Significance
The angiosperm life cycle has alternating diploid (sporophyte) and haploid (gametophyte) generations. The sporophyte generation begins with fertilization of haploid gametes and the gametophyte generation begins after meiosis. In Arabidopsis, the DEMETER (DME) DNA demethylase is essential for reproduction and is expressed in the central cell and vegetative cell of the female and male gametophyte, respectively. Little is known about DME function in the sporophyte. We show that DME activity is required for sporophyte development—seed germination, root hair growth, and cellular proliferation and differentiation during development—and we identify sporophytic genes whose proper expression requires DME activity. Together, our study provides important clues about the genetic circuits regulated by the DME DNA demethylase that control Arabidopsis sporophyte development.
Keywords: DNA demethylation, sprorophytic development, cell proliferation, pluripotency
Abstract
The flowering plant life cycle consists of alternating haploid (gametophyte) and diploid (sporophyte) generations, where the sporophytic generation begins with fertilization of haploid gametes. In Arabidopsis, genome-wide DNA demethylation is required for normal development, catalyzed by the DEMETER (DME) DNA demethylase in the gamete companion cells of male and female gametophytes. In the sporophyte, postembryonic growth and development are largely dependent on the activity of numerous stem cell niches, or meristems. Analyzing Arabidopsis plants homozygous for a loss-of-function dme-2 allele, we show that DME influences many aspects of sporophytic growth and development. dme-2 mutants exhibited delayed seed germination, variable root hair growth, aberrant cellular proliferation and differentiation followed by enhanced de novo shoot formation, dysregulation of root quiescence and stomatal precursor cells, and inflorescence meristem (IM) resurrection. We also show that sporophytic DME activity exerts a profound effect on the transcriptome of developing Arabidopsis plants, including discrete groups of regulatory genes that are misregulated in dme-2 mutant tissues, allowing us to potentially link phenotypes to changes in specific gene expression pathways. These results show that DME plays a key role in sporophytic development and suggest that DME-mediated active DNA demethylation may be involved in the maintenance of stem cell activities during the sporophytic life cycle in Arabidopsis.
The land plant life cycle alternates between two generations, the haploid gametophyte and the diploid sporophyte (1). The DEMETER (DME) DNA glycosylase is required for development of male and female gametophytic generations (2–4). DME is expressed in the vegetative and central cells of the male and female gametophytes, respectively (2), where it demethylates small transposons and edges of long transposons, excising 5-methylcytosine and demethylating DNA via the base excision repair pathway (3–7). In the female gametophyte, this process regulates adjacent gene expression and initiates gene imprinting in the endosperm (8), whereby a mutant maternal dme allele causes seed abortion (2). In the male gametophyte, a mutant paternal dme allele causes altered gene expression and reduces pollen tube germination in certain ecotypes (3).
The sporophytic generation begins when sperm fertilize the haploid egg and homodiploid central cell in the female gametophyte. The fertilized egg develops into a diploid embryo with a vegetative axis, cotyledon leaves, and shoot and root apical meristems (SAM/RAMs). The fertilized central cell generates the triploid endosperm, which provides nutrients for the developing embryo, which together with the maternal seed coat, comprise the seed. Following seed germination, the vegetative growth stage begins with leaf primordia and rosette leaf production by the SAM, and root production by the RAM. Floral transition reprograms the SAM to an inflorescence meristem (IM), producing additional IM and floral meristems (FMs), which generate four floral organs: sepals, petals, stamen, and carpel (9, 10). Approximately 5 wk after Arabidopsis thaliana germination, the IM stops producing FMs, and senescence begins.
We previously demonstrated that the DME gene contains both gametophyte- and sporophyte-specific active enhancers (4). Consistent with this, DME is expressed in the SAM, RAM, and young leaf primordia of the sporophytic generation (4, 11–13). We also showed that rare Arabidopsis plants homozygous for a partial loss-of-function mutant allele, dme-1, displayed sporadic defects in floral organ number, patterning, and development (2). Together, these observations suggest that DME has a role in sporophytic development. By studying Arabidopsis plants homozygous for a strong loss-of-function allele, dme-2, we discovered mutant phenotypes present throughout the sporophytic life cycle, but in general confined to meristematic cell populations, including defective cell division at the RAM, aberrant root and root hair growth, disorganized stomata development, enhanced cellular dedifferentiation, and meristem resurrection. We also compared mRNA transcriptomes primarily from wild-type and homozygous dme-2 seedlings to obtain an initial global view of sporophytic genetic circuits regulated by DME. Linking the mutant phenotypes to changes in specific gene expression pathways, we provide vital preliminary insights into how DME-mediated DNA demethylation plays a significant role in regulating Arabidopsis sporophyte development.
Results
Generating Homozygous dme-2 Lines.
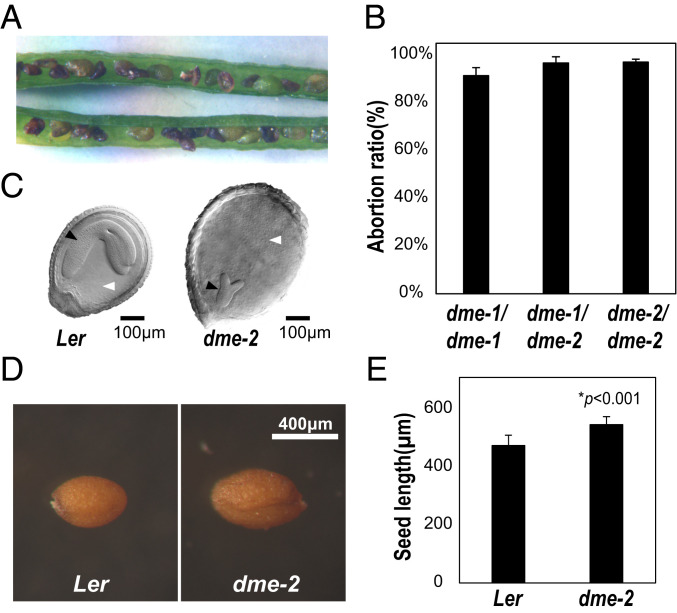
dme alleles with reduced function are recessive to the wild-type DME allele throughout the sporophyte generation with one exception—heterozygous embryo and endosperm that inherit the maternal mutant dme allele are usually inviable due to aberrant reprogramming of DNA demethylation in the gametophyte central cell (2, 14). Therefore, to observe recessive mutant phenotypes, we need plant lines homozygous for dme mutant alleles. That dme-2 is stronger than dme-1 is supported by the insertion of the T-DNA near the translation start site in the DME.1 model for dme-1, versus T-DNA insertion in the middle of gene exons for dme-2, suggesting that transcription and translation of the dme-1 might produce a low level of DME, whereas transcription and translation of the dme-2 allele likely makes no functional DME protein (2) (SI Appendix, Fig. S1 A and B). Our previous study of DME function during sporophytic growth was limited because we could only generate and propagate weak dme-1 homozygous lines (2). Here we generated multiple, independently-isolated F2 homozygous dme-2 mutant lines plant lines, as described in SI Appendix, Fig. S2 and SI Materials and Methods). The dme-1 and dme-2 alleles were initially generated in the Columbia gl (Col-gl) background and were subsequently backcrossed six times to the Landsberg erecta (Ler) wild type for these studies (2). dme-2 homozygous plants showed a higher level of seed abortion, 97.1%, (n = 1,284) than dme-1 lines 91.5% (n = 1,306) (Fig. 1 A–C and SI Appendix, Table S1), and dme-2 homozygous seedlings had less DME RNA than dme-1 seedlings (SI Appendix, Fig. S1C), in accord with dme-2 being a stronger mutant allele than dme-1.
Fig. 1.
dme-2 homozygous seeds are large and dme-2 showed a higher abortion ratio than dme-1 plants. (A) dme-2 homozygous mutant silique with aborted seeds. (B) Comparison of the seed abortion ratio of F3 progenies from dme-1/dme-2 F1 progenitors. The total number of seeds counted for dme-1/dme-1, dme-1/dme-2, and dme-2/dme-2 was 1,306, 1,370, and 1,284, respectively, as shown in SI Appendix, Table S1. (C) Comparison of dme-2 aborting seed with wild-type Ler from the same silique (200x magnification). Black arrowhead, embryo; white arrowhead, endosperm. (Scale bar, 100 µm.) (D) Viable seeds from wild type and dme-2 homozygous mutants. (Scale bar, 400 µm.) (E) Average length of mature seeds from wild type and dme-2 mutants. Individual seed length of the major axis is shown in SI Appendix, Table S7. Data represent means ± SD in B and E.
