.
Fig. 3.
Effect of mRNA cap methylation on PCIF1 modification of VSV mRNA. (A) The indicated 293T cells were infected with VSV-LG1670A at a MOI of 3, and viral RNA was radiolabeled, extracted, and analyzed by 2D-TLC as in Fig. 2A (representative images, n = 3). (B) mRNA synthesized in vitro by VSV-LG1670A mRNA was incubated with purified PCIF1 and analyzed as in Fig. 2C (representative image, n = 3). (C) The indicated 293T cells were infected with VSV-LG4A as in A (representative images, n = 3). (D) mRNA synthesized by VSV in vitro in the presence of 200 µM S-adenosyl-homocysteine was used as substrate for PCIF1 in vitro and analyzed as in B (representative image, n = 3).
Effect of m6Am on Viral Growth, mRNA Stability, and Translation.
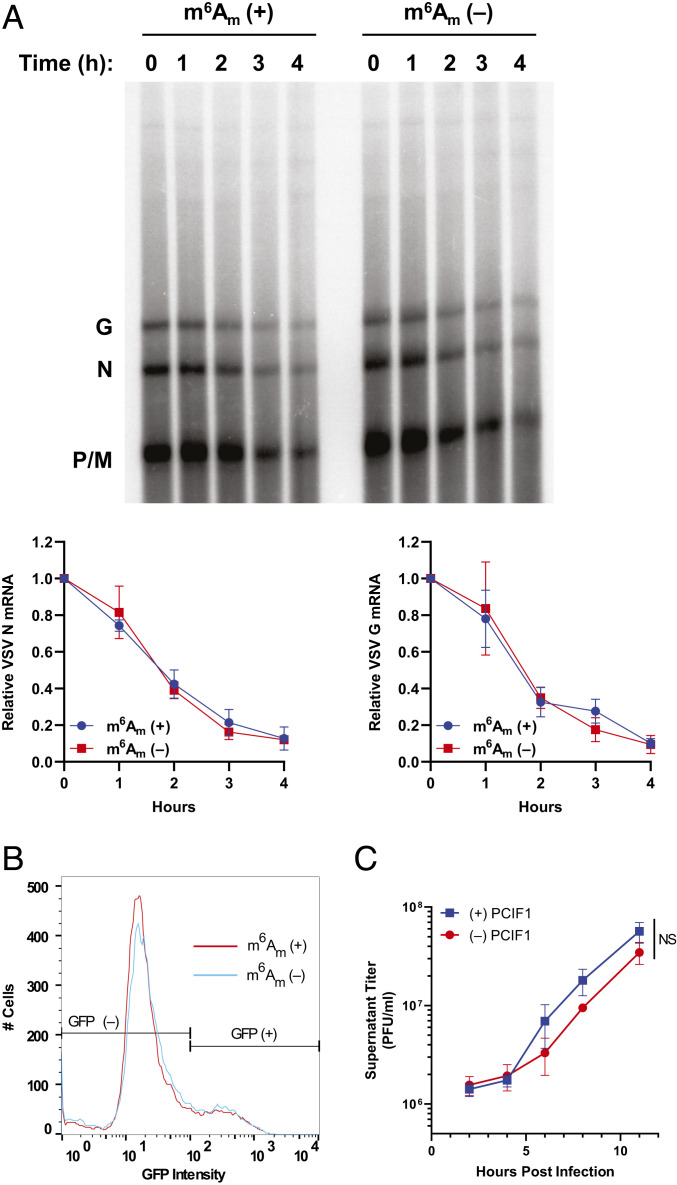
To determine whether PCIF1 modification influences VSV mRNA stability, we compared the decay of modified and unmodified transcripts. Briefly, mRNA was specifically isolated from infected PCIF1 KO cells, in vitro methylated with purified PCIF1 where indicated, transfected into uninfected cells, and isolated at the indicated time points. Analysis of the isolated mRNA by electrophoresis on acid agarose gels demonstrates that viral mRNA stability is unaffected by the presence of m6Am (Fig. 4A, two-way ANOVA, P > 0.4). The stability of each individual mRNA appeared unaffected by PCIF1-dependent modification to m6Am when transfected into either wild-type or PCIF1 KO cells (SI Appendix, Fig. S4).
Fig. 4.
Effect on m6Am on viral mRNA translation and stability. (A and B) VSV mRNA isolated from PCIF1 KO 293T cells and methylated in vitro with PCIF1 prior to transfection of 500 ng RNA into PCIF1 KO HeLa cells and assessed for: (A) mRNA stability by extraction from cells at the indicated time post-transfection and analyzed by electrophoresis on acid agarose gels. A representative image is shown along with quantitative analysis of the abundance of the N and G mRNAs (n = 3, ± SD, two-way ANOVA, P > 0.4). (B) mRNA translation by measurement of GFP-positive cells and their intensity by flow cytometry (n = 3, 0.95 > P > 0.18, Student’s t test). (C) Viral replication assessed in PCIF1 KO HeLa cells expressing 3X-FLAG-PCIF1 or an empty vector following infection with VSV at a MOI of 3. Viral titers were determined by plaque assay on Vero cells at the indicated time post-inoculation. (n = 3, ± SD, NS: P > 0.08, Student’s t test, statistics shown are for the 11 h timepoint).
To measure whether mRNA translation was affected by the presence of m6Am, we measured the expression of a viral encoded eGFP reporter gene following transfection of methylated or unmethylated mRNA into PCIF1 KO cells. The measurement of eGFP by flow cytometry at 7 h post-transfection reveals that neither the fraction of positive cells nor the fluorescence intensity was significantly altered by the presence of m6Am (Fig. 4B and SI Appendix, Fig. S5A). Similar findings on stability and translation were obtained using a luciferase reporter (SI Appendix, Fig. S5 B and C). To further verify that m6Am does not impact the translation of VSV mRNA, we cotransfected mRNA from two VSV reporter viruses encoding firefly (Luc) or renilla (Ren) luciferase with opposing methylations. In this competitive translation experiment, the ratio of firefly and renilla translated in transfected cells was unaffected by the presence of m6Am, irrespective of which reporter virus mRNA was modified (SI Appendix, Fig. S5 D and E). Collectively, these results demonstrate that translation of VSV mRNA is unaffected by the presence of m6Am.
