Abstract
Simple Summary
The presence of an EGFR activating mutation in tumors of non-small-cell lung cancer patients enables effective targeted therapy towards EGFR. Studies that describe a nationwide uptake of EGFR testing, the impact of the switch from single-gene EGFR to multi-gene testing, and the clinical response towards EGFR inhibitors in first-line treatment are limited. From 2013 to 2017 the percentage of patients routinely tested for EGFR mutations increased from 73% to 81% in the Netherlands. A strong shift towards EGFR testing as part of a multi-gene next generation sequencing analysis was observed. However, this did not change the percentage of EGFR mutations that were reported for this patient population, which remained stable at 12%. When treated with EGFR inhibitors that were available in a routine clinical setting prior to 2018, clear differences were observed between the type of EGFR mutation and survival.
Abstract
EGFR mutation analysis in non-small-cell lung cancer (NSCLC) patients is currently standard-of-care. We determined the uptake of EGFR testing, test results and survival of EGFR-mutant NSCLC patients in the Netherlands, with the overall objective to characterize the landscape of clinically actionable EGFR mutations and determine the role and clinical relevance of uncommon and composite EGFR mutations. Non-squamous NSCLC patients diagnosed in 2013, 2015 and 2017 were identified in the Netherlands Cancer Registry (NCR) and matched to the Dutch Pathology Registry (PALGA). Overall, 10,254 patients were included. Between 2013–2017, the uptake of EGFR testing gradually increased from 72.7% to 80.9% (p < 0.001). Multi-gene testing via next-generation sequencing (increased from 7.8% to 78.7% (p < 0.001), but did not affect the number of detected EGFR mutations (n = 925; 11.7%; 95% confidence interval (CI), 11.0–12.4) nor the distribution of variants. For patients treated with first-line EGFR inhibitors (n = 651), exon 19 deletions were associated with longer OS than L858R (HR 1.58; 95% CI, 1.30–1.92; p < 0.001) or uncommon, actionable variants (HR 2.13; 95% CI, 1.60–2.84; p < 0.001). Interestingly, OS for patients with L858R was similar to those with uncommon, actionable variants (HR 1.31; 95% CI, 0.98–1.75; p = 0.069). Our analysis indicates that grouping exon 19 deletions and L858R into one class of ‘common’ EGFR mutations in a clinical trial may mask the true activity of an EGFR inhibitor towards specific mutations.
Keywords: EGFR, non-small cell lung cancer, molecular diagnostics, nationwide, real-world, survival
1. Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of global cancer-related deaths [1]. Activating mutations in EGFR account for the second most common mechanism of malignant transformation in NSCLC in western Europe and the United States [2]. Clinical trials have demonstrated that first-line treatment with an EGFR tyrosine kinase inhibitor (TKI), such as gefitinib [3], erlotinib [4], afatinib [5], dacomitinib [6], or osimertinib [7]. improves survival of advanced NSCLC patients with EGFR exon 19 deletions and L858R point mutations. Therefore, molecular diagnostics to detect these and other EGFR mutations has been standard-of-care in Europe since 2010 [8,9,10]. Molecular testing has since evolved from a single-gene polymerase chain reaction-based approach to the implementation of multiplex analyses such as next-generation sequencing (NGS) in routine diagnostics for the detection of multiple gene variants in a limited amount of tissue [11,12]. A multiplex analysis is particularly useful for NSCLC due to often limited amount of available tissue and the increasing number of predictive markers beyond EGFR (including BRAF, ERBB2 and MET) [13,14,15]. The Dutch National Healthcare Institute does not specify the methodology or commercially available companion diagnostic test that must be used for the evaluation of these biomarkers. Instead, individual molecular laboratories perform local validation of sequencing techniques according to national accreditation guidelines (ISO-NEN-15189:2012 since 2016/2017) [16]. All NGS panels used in the Netherlands cover the full region of interest in EGFR (exons 18–21).
There have been limited evaluations of the real-world, population-level effect of changes in routine EGFR testing and the introduction of multiplex testing on the landscape of EGFR mutations and overall survival rates. Furthermore, the clinical relevance of uncommon and composite EGFR mutations remains elusive. In the Netherlands, all patients diagnosed with cancer are registered in the Netherlands Cancer Registry (NCR), managed by the Netherlands Comprehensive Cancer Organization (IKNL). In addition, the nationwide network and registry of histo- and cytopathology in the Netherlands (PALGA) maintains a database of all pathology reports from pathology departments in the Netherlands [17]. The effects of routine EGFR mutation testing for advanced NSCLC patients on a population-level scale in 2013, 2015 and 2017 were evaluated following a query of both registries. During this period, only gefitinib, erlotinib and afatinib were available for use in first-line treatment.
2. Materials and Methods
2.1. Patient Selection
Testing for EGFR mutations has been standard-of-care in the Netherlands since 2011 [18], and NGS has gradually been implemented into the routine setting for non-squamous NSCLC (ns-NSCLC). Therefore, data were requested from the NCR and PALGA for advanced NSCLC patients diagnosed in 2013, 2015 and 2017. All patients recorded in the NCR who were diagnosed with advanced adenocarcinoma of the lung, adenosquamous carcinoma of the lung and NSCLC not otherwise specified (NOS) were included. Patients were matched with the nationwide PALGA registry to retrieve the pathology reports. Data requests were approved by the scientific and privacy committees of IKNL (application numbers K15.115, K16.264, and K18.311) and PALGA (application numbers LZV1172, LZV2016-91, LZV2018-199).
2.2. Data Extraction and Handling
Variables retrieved from the NCR included sex, age at diagnosis, morphology code (ICD-O 3rd edition), type of first-line treatment (categories including chemotherapy, radiotherapy, surgery, targeted therapy, other therapy, a combination of these, or no therapy), overall survival (OS) (time in days from diagnosis to death or data cut-off) and vital status as of 1 February 2020. Molecular testing results were manually extracted from the pathology reports by dedicated researchers (CCHJK, BNCG, BK). Extracted variables included whether testing was performed or not, the type of molecular test(s) used to detect EGFR mutations, an identifier of the pathology department that requested the test, and the reported EGFR mutation status. The type of molecular test(s) used were corrected to exclude secondary testing on new tissue obtained at progressive disease.
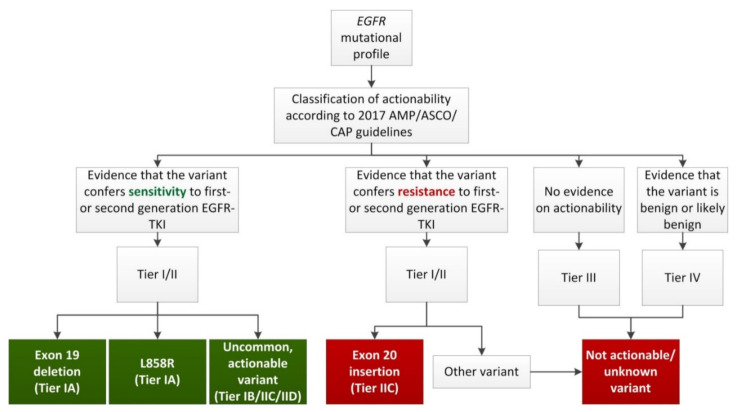
Reported EGFR driver mutations were reannotated in accordance with the Human Genome Variant Society recommendations for the description of sequence variants [19]. Each variant was classified for predicted actionability with EGFR-TKI as ‘sensitive’ or ‘no benefit’, with level of evidence tiers based on the 2017 Association for Molecular Pathology (AMP)/American Society of Clinical Oncology (ASCO)/College of American Pathologists (AMP) guidelines [20]. Patients with EGFR mutations were grouped into one of five nominal categories based on the frequency of the reported EGFR mutations and the AMP/ASCO/CAP level of evidence (Figure 1). These categories included: exon 19 deletion (sensitive, Tier IA); L858R (sensitive, Tier IA); uncommon, actionable (sensitive, Tier IB/IIC/IID); exon 20 insertion (no benefit expected, Tier IIC) or not actionable/unknown (no benefit expected, Tier I–IV, except for exon 20 insertions). In case of a double variant, actionability was assessed for the combination. For example, the combination of uncommon, actionable variants E709A and G719A has been reported to be sensitive to afatinib [21], and the combination is thus tiered IIC (uncommon, actionable). However, if evidence for the combination was lacking, the patient was assigned according to the highest Tier of the individual variants. For instance, patients with a combination of an exon 19 deletion (Tier IA) and a variant of unknown significance (Tier III) were assigned to category ‘exon 19 deletion’. Classification was performed by a certified clinical scientist in molecular pathology (LCvK) [22], and only considered for first- and second-generation EGFR-TKI (gefitinib, erlotinib, afatinib) due to the unavailability of first-line osimertinib treatment in the study period. Clinical data processing was performed in accordance with the General Data Protection Regulation (EU) 2016/679.
Figure 1.
Flowchart used for classification of EGFR mutational profiles. Grouping of EGFR mutational profiles included in this study, based on the frequency of the mutation(s) and the corresponding AMP/ASCO/CAP level of evidence for actionability with first- or second-generation EGFR-TKI. Groups in green (Exon 19 deletion, L858R and Uncommon, actionable variants) are considered sensitive to first- or second-generation EGFR-TKI, groups in red (Exon 20 insertion and Not actionable/unknown variants) are considered resistant to first- or second-generation EGFR-TKI. In case of a double variant, actionability was assessed for the combination. However, if evidence for the combination was lacking, the patient was assigned according to the highest tier of the individual variants. Abbreviations: AMP, Association for Molecular Pathology; ASCO, American Society of Clinical Oncology; CAP, College of American Pathologists; EGFR, epidermal growth factor receptor gene; TKI, tyrosine kinase inhibitor.
2.3. Statistical Analysis
Statistical analysis was performed with SPSS version 23 (SPSS Inc., Chicago, IL, USA). Comparisons were performed for patient characteristics, testing rates and EGFR mutation detection rates between the three years of diagnosis (2013, 2015 and 2017) and between testing modalities (multi-gene assays or single-gene tests). Multi-gene assays were defined as NGS technologies or massARRAY. Proportional differences between years were assessed using Pearson’s Chi-square analysis, and, if significant, a subgroup analysis was performed with Fisher’s exact tests. Differences between testing modalities were assessed using Fisher’s exact tests.
Median OS from date of diagnosis was estimated with the Kaplan-Meier method, including 95% confidence intervals (CI), and tested for significance with the Log-rank test. For EGFR-mutant patients treated with first-line targeted therapy, uni- and multivariate Cox regression analyses were performed to correct for the co-variables age, sex, year of diagnosis and tumor histology. Variables with a p-value < 0.05 in the univariate analysis were included in the multivariate analysis (forward stepwise logistic regression). Hazard ratio (HR) and 95% CI were calculated. Differences were considered statistically significant at a p-value < 0.05.