Generating and Comparing Wild-Type and dme-2 Seedling Transcriptomes.
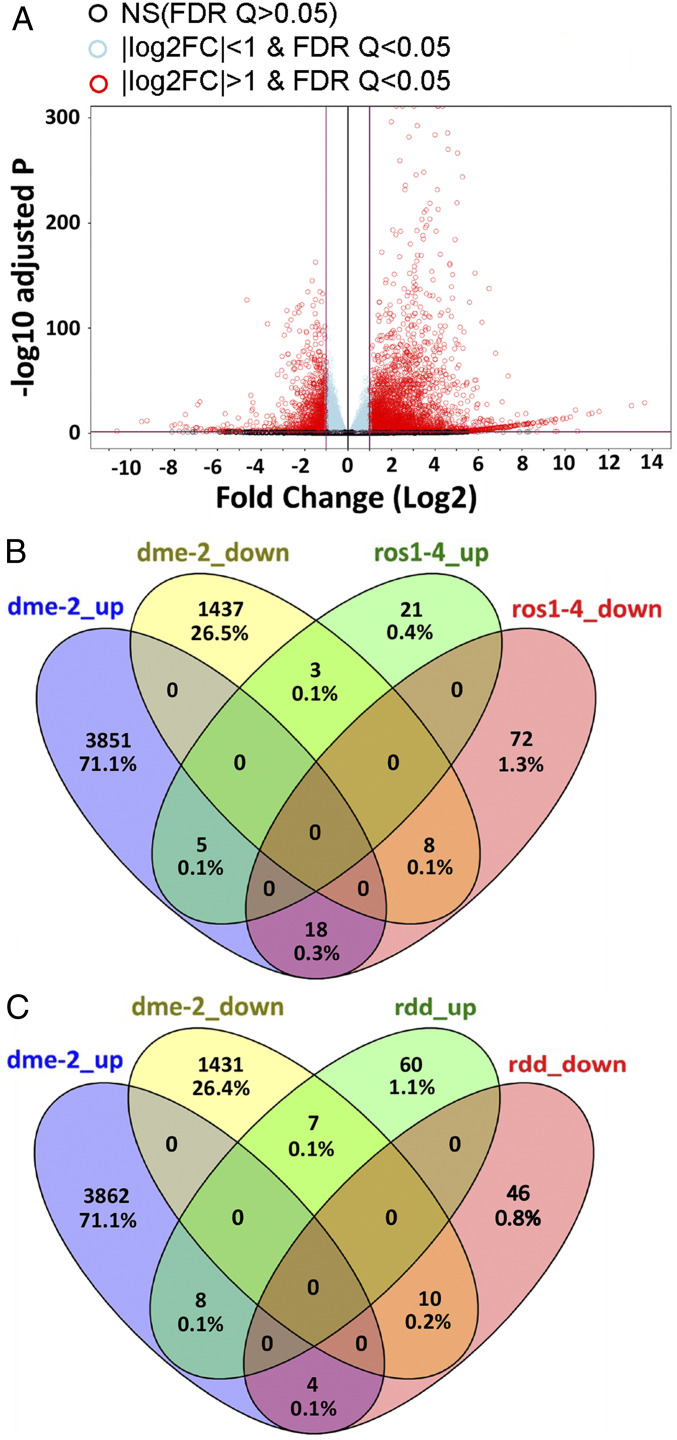
In order to identify genes regulated by DME in the sporophyte, we generated three biological replicas of next generation RNA-sequencing (RNA-seq) data from wild-type and homozygous dme-2 seedlings (Ler ecotype, 100 seedlings per replica) grown on Murashige and Skoog (MS) plates and harvested 7 d after germination (DAG). We found 5,295 genes that displayed significant gene expression differences when we compared the transcriptomes of wild-type and dme-2 genotypes. We identified 1,448 genes down-regulated at least twofold (false discovery rate [FDR] <0.05) in dme-2 mutants compared to wild-type (Fig. 2A and Dataset S1, dme differentially expressed genes [DEG]), consistent with the function of the wild-type DME activity in genome-wide demethylation at gene promoters (6, 14, 15). Gene ontology (GO) analysis (ThaleMine v4.2.0-20200615 GO tool) shows that down-regulated genes were enriched in transcription regulator activity (GO:0140110) and DNA-binding transcription factor activity (GO:0003700) (Dataset S1, dme_GO_down-regulated twofold). This is consistent with a model whereby DME-mediated demethylation promotes DNA accessibility at some promoter elements, and thus increased transcription in wild type (8). We also found a striking number of up-regulated genes (3,847) in the dme-2 mutant sporophytic transcriptome (at least a twofold change, FDR <0.05) (Dataset S1, dme DEG). Up-regulated genes in dme-2 mutants were most enriched in biochemical GO categories: catalytic (GO:0003824) and peroxidase activity (GO:0004601) (Dataset S1, dme_GO_up-regulated twofold).
Fig. 2.
RNA-seq analysis of Ler and dme-2 mutant seedlings. (A) Volcano plot visualizing differentially expressed genes between dme-2 mutant and Ler wild type. The comparison was made using the DESeq2 R package with untransformed RNA-seq read counts. Data from three replicates per sample were used. Totals of 3,874 genes and 1,448 genes are significantly up-regulated and down-regulated, respectively, in dme-2 mutants (log2 fold change >1, FDR <0.05). (B) Venn diagram of genes regulated by DME and ROS1. Genes up-regulated and down-regulated in dme-2 seedlings compared to wild-type Ler are in blue and yellow, respectively. Genes up-regulated and down-regulated in ros1-4 seedlings compared to wild-type Columbia are in green and magenta, respectively. ros1-4 data are from ref. 21. (C) Venn diagram of genes regulated by DME and RDD. Genes up-regulated and down-regulated in dme-2 seedlings compared to wild-type Ler are in blue and yellow, respectively. Genes up-regulated and down-regulated in rdd immature floral buds compared to wild-type Columbia are in green and magenta, respectively. rdd data are from ref. 23. All genes shown in the Venn diagram are twofold changed and FDR <0.05.
DME Regulates Transposable Element Expression.
Since we utilized polyadenylation-enriched RNA sequencing, we were not able to capture the majority of transposable elements (TEs) in our dataset. However, among a subset of TEs that harbor poly(A) sites in A. thaliana (16), we found 39 TEs to be down-regulated in dme-2 seedlings (Dataset S1, TE gene_DEG), suggesting that DME-mediated demethylation may promote their expression.
DME Has Distinct Sporophytic Regulatory Roles to Play Compared to DME-Like Paralog Genes.
There are three DME paralogs encoded in the A. thaliana genome, REPRESSOR OF SILENCING 1 (ROS1), DEMETER-LIKE 2 (DML2), and DML3. During sporophytic growth, ROS1, DML2, and DML3 prevent excessive DNA methylation at genomic regions and transgenes (17–19), mediate the response to sporophytic biotic and abiotic stress (20, 21), and production of stomata precursor cells (22). Using published transcriptome data from ros1-4 12-DAG seedlings (21) as well as ros1/dml2/dml3 (rdd) mutant immature floral buds (23) both in the Columbia ecotype background, we found 127 and 135 genes that displayed significant gene expression differences when we compared transcriptomes of wild-type versus ros1 and rdd genotypes, respectively. Our analysis identified that 29 and 75 genes were up-regulated in the ros1 and rdd mutant background, respectively, compared to wild type, whereas 98 and 60 genes were down-regulated in the ros1 and rdd mutant background, respectively, compared to wild type (twofold change, FDR <0.05) (Fig. 2 B and C) (Dataset S1, ros1 and rdd). We identified 13 and 18 genes that followed the same alteration patterns in dme-2 versus ros1, and dme-2 versus rdd in sporophytic tissues compared to wild type (Fig. 2 B and C and SI Appendix, Table S2). These include AT4G19520, a putative TIR-NBS-LRR gene linked to disease resistance, and AT4G05020/NAD(P)H dehydrogenase B2 (NDB2), required for stress tolerance (24). In summary, there is a 40-fold increase in genes impacted by the dme-2 mutation (5,295) compared to ros1 (127) or rdd (135), and only a few of these genes (18) share common identity and expression pattern (Fig. 2 A–C). This indicates that DME has a broad, independent role for the regulation of gene expression compared to its paralogs in the sporophyte.