As neither viral mRNA translation nor stability were altered by the presence of m6Am, we next compared the kinetics of viral growth in PCIF1 knockout or addback cells. Cells were infected at a MOI of 3 and viral titers determined by plaque assay at various times postinfection. The kinetics of viral growth were also unaffected in cells lacking PCIF1 (Fig. 4C and SI Appendix, Fig. S6). Collectively, these data demonstrate that despite extensive modification of the viral mRNAs by PCIF1, viral replication and gene expression are unaltered by this modification.
Interferon-β Treatment Uncovers a Role for PCIF1 in the Host Response to Infection.
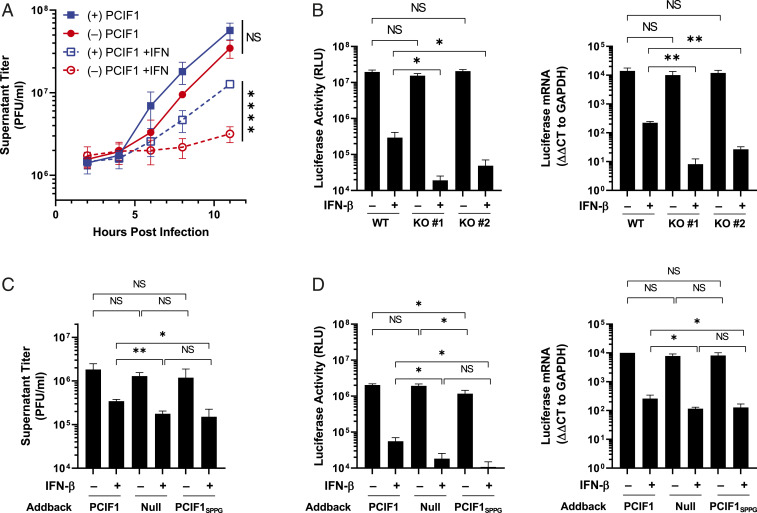
As cap methylation at the 2’-O position of the first nucleotide helps distinguish self from viral RNA during infection (4), we examined whether PCIF1 modification of mRNA plays a similar role. To examine whether cap-proximal m6Am helps counter host cell antiviral responses, we measured how PCIF1 affects the interferon-β (IFN-β)–mediated inhibition of virus growth. The treatment of cells with IFN-β prior to infection uncovered a PCIF1-dependent attenuation of the antiviral effect (Fig. 5A and SI Appendix, Fig. S6). The infection of cells by a VSV reporter virus that expresses firefly luciferase confirmed that PCIF1 attenuates the suppressive effect of IFN-β on viral gene expression at the RNA and protein levels in a single round of infection (SI Appendix, Fig. S7A). We validated this finding in two independent PCIF1-null HeLa cell lines as well as reconstitution with functional PCIF1 (SI Appendix, Fig. S7 A and B). This result suggests that the effects of PCIF1 are restricted to steps of the viral replication cycle up to and including gene expression. To eliminate viral entry as a possible contributor, we transfected ribonucleoprotein cores purified from VSV-Luc into cells, thereby bypassing viral entry. The pretreatment of cells with IFN-β was still accompanied by augmented inhibition of gene expression in cells lacking PCIF1 (SI Appendix, Fig. S7C). This result demonstrates that the IFN-β–mediated suppression of VSV gene expression is enhanced in cells lacking PCIF1.
Fig. 5.
Effect of IFN-β pretreatment of cells on viral infection. (A) PCIF1 KO HeLa cells reconstituted with PCIF1 or an empty vector were pretreated with vehicle (0.1% BSA) or 500 U ⋅ ml−1 of IFN-β for 5 h and infected with VSV at a MOI of 3. Viral titer was determined at the indicated time points by plaque assay on Vero cells (n = 3, ± SD, NS: P > 0.08, ****P < 0.0001, Student’s t test). (B) WT (parental) or PCIF1 KO A549 cells were pretreated with vehicle (0.1% BSA) or 500 U ⋅ ml−1 IFN-β for 5 h prior to infection with VSV expressing a luciferase reporter at a MOI of 5. Cells were lysed 6 hpi and luciferase activity determined (n = 4, NS: 0.50 > P > 0.05, *P < 0.05, Student’s t test) and mRNA levels verified by qRT-PCR (n = 3, NS: 0.47 > P > 0.23, *P < 0.05, **P < 0.01, Student’s t test). (C) PCIF1 KO HeLa cells were reconstituted with PCIF1, PCIF1SPPG, or empty vector and infected with VSV at a MOI of 3, and viral titers were measured at 11 hpi by plaque assay (n = 3, ± SD, NS: 0.83 > P > 0.05, *P < 0.05, **P < 0.01, Student’s t test). (D) As in C, except cells were infected with VSV-Luc and luciferase protein and RNA measured as in B (n = 3, ± SD, NS: 0.83 > P > 0.29, *P < 0.05, Student’s t test).
The antiviral response in HeLa cells is partly attenuated (30), and therefore, we examined whether loss of PCIF1 results in a similar IFN-β–dependent inhibition of viral replication in A549 cells. We confirmed that VSV mRNAs were also N6-methylated by PCIF1 in these cells (SI Appendix, Fig. S8). The pretreatment of A549 cells with IFN-β revealed that, in the absence of PCIF1, viral gene expression was suppressed an additional 10-fold as evident by levels of viral mRNA and protein (Fig. 5B). Measurements of specific viral proteins following metabolic incorporation of [35S]-met and [35S]-cys followed by analysis of proteins on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) demonstrates that the three most abundant viral proteins, N, M, and G, are further suppressed in cells lacking PCIF1, but cellular translation in uninfected cells is unaffected (SI Appendix, Fig. S9).
To rule out the possibility of N6-methylation–independent activities of PCIF1 mediating this effect, we examined infection in cells expressing a catalytically inactive mutant of PCIF1, PCIF1SPPG (Fig. 2B). Both PCIF1SPPG addback and PCIF1 knockout equivalently augmented the effect of IFN-β on viral growth (Fig. 5C) and VSV-luciferase gene expression (Fig. 5D). Collectively, the above experiments reveal that loss of PCIF1 or its ability to synthesize m6Am augments the suppressive effect of IFN-β on VSV gene expression, suggesting that m6Am methylation of viral mRNAs protects against the otherwise antiviral effects of the IFN-mediated innate immune response.