3. Results
3.1. Patients Included in the Analysis
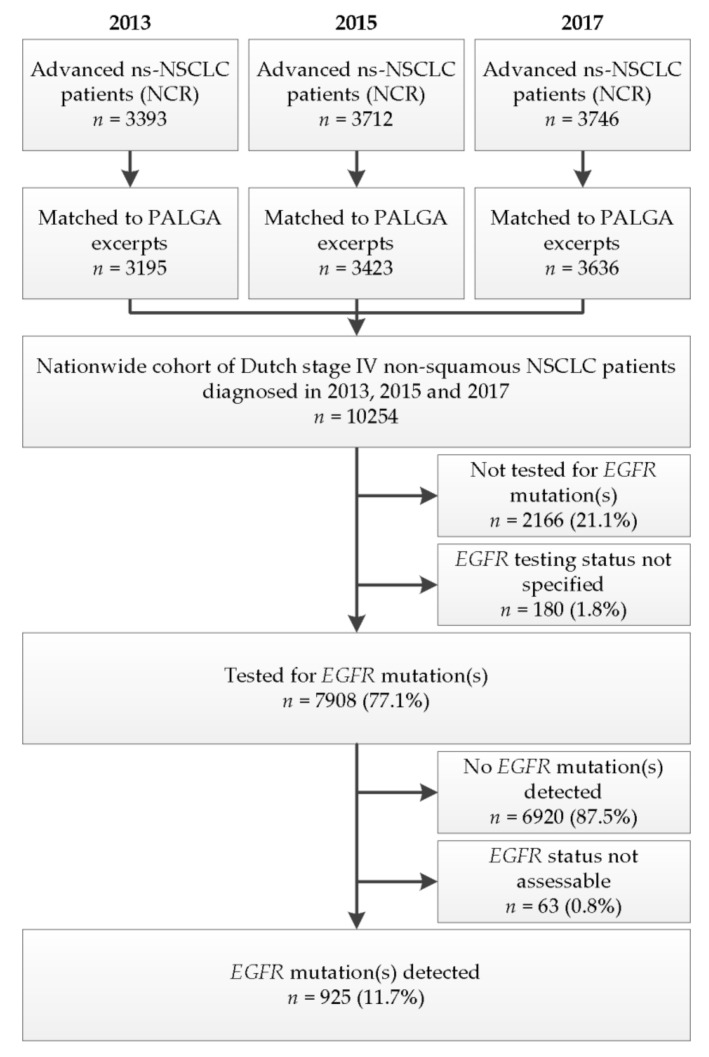
The NCR recorded 3393, 3712 and 3746 advanced ns-NSCLC patients in 2013, 2015 and 2017, respectively. Of these, 3195 (2013), 3423 (2015) and 3636 (2017) could be matched to excerpts in the PALGA registry, creating a population-level cohort of 10,254 Dutch advanced ns-NSCLC patients (Figure 2). Patient characteristics are shown in Table 1. The mean age for the total population was 66.9 years. A small majority of patients was male (53.6%). The majority of patients were diagnosed with adenocarcinoma (86.3%), followed by NSCLC-not otherwise specified (NSCLC-NOS) (13.1%), and adenosquamous carcinoma (0.6%).
Figure 2.
Patient selection. Flowchart depicting the selection of patients for inclusion in this study. Abbreviations: EGFR, epidermal growth factor receptor gene; PALGA, Dutch Pathology Registry; NCR, Netherlands Cancer Registry; ns-NSCLC, non-squamous non-small cell lung cancer.
Table 1.
Patient characteristics, EGFR testing method and mutation status at initial diagnosis of advanced NSCLC.
| Characteristic | Total | 2013 | 2015 | 2017 |
|---|---|---|---|---|
| Cases, n (%) | 10,254 (100) | 3195 (100) | 3423 (100) | 3636 (100) |
| Sex | ||||
| Male | 5498 (53.6) | 1730 (54.1) | 1820 (53.2) | 1947 (53.5) |
| Female | 4757 (46.4) | 1465 (45.9) | 1603 (46.8) | 1689 (46.5) |
| Age | ||||
| Mean (range) | 66.9 (20–101) | 66.2 (23–98) | 67.0 (24–97) | 67.4 (20–101) |
| <65 years | 4133 (40.3) | 1386 (43.4) | 1355 (39.6) | 1392 (38.3) |
| ≥65 years | 6121 (59.7) | 1809 (56.6) | 2068 (60.4) | 2244 (61.7) |
| Tumor histology | ||||
| Adenocarcinoma | 8845 (86.3) | 2724 (85.3) | 2961 (86.5) | 3160 (86.9) |
| Adenosquamous carcinoma | 62 (0.6) | 19 (0.6) | 25 (0.7) | 18 (0.5) |
| NSCLC, NOS | 1347 (13.1) | 452 (14.1) | 437 (12.8) | 458 (12.6) |
| EGFR mutation testing | ||||
| Tested | 7908 (77.1) | 2324 (72.7) | 2643 (77.2) | 2941 (80.9) |
| Not tested a | 2166 (21.1) | 765 (24.0) | 766 (22.4) | 635 (17.5) |
| Testing status not specified | 180 (1.8) | 106 (3.3) | 14 (0.4) | 60 (1.6) |
| EGFR test performed at initial diagnosis (% of n tested) | ||||
| Single-gene test | 2350 (29.7) | 1356 (58.3) | 653 (24.7) | 341 (11.6) |
| Fragment analysis | 8 (0.1) | 8 (0.3) | – | – |
| HRM (±sequencing) | 1215 (15.4) | 693 (29.8) | 382 (14.5) | 140 (4.8) |
| Immunohistochemistry | 20 (0.3) | 20 (0.9) | – | – |
| Mutation-specific PCR | 290 (3.7) | 121 (5.2) | 141 (5.3) | 28 (1.0) |
| PCR screening (±sequencing) b | 259 (3.3) | 232 (10.0) | 27 (1.0) | – |
| Pyrosequencing | 109 (1.4) | 2 (0.1) | 12 (0.5) | 95 (3.2) |
| Sanger sequencing | 257 (3.2) | 115 (4.9) | 67 (2.5) | 75 (2.6) |
| Combination of different single-gene tests | 192 (2.4) | 165 (7.1) | 24 (0.9) | 3 (0.1) |
| Multi-gene assay | 3958 (50.1) | 181 (7.8) | 1463 (55.4) | 2314 (78.7) |
| MassARRAY | 59 (0.7) | 35 (1.5) | 8 (0.3) | 16 (0.5) |
| NGS | 3299 (41.7) | 63 (2.7) | 1111 (42.0) | 2125 (72.3) |
| Combination of single- and multi-gene assays | 600 (7.6) | 83 (2.6) | 344 (13.0) | 176 (5.9) |
| Test not specified | 1600 (20.2) | 787 (33.9) | 527 (19.9) | 286 (9.7) |
| EGFR mutation status (% of n tested) | ||||
| EGFR mutation(s) reported | 925 (11.7) | 256 (11.0) | 328 (12.4) | 341 (11.6) |
| No EGFR mutation(s) | 6920 (87.5) | 2046 (88.0) | 2297 (86.9) | 2577 (87.6) |
| EGFR testing failed | 63 (0.8) | 22 (1.0) | 18 (0.7) | 23 (0.8) |
a Reasons for not performing an EGFR mutation analysis were not systematically captured in the pathology reports used in our study. Therefore, it could not be determined what factors (technical and/or patient-specific) contributed to not testing for EGFR mutations. b Diagnostic algorithm consisting of screening with mutation-specific PCR, followed by EGFR sequencing. Abbreviations: EGFR, epidermal growth factor receptor gene; HRM, high-resolution melting; NGS, next-generation sequencing; NOS, not otherwise specified; NSCLC; non-small cell lung cancer; PCR, polymerase chain reaction.
3.2. EGFR Testing Rates, Inter-Pathology Department Variance and Assays Used
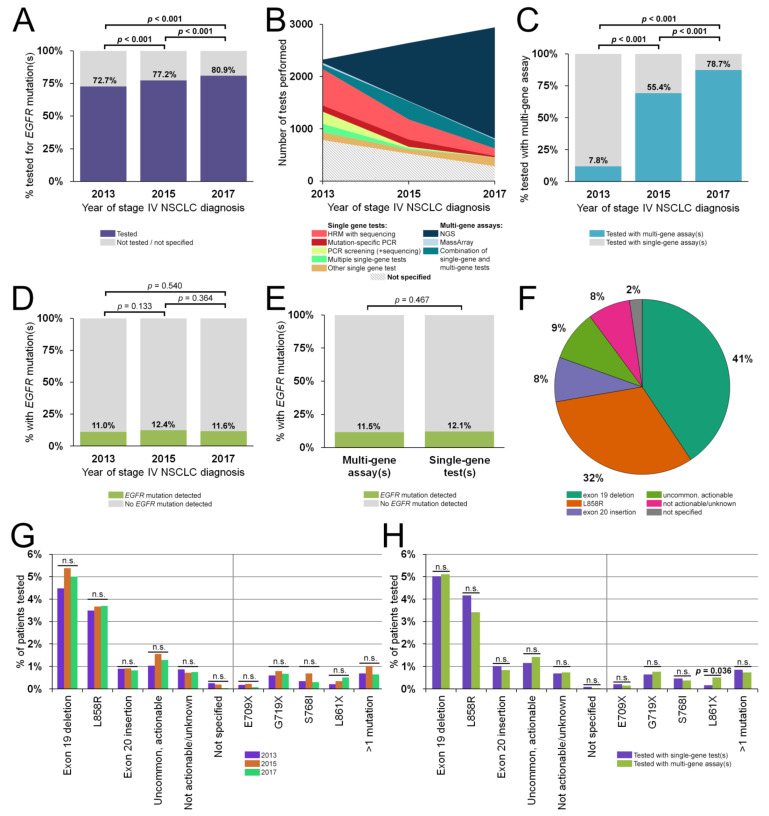
Out of the total population of 10,254 advanced NSCLC patients, 77.1% (95% CI, 76.3–77.9%; n = 7908) were tested for EGFR mutations. The testing rate increased significantly throughout the study period, from 72.7% (95% CI, 71.2–74.2%) in 2013 to 77.2% (95% CI, 75.8–78.6%) in 2015 (p < 0.001), up to 80.9% (95% CI, 79.6–82.2%) in 2017 (p < 0.001) (Figure 3A). A commonly reported reason for not performing an EGFR mutation analysis was the lack of sufficient suitable tissue or a low percentage of tumor cells. However, reasons for not performing an EGFR mutation analysis were not systematically captured in the pathology reports used in our study. Therefore, it could not be determined with high confidence which other technical, patient or infrastructural factors contributed to not testing for EGFR mutations.
Figure 3.
EGFR testing rates, methods and EGFR mutations. (A) Percentage of patients tested for EGFR mutations, by year of diagnosis. (B) Distribution of EGFR testing methods in number of tests by year. (C) Percentage of patients tested with single-gene versus multi-gene assays, by year of diagnosis. (D,E) Percentage of patients tested for EGFR mutations that tested positive for one or more EGFR mutation(s), by year of diagnosis (D) and multi-gene assay versus single-gene assay (E). (F) Distribution of EGFR mutation classes. (G,H) Comparison of distributions of EGFR mutation classes and occurrence of specific uncommon mutations, by year of diagnosis (G) and single-gene assay versus multi-gene assay (H). Abbreviations: EGFR, epidermal growth factor receptor gene; HRM, high-resolution melting; NGS, next-generation sequencing; n.s., not significant; NSCLC, non-small cell lung cancer; PCR, polymerase chain reaction.
Samples from the patients in this study originated from 52 different pathology departments in the Netherlands. Forty-one departments reported EGFR mutation test results for 10 or more patients in each of the three time periods (2013, 2015 and 2017). The majority of departments improved their testing rates and the inter-pathology department variance in requesting EGFR testing gradually decreased from 2013 to 2017 (Figure S1). In 2017, only four out of the 41 departments (9.8%) underperformed compared to the national average. In other words, these departments reported fewer EGFR mutation tests (<60% of the eligible patients) than expected for their respective tumor volumes (based on the lower 99.7% CI limit) and the national average of 80.9% in 2017.
The landscape of tests used to detect EGFR mutations changed in the study years. Whereas the most common tests in 2013 were high-resolution melting (HRM), mutation-specific polymerase chain reaction (PCR), single-gene sequencing (such as Sanger sequencing) or combinations thereof, EGFR testing was mostly performed (78.7%) with multi-gene assays in 2017 (Figure 3B). The use of multi-gene assays as first-line molecular diagnostics surged from 7.8% in 2013 to 55.4% in 2015 (p < 0.001), and further increased to 78.7% in 2017 (p < 0.001) (Figure 3C). Laboratories generally use custom or generic NGS panels validated in accordance with national accreditation guidelines (ISO-NEN-15189:2012 since 2016/2017). Examples of more common NGS assays are custom AmpliSeq-panels on an IonTorrent platform [23], the TruSeq Amplicon Cancer Panel-based NGS using a MiSeq Personal sequencer [24], and the nationally aligned single-molecule Molecular Inversion Probe (smMIP) PATHv2D panel on an Illumina platform [25]. The panels used by Dutch molecular pathology laboratories all covered the full region of interest in EGFR (exons 18–21).