DME Influences Seed Length and Germination Rate.
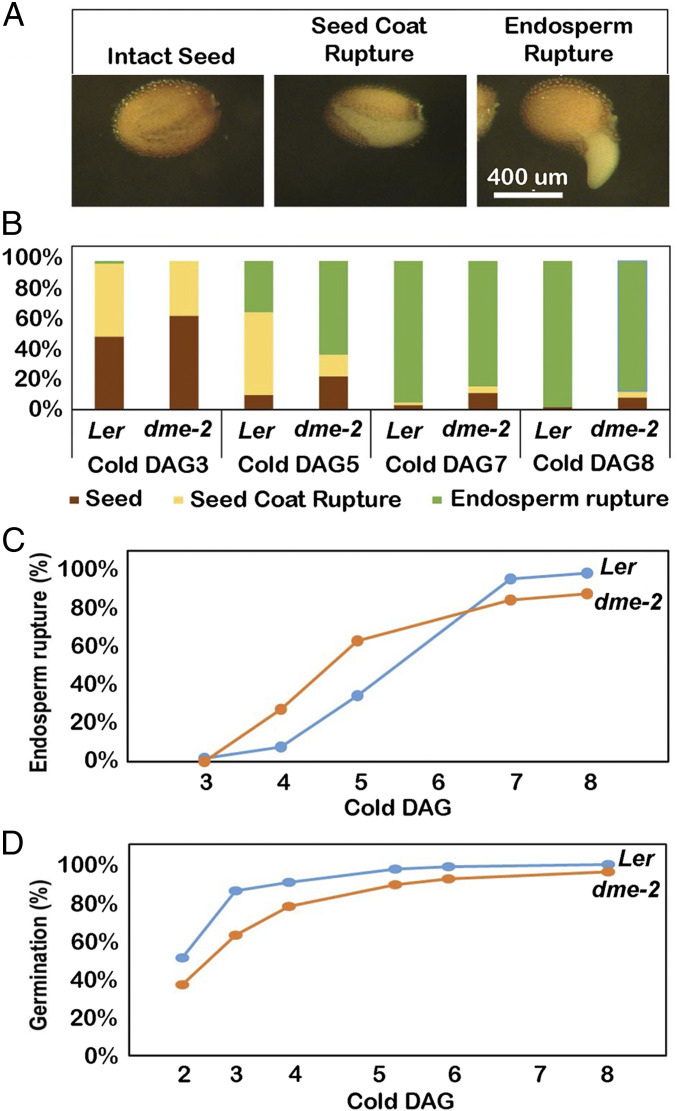
The average length of homozygous dme-2 (Ler ecotype) viable seeds was longer than that of wild-type seeds (Ler ecotype background) by around 70 μm (P < 0.001) (Fig. 1 D and E and SI Appendix, Fig. S3 and Table S3). To examine whether viable homozygous dme-2 mutant seeds exhibit defects during germination, wild-type and dme-2 seeds were sown on MS plates under long-day conditions (16 h light/8 h dark, 22 °C) and under continuous light at 22 °C or 4 °C (Fig. 3A). We did not detect any differences in seed germination at 22 °C; however, germination was slower at 4 °C, and we were able to observe delayed seed-coat rupture in homozygous dme-2 seeds (Fig. 3B, DAG3), and more rapid endosperm rupture in dme-2 seeds compared to wild type. (Fig. 3B, DAG4,and 3C). By 8 DAG, the overall germination ratio, the sum of seed-coat rupture, and endosperm rupture divided by the total number of seeds, was lower in dme-2 seeds (88%, n = 161) compared to wild-type seeds (98%, n = 177), mainly due to a larger number of nongerminating dme-2 seeds compared to wild-type seeds (Fig. 3 B and D, DAG8, and SI Appendix, Table S4).
Fig. 3.
Seed germination comparison between wild type and dme-2 mutant in continuous cold conditions. (A) Sequential stages of wild-type seed germination. (Scale bar, 400 µm.) (B) Distribution ratios of germination stages of wild-type and dme-2 seeds in continuous light with cold condition. Brown, nongerminated seed; yellow, seed-coat rupture; green, endosperm rupture. The counted numbers are shown in SI Appendix, Table S8. (C) Comparison of endosperm rupture. X = DAG, Y = (endosperm ruptured seeds/all seeds) x 100. (D) Comparison of overall germination ratio. X = DAG, Y = (ruptured seeds/all seeds) x 100.
DME Is Necessary for Proper Cell Division in Roots.
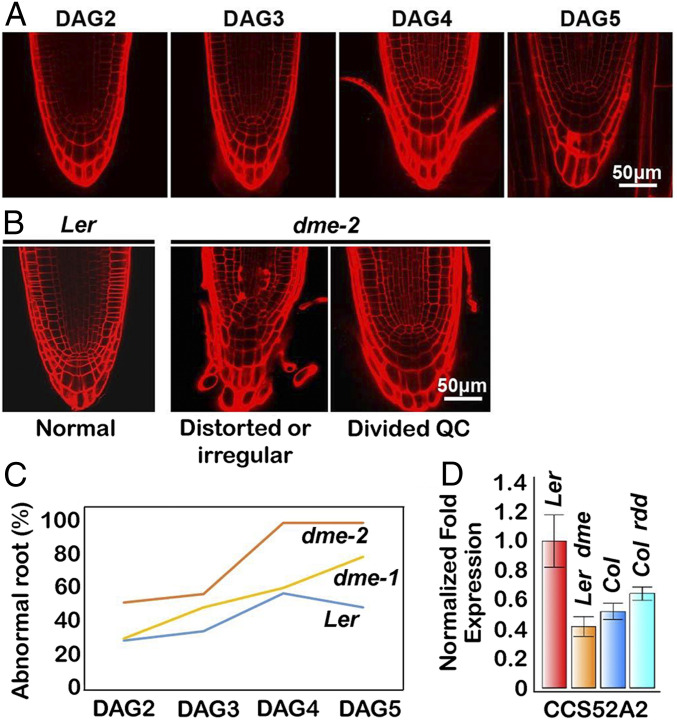
DME is highly expressed in actively dividing regions of the RAM and their adjacent cells (4, 11). Here, we used a DME:GFP (GREEN FLUORESCENT PROTEIN) transgene (2) coupled with confocal microscopy to visualize the internal patterns of DME expression during root development (SI Appendix, Fig. S4A). The root quiescent center (QC) is a small group of organizing cells that maintains the RAM stem cell niche, defined by mitotic quiescence, critical for its function (25–27). We visualized DME:GFP expression in 2 DAG to 5 DAG root tips under the confocal microscope following propidium iodide (PI) staining that delineates cell walls. Wild-type root tips (Ler ecotype) display a well-defined organized cell pattern whereby cellular lineage and identity are tightly established and maintained early after germination (Fig. 4A, 2 DAG). In contrast, dme-2 root tips (Ler ecotype) display an increased frequency of irregular cell patterning, often visualized as precocious and aberrant cell divisions in the QC compared to wild type (Fig. 4 B and C and SI Appendix, Table S5). More than half of the roots showed increased QC division in dme-2 roots at DAG2, compared to only 30% in wild-type roots at 2 DAG. The proportion of abnormal root morphology in dme-2 root tips increased to almost 100% at 4 DAG, compared to 50% in the wild type (Fig. 4C).
Fig. 4.
dme mutants show an abnormal RAM with a disorganized quiescent center. (A) Wild-type Ler root development from DAG2 to DAG5, showing natural QC division onset. (B) Major phenotype observed in dme mutant roots. (C) Frequency of abnormal roots. The numbers of primary roots displayed abnormally distorted QC versus normal roots are in SI Appendix, Table S5. (D) Reduced expression of CCS52A gene in dme-2 mutants. Data represent means ± SD.