Effect of PCIF1 on Innate Immune Sensing and IFIT Proteins.
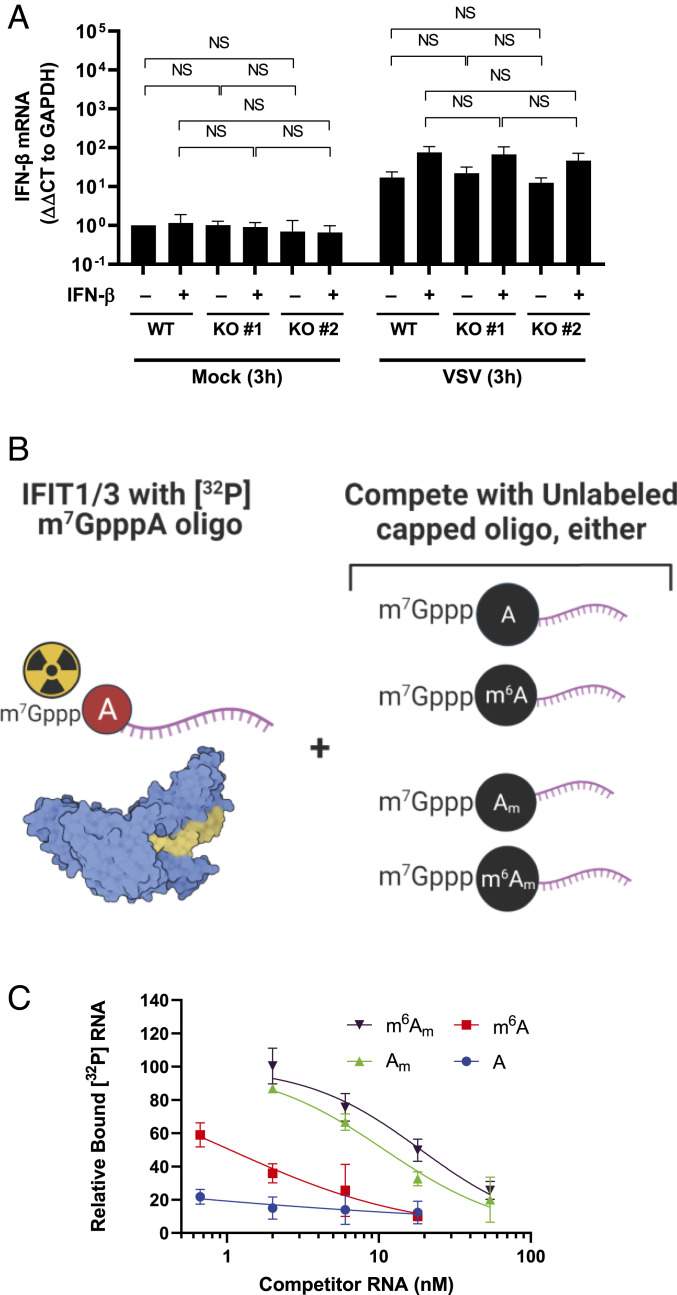
We next examined whether m6Am functionally impacts known discriminators of the cap-proximal nucleotide in innate immunity, specifically RIG-I and IFIT1/3 (4). To determine whether sensing of VSV mRNA, for which RIG-I is essential (31), was affected by PCIF1 modification of viral mRNA, we measured induction of IFN-β mRNA during infection. IFN-β mRNA levels were indistinguishable in cells independent of the presence of PCIF1 at 3 (Fig. 6A) and 6 h post infection (SI Appendix, Fig. S10). This result demonstrates that loss of PCIF1 does not alter the RIG-I–dependent sensing of VSV RNA in cells.
Fig. 6.
PCIF1 does not regulate VSV mRNA sensing or IFIT1/3 binding. (A) The indicated A549 cells were pretreated with vehicle (0.1% BSA) or 500 U ⋅ ml−1 IFN-β for 5 h prior to infection with VSV at a MOI of 5 for 3 h. Cells were lysed and IFN-β mRNA levels determined by qRT-PCR (n = 4, ±SD, NS: 0.08 < P < 0.80, Student’s t test). (B) Schematic for testing IFIT1/3 affinity for differentially methylated cap structures [structure shown from Protein Data Bank 6C6K (32)]. IFIT1-IFIT3 complexes (1 µM) were bound to 1 nM radiolabeled RNA with a m7GpppA cap in the presence of unlabeled competitor RNAs at the indicated concentration with the indicated cap-proximal nucleotide. A high affinity of the competitor for IFIT1/3 will result in displacement of the radiolabeled substrate and loss of signal. A low affinity will allow the radiolabeled substrate to remain bound. (C) Radiolabeled RNA bound to IFIT1/3 was measured by filter-binding assay and phosphorimaging and normalized to a control without competitor RNA (n = 3, ± SD, least squares regression line shown).
We next examined whether IFIT1, which represses translation of mRNAs lacking cap-proximal 2’O methylation (7), also discriminates against N6-methylation. To do this, we established a competitive binding assay for capped RNA substrates with a complex of purified IFIT1 and IFIT3 (32). Briefly, we used as binding substrate a radiolabeled and capped 27-nt (nucleotide) synthetic RNA corresponding to the first 27 nucleotides of the VSV genome in competition with unlabeled RNAs containing a cap-proximal A, m6A, Am, or m6Am (Fig. 6 B and C). As expected, the binding of the radioactive 27-nt capped RNA was efficiently out competed by the identical cold competitor (half maximal inhibitory concentration [IC50]: < 0.66 nM; Fig. 6C), confirming that IFIT1/3 efficiently binds m7GpppA capped transcripts (7, 32). The modification of the competitor RNA to contain a m7GpppAm cap structure decreased its effectiveness >16-fold, consistent with published results (IC50: 10.89 nM; Fig. 6C) (7, 32), and further N6-methylation to m7Gpppm6Am resulted in a slight further reduction (IC50: 18.88 nM; Fig. 6C) that failed to rise to the level of statistical significance. At high concentrations of PCIF1, it is possible to N6-methylate cap structures in vitro that are not 2’-O methylated. Such an m7Gpppm6A cap structure competes for IFIT1/3 binding slightly better than m7GpppA (IC50: 1.05 nM; Fig. 6C). As such RNAs require 2’O methylation to be substrates for PCIF1 in VSV and cellular mRNA, we conclude that the PCIF1-dependent modification of viral RNA does not significantly alter its recognition by IFIT1/3.