3.3. Landscape of EGFR Mutations in Untreated Dutch NSCLC Patients
Of the 7908 patients tested for EGFR mutations at initial diagnosis, one or more mutations were reported in 11.7% of all cases (95% CI, 11.0–12.4%; n = 925) (Table 2). Female patients were more likely to harbor EGFR mutations (16.0%; 95% CI, 14.8–17.2%; n = 600) than male patients (7.8%; 95% CI, 7.0–8.6; n = 325; p < 0.001). There were no significant differences in EGFR positivity between the three study years (Figure 3D) nor between the use of single-gene or multi-gene assays (Figure 3E). The majority of patients harbored a classical, actionable exon 19 deletion (41% of 925 cases) or L858R (32%), whereas uncommon but actionable mutations (9%), exon 20 insertions (8%), and other not actionable/unknown variants (8%) constituted a much smaller proportion of the EGFR-mutant patients (Figure 3F). On a variant class- or single mutant-level, no significant distribution differences were observed for the three study years (Figure 3G) nor for the use of single-gene or multi-gene assays (Figure 3H), except for EGFR L861X. This variant (either alone or in combination with a second EGFR variant) was slightly more common in patients tested with a multi-gene assay (n = 20; 0.5% of those tested for EGFR mutations) compared to those tested with single-gene tests (n = 5; 0.2%; p = 0.036).
Table 2.
EGFR mutations in the Dutch population of advanced non-squamous NSCLC patients by year of diagnosis (A) and molecular diagnostic modality (B).
| A | By Year of Advanced NSCLC Diagnosis | ||||||
| Mutation(s) | Total | 2013 | 2015 | 2017 | p a | ||
| Tested for Mutations, n (%) | 7908 (100) | 2324 (100) | 2643 (100) | 2941 (100) | |||
| Any EGFR mutation | 925 (11.7) | 256 (11.0) | 328 (12.4) | 341 (11.6) | 0.303 | ||
| Distribution of predefined classes of sensitivity | |||||||
| Exon 19 deletion | 393 (5.0) | 104 (4.5) | 142 (5.4) | 147 (5.0) | 0.347 | ||
| L858R | 287 (3.6) | 81 (3.5) | 97 (3.7) | 109 (3.7) | 0.905 | ||
| Exon 20 insertion | 69 (0.9) | 21 (0.9) | 24 (0.9) | 24 (0.8) | 0.917 | ||
| Uncommon, actionable | 103 (1.3) | 24 (1.0) | 41 (1.6) | 38 (1.3) | 0.274 | ||
| Not actionable/unknown | 61 (0.8) | 20 (0.9) | 19 (0.7) | 22 (0.7) | 0.836 | ||
| Not specified | 12 (0.2) | 6 (0.3) | 5 (0.2) | 1 (0.0) | 0.097 | ||
| Distribution of uncommon EGFR mutation(s) | |||||||
| >1 EGFR mutation | 62 (0.8) | 16 (0.7) | 27 (1.0) | 19 (0.6) | 0.233 | ||
| E709X | 12 (0.2) | 4 (0.2) | 6 (0.2) | 2 (0.1) | 0.299 | ||
| G719X | 55 (0.7) | 14 (0.6) | 21 (0.8) | 20 (0.7) | 0.713 | ||
| S768I | 35 (0.4) | 8 (0.3) | 18 (0.7) | 9 (0.3) | 0.076 | ||
| L861X | 29 (0.4) | 5 (0.2) | 9 (0.3) | 15 (0.5) | 0.206 | ||
| B | By Molecular Diagnostic Modality | ||||||
| Mutation(s) | Total | Single-Gene Test(s) | Multi-Gene Assay | p c | |||
| Cases, n (%) | 6308 (100) b | 2350 (100) | 3958 (100) | ||||
| Distribution of predefined classes of sensitivity | |||||||
| Exon 19 deletion | 320 (5.1) | 118 (5.0) | 202 (5.1) | 0.906 | |||
| L858R | 233 (3.7) | 98 (4.2) | 135 (3.4) | 0.129 | |||
| Exon 20 insertion | 57 (0.9) | 24 (1.0) | 33 (0.8) | 0.492 | |||
| Uncommon, actionable | 83 (1.3) | 27 (1.1) | 56 (1.4) | 0.424 | |||
| Not actionable/unknown | 45 (0.7) | 16 (0.7) | 29 (0.7) | 0.878 | |||
| Not specified | 3 (0.0) | 2 (0.1) | 1 (0.0) | 0.560 | |||
| Distribution of individual uncommon EGFR mutation(s) | |||||||
| >1 EGFR mutation | 49 (0.8) | 20 (0.9) | 29 (0.7) | 0.657 | |||
| E709X | 11 (0.2) | 5 (0.2) | 6 (0.2) | 0.551 | |||
| G719X | 45 (0.7) | 15 (0.6) | 30 (0.8) | 0.645 | |||
| S768I | 26 (0.4) | 11 (0.5) | 15 (0.4) | 0.685 | |||
| L861X | 24 (0.4) | 4 (0.2) | 20 (0.5) | 0.036 | |||
Values in bold indicate significant differences. a Level of significance tested with Pearson’s Chi-square test, considered significant at p < 0.05. b Does not include patients for whom the testing modality was not reported (n = 1600). c Level of significance tested with Fisher’s exact test, considered significant at p < 0.05. Abbreviations: EGFR, epidermal growth factor receptor gene; HRM, high-resolution melting; NGS, next-generation sequencing; NOS, not otherwise specified; NSCLC; non-small cell lung cancer; PCR, polymerase chain reaction.
3.4. Impact of EGFR Testing on First-Line Targeted Therapy and Subsequent Overall Survival
Treatment and survival data were available for 10,237 out of 10,254 patients (99.8%). This included 390 patients with an exon 19 deletion, 287 patients with L858R, 103 patients with an uncommon, actionable variant, 69 patients with an exon 20 insertion, 61 patients with a not actionable/unknown variant and 6972 patients without EGFR mutation. Patients who were not tested for EGFR mutations (n = 2343) were excluded from the analysis, as were patients reported to be EGFR mutation positive but for whom the exact variant was not reported (and could thus not be classified) (n = 12).
3.4.1. Overall Survival of EGFR-Mutant Patients Versus Those without EGFR Mutations
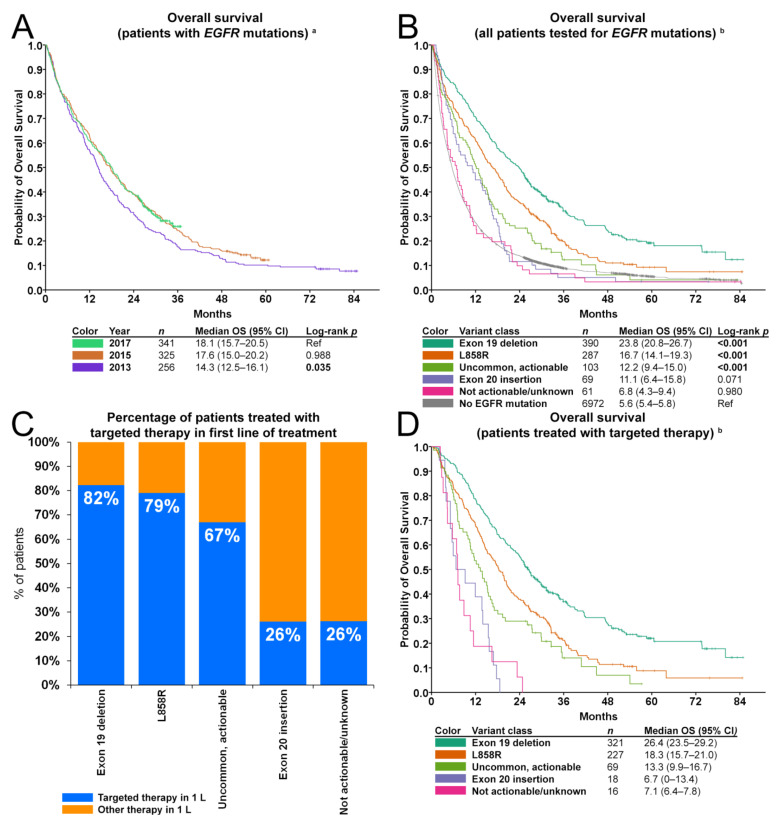
OS in patients with any EGFR mutation was higher for those diagnosed in 2017 (median 18.1 months; 95% CI, 15.7–20.5) compared to 2013 (median 14.3 months; 95% CI, 12.5–16.1; p = 0.035), but similar to 2015 (median 17.6 months; 95% CI, 15.0–20.2; p = 0.988) (Figure 4A). Irrespective of first-line treatment, distinct survival patterns were observed in the different EGFR mutation subclasses (Figure 4B). Median OS in patients without EGFR mutations was 5.6 months (95% CI, 5.4–5.8) in the period 2013–2017. Evaluation of OS for the various EGFR mutation classes revealed a favorable OS for patients with exon 19 deletions (23.8 months; 95% CI, 20.8–26.7; p < 0.001), L858R (16.7 months; 95% CI, 14.1–19.3; p < 0.001) and uncommon, actionable variants (12.2 months; 95% CI, 9.4–15.0; p < 0.001). In contrast, median OS of patients with exon 20 insertions (11.1 months; 95% CI, 6.4–15.8; p = 0.071) or with not actionable/unknown variants (6.8 months; 95% CI, 4.3–9.4; p = 0.980) was comparable to the OS of patients without an EGFR mutation.
Figure 4.
First-line targeted therapy and overall survival. (A) Kaplan-Meier curve depicting overall survival by year of diagnosis, irrespective of first-line treatment. Censored cases are indicated with a +. (B) Kaplan-Meier curve depicting overall survival by mutation class, irrespective of first-line treatment. Censored cases are indicated with a +. (C) Percentage of patients treated with targeted therapy in first-line of treatment, by EGFR mutation class. (D) Kaplan-Meier curve depicting overall survival of EGFR-mutant patients treated with targeted therapy in first-line of treatment. Censored cases are indicated with a +. a Excluding patients with missing follow-up data (n = 3). b Excluding patients with missing follow-up data (n = 3) and patients reported to have a non-specified EGFR mutation (n = 12). Abbreviations: 1 L; first-line of treatment; CI, confidence interval; EGFR, epidermal growth factor receptor gene; p, level of significance; OS, overall survival.
3.4.2. Overall Survival in Patients Treated with First-Line Targeted Therapy
Out of the 910 patients with EGFR mutation(s) and available follow-up, 651 (72%) received first-line treatment with targeted therapy. Of note, this included patients in the not actionable/unknown group (Tier III–IV), which today would likely not be treated with targeted therapy. The proportion receiving targeted therapy was not affected by year of diagnosis (70%, 70% and 74% in 2013, 2015 and 2017, respectively; p = 0.332) (Figure S2). There was no information on which inhibitor was used. However, in 2013–2017, gefitinib, erlotinib and afatinib were the only EGFR inhibitors used in first-line therapy [26]. The remaining patients (representing 29% of all EGFR-mutated patients) were either treated with a non-targeted modality (n = 171; 19%) or did not receive treatment (n = 88; 10%). The NCR did not register reasons for not treating patients with targeted therapy, nor was this information available in the pathology reports. Therefore, reasons for not treating patients with targeted therapy could not be investigated.
The proportion of patients receiving first-line targeted therapy was highest for those with an exon 19 deletion (321/390; 82%) or L858R mutation (227/287; 79%), lower for those with uncommon, actionable variants (69/103; 67%) and smallest for those with exon 20 insertions (18/69; 26%) and not actionable/unknown variants (16/61; 26%) (Figure 4C). These proportions were not affected by year of diagnosis (Figure S2). Distinct differences in survival were observed between the predefined categories of EGFR mutations (Figure 4D). Median OS in patients treated with first-line targeted therapy was 26.4 months for patients with exon 19 deletions (95% CI, 23.5–29.2), 18.3 months for patients with L858R (95% CI, 15.7–21.0), 13.3 months for patients with uncommon, actionable variants (95% CI, 9.9–16.7), 6.7 months for patients with exon 20 insertions (95% CI, 0–13.4), and 7.1 months for patients with not actionable/unknown variants (95% CI, 6.4–7.8).