QC cell proliferation is regulated by SCARECROW (SCR)/RETINOBLASTOMA-RELATED (RBR) protein signaling, as well as CYCLIN-D3-3 (CYCD3;3) and CYCD1;1 proteins, which promote cell proliferation in the QC (28), and the CELL CYCLE SWITCH 52 A2 (CCS52A2) subunit of the Anaphase Promoting Complex/Cyclosome (APC/CCCS52A2). APC/CCS52A2 is thought to act by targeting the Ethylene Response Factor 115 (ERF115) transcription factor to the proteosome, restraining QC cell division, and prematurely divided QC are observed in CCS52A2 mutant roots as well as in ERF115 overexpression mutants (29, 30). RBR was not differentially expressed in dme-2 mutant seedlings. In contrast, CYCD3;3 and CYCD1;1 levels were both increased, and SCR was decreased, consistent with the QC division phenotype we observed, but these changes did not reach our DEG cutoff (see Fig. 8D). For CCS52A2 and ERF115 we measured expression of 7-DAG wild-type and dme-2 (Ler ecotype) roots using qRT-PCR. CCS52A2 was significantly decreased in dme-2 mutant seedlings (Fig. 4D). The reduction of CCS52A2 gene expression was specific to the dme mutant, since expression changes were not identified in rdd mutants (Fig. 4D). The expression of ERF115 was very difficult to detect due to its specific and transient expression in the QC (30). However, we did identify a slight increase in expression in dme-2 mutant roots (SI Appendix, Fig. S5), consistent with its negative regulation by CCS52A2. Our result suggests that DME functions in maintenance of QC and RAM activity, at least in part, by influencing the expression of QC regulatory genes described above.
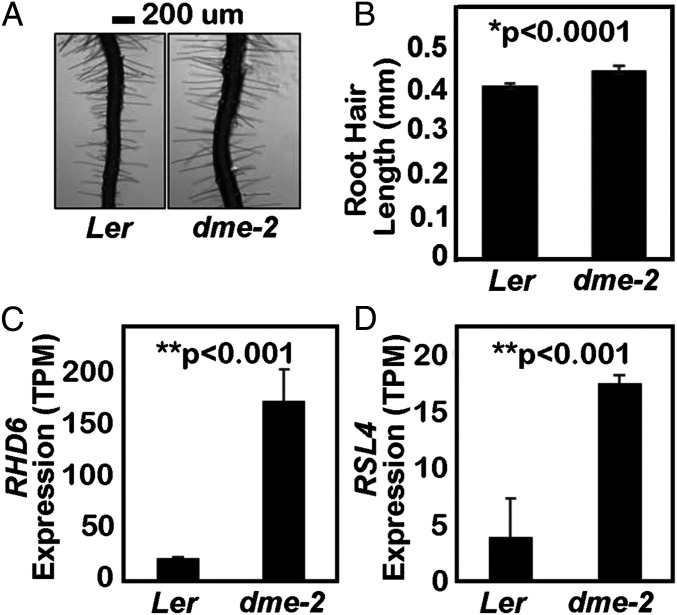
DME Plays a Role in Root Hair Growth.
We measured root hair length in wild type and dme-2 mutants in the Ler ecotype using a stereomicroscope. On 6 DAG, dme-2 root hair length was significantly longer than that of wild type (Fig. 5 A and B), suggesting that DME-mediated DNA demethylation inhibits root hair tip growth. We examined our dme-2 transcriptome data for genes associated with root hair growth and found that ROOT HAIR DEFECTIVE 6 (RHD6) expression was significantly increased in dme-2 seedlings (Fig. 5C). RHD6 is a key basic helix–loop–helix (bHLH) family transcription factor that positively regulates root hair initiation and elongation (31–33). Expression of RHD6’s primary downstream target gene, ROOT HAIR DEFECTIVE SIX‐RHD6LIKE4 (RSL4), which is necessary and sufficient for root hair growth (32, 34), was also increased in dme-2 mutant seedlings (Fig. 5D). Expression of these genes was not altered in published ros1 seedling or rdd mutant immature floral bud transcriptomes. Thus, the negative regulation of root hair growth is specific to DME and may be mediated by transcriptional suppression of RHD6 and RSL4 gene loci.
Fig. 5.
DME suppresses root hair growth. (A and B) Comparison of root hair length in Ler and dme-2 mutants. Data represent mean ± SEM, n = 517 root hairs for Ler and n = 355 root hairs for dme-2 from approximately 20 ∼ 29 roots. The values are from two biological replicates and are significantly different (*P < 0.0001; Student’s t test). (Scale bar, 100 μm.) (C) Relative expression levels of the RHD6 gene from RNA-seq, plotted as TPM value (transcripts per kilobase million, number of transcripts scaled using the average transcript length over samples and then the library size). (D) Relative expression levels of RSL4 from the RNA-seq plotted as TPM value. ** in C and D, FDR-adjusted P value < 0.001 by DESeq2. Data represent means ± SD.
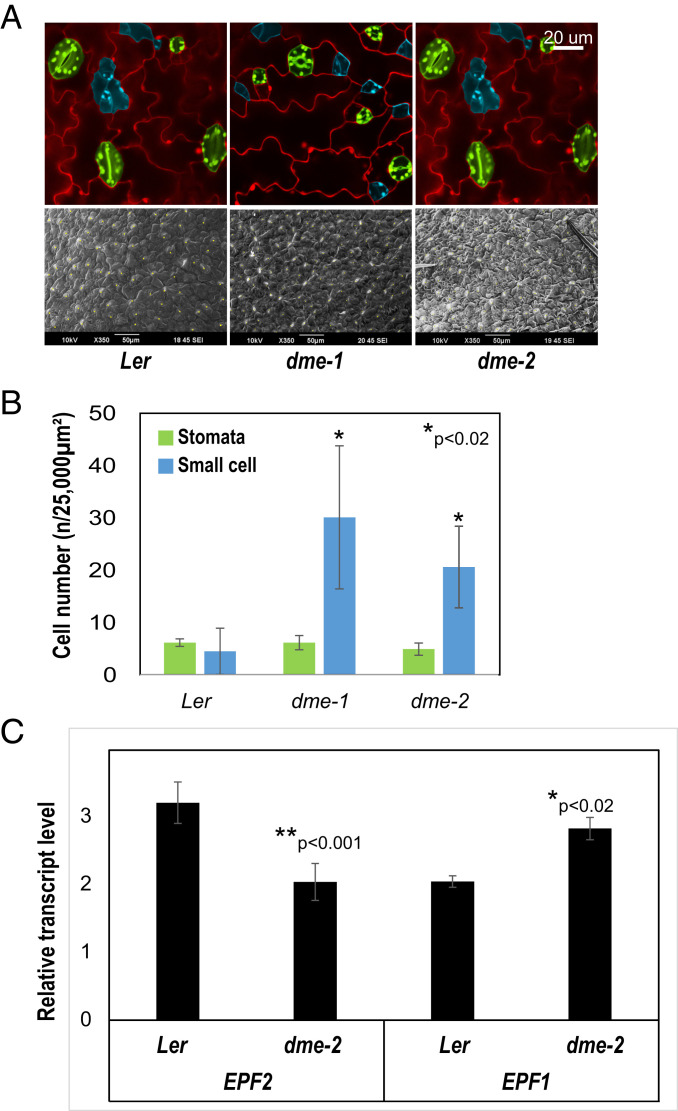
DME Influences Stomatal Precursor Cell Number.
Stomata are regularly spaced and distributed throughout the leaf epidermis and are formed by a series of asymmetric cell divisions by meristemoid mother cells (MMCs). The larger cell produced following MMC division is a stomatal lineage ground cell (SLGC), while the smaller is a precursor meristemoid. Precursor meristemoids undergo asymmetric divisions to produce further meristemoids and SLGCs, and eventually differentiate into guard mother cells, which produce guard cell pairs (35–37). Here we show that dme-2 mutant leaves (Ler ecotype) displayed a significantly increased number of characteristic small and triangular meristemoid precursor cells compared to wild type (Fig. 6 A and B, blue). This phenotype was also observed in DME-paralog mutant leaves, (ros1 and rdd), which overproduce stomata precursors, but not stomata, in the leaf epidermis (22). This is, at least in part, due to the down-regulation of EPIDERMAL PATTERNING FACTOR 2 (EPF2), a negative regulator of stomata formation, and epf2 mutant plants exhibit the same phenotype (22, 38–40). Given the phenotypic similarities in epidermal patterning between in dme-2, ros1, rdd, and epf2, we compared EPF2 expression in 3 DAG wild-type and dme-2 seedlings. qRT-PCR showed that EPF2 expression was reduced in dme-2 mutants (Fig. 6C), supporting the idea that the mechanism of meristemoid cell increase is mirrored between rdd and dme mutant leaves. The final number of mature stomata was not increased (Fig. 6B, green) as also seen in ros1 mutants (22). Interestingly, EPF1, another negative regulator of stomata formation which is related to EPF2 (41), showed increased expression in dme-2 and rdd mutants (Fig. 6C) (22), perhaps compensating for the down-regulation of EPF2 in DNA demethylase mutants, and providing an explanation for the maintenance of stomata number. The commonality of dme and ros1/rdd phenotypes, and the shared regulation of the EPF2 gene, provides strong evidence that DME and RDD-mediated DNA demethylation is involved in the regulation of the stomatal lineage in A. thaliana.