Effect of PCIF1 on mRNA From Other Negative-Sense RNA Viruses.
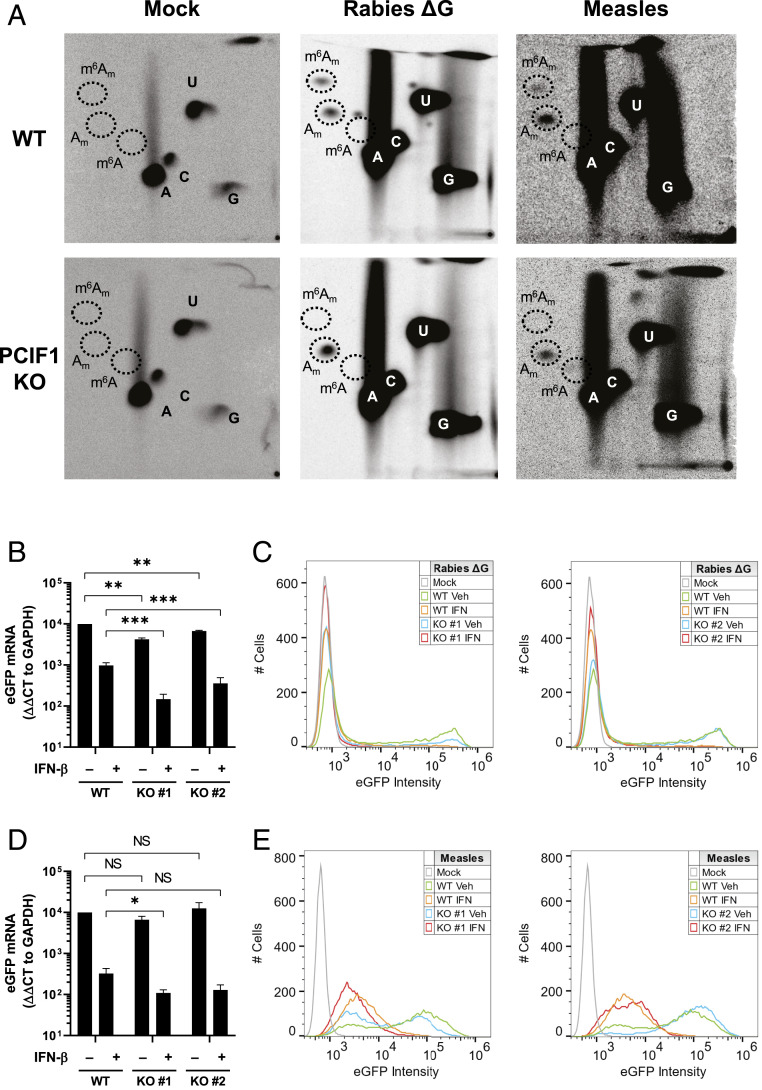
We next examined whether PCIF1 modifies the mRNA of other negative-sense RNA viruses that replicate in the cytoplasm. For this purpose, we selected rabies virus from the same family as VSV, and whose mRNAs start with the same sequence, and measles virus from the family paramyxoviridae. Both viruses synthesize m7GpppAm cap structures (33–35) that were further modified in a PCIF1-dependent manner to m7Gpppm6Am (Fig. 7A). We note, however, that the PCIF1-dependent methylation of measles virus RNA appears inefficient (Fig. 7A). We also observed a PCIF1-dependent attenuation of the effect of interferon-β pretreatment of cells on rabies virus gene expression at both the RNA and protein level (Fig. 7 B and C and SI Appendix, Fig. S11A). Reflecting the inefficient modification of measles virus mRNA, we observed an approximately ∼2.5-fold PCIF1-dependent attenuation of the suppressive effect of interferon-β on measles virus mRNA levels (clone #1, P = 0.048; clone #2, P = 0.072) (Fig. 7 D and E and SI Appendix, Fig. S11B).Collectively, this data demonstrates that PCIF1 N6-methylates other negative-sense RNA virus mRNAs that can serve to attenuate the interferon-β–dependent antiviral response.
Fig. 7.
Effect of PCIF1 on other NNS RNA viruses. (A) The indicated A549 cells were infected with the indicated viruses at a MOI of 3, cellular transcription was halted by adding 10 µg ⋅ ml−1 actinomycin D at 5.5 hpi, and viral RNA was labeled by metabolic incorporation of 100 µCi ⋅ ml−1 [32P] phosphoric acid from 6 to 24 hpi. Viral RNA was extracted and analyzed by 2D-TLC as in Fig. 1B. (B) The indicated A549 cells were pretreated with vehicle (0.1% BSA) or 500 U ⋅ ml−1 IFN-β concurrent with infection with rabies virus ΔG at a MOI of 1 for 24 h. Cells were lysed and eGFP mRNA levels determined by qRT-PCR (n = 3, ± SD, **P < 0.01, ***P < 0.001, Student’s t test). (C) Same as B, except cells were fixed in formaldehyde and flow cytometry performed for eGFP expression (n = 3, representative histograms shown). (D) The indicated A549 cells were pretreated with vehicle (0.1% BSA) or 500 U ⋅ ml−1 IFN-β for 5 h prior to infection with measles virus at a MOI of 3 for 24 h. Cells were lysed and eGFP mRNA levels determined by qRT-PCR (n = 3, ± SD, NS: 0.06 < P < 0.47, *P < 0.05, Student’s t test). (E) Same as D, except cells were fixed in formaldehyde and flow cytometry performed for eGFP expression (n = 3, representative histograms shown).
Discussion
The major finding of this study is the identification of a role for PCIF1-mediated m6Am methylation in the type I interferon response to VSV infection. We demonstrate that loss of PCIF1 enhances the sensitivity of viral replication to the pretreatment of cells with IFN-β by affecting VSV gene expression. PCIF1 is necessary and sufficient for modification of VSV mRNA to yield cap-proximal m6Am, and in contrast to cellular mRNA modification, viral mRNAs require prior ribose-2’-O but not guanine-N7 methylation of the cap structure. The most parsimonious explanation of our results is that the PCIF1-dependent modification of viral mRNA cap structures to m6Am serves to dampen an IFN-β–mediated suppression of gene expression. Mechanistically, how this occurs was not resolved by the present study, but we posit that this requires discrimination of modified from unmodified RNA by an interferon-stimulated gene (ISG).