Uni- and multivariable Cox regression analyses were performed to evaluate whether the type of EGFR mutation is associated with OS (Table 3). Because OS for patients with EGFR exon 20 insertions and patients with not actionable/unknown variants treated with first- and second-generation EGFR inhibitors was comparably poor (Figure 4D), these groups were pooled into one category dubbed as “resistant/unknown variants”. A higher age (p < 0.001), male sex (p < 0.008) and diagnosis in 2013 (p = 0.011) were significantly associated with worse outcome in the univariate analyses. When corrected for these factors, patients with EGFR exon 19 deletions showed superior outcomes compared to those with L858R (hazard ratio (HR) 1.6; 95% CI, 1.3–1.9; p < 0.001), uncommon, actionable variants (HR 2.1; 95% CI, 1.5–2.7; p < 0.001) and resistant/unknown variants (HR 4.7; 95% CI, 3.2–6.8; p < 0.001). OS of patients with EGFR L858R when corrected for age, sex and year of diagnosis was not different from patients with uncommon actionable EGFR mutations (HR 1.31; 95% CI, 1.0–1.8; p = 0.069). Furthermore, patients with resistant/unknown variants had a worse outcome compared to those with EGFR L858R (HR 3.0; 95% CI, 2.1–4.3; p < 0.001) or those with uncommon actionable variants (HR 2.4; 95% CI, 1.6–3.6; p < 0.001).
Table 3.
Uni- and multivariate Cox regression overall survival analyses for EGFR mutation-positive NSCLC patients treated with first-line targeted therapy (n = 651).
| Factor | Univariate Analysis | Multivariate Analysis a | |||||
|---|---|---|---|---|---|---|---|
| n | HR | 95% CI | p | HR | 95% CI | p | |
| Age (cont.) | 651 | 1.02 | 1.01–1.03 | <0.001 | |||
| Age (in years) | |||||||
| <65 years | 258 | Ref | Ref | Ref | |||
| ≥65 years | 393 | 1.26 | 1.06–1.51 | 0.011 | |||
| Sex | |||||||
| Male | 223 | Ref | Ref | Ref | |||
| Female | 428 | 0.78 | 0.65–0.94 | 0.008 | |||
| Tumor histology | |||||||
| Adenocarcinoma | 630 | Ref | Ref | Ref | |||
| NSCLC, NOS | 19 | 1.45 | 0.88–2.39 | 0.142 | |||
| Adenosquamous carcinoma | 2 | 2.23 | 0.56–8.96 | 0.258 | |||
| Year of diagnosis | |||||||
| 2013 | 175 | 1.32 | 1.07–1.63 | 0.011 | |||
| 2015 | 223 | Ref | Ref | Ref | |||
| 2017 | 253 | 1.01 | 0.82–1.26 | 0.902 | |||
| EGFR mutation class (Ref: Exon 19 deletion) | |||||||
| Exon 19 deletion | 321 | Ref | Ref | Ref | Ref | Ref | Ref |
| L858R | 227 | 1.58 | 1.30–1.92 | <0.001 | 1.57 | 1.29–1.90 | <0.001 |
| Uncommon, actionable variant | 69 | 2.13 | 1.60–2.84 | <0.001 | 2.05 | 1.54–2.74 | <0.001 |
| Resistant/unknown variants b | 34 | 5.05 | 3.48–7.33 | <0.001 | 4.67 | 3.21–6.80 | <0.001 |
| EGFR mutation class (Ref: L858R) | |||||||
| L858R | 227 | Ref | Ref | Ref | Ref | Ref | Ref |
| Uncommon, actionable variant | 69 | 1.35 | 1.01–1.80 | 0.046 | 1.31 | 0.98–1.76 | 0.069 |
| Resistant/unknown variants b | 34 | 3.19 | 2.19–4.63 | <0.001 | 2.98 | 2.05–4.34 | <0.001 |
| EGFR mutation class (Ref: uncommon, actionable variant) | |||||||
| Uncommon, actionable variant | 69 | Ref | Ref | Ref | Ref | Ref | Ref |
| Resistant/unknown variants b | 34 | 2.37 | 1.55–3.63 | <0.001 | 2.27 | 1.48–3.49 | <0.001 |
Values in bold indicate significant differences. a Corrected for age (continuous), sex, and year of diagnosis. b Because patients with EGFR exon 20 insertions and patients with not actionable/unknown variants had comparably poor outcomes (Figure 3D), these groups were pooled into one category named “resistant/unknown variants”. Abbreviations: CI, confidence interval; cont., continuous variable; EGFR, epidermal growth factor receptor gene; HR, hazard ratio; NOS, not otherwise specified; NSCLC; non-small cell lung cancer; p, level of significance; Ref, reference group.
In 2017, the third-generation EGFR inhibitor osimertinib was approved for use in patients who developed the EGFR T790M mutation as a mechanism of resistance. In this year, of the 254 patients who received an EGFR inhibitor, 27 (11%) developed the T790M within one year after diagnosis. An additional 16 patients (6%) developed this mutation in the remainder of 2018. The T790M mutation was detected on the background of an exon 19 deletion (27 out of 321 (8.4%) treated with targeted therapy), the L858R mutation (15/227; 6.6%) and the composite G719S/L861Q mutation (n = 1). Because the reports were limited to 2018, the number of long-term responders cannot be estimated. The NCR data did not contain information about the type of therapy in second-line nor the duration of response to second-line treatment. Therefore, a survival analysis to investigate the impact of resistance-targeted treatment options could not be performed. There is insufficient data to investigate whether the treatment-induced T790M is more prevalent on the background of an exon 19 deletion than L858R.
4. Discussion
In this study, the impact of routine testing for EGFR mutations was investigated in the Dutch population of patients with newly diagnosed advanced ns-NSCLC in 2013, 2015 and 2017. This analysis revealed that, despite a significant increase in testing rate and a nationwide shift from single-gene testing to multi-gene assays such as NGS, the EGFR positivity rate and distributions of specific (common and uncommon) alterations that can be observed at primary diagnosis remained comparable. In addition, the analysis of real-world treatment outcome data in the Netherlands revealed distinct survival patterns for patients with different classes of EGFR mutations when treated in first-line with first- (gefitinib, erlotinib) or second-generation (afatinib) EGFR-TKI. The results of this population-based analysis hold not only value for policy makers and healthcare providers, but also offer insight into the frequency and consequences of reported uncommon and composite variants for clinicians and scientists.
4.1. Impact of Routine EGFR Testings on Detection of EGFR Mutations and Overall Survival
Testing for EGFR mutations at primary diagnosis has been standard-of-care for advanced NSCLC patients in the Netherlands since 2011 [9,27]. Our analysis showed that the testing rate has since then gradually but significantly increased from 72.7% in 2013 to 80.2% in 2017 (p < 0.001). Whether nationwide testing rates further increased thereafter will be the subject of future studies. Reasons for not performing an EGFR mutation analysis were not systematically captured in the pathology reports. The availability and suitability of sufficient tissue, patient factors negating the need for testing (death prior to testing, poor condition, or personal preference provided at shared decision-making visits) are likely important factors. However, it cannot be excluded that a percentage of eligible NSCLC patients are not offered predictive testing despite the availability of tissue and increasing number of registered (EGFR-)targeted therapies.
The prevalence of EGFR mutations in the Dutch population of ns-NSCLC is 11.7% (95% CI, 11.0–12.4%). This was comparable to the prevalence found in a smaller Dutch cohort in 2012 (10.6% in adenocarcinoma of the lung) [28], though slightly lower than previously reported in a pooled prevalence analysis of studies with European subjects (14.1%) [29]. As expected, EGFR mutations associated with EGFR inhibitor-induced on-target resistance mutations were not observed in this untreated cohort, except for one case. This single case of EGFR T790M without another mutation might suggests a germline event, but cannot be concluded without additional testing.
The uptake of NGS as the method of choice for EGFR analysis (from 8% in 2013 to 79% in 2017) did not impact the nationwide detection rate of EGFR mutations nor the distribution of uncommon mutations. This is likely because the majority of single-gene tests in this study were sequencing techniques (such as pyrosequencing, Sanger sequencing or HRM in combination with sequencing) that covered the full region of interest in EGFR (exons 18–21). The only variant impacted by the shift to multi-gene testing was the uncommon EGFR L861X mutation in exon 21. This variant was slightly more common in patients tested with a multi-gene assay compared to single-gene tests (p = 0.036). The first reports on the actionability of this variant with first-generation EGFR-TKI emerged in 2018 [30]. It is conceivable that this continuously increasing knowledge resulted in a more frequent reporting of previously unrecognized uncommon yet actionable EGFR variants. In addition, single-gene tests available in 2013—such as cobas® EGFR Mutation Test v1 (Roche Holding AG, Basel, Switzerland)—might not have been designed to capture an L861X mutation due to its previously unknown clinical significance. As such, the introduction of EGFR mutation analysis by NGS likely contributed to the discovery of treatment options for the current uncommon EGFR mutations. Our results indicate that the utilization of NGS did not adversely impact the detection rate for EGFR mutations underscoring the accurate detection of mutations.
Median OS in the Dutch population of advanced ns-NSCLC patients with EGFR mutations was 18.1 months in 2017, which is comparable to other real-world studies with European subjects [31]. Although median OS in patients diagnosed in 2015 was comparable (17.6 months), patients diagnosed in 2013 survived shorter (14.3 months, p = 0.035). This may be because patients diagnosed in 2013 generally did not have access to second-line testing and novel treatment options. For example, a multitude of potential resistance mechanisms to first- and second-generation EGFR-TKIs therapies have been reported, most commonly the T790M resistance mutation [32]. The activity of third-generation inhibitors, such as osimertinib, in first and later lines of therapy, is unaffected by this mutation [33]. In addition, checkpoint inhibition has been approved for patients with tumors expressing PD-L1. This has improved survival rates in advanced NSCLC patients [34]. Checkpoint inhibitors are not used in first- or second-line treatment of EGFR-mutant patients as they do not improve OS in these patients [35]. Unfortunately, assessment of the impact of the introduction of the new treatment modalities could not be performed as the NCR database lacks information on second and later lines of treatment.
4.2. Beyond EGFR
The number of options for targeted therapy for NSCLC patients beyond EGFR has strongly increased. Different therapies are now registered or being tested in trials for patients with an activating mutation ALK [36], BRAF [13], ERBB2 [15], MET [14], NTRK1-3 [37], RET [38], and ROS1 [39]. In addition, trials with specific KRAS G12C inhibitors demonstrate strong clinical benefit with registration expected by the end of 2021 [40]. As such, the panel of therapeutically relevant biomarkers in NSCLC is continuously increasing. Testing patients for all these predictive markers using consecutive single-gene tests is inefficient considering the often limited amount of available tissue. Therefore, NGS has become standard-of-care in the Netherlands to simultaneously test for all relevant actionable mutations also beyond EGFR as an efficient diagnostic work-up [25]. Although the content of the panels has changed since 2017, the current core set of genes relevant for NSCLC was already analyzed in 2017. Due to higher volumes of NGS tests, the turnaround times and costs per sample have decreased. Improvements of library preparation methods and optimization of sample procurement has decreased the number of samples that could not be analyzed due to low tissue volume.
Publicly available cohort databases such as the AACR project GENIE database indicate that EGFR-activating mutations are mutually exclusive with other known driver mutations [2]. However, co-occurrence of mutations in other genes may affect survival and response to EGFR-TKI [41]. Notably, the prognostic value of TP53 mutations in combination has been demonstrated [42]. Patients with concurrent EGFR and TP53 mutations may benefit from combining EGFR-TKI with anti-VEGF therapies such as bevacizumab [43], ramucirumab [44], or apatinib [45]. Because TP53 was not part of the list of genes that must be sequenced in the context of NSCLC in the Netherlands in the study period [18], most laboratories did not report mutations in these genes. As such, our results could not be stratified for co-occurrence of EGFR and TP53 mutations.