Fig. 6.
DME affects the determination of stomatal precursor cells. (A) Stomata (green) and stomatal precursor cell (light blue) of adaxial leaf and overall SEM visualization (white spots show stomata). (Scale bar, 20 µm.) (B) Normalized counted cell number of stomata and precursor cell in DAG3 seedlings. Pictures in A were used for analysis, n = 4 (*P < 0.02, Student’s t test). (C) qRT-PCR of stomatal regulatory genes, EPF2 and EPF1 (*P < 0.02, **P < 0.001, Student’s t test). Data represent means ± SD.
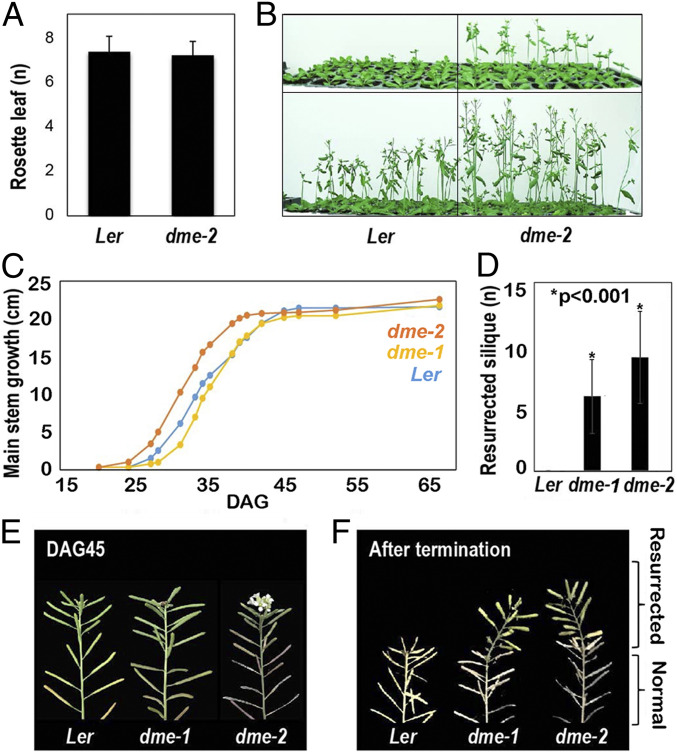
DME Regulates Aerial Growth Rate.
DME is expressed in the vegetative shoot apical meristem (SAM), inflorescence meristem (IM) and floral meristem (FM) regions (SI Appendix, Fig. S6) (4, 11). Hence, we investigated the aerial growth of dme mutant seedlings. We planted dme-2 mutant and wild-type (Ler ecotype) seeds at the same time and counted the number of leaves present at the time of SAM to IM transition to measure flowering time. No differences were observed in leaf number at the SAM to IM transition when we compared wild-type and dme-2 mutant plants (Fig. 7A). However, we observed that dme-2 plants produced an IM earlier than wild-type plants (Fig. 7B) and remained developmentally ahead until cessation of growth at 40 DAG. By contrast, wild-type Ler ceased growth at 47 DAG, although the terminal height of dme-2 and wild-type primary inflorescences were the same (∼21 cm) (Fig. 7C).
Fig. 7.
Earlier bolting and resurrection after termination in dme mutants. (A) Rosette leaf counting on flowering (n = 25 for both Ler and dme-2 plants). (B) Flowering time and growth differences between Ler and dme-2 mutants. (C) Growth plot of main stem (n = 16) of each genotype. Resurrection (bloom after the first termination) makes a difference in the final height of the main stem at DAG65. (D) Number of siliques generated from the resurrection (n = 16, *P < 0.001, Student’s t test). (E) Resurrected green siliques are shown on top of the dried terminated IM in dme mutants. Data in A and D represent means ± SD.
dme Mutant Plants Display a “Resurrection” Phenotype.
Following cessation of inflorescence stem growth, Arabidopsis plants change color from green to yellow, begin to dessicate, seeds mature in the siliques, and ultimately wild-type plants all die. However, after a pause of about 10 d, we found that dme-2 plants (Ler ecotype) surprisingly began to make additional flowers that produced fertile siliques (Fig. 7D). That is, in dme-2 mutants, the inflorescence meristem reinitiated the formation of flowers, resulting in elongating green inflorescence shoots above the yellow drying stems on the same plant (Fig. 7 E and F). Due to the resurrection of the IM, the dme-2 plants tended to reach a final height that was taller than wild type at DAG65 (Fig. 7C), displaying siliques with viable seeds atop the dried yellow stems below (Fig. 7E). We call this flowering phenotype "resurrection" because the plants produce new siliques and seeds after the initial termination of flowering appears to have occurred. This indicates that DME may play a role in the maintenance of SAM activity and IM termination, that manifests in the appropriate cessation of sporophytic growth, to allow resources to be deferred to the next generation.
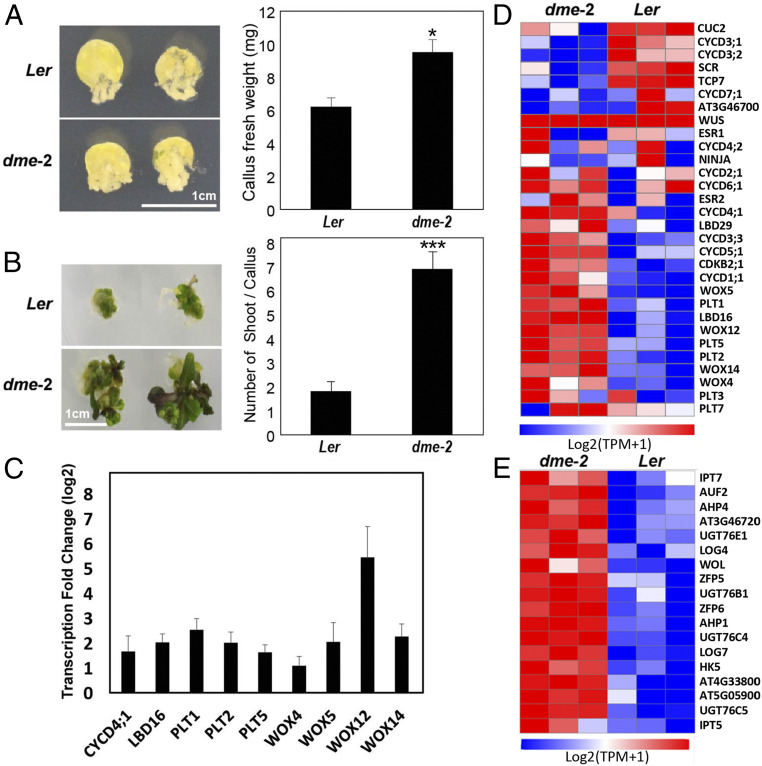
DME Plays a Role in Inhibiting Dedifferentiation and De Novo Shoot Formation.
The observation that the dme-2 mutation allowed previously senescent plants to reenter an active phase of growth led us to further explore the capacity of dme-2 mutant tissues for regeneration, using an in vitro callus culture system, where a mass of undifferentiated plant callus cells can differentiate into mature tissues or entire new individuals in appropriate conditions (42, 43). Plant regeneration usually involves a two-step process, including callus formation and de novo shoot organogenesis (44). To examine cell proliferation activity in callus, leaf explants of 2-wk-old seedlings were incubated on callus-inducing medium (CIM) for 2 wk. Measurement of fresh weight revealed that callus proliferation was increased by 60% in dme-2 compared with wild type (Fig. 8A). This result is consistent with a recent report by Shim et al. (45) that DME expression is dramatically reduced during wild-type Arabidopsis leaf-to-callus formation.
Fig. 8.