The precedent for a role of mRNA cap modifications in the antiviral response already exists. The RIG-I–dependent recognition of a 5′ triphosphate is suppressed by the presence of an mRNA cap structure (4, 6), and ribose 2’-O methylation of the cap structure inhibits the ability of an ISG, IFIT1, to suppress translation of mRNA (4, 7). The PCIF1-dependent modification of VSV mRNA cap structures may work by a similar mechanism by helping viral mRNA appear more host-like, but this modification has no significant effect on the RIG-I– or IFIT1–dependent recognition of viral mRNA (Fig. 6). This finding is entirely consistent with the known requirements of a 5′ triphosphate for RIG-I–dependent recognition. Although we observed that generation of an artificial m7Gpppm6A cap structure in vitro reduced binding to IFIT1, it seems unlikely that this ISG plays a major role in this discrimination. This conclusion is based on the known recognition of m7GpppA cap structures by IFIT1 (4, 7, 32) and the requirement for 2’-O modification of mRNAs for their subsequent PCIF1 modification in cells (11–13). Additional work will be required to define whether an ISG is required to discriminate between m6Am modified and unmodified cap structures.
Although we establish a role of PCIF1 and m6Am in the IFN-β–mediated suppression of VSV gene expression and demonstrate that viral mRNAs are modified by PCIF1, the modification of cellular mRNA may also play a role. Cap-proximal m6Am inhibits the host mRNA decapping enzyme DCP2 (17), which may alter the stability of cellular mRNAs including those induced on the treatment of cells with IFN-β (36). This seems unlikely to account for the effects we observe on VSV infection, as the DCP2-dependent decapping and degradation of cellular mRNA would be expected to increase in cells lacking PCIF1, likely dampening rather than augmenting the effect of IFN-β treatment. If the antiviral response is due to m6Am modification of cellular mRNA, this contrasts with the consequences of 2’-O methylation of the mRNA cap structure of ISG mRNAs, which enhances their expression (37). We are also mindful of the possibility that PCIF1 may have unknown functions in the cell beyond N6-methylation of mRNA. Insects, including Drosophila, express an ortholog of PCIF1 that associates with the phosphorylated CTD of PolII but is catalytically inactive as an RNA N6-methyltransferase (38). As the catalytic activity of PCIF1 is required for the attenuation of the antiviral response, we also find this explanation unlikely.
The substrate requirements for PCIF1 modification of VSV mRNA differ to those previously shown in cellular mRNA (11, 14). Specifically, we found that guanine-N7-methylation was dispensable for N6-methylation and that ribose 2’-O methylation was required in its absence. Although we do not understand why this is the case, we recapitulate the substrate specificity in vitro, making it unlikely that this distinction reflects the cytoplasmic modification of viral mRNA rather than the nuclear modification of cellular mRNA. This altered specificity for the modification of the VSV mRNA coupled with the altered recognition specificity of the VSV cap methylation machinery—which requires 2’-O methylation prior to guanine-N7 methylation—raises the possibility that the structure of the 5′ end of VSV mRNAs leads to the altered specificity (29).
The finding that PCIF1 and m6Am affect the antiviral response raises the question of why modify cap-proximal A and not other cap-proximal bases. More cellular mRNAs initiate with G than A (16), but guanosine is typically only methylated at the 2’O position in mRNA (39). This is likely because O6-methylation of guanosine (m6G) has been shown to have a large fitness cost. Its presence in DNA is highly mutagenic, though pairing with thymidine during DNA replication, and when present in RNA, it causes incorrect ribosome decoding (40) and a 1,000-fold decrease in the peptide bond formation rate (41). N6-methylation in adenosine, by contrast, has a minimal effect on these processes (40, 41).
The importance of m6Am for most viruses has not been examined. As expected, the mRNAs of DNA viruses which rely on host RNA polymerases for transcription, including adenoviruses (42, 43), simian virus 40 (44), herpes simplex virus 1 (45), and polyomaviruses (46), contain m6Am (1, 3). Vaccinia virus, which replicates in the cytoplasm, also produces m6Am-containing mRNA (1, 3, 47), likely through modification by PCIF1. Whether such modifications also attenuate the antiviral response remains to be determined. We observed a PCIF1–dependent attenuation of the suppressive effect of interferon-β on rabies virus and, to a lesser extent, measles virus. What impacts the magnitude of PCIF1-dependent attenuation of the suppressive effect of interferon-β between different viruses was not determined. We suggest that this may be related to the different extent of PCIF1 modification of viral RNA, the different kinetics with which VSV, rabies, and measles replicate, and differences between the viruses in how they regulate antiviral signaling. The evolution by many viruses of their own capping machinery also begs the question of whether viruses have evolved a PCIF1-like cap modifying enzyme, particularly given the potential advantage in the face of an antiviral response. Additional studies with VSV and other viruses will be required to fully define the role of PCIF1 in the host response to infection.
Materials and Methods
Cells.
Human embryonic kidney 293T (HEK293T), HeLa, A549, Vero CCL81, and BsrT7/5 cells were maintained in humidified incubators at 37 °C and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with L-glutamine, sodium pyruvate, glucose, (Corning no. 10013CV), and 10% fetal bovine serum (FBS; Tissue Culture Biologicals no. 101). The generation of HEK293T and HeLa PCIF1 KO cell lines and addbacks were previously described (12), and A549 PCIF1 KO cell lines were generated and verified using these same methods. Cells were tested regularly using the e-Myco PLUS PCR Kit (Bulldog Bio no. 2523348).
Viruses.
VSV (as rescued from an infectious complementary DNA clone of VSV, pVSV (1)+), VSV-LG1670A, VSV-LG4A, VSV-luciferase, VSV-RenillaP, VSV-IRES, and VSV-eGFP have been described previously (29, 48–52). VSV viruses were propagated in BsrT7/5 cells. Rabies ΔG expressing eGFP was kind gift of E.M. Callaway, The Salk Institute for Biological Studies, La Jolla, CA (53) and propagated in BsrT7/5 cells expressing SAD B19 rabies virus G. Measles expressing eGFP was propagated in Vero cells.