4.3. A Clinical Evidence-Driven Reclassification of EGFR Mutations
Previous reports on nationwide real-world data regarding the effect of EGFR mutation status and response to EGFR inhibitors generally lack detailed information with respect to EGFR mutation type [26,31,46,47,48]. In smaller cohorts, EGFR mutation status and response rates have been reported, but irrespective of evidence of response to first- and second-generation EGFR-TKI, often including T790M resistance mutations, exon 20 insertions and variants of unknown significance [49,50,51,52,53,54]. Furthermore, the EGFR L858R mutation and exon 19 deletions are often pooled together as ‘common’ mutations [53,55], due to their frequency but despite a difference in response in the Asian population [56]. In a recent, unpublished study, Robichaux et al. suggested four subgroups: classical-like, T790M-like, exon 20 insertions and ATP binding-pocket volume-reducing (PVR) mutations [57]. However, also in this study exon 19 deletions and L858R were pooled into the classical-like group, and included combinations between a common and an uncommon mutation in the ‘PVR’ group. In contrast, it may be more appropriate to classify variants based on their respective sensitivity to individual inhibitors in terms of survival [58]. Thus, we differentiated EGFR variants in terms of frequency and actionability with first- and second-generation EGFR-TKI. The classification scheme proposed by the 2017 AMP/ASCO/CAP guidelines [20], was used to individually classify all detected variants into EGFR “exon 19 deletion” (Tier I), “L858R” (Tier I) and into three groups of uncommon variants: “exon 20 insertions” (Tier II), “uncommon, actionable variants” including known double mutations such as S768I/L861Q and G719A/S768I (Tier I or II) and “not actionable/unknown variants” (Tier III or IV) (Figure 1 and Table 4). This classification scheme revealed distinct survival characteristics in patients treated with first-line targeted therapy.
Table 4.
Rationale for classification of expected first- and second-generation EGFR-TKI sensitivity for EGFR variants reported in the Dutch population in 2013, 2015 and 2017.
| Variant(s) | n (%) a | Rationale | Prediction | LoE | Group |
|---|---|---|---|---|---|
| R108K | 1 (0.11%) | Known gain of function in brain tumors [59], but actionability unknown in NSCLC | no benefit | III | not actionable/unknown |
| C595F | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| Q701K | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| L703F | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| R705S | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| E709_T710delinsD | 3 (0.32%) | Clinical sensitivity to EGFR-TKI reported [60,61] | sensitive | IID | uncommon, actionable |
| E709A/G719A | 4 (0.43%) | Clinical sensitivity to EGFR-TKI reported [21] | sensitive | IIC | uncommon, actionable |
| E709A/G719R | 1 (0.11%) | Considered comparable to E709A/G719A | sensitive | IIC | uncommon, actionable |
| E709A/G719S | 1 (0.11%) | Considered comparable to E709A/G719A | sensitive | IIC | uncommon, actionable |
| E709D | 1 (0.11%) | No evidence on pathogenicity or actionability. Similar amino acid properties between Glu (E) and Asp (D), thus no effect expected | no benefit | III | not actionable/unknown |
| E709K/G719S | 1 (0.11%) | Clinical sensitivity to EGFR-TKI reported [62] | sensitive | IIC | uncommon, actionable |
| I715fs* | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| G719_S720delinsAF | 1 (0.11%) | Considered comparable to G719A | sensitive | IID | uncommon, actionable |
| G719A | 18 (1.95%) | Clinical sensitivity to EGFR-TKI reported [63] | sensitive | IIC | uncommon, actionable |
| G719A/D761Y | 1 (0.11%) | Considered comparable to G719A | sensitive | IIC | uncommon, actionable |
|
G719A +
Exon 20 insertion, NOS |
1 (0.11%) | No EGFR-TKI sensitivity expected due to exon 20 insertion, grouped accordingly | no benefit | IIC | exon 20 insertion |
| G719A/L861Q | 3 (0.32%) | Clinical sensitivity to EGFR-TKI reported [64] | sensitive | IIC | uncommon, actionable |
| G719A/R776H | 1 (0.11%) | Considered comparable to G719A | sensitive | IIC | uncommon, actionable |
| G719A/S768I | 5 (0.54%) | Clinical sensitivity to EGFR-TKI reported [65] | sensitive | IIC | uncommon, actionable |
| G719A/V769M | 1 (0.11%) | Considered comparable to G719A | sensitive | IIC | uncommon, actionable |
| G719C | 2 (0.22%) | Clinical sensitivity to EGFR-TKI reported [66] | sensitive | IIC | uncommon, actionable |
| G719C/S768I | 6 (0.65%) | Clinical sensitivity to EGFR-TKI reported [67] | sensitive | IIC | uncommon, actionable |
| G719S | 1 (0.11%) | Clinical sensitivity to EGFR-TKI reported [66] | sensitive | IIC | uncommon, actionable |
| G719S/L747S | 1 (0.11%) | Considered comparable to G719S | sensitive | IIC | uncommon, actionable |
| G719S/L861Q | 1 (0.11%) | Considered comparable to G719A/L861Q | sensitive | IIC | uncommon, actionable |
| G719S/S768I | 1 (0.11%) | Considered comparable to G719A/S768I and G719C/S768I | sensitive | IIC | uncommon, actionable |
| G719X, NOS | 2 (0.22%) | Similar grouping as other G719 substitutions | sensitive | IIC | uncommon, actionable |
| G719X, NOS/S768I | 3 (0.32%) | Similar grouping as other G719X/S768I variants | sensitive | IIC | uncommon, actionable |
| G724A/S768I | 1 (0.11%) | Considered comparable to G724S/S768I | sensitive | IIC | uncommon, actionable |
| G724S/S768I | 1 (0.11%) | Clinical sensitivity to EGFR-TKI reported [68] | sensitive | IID | uncommon, actionable |
| c.2184 + 19G > A | 1 (0.11%) | Likely a SNP due to high allele frequency in general population (3.5%; GnomAD) b | no benefit | IV | not actionable/unknown |
| L730fs*1 | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| I740_K745dup | 2 (0.22%) | Clinical sensitivity to osimertinib reported [69], but no evidence on first- or second-generation TKI | no benefit | III | not actionable/unknown |
| A743S | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| I744_P753delinsSNISG | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_A750del | 210 (22.7%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_A750del/G873E | 1 (0.11%) | Considered comparable to E746_A750del | sensitive | IA | exon 19 deletion |
| E746_A750del/K754Q | 1 (0.11%) | Considered comparable to E746_A750del | sensitive | IA | exon 19 deletion |
| E746_A750del/P848L | 1 (0.11%) | Considered comparable to E746_A750del | sensitive | IA | exon 19 deletion |
| E746_A750del/V765M | 1 (0.11%) | Considered comparable to E746_A750del | sensitive | IA | exon 19 deletion |
| E746_A750delinsEP | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_A750delinsIP | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_A750dup | 1 (0.11%) | Considered comparable to I740_K745dup | no benefit | III | not actionable/unknown |
| E746_K754delinsVSR | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_L747delinsIP | 1 (0.11%) | Considered comparable to L747P | sensitive | IID | uncommon, actionable |
| E746_P753delinsANKE | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_P753delinsIS | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_P753delinsVS | 2 (0.22%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_S752delinsI | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_S752delinsV | 20 (2.16%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751del | 2 (0.22%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsA | 3 (0.32%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsAA | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsI | 2 (0.22%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsK | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsL | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsP | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsS | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| E746_T751delinsVP | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_A750delinsP | 10 (1.08%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_A750delinsP/V845L | 1 (0.11%) | Considered comparable to L747_A750delinsP | sensitive | IA | exon 19 deletion |
| L747_A755delinsSKD | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_E749del | 3 (0.32%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_K754delinsATSPE | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_K754delinsG | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_K754delinsQPN | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_P753delinsQ | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_P753delinsS | 25 (2.70%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_S752del | 7 (0.76%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_S752del/K754R | 1 (0.11%) | Considered comparable to L747_S752del | sensitive | IA | exon 19 deletion |
| L747_S752del/L777Q | 1 (0.11%) | Considered comparable to L747_S752del | sensitive | IA | exon 19 deletion |
| L747_S752delinsQ | 2 (0.22%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_T751del | 16 (1.73%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747_T751del/S768I | 1 (0.11%) | Considered comparable to L747_T751del | sensitive | IA | exon 19 deletion |
| L747_T751delinsP | 6 (0.65%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| L747P | 4 (0.43%) | Clinical sensitivity to EGFR-TKI reported [70] | sensitive | IID | uncommon, actionable |
| E749_A755delinsD | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| A750_E758delinsP | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| T751_I759delinsN | 1 (0.11%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| S752_I759del | 4 (0.43%) | Net loss of amino acids on exon 19 (deletion) | sensitive | IA | exon 19 deletion |
| P753L | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| K757R | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| A763S | 2 (0.22%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| A763_Y764insFQEA | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| V765M | 2 (0.22%) | Only reported in combination with other variants, no evidence on individual variant | no benefit | III | not actionable/unknown |
| A767_V769dup | 16 (1.73%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| A767T | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| S768_D770dup | 9 (0.97%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| S768_V769delinsIL | 4 (0.43%) | Considered comparable to S768I | sensitive | IIC | uncommon, actionable |
| S768I | 5 (0.54%) | Clinical sensitivity to EGFR-TKI reported [49] | sensitive | IIC | uncommon, actionable |
| S768I/L861Q | 2 (0.22%) | Clinical sensitivity to EGFR-TKI reported [65] | sensitive | IIC | uncommon, actionable |
| S768I/V774M | 2 (0.22%) | Considered comparable to S768I | sensitive | IIC | uncommon, actionable |
| V769L | 2 (0.22%) | Only reported in combination with other variants, no evidence on individual variant | no benefit | III | not actionable/unknown |
| V769M | 2 (0.22%) | Possible germline variant [71], no evidence sensitivity to EGFR-TKI | no benefit | III | not actionable/unknown |
| D770_H773dup | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770_N771insG | 3 (0.32%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770_N771insGF | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770_N771insSVA | 2 (0.22%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770_N771insT | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770_N771insY | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770_P772dup | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770delinsEQPP | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770delinsGY | 3 (0.32%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| D770Y | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| N771_H773dup | 2 (0.22%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| N771_P772insH | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| N771_P772insR | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| N771delinsGF | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| N771delinsGY | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| N771delinsKG | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| N771delinsTH | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| N771L | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| P772_C775dup | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| P772_H773dup | 3 (0.32%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| P772_H773insANP | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| H773_V774dup | 2 (0.22%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| H773_V774insAH | 2 (0.22%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| H773delinsYNPY | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| H773dup | 5 (0.54%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| V774delinsHC | 1 (0.11%) | Net gain of amino acids on exon 20 (insertion) | no benefit | IIC | exon 20 insertion |
| V774M | 3 (0.32%) | Only reported in combination with other variants, no evidence on individual variant | sensitive | III | not actionable/unknown |
| V774M/L861R | 1 (0.11%) | Considered comparable to L861R | sensitive | IIC | uncommon, actionable |
| C775F | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| R776H | 1 (0.11%) | Only reported in combination with other variants, no evidence on individual variant | no benefit | III | not actionable/unknown |
| R776L | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| G779F | 3 (0.32%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| G779S | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| C781F | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| Q787E | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| T790M | 1 (0.11%) | Known resistance-inducing mutation, but not transforming without a driver mutationg [72] | no benefit | III | not actionable/unknown |
| G796C | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| L799M | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| D830Y | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| L832T | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| L833V/H835L | 2 (0.22%) | Clinical sensitivity to EGFR-TKI reported [73] | sensitive | IID | uncommon, actionable |
| R836L | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| R836R | 4 (0.43%) | Likely a SNP due to high allele frequency in general population (1.7%; GnomAD) b | no benefit | IV | not actionable/unknown |
| A840T | 4 (0.43%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| P848L | 5 (0.54%) | No evidence on pathogenicity or actionability, may be a rare SNP (AF 0.03; GnomAD) b | no benefit | IV | not actionable/unknown |
| L858_K860delinsRTI | 1 (0.11%) | Considered comparable to L858R | sensitive | IID | L858R |
| L858R | 274 (29.6%) | Classical, well known activating mutation sensitive to first- and second-generation EGFR-TKI | sensitive | IA | L858R |
| L858R/A871E | 1 (0.11%) | Considered comparable to L858R | sensitive | IA | L858R |
| L858R/A871G | 1 (0.11%) | Considered comparable to L858R | sensitive | IA | L858R |
| L858R/E709G | 1 (0.11%) | Considered comparable to L858R | sensitive | IA | L858R |
| L858R/L718M | 1 (0.11%) | Considered comparable to L858R | sensitive | IA | L858R |
| L858R/L747V | 1 (0.11%) | Considered comparable to L858R | sensitive | IA | L858R |
| L858R/R776H | 2 (0.22%) | Considered comparable to L858R | sensitive | IA | L858R |
| L858R/S768I | 4 (0.43%) | Considered comparable to L858R | sensitive | IA | L858R |
| L858R/V834L | 1 (0.11%) | Considered comparable to L858R | sensitive | IA | L858R |
| A859T | 1 (0.11%) | Clinical sensitivity to EGFR-TKI reported [74] | sensitive | IID | uncommon, actionable |
| L861Q | 19 (2.05%) | Clinical sensitivity to EGFR-TKI reported [75] | sensitive | IIC | uncommon, actionable |
| L861R | 3 (0.32%) | Clinical sensitivity to EGFR-TKI reported [75] | sensitive | IIC | uncommon, actionable |
| G863D | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| A864T | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| A864V | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| E866K | 1 (0.11%) | No evidence on pathogenicity or actionability | no benefit | III | not actionable/unknown |
| EGFR variant, NOS | 4 (0.43%) | Exact variant and exon not reported | unknown | NA | unspecified |
| Exon 18 variant, NOS + Exon 20 variant, NOS | 2 (0.22%) | Exact variant not reported | unknown | NA | unspecified |
| Exon 19 deletion, NOS | 52 (5.62%) | Exact variant not reported but confirmed net loss of amino acids on exon 19 (deletion) | unknown | NA | exon 19 deletion |
| Exon 19 variant, NOS | 3 (0.32%) | Exact variant not reported (could be a deletion or a single-nucleotide variant) | sensitive | IA | unspecified |
| Exon 20 insertion, NOS | 3 (0.32%) | Exact variant not reported but confirmed net gain of amino acids on exon 21 (insertion) | unknown | NA | exon 20 insertion |
| Exon 20 SNV (silent) | 1 (0.11%) | Silent mutation, no change in amino acids | no benefit | IIC | not actionable/unknown |
| Exon 21 variant, NOS | 3 (0.32%) | Exact variant not reported (could be L858R or a different, non-classical mutation) | no benefit | IV | unspecified |
a Amount of tumors reported to harbor this mutation, with percentage of all EGFR-mutant tumors between brackets. b According to GnomAD [76], query on 18 December 2020. Abbreviations: AF, allele frequency; DOI, digital object identifier; EGFR, epidermal growth factor receptor; LoE, AMP/ASCO/CAP Level of evidence for actionability [20]; NOS, not otherwise specified; NSCLC, non-small cell lung cancer; SNP, single nucleotide polymorphism; TKI, tyrosine kinase inhibitor.