Increased callus formation and de novo shoot regeneration in dme-2. (A) Callus formation. Leaf explants from the third leaves of 2‐wk‐old plants were used to induce callus on callus‐inducing medium (CIM) (n > 30). Plates were incubated for 2 wk under continuous dark conditions and photographed (Left). Thirty calli dissected from leaf explants were collected to measure fresh weight (Right). (B) De novo shoot regeneration. Calli preincubated for 6 d on CIM were used to induce shoot regeneration on SIM (n > 30). Plates were incubated for 3 wk under continuous light conditions and photographed (Left). The number of regenerated shoots from calli was measured (Right). In A and B, three independent measurements were averaged. Statistically significant differences between wild‐type and mutant calli are indicated by asterisks (Student’s t test, *P < 0.05, ***P < 0.001). Bars indicate ± SEM. (C) Some key regulatory genes involved in pluripotency and shoot formation are overexpressed in dme-2 mutant compared to wild type. RNA-seq analyses were performed as described in Materials and Methods and differentially expressed genes analyzed with DESeq2. The bars represent the average fold expression changes with SEs. (D) A heatmap of genes involved in callus regeneration, pluripotency, and de novo shoot formation in dme-2 mutants and Ler wild type. TPM (transcripts per kilobase million) counts were log2(x + 1) transformed. The Far Left is Min(logTPM + 1) and the Far Right is Max(logTPM + 1). Min and Max mean the minimum and maximum value of log TPM(normalized read count) of the total genes in the figure. Blue color corresponds to a decreasing transcript abundance, while red color corresponds to an increasing transcript abundance. (E) A heatmap of significant gene expression (DESeq2, padj of 0.05) that are up-regulated in dme-2 mutants involved in cytokinin biosynthesis and signaling. Transcriptome reads were log2(x + 1) transformed.
Furthermore, de novo shoot organogenesis from leaf-derived callus was strongly enhanced in dme-2 on shoot-inducing medium (SIM). While few leaves were produced in wild-type callus on SIM, we observed a more than threefold increase in shoot regeneration capacity in dme-2 callus (Fig. 8B).
To examine which transcriptional processes may contribute to the resurrection and dedifferentiation phenotypes observed in dme-2 mutant plants, we focused on pathways of regeneration and cell proliferation. CYCD genes are associated with cell proliferation during whole-plant regeneration processes (46, 47). CYCD4;1, which is expressed early during germination and whose loss of function leads to decreased cell division and germination rate (48) was up-regulated in the dme-2 mutant (Fig. 8C). Moreover, CYCD2;1, CYCD3;3, CYCD5;1, and CYCD6;1 were likewise up-regulated, although they did not meet our DEG criteria (Fig. 8D). We examined expression of A. thaliana meristem and regeneration regulatory genes and found that over half were significantly up-regulated in dme-2 mutants, including LATERAL ORGAN BOUNDARIES-DOMAIN 16 (LBD16), PLETHORA1 (PLT1), PLT2, PLT5, WUSCHEL RELATED HOMEOBOX4 (WOX4), WOX5, WOX12, and WOX14 (Fig. 8 C and D).
As described by Shim et al. (45), two myb-related transcriptional repressors, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL 1 (LHY1), are key upstream negative regulators of callus proliferation and plant regeneration. Consistent with this, we found that CCA1 and LHY1 expression are significantly reduced in our dme mutant seedling transcriptome (SI Appendix, Fig. S7). These results suggest that DME may function as one of the master regulators for maintenance of the differentiated state and a negative regulator of plant reprogramming and dedifferentiation.
The decision to initiate shoot regeneration is greatly influenced by cytokinin biosynthesis and signaling (49–51). Several key cytokinin-pathway genes such as ISOPENTENYLTRANSFERASE (IPT)5 and 7 (52, 53), Arabidopsis HISTIDINE PHOSPHOTRANSFER PROTEIN (AHP)1 and 4 (54), and LONELY GUY (LOG)4 and 7 (55) were up-regulated in the dme-2 mutant and formed pathway interconnections consistent with their promotion of de novo shoot formation following the transcriptional dysregulation resulting from a loss of dme-2 (Fig. 8E).
Discussion
Previous work has focused on the function of DME during the reproductive stage; however, DME is highly expressed during sporophytic development (11). We detected certain sporophytic mutant phenotypes by analyzing homozygous lines for the weaker dme-1 allele (2). To broaden our understanding of DME function in the sporophyte, we generated loss-of-function dme-2 homozygous lines, discovered additional DME mutant phenotypes throughout the A. thaliana life cycle, and used our analyses with RNA-seq to evaluate the transcriptomic changes accompanying the loss of sporophytic DME function.
The rationale for the method we used to generate homozygous dme-2 lines is based on our observation that dme-1 seed viability increased slightly, but continuously, as generations went by. We speculated that this is due to accumulative epigenetic phenomena. Therefore, we were able to use the slightly increased viability of the later generation of dme-1 homozygous plants as female donors to introduce the male dme-2 allele to make hybrid dme-1/dme-2 mutants. Since maternal dme-1 seed viability has been already slightly increased, we speculated that dme-2 homozygous seeds could be generated from the dme-1/dme-2 hybrid plants, albeit at low frequency.
dme-2 mutant plants exhibit a number of developmental defects: precocious inflorescent stem formation (Fig. 7), aberrant root patterning and development (Fig. 4), excess root hair growth (Fig. 5), overproduction of stomatal lineage stem cells (Fig. 6), and a resurrection of the inflorescence meristem in previously terminated and desiccating plants (Fig. 7). In general, these phenotypes correspond with the sites of DME sporophytic expression: the SAM, RAM, and leaf primordia (4, 11).
Most homozygous dme-2 seeds abort their development, but those that survive are larger than wild-type seeds (Fig. 1). This is likely due to excess endosperm compared to wild-type, visible in aborting dme-2 seeds (Fig. 1C), as well as in aborting heterozygous seeds that inherited a maternal mutant dme allele (2). We also observed an increase in the rate of endosperm rupture (Fig. 3). One possibility is that endosperm overproliferation during dme-2 seed development may influence subsequent endosperm rupture during germination.
The earlier transition of the SAM to the IM (bolting), reflected in the early development of inflorescence stems in dme-2, did not accompany a decrease in the number of rosette leaves in dme mutant plants (Fig. 7). This indicates that DME is not involved in the FLC and FT-associated floral-induction pathway (56, 57). DME is highly expressed in the SAM and in leaf primordia, so it is possible that in dme mutant plants, the more rapid inflorescent stem induction may be the result of more rapid growth and development influenced by the loss of DME.
A. thaliana stomata are two-cell valves controlling epidermal pores for gas exchange. Stomatal formation and patterning are highly regulated by the frequency and the placement of asymmetric cell divisions of the meristemoid mother cell and stem-like derivatives (36, 37). In dme mutant seedlings we observed increased small precursor (meristemoid) cells, which retain stem-like features (Fig. 6). This phenotype is also observed in ros1/rdd and epf mutant leaves (22) and was found in this case to be caused by RDD-mediated DNA demethylation at the EPF2 promoter. The same stomatal cell specification phenotype in dme-2 mutant leaves strongly suggests that DME function overlaps with other RDD proteins in this pathway, regulating the stomatal stem cell niche by DNA demethylation at the EPF locus.
DME also affects the structure and function of the RAM. The early QC cell divisions in dme-2 roots suggest that the stereotyped cell divisions of the QC in the dme-2 RAM are not well established (Fig. 4). Despite being surrounded by actively proliferating mitotic cells, QC cells self-renew at a low rate, key to maintaining the root stem cell niche (30). We found that earlier QC division and irregular RAM cell patterning in dme-2 mutants phenocopies that in cycd3;3/cycd1;1 (28) and ccs52a2 loss-of-function mutants as well as in ERF115 overexpression mutants (29, 30). Of note, previous studies have shown that ERF115 promoter activity is detected only in QC cells that are associated with QC division (30). CCS52A2, a ubiquitin ligase that targets ERF115 in the QC, is involved in maintaining the QC. Our data indicate that DME may directly or indirectly impact this pathway, since CYCD3;3, CYCD1;1, and CCS52A2 expression was reduced in dme-2 mutants, (Figs. 4 and 8), whereas ERF115 expression was slightly increased (SI Appendix, Fig. S6). It should be noted that other pathways increase QC division, such as responses to stress, which are unrelated to potential transcriptional regulation of specific pathways by DME (58).