Radiolabeling of mRNA.
Cap-proximal nucleotides were radiolabeled as previously described (17). For specific radiolabeling of VSV mRNA, cells were infected with VSV at a MOI of 3 in serum/phosphate-free DMEM (Gibco no. 11971–025). A total of 10 µg ⋅ ml−1 Actinomycin D (Sigma no. A5156) was added to halt cellular transcription at 2.5 hpi, and at 3 h postinfection, 100 µCi ⋅ ml−1 [32P] phosphoric acid was added to label newly synthesized RNA (Perkin-Elmer no. NEX053H). RNA was harvested in TRIzol (Thermo Fisher no. 15596018) and poly(A)+ mRNA selected using the NEB Magnetic mRNA Isolation Kit (NEB S1550S). The radiolabeling of measles and rabies mRNA was performed similarly, except ActD was added at 5.5 h post infection, and radiolabeling continued until 24 h postinfection.
Identification of Methylated Nucleotide Levels by 2D-TLC.
A total of 2 µg mRNA suspended in 6 µl RNase-free H2O was digested with 2 units of nuclease P1 (Sigma N8630) for 3 h at 37 °C. The volume was then increased to 20 µl and RNA further digested with 2 units of Cap-Clip acid pyrophosphatase for 3 h (Cell Script no. C-CC15011H) in the manufacturer’s buffer. 2D-TLC was performed as previously described (27). Plates were developed in the first dimension with five parts isobutyric acid (Sigma no. I1754) to three parts 0.5 M ammonia (VWR no. BDH153312K) for 14 h and in the second dimension with a solvent of 70 parts isopropanol, 15 parts hydrochloric acid, and 15 parts water for 20 h. RNA species were positively identified by ultraviolet shadowing (254 nm) of co-spotted (nonradioactive) commercially available standards (5′ monophosphate forms). The standards Am and m6Am 5′ monophosphate were generated by digesting their triphosphate forms (TriLink N-1015 and N-1112) with 1 unit Apyrase (NEB M0398).
In Vitro Transcription of VSV mRNA and Methylation with PCIF1.
VSV or VSV-LG1670A mRNA was synthesized in vitro as previously described with 30 µCi [32P]-α-ATP per reaction (29, 54). RNA was extracted in TRIzol, poly(A) selected, and extracted in TRIzol again to concentrate the samples. Purified recombinant GST-PCIF1 was generated and used to in vitro methylate 225 ng of this mRNA as previously described (12).
Purification, Selection, and In Vitro Methylation of VSV mRNA From Cells.
HEK293T PCIF1 KO cells were infected at a MOI of 10 for 7 h. RNA was extracted in TRIzol and VSV mRNAs selected using a biotinylated oligo against the conserved stop and poly(A) sequence present at the 3′ end of VSV mRNAs (“Biotin-VSVstop”). A total of 1.5 nmol oligo was annealed to this mRNA by incubating at 65 °C for 5 min followed by cooling on ice for 5 min, and complexes were isolated by pulldown with NEB Streptavidin Magnetic Beads (NEB no. S1420, manufacturer’s protocol). Following cleanup by TRIzol extraction, mRNA was purified further using the NEB poly(A) magnetic kit as above, and TRIzol was extracted again. This stock of RNA from PCIF1 KO cells was then in vitro methylated (or mock methylated) with purified PCIF1 as above.
Determination of VSV mRNA Stability.
Biotin-VSVstop–selected mRNAs were transfected into HeLa wild-type (WT) or PCIF1 KO cells. RNA was transfected into separate wells of a 24-well plate (2 × 105 cells; 500 ng RNA per well) using Lipofectamine 2000 (Thermo Fisher no. 11668019), media was changed at 3 h post-transfection, and wells were harvested in TRIzol at 1 h intervals for 4 h. Extracted mRNAs were separated by electrophoresis on acid agarose gels, which were dried and exposed to a phosphor screen. VSV mRNAs levels were quantified using ImageQuant version 8.2 and normalized to the 0 h timepoint.
Transfection and Flow Cytometry Analysis of Translation of VSV mRNA.
A total of 500 ng Biotin-VSVstop–selected VSV mRNAs were transfected into 2 × 105 HeLa WT or PCIF1 KO cells using Lipofectamine 2000. At 6 h post-transfection, cells were trypsinized and washed and resuspended in phosphate-buffered saline. Half the cells were analyzed for GFP expression by flow cytometry (BD FACS Calibur); GFP-positive cells and the mean fluorescence intensity of GFP were calculated in FlowJo (20,000 cells analyzed per replicate). Studies of gene expression with GFP-expressing rabies and measles virus were performed similarly (24 h post infection), except flow cytometry was performed on a Beckman Coulter Cytoflex S.
Determination of VSV mRNA Gene Expression by Luciferase Luminescence and RT-qPCR.
Either 2 × 105 cells (24-well format; transfection experiments) or 4 × 105 cells (12-well format; IFN experiments) were lysed in 120 µl passive lysis buffer (Promega no. E1941). Half the lysate was used to quantify luciferase protein (Promega Luciferase Assay System no. E1501) using a SpectraMax L luminometer with reagent injectors in technical triplicate. RNA was extracted from the other half of cells in TRIzol, and 1 µg was reverse transcribed using SuperScript III (Invitrogen no. 18080044), oligo-dT primers (IDT no. 51011501), and RNase inhibitors (Promega no. N2515). Real-time qPCR was performed using Fast SYBR Green (Thermo Fisher no. 4385612) in technical duplicate. Relative RNA was calculated as ΔΔCT (normalized to GAPDH) × 104. RT-qPCR was performed similarly for rabies and measles virus–expressing eGFP.
Detection of Radiolabeled Samples.
Gels were fixed in 30% methanol and 10% acetic acid, washed twice in methanol, and dried using a vacuum pump gel dryer. TLC plates were air dried. Dried gels or plates were then exposed to a phosphor screen and scanned on a Typhoon scanner.
Growth Curve with IFN Pretreatment.