Patients with an EGFR exon 19 deletion, L858R or uncommon actionable mutation benefit from first- and second-generation EGFR inhibitors, albeit to a different extent (median OS 95% CI: 20.8–26.7, 14.1–19.3, 9.4–15.1 months, respectively). OS for patients with EGFR exon 20 insertions and not actionable/unknown variants was comparable to patients without an EGFR mutation who were not treated with an inhibitor. The current study confirmed the superior OS for patients with an EGFR exon 19 deletions compared the L858R mutation (adjusted HR of L858R versus exon 19 deletion was 1.57; p < 0.001) [56]. When corrected for covariates such as age, sex and year of diagnosis, the difference in OS for patients with an EGFR L858R or uncommon actionable mutation was not significant (HR uncommon, EGFR actionable variants versus L858R: 1.311; p = 0.069). It should be noted that the majority of these patients were treated with first-generation EGFR-TKIs gefitinib or erlotinib [26], which may be less effective in patients with uncommon EGFR mutations than afatinib [49,54], or osimertinib [77]. Afatinib was likely used in a minority of patients [26], and osimertinib was not available for first-line treatment in the study period. Thus, survival rates for L858R and uncommon, actionable variants may be even more similar with newer agents but has to be investigated. Collectively, these results reinforce the notion that future clinical trials need to avoid pooling EGFR L858R and exon 19 deletions into a single category, and warrants further investigation into the similarities between L858R and ‘uncommon, actionable’ variants.
Both patients with EGFR exon 20 insertions and patients with variants classified as ‘not actionable/unknown’ showed dismal survival when treated with first- or second-generation EGFR-TKI in first-line. Both categories were considered as ‘resistant’ to first- and second-generation EGFR-TKI: the former category because these are known to be non-responsive to first- and second-generation TKI [78], and the latter because (pre)clinical evidence on actionability is lacking. However, these perspectives might change with new evidence: for example, there are promising novel agents and combination therapies currently under investigation that may be effective against EGFR exon 20 insertions, including amivantamab [79], poziotinib [80], and mobocertinib [81], or combining osimertinib or afatinib with cetuximab [82]. In the Netherlands, all Tier II and III EGFR variants are nowadays commonly discussed in Molecular Tumor Boards [83], resulting in favorable treatment outcomes [61].
4.4. Limitations of the Study
Although the PALGA registry offers nationwide coverage on pathology testing in the Netherlands, the collected information in the study period represented narrative pathology reports in routine diagnostics. Thus, if patients were tested for EGFR mutations for research purposes or if the pathologist in charge did not include testing results in the pathology report, patients may have been missed and the testing rate may be underestimated.
Similarly, the NCR, which also has nationwide coverage and therefore allows for unique real-world analyses, has several limitations. First, it does not include treatment-specific information such as best overall response or progression-free survival and is limited to first-line treatment. Second, the NCR did not register reasons for not treating patients with targeted therapy; thus, we could not investigate why circa 20% of patients with common EGFR mutations were not treated with EGFR-targeted therapy. Finally, information on which EGFR-TKI was specifically used for each patient was not provided, but was restricted to first- (gefitinib, erlotinib) and second-generation (afatinib) inhibitors that could have been used in the period 2013–2017. In 2017, osimertinib became available which has made a positive impact on overall survival. Due to the limitations of the NCR database, this impact could not be quantified. The clinical impact of currently available third-generation inhibitors on EGFR variants in second-line testing and multiple lines of treatment was therefore not the subject of this study.
Despite the increase in the use of NGS, the current study did not capture mutations in genes other than EGFR. As such, frequencies of other driver mutations are not reported and a complete landscape of mutations in the known drivers is not presented.
5. Conclusions
In the period 2013–2017, EGFR mutation testing in the Netherlands has transformed from a single-gene approach to the nationwide implementation of NGS using a multigene panel for predictive biomarker testing including EGFR according to the current Dutch national guideline for lung cancer [10]. This shift did not affect the overall detection rate of EGFR mutations (11.7%) nor the distribution of mutations. In patients treated with first-line targeted therapy, EGFR exon 19 deletions, L858R, and uncommon, actionable variants were all independently associated with improved survival compared to patients with EGFR exon 20 insertions or not actionable/unknown variants. Furthermore, we demonstrated that overall survival in patients with L858R is more comparable to those with uncommon, actionable mutations than to those with EGFR exon 19 deletions. These results indicate that distinguishing between EGFR exon 19 deletions and L858R mutations, as well as classifying actionability of uncommon variants using the 2017 AMP/ASCO/CAP guidelines, is a more appropriate method to predict actionability and survival than the grouping of mutations into ‘common’ and ‘uncommon’ variants based on their frequencies in the patient population.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13143641/s1, Figure S1: Inter-pathology department variance in EGFR testing request rates, Figure S2: Changes in percentage of patients treated with targeted therapy in first-line of treatment.
Author Contributions
Conceptualization, B.K., H.J.M.G., E.S., S.M.W. and L.C.v.K.; Data curation, B.K., B.N.C.G. and C.C.H.J.K.; Formal analysis, B.K.; Funding acquisition, S.M.W.; Methodology, B.K., H.J.M.G., E.S., S.M.W. and L.C.v.K.; Project administration, L.C.v.K.; Resources, C.C.H.J.K., R.A.M.D. and A.J.v.d.W.; Supervision, H.J.M.G., E.S., S.M.W. and L.C.v.K.; Validation, L.C.v.K.; Visualization, B.K.; Writing—original draft, B.K. and L.C.v.K.; Writing—review and editing, B.N.C.G., C.C.H.J.K., R.A.M.D., A.J.v.d.W., H.J.M.G., E.S. and S.M.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by AstraZeneca [grant number AZNL202000093], but had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and data requests were approved by the scientific and privacy committees of IKNL (application numbers K15.115, K16.264, and K18.311) and PALGA (application numbers LZV1172, LZV2016-91, LZV2018-199).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
A.J.v.d.W. reports grants from AstraZeneca, Boehringer-Ingelheim, Pfizer, Roche and Takeda, and reports advisory board presence from AstraZeneca, Bayer, Boehringer-Ingelheim, Pfizer, Roche and Takeda, all outside the submitted work and money to UMCG. H.J.M.G. reports advisory board presence from Novartis and Eli Lilly, outside the submitted work. E.S. reports lectures for Bio-Rad, Novartis, Roche, Biocartis, Illumina, Lilly, Pfizer, AstraZeneca, and Agena Bioscience, is consultant in advisory boards for AstraZeneca, Roche, Pfizer, Novartis, Bayer, Lilly, BMS, Amgen, Biocartis, Illumina, Agena Bioscience and MSD/Merck, and received research grants from Pfizer, Biocartis, Invitae-ArcherDX, AstraZeneca, Agena Bioscience, BMS, Bio-Rad, Roche, Boehringer Ingelheim. S.M.W reports grants from Roche, Pfizer, Bayer, MSD, AstraZeneca and Amgen, outside the submitted work. L.C.v.K. reports grants, non-financial support from Roche, advisory board presence for AstraZeneca, Novartis, Merck, Janssen-Cilag, Bayer, BMS, nanoString and Pfizer, grants and non-financial support from Invitae, non-financial support from Biocartis, grants from Bayer, non-financial support from nanoString, outside the submitted work. All remaining authors (B.K., B.N.C.G., C.C.H.J.K., R.A.M.D.) have declared no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.AACR Project GENIE Consortium AACR Project GENIE: Powering precision medicine through an international consortium. Cancer Discov. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sequist L.V., Martins R.G., Spigel D., Grunberg S.M., Spira A., Jänne P.A., Joshi V.A., McCollum D., Evans T.L., Muzikansky A., et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J. Clin. Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y.-L., Zhou C., Liam C.-K., Wu G., Liu X., Zhong Z., Lu S., Cheng Y., Han B., Chen L., et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: Analyses from the phase III, randomized, open-label, ENSURE study. Ann. Oncol. 2015;26:1883–1889. doi: 10.1093/annonc/mdv270. [DOI] [PubMed] [Google Scholar]
- 5.Park K., Tan E.-H., O’Byrne K., Zhang L., Boyer M., Mok T., Hirsh V., Yang J.C.-H., Lee K.H., Lu S., et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): A phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17:577–589. doi: 10.1016/S1470-2045(16)30033-X. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y.-L., Cheng Y., Zhou X., Lee K.H., Nakagawa K., Niho S., Tsuji F., Linke R., Rosell R., Corral J., et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18:1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 7.Soria J.-C., Ohe Y., Vansteenkiste J., Reungwetwattana T., Chewaskulyong B., Lee K.H., Dechaphunkul A., Imamura F., Nogami N., Kurata T., et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 8.D’Addario G., Früh M., Reck M., Baumann P., Klepetko W., Felip E., ESMO Guidelines Working Group Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010;21(Suppl. 5):v116–v119. doi: 10.1093/annonc/mdq189. [DOI] [PubMed] [Google Scholar]
- 9.Planchard D., Popat S., Kerr K., Novello S., Smit E.F., Faivre-Finn C., Mok T.S., Reck M., van Schil P.E., Hellmann M.D., et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018;29:iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 10.Hendriks L.E.L., Dingemans A.-M.C., de Ruysscher D.K.M., Aarts M.J., Barberio L., Cornelissen R., Hartemink K.J., van den Heuvel M., Schuuring E., Smit H.J.M., et al. Lung cancer in the Netherlands. J. Thorac. Oncol. 2021;16:355–365. doi: 10.1016/j.jtho.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin M.-T., Mosier S.L., Thiess M., Beierl K.F., Debeljak M., Tseng L.-H., Chen G., Yegnasubramanian S., Ho H., Cope L., et al. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am. J. Clin. Pathol. 2014;141:856–866. doi: 10.1309/AJCPMWGWGO34EGOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planchard D., Besse B., Groen H.J.M., Souquet P.-J., Quoix E., Baik C.S., Barlesi F., Kim T.M., Mazieres J., Novello S., et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: An open-label, multicentre phase 2 trial. Lancet Oncol. 2016;17:984–993. doi: 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf J., Seto T., Han J.-Y., Reguart N., Garon E.B., Groen H.J.M., Tan D.S.W., Hida T., de Jonge M., Orlov S.V., et al. Capmatinib in MET Exon 14-mutated or MET-amplified non-small-cell lung cancer. N. Engl. J. Med. 2020;383:944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 15.Li B.T., Shen R., Buonocore D., Olah Z.T., Ni A., Ginsberg M.S., Ulaner G.A., Offin M., Feldman D., Hembrough T., et al. Ado-trastuzumab emtansine for patients with HER2-mutant lung cancers: Results from a phase II basket trial. J. Clin. Oncol. 2018;36:2532–2537. doi: 10.1200/JCO.2018.77.9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.UMCG Pathologie en Medische Biologie Moleculaire Diagnostiek. [(accessed on 19 May 2020)]; Available online: https://www.umcg.nl/NL/UMCG/Afdelingen/Pathologie/Professionals/moleculaire-diagnostiek/Paginas/default.aspx.