A striking phenotype that we observed in dme-2 mutant sporophytes was the resurrection of inflorescent stems, where the dme-2 shoot apical region reacquired IM activity following a temporary termination process. This resulted in the production of an average of 10 more siliques (Fig. 7). Such a phenotype indicates a role for DME in the regulation of cellular pluripotency, perhaps preventing proliferation or dedifferentiation in differentiated tissues. This hypothesis is supported by the phenotype that we observed in dme mutant callus culture, whereby there was a significant increase in cell proliferation activity in dme mutant callus compared to wild type. In addition, dme mutant callus produced significantly more de novo shoots compared to wild type (Fig. 8). Interestingly, plants homozygous for the met1 mutation exhibited reduced callus formation and a low proliferation rate and maintained a differentiation-associated feature (i.e., green pigmentation) compared to wild type (59, 60). These met1 phenotypes are all in opposition to those in dme homozygous mutant plants (Fig. 8), consistent with what we observed during seed development, that DME-mediated demethylation and MET-mediated methylation are antagonistic to each other, promoting and repressing endosperm cell proliferation, respectively (2, 61, 62). We therefore suggest that the respective gain and loss of DNA methylation in dme and met1 mutant plants directly and antagonistically influences sporophyte cellular pluripotency in A. thaliana.
Dedifferentiation is the transformation of cells from a given differentiated state to a less differentiated or stem-cell–like state, associated with reentry into the cell cycle and reacquisition of pluripotency or trans/redifferentiation (63, 64). In dme-2 mutant seedlings a number of cell cycle regulators, pluripotency genes, and members of cytokinin biosynthesis pathways were up-regulated. Of note, LBD16 and LBD29, which regulate the root cell cycle, a number of D-type cyclins (Fig. 8), as well as PLT1, PLT2, and PLT5, which encode AP2 class putative transcription factors and act to maintain stem cell activity (65), were all significantly up-regulated. Also up-regulated were WOX5, 12, and 14, which regulate stem cell maintenance (Fig. 8). The up-regulation of these genes in dme mutant sporophytes suggests that DME activity may correlate with their repression, implicating these transcripts in the gain of pluripotency and resurrection we observe here.
CCA1 and LHY1 have long been known as key myb transcriptional repressors that can form heterodimers in vivo and function synergistically in the circadian clock of Arabidopsis (66, 67). However, a recent report showed they function independently of the circadian clock where their gene expression is suppressed resulting in activation of cell cycle genes that promote cell proliferation and pluripotency acquisition during callus formation (45). Consistent with this hypothesis, they showed more callus is formed in cca1 single mutants and cca1; lhy1 double mutants compared to wild-type plants (45). Likewise, in dme-2 mutant plants, we observed decreased CCA1 and LHY1 expression (SI Appendix, Fig. S7) and increased callus and shoot formation (Fig. 8 A and B) compared to wild-type plants. Finally, DME expression was shown to be dramatically reduced during wild-type leaf-to-callus formation (45). Taken together, these results suggest that DME functions as a key negative upstream regulator for cell proliferation and acquisition of pluripotency in Arabidopsis.
Genomic targets of DME-mediated DNA demethylation in gametophytes have been identified and include several endosperm-imprinted gene regulatory regions (2, 3, 5–8, 68). We compared our expression data with genes impacted by hypermethylation of maternal DNA at imprinting control regions in dme-2 mutant endosperm (8). Of the seven genes known to depend on DME for maternal-specific expression in endosperm, two genes were also significantly down-regulated in dme-2 seedling mutant sporophytic tissue (AT1G77960 and AT4G18150), indicating that expression of these genes is shared between the central cell and the sporophyte (SI Appendix, Table S2). Of the remaining five genes, three did not change significantly and two were significantly up-regulated in the sporophyte, indicating that the transcriptional regulation of these genes may differ between the central cell and the sporophyte (SI Appendix, Table S2). It is possible that the distinct gene expression patterns in the central cell versus the sporophyte might reflect different cell-specific epigenetic profiles (7, 69) or different transcriptional complexes required to express the same DME target genes.
During reproduction, aside from mediating genomic imprinting, DME is thought to function in TE silencing. The majority of DME targets are small, euchromatic TEs, and it is suggested that DME may contribute to TE silencing in vital cells, such as gametes, by initiating RNA-directed DNA methylation (RdDM) in gamete companion cells (6, 70, 71). It has been suggested that RdDM in A. thaliana meristems occurs in a similar manner, to reinforce TE silencing during vegetative growth (72). In support of this, the transient expression of TEs was recently reported in meristems prior to flower induction, in conjunction with increases in CHG and decreases in CHH methylation, indicative of epigenetic reprogramming, which contributes to the protection of the stem cell genome (73). RdDM-mediated TE silencing is characterized by CHH methylation, and during the rice SAM developmental transition from the vegetative to reproductive state, methylation at CHH sites is kept high, particularly at TEs, via RdDM (74). The observation that, in addition to gamete companion cells, DME is expressed in meristematic and newly developing tissues (SI Appendix, Fig. S7) (4) therefore leads to the intriguing possibility that DME, and perhaps RDD paralogs, may act in the stem cell compartments of the sporophyte to demethylate TEs in certain cell compartments, initiating RdDM and thus reinforcing TE silencing in stem cells. Silencing of TEs in stem cells throughout the sporophyte life cycle might therefore also be essential for the generation of the germline, distinguishing plant developmental biology from animal developmental biology where the germline is specified early in embryogenesis. The precise cells in which DNA demethylases may act in order to initiate RdDM in the sporophyte are unknown, although in A. thaliana root apical meristem, excess 24-nt smRNAs are thought to be produced in the columella and transported to the nearby QC, reinforcing TE silencing (75). Future work to profile the DNA methylation and TE expression landscape of dme mutant vegetative tissues will provide important clues as to the mechanism of TE regulation and its potential link to active DNA demethylation.
Here, we report the generation of homozygous dme-2 mutant plants and examination of their developmental phenotypes and transcriptome. We find profound alterations in the dme mutant sporophytic transcript landscape, and in several cases found evidence of links to the loss of DME-mediated DNA demethylation activity. In addition, we identify dysregulation of a number of genetic pathways that correlated with the observed phenotypes, including precocious inflorescent stem formation, aberrant root patterning and development, overproduction of stomatal lineage stem cells and the resurrection of the SAMs in terminated bolts and enhanced regeneration. Our data show that DME is required for normal sporophytic development and may play an important role in the regulation of pluripotency and cellular differentiation.
Materials and Methods
Please see SI Appendix, SI Materials and Methods for full details.
Transcriptomics Data Acquisition.
We sequenced the cDNA of three biological replicates of DAG7 Ler wild-type and dme-2 mutant seedlings grown in MS plates on the DNBseq platform at the Beijing Genomics Institute, generating on average 4.40 Gb of data per sample. Fragments with low-quality reads or reads with adaptors were filtered using Trimmomatic (v.0.36) (76). We mapped filtered reads onto the reference genome using HISAT2 (v2.1.0) identifying a total of 24,739 genes (77). Reads were assembled into transcripts using StringTie (v2.1.3) (78). Annotation was conducted using the TAIR10 FASTA sequence and the TAIR10 genome GTF annotation file (http://www.arabidopsis.org/). Published transcriptome data discussed in this publication, as accession numbers GSE65016 (21) and GSE10877 (23), were obtained from the National Center for Biotechnology Information Gene Expression Omnibus.
Differentially Expressed Gene Analysis and Data Visualization.
Gene-level read count estimates were calculated with transcript-level count data from StringTie with Tximport (v.1.16.1) (79). We used DESeq2 (80) to find DEGs between dme-2 mutants and Ler wild type. Unless otherwise stated, statistical significance of DEGs were determined at α = 0.05 and corrected for multiple hypothesis testing using the FDR method. Relative expression levels of the genes from RNA-seq were plotted with FPKM values (fragments per kilobase of transcript per million). Volcano plots were plotted to visualize distribution of DEGs, using the R package ggplot2 (81). Heatmaps were built to show significant (FDR-adjusted P value of 0.01) up-regulation of genes involved in cytokinin biosynthesis and signaling in dme-2 mutants. Transcriptome reads were log2(× + 1) transformed before visualization. Hierarchical clustering was done by row-wise Pearson correlation values and visualized on the Morpheus web platform (https://software.broadinstitute.org/morpheus). Venn diagrams were constructed using Venny (v. 2.1) developed by J. C. Oliveros (https://bioinfogp.cnb.csic.es/tools/venny_old/venny.php).