HeLa cells were pretreated with 500 U ⋅ ml−1 IFN-β (Tonbo Biosciences 21–8699) or vehicle (0.1% bovine serum albumin [BSA]) for 5 h in serum-free DMEM. Cells were washed and infected with VSV at a MOI of 3 for 1 h in serum-free DMEM. After 1 h, the inoculum was removed, and cells were washed and supplemented with 2% FBS. At 2, 4, 6, 8, and/or 11 h postinfection, 1% of the supernatant was removed and frozen at −80 °C. After all samples had been collected, viral titers were determined by plaque assay on Vero cells.
Gene Expression with IFN Pretreatment.
HeLa or A549 cells were pretreated with 500 U ⋅ ml−1 IFN-β as above for 5 h, infected with VSV-Luc at a MOI of 3, and at 6 h post infection, were lysed and processed for luciferase protein and mRNA quantitation. Alternatively, VSV-Luc ribonucleoprotein particles (RNPs) were purified as previously described, and 50 ng was transfected into HeLa cells instead of infection with virus.
Metabolic Radiolabeling of Protein.
For 5 h, 4 × 105 A549 cells were pretreated with 500 U ⋅ ml−1 IFN-β as above and infected (or uninfected control) with WT VSV at MOI 5. At 5 h postinfection, cells were washed, and the media was changed to methionine/cysteine-free DMEM (Gibco no. 21013–024). After 40 min of starvation, cells were pulse labeled with 30 µCi ⋅ ml−1 [35S] methionine (Perkin-Elmer no. NEG009T) and 30 µCi ⋅ ml−1 [35S] cysteine (Perkin-Elmer no. NEG022T) for 20 min. Cells were then lysed in SDS sample buffer and run on a low-bis 10% SDS-PAGE gel. Protein translation was determined by phosphorimaging as above. Equal loading was determined by staining with 0.25% Coomassie Brilliant Blue G-250.
IFIT1-IFIT3CTD Complex Purification and Filter-Binding Assay.
Protein complexes were purified as previously described (32). Maltose-binding protein (MBP) and His-tagged IFIT1 and the terminal 87 amino acids of IFIT3 (IFIT3CTD) were expressed in Rosetta 2 Escherichia coli cells (Novagen; the plasmids were a kind gift of G. Amarasinghe, Washington University, Saint Louis, MO), cultured in Luria Broth at 37 °C, induced at an optical density at wavelength 600 nm (OD600) of 0.6 with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3 h, resuspended in lysis buffer with 25 mM sodium phosphate (pH 7.5), 250 mM NaCl, and 5 mM 2-mercaptoethol, lysed using a Constant Systems CF1 Cell Disrupter, clarified by centrifugation at 20,000 × g at 4 °C for 1 h, and purified by an imidazole gradient elution from a His-trap purification column (GE) using an AKTA chromatography system (GE). Purified IFIT1 and IFIT3CTD were combined and tags cleaved using tobacco etch virus (TEV) protease at 4 °C for 14 h. IFIT1-IFIT3CTD complexes were selected by size exclusion using an S200 column (GE), and the concentration was determined by A280 absorbance.
RNA substrates were prepared as follows. Triphosphate RNA with the 27 nt VSV leader sequence (5′(ppp)ACGAAGACAAACAAACCAUUAUUAUCA 3′) were purchased from Trilink Biosciences. Radiolabeled substrate with a m7GpppA cap was prepared by capping with a vaccinia capping system (NEB) and [32P]-GTP (Perkin-Elmer) according to manufacturer’s instructions. Unlabeled substrate with a m7GpppA or m7GpppAm cap were prepared by capping with a vaccinia capping system (NEB) and/or 2’O methyltransferase (NEB). m7Gpppm6A or m7Gpppm6Am substrates were prepared by further methylation with purified PCIF1 as described above. To eliminate contaminating uncapped (triphosphate) RNA, samples were further digested by sequential incubation with shrimp alkaline phosphatase (NEB), polynucleotide kinase (NEB), and terminator exonuclease (Lucigen). Samples were TRIzol extracted into nuclease-free water before use, and concentrations were measured by Qubit RNA High Sensitivity Kit (Invitrogen).
Filter-binding assay was performed as previously described (32, 55). IFIT1-IFIT3CTD complex (1 µM) was incubated with 1 nM radiolabeled m7GpppA RNA and the indicated concentrations of cold RNA in a total reaction volume of 20 µl in a 96-well plate, including controls with no cold RNA. The binding buffer was 20 mM Tris pH 7.4, 200 mM NaCl, 1 mM DTT, 50 µg/mL BSA, and 10% glycerol. Reactions were incubated for 20 min at 37 °C followed by 40 min at 4 °C, applied to nitrocellulose and nylon membranes using a 96-well dot blot apparatus (Bio-Rad), and exposed to a phosphor screen. Relative bound RNA was calculated by dividing the signal from competitor RNA samples from the control reaction without competitor (ImageQuant TL 8.2 software). Least squares fit curves were calculated using Prism 9.
Data Analysis and Replicates.
All experiments were performed with n = 3 or n = 4 biological replicates (as indicated). Each qPCR biological replicate is the average of technical duplicates from the same sample. All qPCR biological replicates (except Figs. 5D, 6A, and 7 B–D) were run and analyzed on the same plate, enabling an SD to be calculated for all samples. Each luciferase biological replicate is the average of technical triplicates from the same sample. Statistical tests were performed in Microsoft Excel, and graphs were generated in GraphPad Prism 9. Schematics were created using https://Biorender.com.