- 17.Casparie M., Tiebosch A.T.M.G., Burger G., Blauwgeers H., van de Pol A., van Krieken J.H.J.M., Meijer G.A. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell. Oncol. 2007;29:19–24. doi: 10.1155/2007/971816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NVALT Niet-Kleincellig Longcarcinoom: Landelijke Richtlijn. [(accessed on 8 July 2021)]; Available online: https://richtlijnendatabase.nl/richtlijn/niet_kleincellig_longcarcinoom/startpagina_-_niet-kleincelling_longcarcinoom.html.
- 19.Den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.-F., Smith T., Antonarakis S.E., Taschner P.E.M. HGVS Recommendations for the description of sequence variants: 2016 update. Hum. Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 20.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heigener D.F., Schumann C., Sebastian M., Sadjadian P., Stehle I., Märten A., Lüers A., Griesinger F., Scheffler M., Abdollahi A., et al. Afatinib in non-small cell lung cancer harboring uncommon EGFR mutations pretreated with reversible EGFR inhibitors. Oncologist. 2015;20:1167–1174. doi: 10.1634/theoncologist.2015-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deans Z.C., Costa J.L., Cree I., Dequeker E., Edsjö A., Henderson S., Hummel M., Ligtenberg M.J., Loddo M., Machado J.C., et al. Integration of next-generation sequencing in clinical diagnostic molecular pathology laboratories for analysis of solid tumours; an expert opinion on behalf of IQN Path ASBL. Virchows Arch. 2017;470:5–20. doi: 10.1007/s00428-016-2025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boonstra P.A., ter Elst A., Tibbesma M., Bosman L.J., Mathijssen R., Atrafi F., van Coevorden F., Steeghs N., Farag S., Gelderblom H., et al. A single digital droplet PCR assay to detect multiple KIT exon 11 mutations in tumor and plasma from patients with gastrointestinal stromal tumors. Oncotarget. 2018;9:13870–13883. doi: 10.18632/oncotarget.24493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sie D., Snijders P.J.F., Meijer G.A., Doeleman M.W., van Moorsel M.I.H., van Essen H.F., Eijk P.P., Grünberg K., van Grieken N.C.T., Thunnissen E., et al. Performance of amplicon-based next generation DNA sequencing for diagnostic gene mutation profiling in oncopathology. Cell. Oncol. 2014;37:353–361. doi: 10.1007/s13402-014-0196-2. [DOI] [PubMed] [Google Scholar]
- 25.Steeghs E.M.P., Kroeze L.I., Tops B.B.J., van Kempen L.C., ter Elst A., Kastner-van Raaij A.W.M., Hendriks-Cornelissen S.J.B., Hermsen M.J.W., Jansen E.A.M., Nederlof P.M., et al. Comprehensive routine diagnostic screening to identify predictive mutations, gene amplifications, and microsatellite instability in FFPE tumor material. BMC Cancer. 2020;20:291. doi: 10.1186/s12885-020-06785-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gijtenbeek R.G.P., Damhuis R.A.M., Groen H.J.M., van der Wekken A.J., van Geffen W.H. Nationwide real-world cohort study of first-line tyrosine kinase inhibitor treatment in epidermal growth factor receptor-mutated non-small-cell lung cancer. Clin. Lung Cancer. 2020;21:e647–e653. doi: 10.1016/j.cllc.2020.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Peters S., Adjei A.A., Gridelli C., Reck M., Kerr K., Felip E., ESMO Guidelines Working Group Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2012;23(Suppl. 7):vii56–vii64. doi: 10.1093/annonc/mds226. [DOI] [PubMed] [Google Scholar]
- 28.Smits A.J.J., Kummer J.A., Hinrichs J.W.J., Herder G.J.M., Scheidel-Jacobse K.C., Jiwa N.M., Ruijter T.E.G., Nooijen P.T.G.A., Looijen-Salamon M.G., Ligtenberg M.J.L., et al. EGFR and KRAS mutations in lung carcinomas in the Dutch population: Increased EGFR mutation frequency in malignant pleural effusion of lung adenocarcinoma. Cell. Oncol. 2012;35:189–196. doi: 10.1007/s13402-012-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y.-L., Yuan J.-Q., Wang K.-F., Fu X.-H., Han X.-R., Threapleton D., Yang Z.-Y., Mao C., Tang J.-L. The prevalence of EGFR mutation in patients with non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget. 2016;7:78985–78993. doi: 10.18632/oncotarget.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brindel A., Althakfi W., Barritault M., Bringuier P.-P., Watkin E., Maury J.-M., Girard N., Brevet M. Uncommon EGFR mutations in lung adenocarcinomas: Clinical features and response to tyrosine kinase inhibitors. Ann. Oncol. 2018;29:viii747–viii748. doi: 10.1093/annonc/mdy424.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leduc C., Merlio J.P., Besse B., Blons H., Debieuvre D., Bringuier P.P., Monnet I., Rouquette I., Fraboulet-Moreau S., Lemoine A., et al. Clinical and molecular characteristics of non-small-cell lung cancer (NSCLC) harboring EGFR mutation: Results of the nationwide French Cooperative Thoracic Intergroup (IFCT) program. Ann. Oncol. 2017;28:2715–2724. doi: 10.1093/annonc/mdx404. [DOI] [PubMed] [Google Scholar]
- 32.Yun C.-H., Mengwasser K.E., Toms A.V., Woo M.S., Greulich H., Wong K.-K., Meyerson M., Eck M.J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross D.A.E., Ashton S.E., Ghiorghiu S., Eberlein C., Nebhan C.A., Spitzler P.J., Orme J.P., Finlay M.R.V., Ward R.A., Mellor M.J., et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reck M., Rodríguez-Abreu D., Robinson A.G., Hui R., Csőszi T., Fülöp A., Gottfried M., Peled N., Tafreshi A., Cuffe S., et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 35.Lee C.K., Man J., Lord S., Links M., Gebski V., Mok T., Yang J.C.-H. Checkpoint inhibitors in metastatic EGFR-mutated non-small cell lung cancer—A meta-analysis. J. Thorac. Oncol. 2017;12:403–407. doi: 10.1016/j.jtho.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Kwak E.L., Bang Y.-J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., Ou S.-H.I., Dezube B.J., Jänne P.A., Costa D.B., et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drilon A., Siena S., Ou S.-H.I., Patel M., Ahn M.J., Lee J., Bauer T.M., Farago A.F., Wheler J.J., Liu S.V., et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: Combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7:400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drilon A., Oxnard G.R., Tan D.S.W., Loong H.H.F., Johnson M., Gainor J., McCoach C.E., Gautschi O., Besse B., Cho B.C., et al. Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. N. Engl. J. Med. 2020;383:813–824. doi: 10.1056/NEJMoa2005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw A.T., Riely G.J., Bang Y.-J., Kim D.-W., Camidge D.R., Solomon B.J., Varella-Garcia M., Iafrate A.J., Shapiro G.I., Usari T., et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): Updated results, including overall survival, from PROFILE 1001. Ann. Oncol. 2019 doi: 10.1093/annonc/mdz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burns T.F., Borghaei H., Ramalingam S.S., Mok T.S., Peters S. Targeting KRAS-mutant non-small-cell lung cancer: One mutation at a time, with a focus on KRAS G12C mutations. J. Clin. Oncol. 2020;38:4208–4218. doi: 10.1200/JCO.20.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gristina V., la Mantia M., Galvano A., Cutaia S., Barraco N., Castiglia M., Perez A., Bono M., Iacono F., Greco M., et al. Non-small cell lung cancer harboring concurrent EGFR genomic alterations: A systematic review and critical appraisal of the double dilemma. J. Mol. Pathol. 2021;2:173–196. doi: 10.3390/jmp2020016. [DOI] [Google Scholar]
- 42.Canale M., Petracci E., Delmonte A., Chiadini E., Dazzi C., Papi M., Capelli L., Casanova C., de Luigi N., Mariotti M., et al. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin. Cancer Res. 2017;23:2195–2202. doi: 10.1158/1078-0432.CCR-16-0966. [DOI] [PubMed] [Google Scholar]
- 43.Horinouchi H. To combine or not to combine: Anti-vascular endothelial growth factor therapies in EGFR mutation positive non-small cell lung cancer. Ann. Transl. Med. 2020;8:554. doi: 10.21037/atm.2020.01.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa K., Garon E.B., Seto T., Nishio M., Ponce Aix S., Paz-Ares L., Chiu C.-H., Park K., Novello S., Nadal E., et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20:1655–1669. doi: 10.1016/S1470-2045(19)30634-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhao H., Yao W., Min X., Gu K., Yu G., Zhang Z., Cui J., Miao L., Zhang L., Yuan X., et al. Apatinib plus gefitinib as first-line treatment in advanced EGFR-mutant NSCLC: The phase III ACTIVE study (CTONG1706) J. Thorac. Oncol. 2021 doi: 10.1016/j.jtho.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Suda K., Mitsudomi T., Shintani Y., Okami J., Ito H., Ohtsuka T., Toyooka S., Mori T., Watanabe S.-I., Asamura H., et al. Clinical impacts of EGFR mutation status: Analysis of 5780 surgically resected lung cancer cases. Ann. Thorac. Surg. 2021;111:269–276. doi: 10.1016/j.athoracsur.2020.05.041. [DOI] [PubMed] [Google Scholar]
- 47.Bergqvist M., Christensen H.N., Wiklund F., Bergström S. Real world utilization of EGFR TKIs and prognostic factors for survival in NSCLC during 2010–2016 in Sweden: A nationwide observational study. Int. J. Cancer. 2020;146:2510–2517. doi: 10.1002/ijc.32596. [DOI] [PubMed] [Google Scholar]
- 48.Aye P.S., McKeage M.J., Tin Tin S., Khwaounjoo P., Elwood J.M. Factors associated with overall survival in a population-based cohort of non- squamous NSCLC patients from northern New Zealand: A comparative analysis by EGFR mutation status. Cancer Epidemiol. 2020;69:101847. doi: 10.1016/j.canep.2020.101847. [DOI] [PubMed] [Google Scholar]
- 49.Yang J.C.-H., Sequist L.V., Geater S.L., Tsai C.-M., Mok T.S.K., Schuler M., Yamamoto N., Yu C.-J., Ou S.-H.I., Zhou C., et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 50.Passaro A., Prelaj A., Bonanno L., Tiseo M., Tuzi A., Proto C., Chiari R., Rocco D., Genova C., Sini C., et al. Activity of EGFR TKIs in Caucasian patients with NSCLC harboring potentially sensitive uncommon EGFR mutations. Clin. Lung Cancer. 2019;20:e186–e194. doi: 10.1016/j.cllc.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Pilotto S., Rossi A., Vavalà T., Follador A., Tiseo M., Galetta D., Morabito A., di Maio M., Martelli O., Caffo O., et al. Outcomes of first-generation EGFR-TKIs against non-small-cell lung cancer harboring uncommon EGFR mutations: A post hoc analysis of the BE-POSITIVE study. Clin. Lung Cancer. 2018;19:93–104. doi: 10.1016/j.cllc.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Yasuda H., Park E., Yun C.-H., Sng N.J., Lucena-Araujo A.R., Yeo W.-L., Huberman M.S., Cohen D.W., Nakayama S., Ishioka K., et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci. Transl. Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arrieta O., Cardona A.F., Corrales L., Campos-Parra A.D., Sánchez-Reyes R., Amieva-Rivera E., Rodríguez J., Vargas C., Carranza H., Otero J., et al. The impact of common and rare EGFR mutations in response to EGFR tyrosine kinase inhibitors and platinum-based chemotherapy in patients with non-small cell lung cancer. Lung Cancer. 2015;87:169–175. doi: 10.1016/j.lungcan.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Chang L.-C., Lim C.-K., Chang L.-Y., Chen K.-Y., Shih J.-Y., Yu C.-J. Non-small cell lung cancer harbouring non-resistant uncommon EGFR mutations: Mutation patterns, effectiveness of epidermal growth factor receptor-tyrosine kinase inhibitors and prognostic factors. Eur. J. Cancer. 2019;119:77–86. doi: 10.1016/j.ejca.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 55.Shi J., Yang H., Jiang T., Li X., Zhao C., Zhang L., Zhao S., Liu X., Jia Y., Wang Y., et al. Uncommon EGFR mutations in a cohort of Chinese NSCLC patients and outcomes of first-line EGFR-TKIs and platinum-based chemotherapy. Chin. J. Cancer Res. 2017;29:543–552. doi: 10.21147/j.issn.1000-9604.2017.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackman D.M., Yeap B.Y., Sequist L.V., Lindeman N., Holmes A.J., Joshi V.A., Bell D.W., Huberman M.S., Halmos B., Rabin M.S., et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin. Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 57.IASCLC Deeper Understanding of EGFR Mutation Subgroups Will Further Personalized Treatment for NSCLC. [(accessed on 16 February 2021)]; Available online: https://www.iaslc.org/iaslc-news/ilcn/deeper-understanding-egfr-mutation-subgroups-will-further-personalize-treatment.