Plant Regeneration.
For callus induction, leaf explants of 2-wk-old plants were placed on CIM (MS medium supplemented with 0.5 mg/l 2,4-dichlorophenoxyacetic acid [2,4-D] and 0.05 mg/l kinetin), followed by incubation at 22 °C in the dark for 2 wk in order to estimate cell proliferation (82). For shoot regeneration, leaf-derived callus preincubated on CIM for 6 d was incubated on SIM (MS medium supplemented with 0.9 μmol/l 3-indoleacetic acid, 2.5 μmol/l 2-isopentenyladenine), followed by incubation at 25 °C under continuous light conditions for 3 wk (82). The number of regenerated leaves was counted to measure shoot regeneration capacity.
Supplementary Material
Acknowledgments
The work was supported by the National Research Foundation of Korea (2020R1A2C2009382) (to Y.C.). S.K., J.-S.P., and J.L. were supported by the Stadelmann-Lee Scholarship Fund, Seoul National University.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026806118/-/DCSupplemental.
Data Availability
The RNA data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession no. GSE164217).
References
- 1.Hofmeister W. F. B., Currey F., On the Germination, Development, and Fructification of the Higher Cryptogamia, and On the Fructification of the Coniferæ;, The Ray society (Publication for the Ray society by R. Hardwicke, London, 1862).
- 2.Choi Y., et al., DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 110, 33–42 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Schoft V. K., et al., Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. U.S.A. 108, 8042–8047 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J. S., et al., Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, 2078–2083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gehring M., et al., DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibarra C. A., et al., Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frost J. M., et al., FACT complex is required for DNA demethylation at heterochromatin during reproduction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E4720–E4729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh T. F., et al., Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108, 1755–1762 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weigel D., Jürgens G., Stem cells that make stems. Nature 415, 751–754 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Pidkowich M. S., Klenz J. E., Haughn G. W., The making of a flower: Control of floral meristem identity in Arabidopsis. Trends Plant Sci. 4, 64–70 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Kim M., et al., Temporal and spatial downregulation of Arabidopsis MET1 activity results in global DNA hypomethylation and developmental defects. Mol. Cells 26, 611–615 (2008). [PMC free article] [PubMed] [Google Scholar]
- 12.Ortega-Galisteo A. P., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67, 671–681 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Schumann U., et al., DEMETER plays a role in DNA demethylation and disease response in somatic tissues of Arabidopsis. Epigenetics 14, 1074–1087 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park K., et al., DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. U.S.A. 113, 15138–15143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh T. F., et al., Genome-wide demethylation of Arabidopsis endosperm. Science 324, 1451–1454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo C., Spinelli M., Liu M., Li Q. Q., Liang C., A genome-wide study of “non-3UTR” polyadenylation sites in Arabidopsis thaliana. Sci. Rep. 6, 28060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Z., et al., ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Penterman J., Uzawa R., Fischer R. L., Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 145, 1549–1557 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stroud H., Greenberg M. V., Feng S., Bernatavichute Y. V., Jacobsen S. E., Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152, 352–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu A., et al., Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. U.S.A. 110, 2389–2394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J. S., et al., ROS1-dependent DNA demethylation is required for ABA-inducible NIC3 expression. Plant Physiol. 179, 1810–1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamuro C., et al., Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat. Commun. 5, 4062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lister R., et al., Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sweetman C., et al., AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress. Plant Physiol. 181, 774–788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Berg C., Willemsen V., Hendriks G., Weisbeek P., Scheres B., Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Ortega-Martínez O., Pernas M., Carol R. J., Dolan L., Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317, 507–510 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Sarkar A. K., et al., Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Forzani C., et al., WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 24, 1939–1944 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanstraelen M., et al., APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 106, 11806–11811 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heyman J., et al., ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342, 860–863 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Masucci J. D., Schiefelbein J. W., The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 106, 1335–1346 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pires N. D., et al., Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. U.S.A. 110, 9571–9576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han X., Zhang M., Yang M., Hu Y., Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 32, 1049–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi K., Menand B., Bell E., Dolan L., A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42, 264–267 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Casson S., Gray J. E., Influence of environmental factors on stomatal development. New Phytol. 178, 9–23 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Pillitteri L. J., Torii K. U., Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63, 591–614 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Le J., Zou J., Yang K., Wang M., Signaling to stomatal initiation and cell division. Front Plant Sci 5, 297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hara K., et al., Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Hunt L., Gray J. E., The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864–869 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Lee J. S., et al., Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522, 439–443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hara K., Kajita R., Torii K. U., Bergmann D. C., Kakimoto T., The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birnbaum K. D., Sánchez Alvarado A., Slicing across kingdoms: Regeneration in plants and animals. Cell 132, 697–710 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugimoto K., Gordon S. P., Meyerowitz E. M., Regeneration in plants and animals: Dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 21, 212–218 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Kareem A., et al., PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25, 1017–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shim S., et al., Dynamic changes in DNA methylation occur in TE regions and affect cell proliferation during leaf-to-callus transition in Arabidopsis. Epigenetics 15, 1–18, 10.1080/15592294.2021.1872927 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dewitte W., et al., Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14537–14542 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishikawa M., et al., Physcomitrella cyclin-dependent kinase A links cell cycle reactivation to other cellular changes during reprogramming of leaf cells. Plant Cell 23, 2924–2938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masubelele N. H., et al., D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 15694–15699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riou-Khamlichi C., Huntley R., Jacqmard A., Murray J. A., Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Rosspopoff O., et al., Direct conversion of root primordium into shoot meristem relies on timing of stem cell niche development. Development 144, 1187–1200 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Zhang T. Q., et al., A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29, 1073–1087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kieber J. J., Tribute to Folke Skoog: Recent advances in our understanding of cytokinin biology. J. Plant Growth Regul. 21, 1–2 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Miyawaki K., et al., Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 16598–16603 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lomin S. N., et al., Studies of cytokinin receptor-phosphotransmitter interaction provide evidences for the initiation of cytokinin signalling in the endoplasmic reticulum. Funct. Plant Biol. 45, 192–202 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Kuroha T., et al., Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21, 3152–3169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hepworth J., Dean C., Flowering Locus C’s lessons: Conserved chromatin switches underpinning developmental timing and adaptation. Plant Physiol. 168, 1237–1245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andrés F., Coupland G., The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Zhang W., Swarup R., Bennett M., Schaller G. E., Kieber J. J., Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 23, 1979–1989 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Berdasco M., et al., Promoter DNA hypermethylation and gene repression in undifferentiated Arabidopsis cells. PLoS One 3, e3306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W., et al., DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 7, e1002243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao W., et al., Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev. Cell 5, 891–901 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Hehenberger E., Kradolfer D., Köhler C., Endosperm cellularization defines an important developmental transition for embryo development. Development 139, 2031–2039 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Grafi G., How cells dedifferentiate: A lesson from plants. Dev. Biol. 268, 1–6 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Jiang F., Feng Z., Liu H., Zhu J., Involvement of plant stem cells or stem cell-like cells in dedifferentiation. Front Plant Sci 6, 1028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galinha C., et al., PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Lu S. X., Knowles S. M., Andronis C., Ong M. S., Tobin E. M., CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150, 834–843 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alabadí D., et al., Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Jullien P. E., Kinoshita T., Ohad N., Berger F., Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18, 1360–1372 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang C., et al., The catalytic core of DEMETER guides active DNA demethylation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 17563–17571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slotkin R. K., et al., Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim M. Y., et al., DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc. Natl. Acad. Sci. U.S.A. 116, 9652–9657 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baubec T., Finke A., Mittelsten Scheid O., Pecinka A., Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 15, 446–452 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutzat R., et al., Arabidopsis shoot stem cells display dynamic transcription and DNA methylation patterns. EMBO J. 39, e103667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Higo A., et al., DNA methylation is reconfigured at the onset of reproduction in rice shoot apical meristem. Nat. Commun. 11, 4079 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawakatsu T., et al., Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2, 16058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pertea M., et al., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000 Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickham H., Ggplot2: Elegant Graphics for Data Analysis (Springer Science+Business Media, LLC, New York, 2016). [Google Scholar]
- 82.Valvekens D., Van Montegu M., Van Lijsebettens M., Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. U.S.A. 85, 5536–5540 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession no. GSE164217).