Supplementary Material
Acknowledgments
Thanks to members of the S.P.J.W. laboratory who provided insightful discussions and advice. Thanks also to S. Hur and G. Amarasinghe for advice on RIG-I and IFIT1/3. This project was supported by NIH Grant Nos. F31AI138448 (to M.A.T), T32AI007245 (to S.P.J.W.), AI059371 (to S.P.J.W.), DP2AG055947 (to E.L.G), and R21HG010066 (to E.L.G).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025769118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Furuichi Y., Discovery of m(7)G-cap in eukaryotic mRNAs. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 91, 394–409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuichi Y., LaFiandra A., Shatkin A. J., 5′-terminal structure and mRNA stability. Nature 266, 235–239 (1977). [DOI] [PubMed] [Google Scholar]
- 3.Furuichi Y., Shatkin A. J., Viral and cellular mRNA capping: Past and prospects. Adv. Virus Res. 55, 135–184 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung D. W., Amarasinghe G. K., When your cap matters: Structural insights into self vs non-self recognition of 5′ RNA by immunomodulatory host proteins. Curr. Opin. Struct. Biol. 36, 133–141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shuman S., Capping enzyme in eukaryotic mRNA synthesis. Prog. Nucleic Acid Res. Mol. Biol. 50, 101–129 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Hornung V., et al., 5′-triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Hyde J. L., Diamond M. S., Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology 479–480, 66–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer K. D., Jaffrey S. R., The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer K. D., Jaffrey S. R., Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keith J. M., Ensinger M. J., Moss B., HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 253, 5033–5039 (1978). [PubMed] [Google Scholar]
- 11.Akichika S., et al., Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Boulias K., et al., Identification of the m6Am Methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell 75, 631–643.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sendinc E., et al., PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell 75, 620–630.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H., Zhang M., Li K., Bai D., Yi C., Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 29, 80–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei C., Gershowitz A., Moss B., N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253 (1975). [DOI] [PubMed] [Google Scholar]
- 16.Wei C. M., Gershowitz A., Moss B., 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401 (1976). [DOI] [PubMed] [Google Scholar]
- 17.Mauer J., et al., Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei J., et al., Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baltimore D., Huang A. S., Stampfer M., Ribonucleic acid synthesis of vesicular stomatitis virus, II. An RNA polymerase in the virion. Proc. Natl. Acad. Sci. U.S.A. 66, 572–576 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang B., et al., Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr J. N., Whelan S. P., Wertz G. W., cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 71, 8718–8725 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stillman E. A., Whitt M. A., Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J. Virol. 71, 2127–2137 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J. T., McElvain L. E., Whelan S. P., Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J. Virol. 81, 11499–11506 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyer S. A., Banerjee A. K., In vivo methylation of vesicular stomatitis virus and its host-cell messenger RNA species. Virology 70, 339–351 (1976). [DOI] [PubMed] [Google Scholar]
- 25.Neidermyer W. J. Jr, Whelan S. P. J., Global analysis of polysome-associated mRNA in vesicular stomatitis virus infected cells. PLoS Pathog. 15, e1007875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knipe D. M., Howley P., Fields Virology (Wolters Kluwer, Lippincott Williams & Wilkins, Philadelphia, 6 ed., 2013). [Google Scholar]
- 27.Kruse S., et al., A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci. Rep. 1, 126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahmeh A. A., Li J., Kranzusch P. J., Whelan S. P., Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 83, 11043–11050 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Wang J. T., Whelan S. P., A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 103, 8493–8498 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G., Kouwaki T., Okamoto M., Oshiumi H., Attenuation of the innate immune response against viral infection due to ZNF598-promoted binding of FAT10 to RIG-I. Cell Rep. 28, 1961–1970.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Kato H., et al., Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Johnson B., et al., Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity 48, 487–499.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz J. A., Jenni S., Harrison S. C., Whelan S. P. J., Structure of a rabies virus polymerase complex from electron cryo-microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 2099–2107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noton S. L., Fearns R., Initiation and regulation of paramyxovirus transcription and replication. Virology 479–480, 545–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino M., Ito N., Sugiyama M., Ogino T., The rabies virus L protein catalyzes mRNA capping with GDP polyribonucleotidyltransferase activity. Viruses 8, E144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Dai J., Song M., Fitzgerald-Bocarsly P., Kiledjian M., Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7. Mol. Cell. Biol. 32, 1164–1172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams G. D., Gokhale N. S., Snider D. L., Horner S. M., The mRNA Cap 2′-O-methyltransferase CMTR1 regulates the expression of certain interferon-stimulated genes. mSphere 5, e00202-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey R. R., et al., The mammalian cap-specific m6Am RNA methyltransferase PCIF1 regulates transcript levels in mouse tissues. Cell Rep. 32, 108038 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Limbach P. A., Crain P. F., McCloskey J. A., Summary: The modified nucleosides of RNA. Nucleic Acids Res. 22, 2183–2196 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You C., Dai X., Wang Y., Position-dependent effects of regioisomeric methylated adenine and guanine ribonucleosides on translation. Nucleic Acids Res. 45, 9059–9067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson B. H., Zaher H. S., O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity. RNA 21, 1648–1659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer S., et al., The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 3, 749–765 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss B., Koczot F., Sequence of methylated nucleotides at the 5′-terminus of adenovirus-specific RNA. J. Virol. 17, 385–392 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haegeman G., Fiers W., Characterization of the 5′-terminal capped structures of late simian virus 40-specific mRNA. J. Virol. 25, 824–830 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moss B., Gershowitz A., Stringer J. R., Holland L. E., Wagner E. K., 5′-terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 23, 234–239 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flavell A. J., Cowie A., Legon S., Kamen R., Multiple 5′ terminal cap structures in late polyoma virus RNA. Cell 16, 357–371 (1979). [DOI] [PubMed] [Google Scholar]
- 47.Boone R. F., Moss B., Methylated 5′-terminal sequences of vaccinia virus mRNA species made in vivo at early and late times after infection. Virology 79, 67–80 (1977). [DOI] [PubMed] [Google Scholar]
- 48.Chandran K., Sullivan N. J., Felbor U., Whelan S. P., Cunningham J. M., Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cureton D. K., Burdeinick-Kerr R., Whelan S. P., Genetic inactivation of COPI coatomer separately inhibits vesicular stomatitis virus entry and gene expression. J. Virol. 86, 655–666 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cureton D. K., Massol R. H., Saffarian S., Kirchhausen T. L., Whelan S. P., Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5, e1000394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whelan S. P., Ball L. A., Barr J. N., Wertz G. T., Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. U.S.A. 92, 8388–8392 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franz K. M., Neidermyer W. J., Tan Y. J., Whelan S. P. J., Kagan J. C., STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl. Acad. Sci. U.S.A. 115, E2058–E2067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickersham I. R., Finke S., Conzelmann K. K., Callaway E. M., Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4, 47–49 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whelan S. P., Wertz G. W., Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc. Natl. Acad. Sci. U.S.A. 99, 9178–9183 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rio D. C., Filter-binding assay for analysis of RNA-protein interactions. Cold Spring Harb. Protoc. 2012, 1078–1081 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.