- 58.Gristina V., Malapelle U., Galvano A., Pisapia P., Pepe F., Rolfo C., Tortorici S., Bazan V., Troncone G., Russo A. The significance of epidermal growth factor receptor uncommon mutations in non-small cell lung cancer: A systematic review and critical appraisal. Cancer Treat. Rev. 2020;85:101994. doi: 10.1016/j.ctrv.2020.101994. [DOI] [PubMed] [Google Scholar]
- 59.Lee J.C., Vivanco I., Beroukhim R., Huang J.H.Y., Feng W.L., DeBiasi R.M., Yoshimoto K., King J.C., Nghiemphu P., Yuza Y., et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ackerman A., Goldstein M.A., Kobayashi S., Costa D.B. EGFR delE709_T710insD: A rare but potentially EGFR inhibitor responsive mutation in non–small-cell lung cancer. J. Thorac. Oncol. 2012;7:e19–e20. doi: 10.1097/JTO.0b013e3182635ab4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koopman B., van der Wekken A.J., ter Elst A., Hiltermann T.J.N., Vilacha J.F., Groves M.R., van den Berg A., Hiddinga B.I., Hijmering-Kappelle L.B.M., Stigt J.A., et al. Relevance and effectiveness of molecular tumor board recommendations for patients with non–small-cell lung cancer with rare or complex mutational profiles. JCO Precis. Oncol. 2020;4:393–410. doi: 10.1200/PO.20.00008. [DOI] [PubMed] [Google Scholar]
- 62.Cappuzzo F., Soo R., Hochmair M., Schuler M., Lam K.C., Stehle G., Cseh A., Lorence R.M., Linden S., Forman N.D., et al. Global named patient use program of afatinib in advanced non-small-cell lung carcinoma patients who progressed following prior therapies. Future Oncol. 2018;14:1477–1486. doi: 10.2217/fon-2017-0666. [DOI] [PubMed] [Google Scholar]
- 63.Sehgal K., Rangachari D., VanderLaan P.A., Kobayashi S.S., Costa D.B. clinical benefit of tyrosine kinase inhibitors in advanced lung cancer with EGFR -G719A and other uncommon EGFR mutations. Oncologist. 2021;26:281–287. doi: 10.1002/onco.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka I., Morise M., Kodama Y., Matsui A., Ozawa N., Ozone S., Goto D., Miyazawa A., Hase T., Hashimoto N., et al. Potential for afatinib as an optimal treatment for advanced non-small cell lung carcinoma in patients with uncommon EGFR mutations. Lung Cancer. 2019;127:169–171. doi: 10.1016/j.lungcan.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 65.Liang S.-K., Hsieh M.-S., Lee M.-R., Keng L.-T., Ko J.-C., Shih J.-Y. Real-world experience of afatinib as a first-line therapy for advanced EGFR mutation-positive lung adenocarcinoma. Oncotarget. 2017;8:90430–90443. doi: 10.18632/oncotarget.19563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brueckl W.M., Reck M., Schäfer H., Kortsik C., Gaska T., Rawluk J., Krüger S., Kokowski K., Budweiser S., Hoffmann C., et al. Efficacy of afatinib in the clinical practice: Final results of the GIDEON study in EGFR mutated non-small cell lung cancer (NSCLC) in Germany. J. Clin. Oncol. 2020;38:e21636. doi: 10.1200/JCO.2020.38.15_suppl.e21636. [DOI] [Google Scholar]
- 67.Caliman E., Petreni P., Brugia M., Antonuzzo L., Mazzoni F. In regard to “Activity of EGFR TKIs in Caucasian Patients with NSCLC harbouring potentially sensitive uncommon EGFR mutations”. Clin. Lung Cancer. 2020;21:e363–e365. doi: 10.1016/j.cllc.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 68.Zhang C., Lin L., Zuo R., Wang Y., Chen P. Response to tyrosine kinase inhibitors in lung adenocarcinoma with the rare epidermal growth factor receptor mutation S768I and G724S: A case report and literature review. Thorac. Cancer. 2020;11:2743–2748. doi: 10.1111/1759-7714.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu J., Jiang Q., Xu H., Liu A., Huang L. Two patients having NSCLC with novel duplication mutation in their EGFR gene (p.I740_K745dupIPVAIK) and their response to osimertinib. J. Thorac. Oncol. 2020;15:e49–e51. doi: 10.1016/j.jtho.2019.11.026. [DOI] [PubMed] [Google Scholar]
- 70.Liang S.-K., Ko J.-C., Yang J.C.-H., Shih J.-Y. Afatinib is effective in the treatment of lung adenocarcinoma with uncommon EGFR p.L747P and p.L747S mutations. Lung Cancer. 2019;133:103–109. doi: 10.1016/j.lungcan.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 71.Hellmann M.D., Hayashi T., Reva B., Yu H.A., Riely G.J., Adusumilli P.S., Travis W.D., Wilkins O., Bramletta N., Chandramohan R., et al. Identification and functional characterization of EGFR V769M, a novel germline variant associated with multiple lung adenocarcinomas. JCO Precis. Oncol. 2017:1–10. doi: 10.1200/PO.16.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huo R., Li J., Li X., Shi J., Wang K., Jiao J., Shang Y. Significant benefits of osimertinib against adenosquamous carcinoma harboring germline T790M mutation. Oncologist. 2020;25:826–832. doi: 10.1634/theoncologist.2019-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Long X., Qin T., Lin J. Great efficacy of afatinib in a patient with lung adenocarcinoma harboring EGFR L833V/H835L mutations: A case report. OncoTargets Ther. 2020;13:10689–10692. doi: 10.2147/OTT.S260157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le Maignan L., Mirebeau-Prunier D., Vervueren L., Jeanfaivre T., Urban T., Hureaux J. First case of A859T epidermal growth factor receptor mutation responding to erlotinib. J. Thorac. Oncol. 2011;6:639–640. doi: 10.1097/JTO.0b013e3182037c0c. [DOI] [PubMed] [Google Scholar]
- 75.Wu J.-Y., Yu C.-J., Chang Y.-C., Yang C.-H., Shih J.-Y., Yang P.-C. Effectiveness of Tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non–small cell lung cancer. Clin. Cancer Res. 2011;17:3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 76.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cho J.H., Lim S.H., An H.J., Kim K.H., Park K.U., Kang E.J., Choi Y.H., Ahn M.S., Lee M.H., Sun J.-M., et al. Osimertinib for patients with non-small-cell lung cancer harboring uncommon EGFR mutations: A multicenter, open-label, phase II trial (KCSG-LU15-09) J. Clin. Oncol. 2020;38:488–495. doi: 10.1200/JCO.19.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yasuda H., Kobayashi S., Costa D.B. EGFR exon 20 insertion mutations in non-small-cell lung cancer: Preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 79.Yun J., Lee S.-H., Kim S.-Y., Jeong S.-Y., Kim J.-H., Pyo K.-H., Park C.-W., Heo S.G., Yun M.R., Lim S., et al. Antitumor activity of amivantamab (JNJ-61186372), an EGFR-MET bispecific antibody, in diverse models of EGFR exon 20 insertion-driven NSCLC. Cancer Discov. 2020;10:1194–1209. doi: 10.1158/2159-8290.CD-20-0116. [DOI] [PubMed] [Google Scholar]
- 80.Robichaux J.P., Elamin Y.Y., Tan Z., Carter B.W., Zhang S., Liu S., Li S., Chen T., Poteete A., Estrada-Bernal A., et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat. Med. 2018;24:638–646. doi: 10.1038/s41591-018-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Riely G.J., Neal J.W., Camidge D.R., Spira A.I., Piotrowska Z., Costa D.B., Tsao A.S., Patel J.D., Gadgeel S.M., Bazhenova L., et al. Activity and safety of mobocertinib (TAK-788) in previously treated non-small cell lung cancer with EGFR exon 20 insertion mutations from a phase 1/2 trial. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hasegawa H., Yasuda H., Hamamoto J., Masuzawa K., Tani T., Nukaga S., Hirano T., Kobayashi K., Manabe T., Terai H., et al. Efficacy of afatinib or osimertinib plus cetuximab combination therapy for non-small-cell lung cancer with EGFR exon 20 insertion mutations. Lung Cancer. 2019;127:146–152. doi: 10.1016/j.lungcan.2018.11.039. [DOI] [PubMed] [Google Scholar]
- 83.Koopman B., Groen H.J.M., Ligtenberg M.J.L., Grünberg K., Monkhorst K., Langen A.J., Boelens M.C., Paats M.S., Thüsen J.H., Dinjens W.N.M., et al. Multicenter comparison of molecular tumor boards in The Netherlands: Definition, composition, methods, and targeted therapy recommendations. Oncologist. 2020 doi: 10.1002/onco.13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.