Significance
Rice breeding programs aim to develop cultivars with improved traits, including high grain yield and superior quality. In rice, OsPDCD5 encodes a programmed cell death 5 protein. Targeted mutagenesis of OsPDCD5 enhanced grain yield and plant architecture. Statistical analysis indicated that plot grain yield of OsPDCD5 knockout lines was enhanced by 6.25 to 20.13% in 11 popular or newly bred rice cultivars compared with the corresponding wild types. The OsPDCD5 knockout lines showed increases in milled rice percentage and gel consistency, and a decrease in amylose content. Our results provide insight into the molecular mechanism by which OsPDCD5 influences grain yield and plant architecture, and highlight a promising candidate gene for use in breeding programs designed to develop super rice cultivars.
Keywords: OsPDCD5, grain yield, plant architecture, programmed cell death, polar auxin transport
Abstract
Plant architecture is an important agronomic trait that affects crop yield. Here, we report that a gene involved in programmed cell death, OsPDCD5, negatively regulates plant architecture and grain yield in rice. We used the CRISPR/Cas9 system to introduce loss-of-function mutations into OsPDCD5 in 11 rice cultivars. Targeted mutagenesis of OsPDCD5 enhanced grain yield and improved plant architecture by increasing plant height and optimizing panicle type and grain shape. Transcriptome analysis showed that OsPDCD5 knockout affected auxin biosynthesis, as well as the gibberellin and cytokinin biosynthesis and signaling pathways. OsPDCD5 interacted directly with OsAGAP, and OsAGAP positively regulated plant architecture and grain yield in rice. Collectively, these findings demonstrate that OsPDCD5 is a promising candidate gene for breeding super rice cultivars with increased yield potential and superior quality.
Rice (Oryza sativa L.) is among the most important food crops worldwide. It is a staple food for half of the world’s population. Rice productivity has more than doubled primarily because of two quantum leaps triggered by the Green Revolution in the late 1950s and exploitation of heterosis in the late 1970s. The former advance is largely attributable to the discovery and subsequent widespread utilization of a mutant gene, sd1, in worldwide rice breeding programs, resulting in the development of modern semidwarf lodging-resistant cultivars that are responsive to high-input modern agricultural systems (1). The latter case is clearly attributable to the discovery of the wild-abortive cytoplasmic male sterility gene, WA, and the development and large-scale adoption of the three-line system of hybrid rice breeding in China (2). Subsequently, rice productivity in most countries has eventually attained respective plateaus, despite extensive efforts to exploit diverse strategies to further increase rice yield potential (3). One successful strategy has been to improve plant architecture through ideotype breeding (4) because plant architecture is known to influence the yield of grain crops through improved photosynthetic efficiency at the population level, as well as through the harvest index (5–7).
The efforts to discover large-effect genes that can increase rice productivity have been greatly facilitated by the recent progress in global rice functional genomics research, which has resulted in the cloning and characterization of more than 3,600 genes (8). The cloned rice genes include many quantitative trait loci that control plant architecture and grain yield. For example, PLANT ARCHITECTURE AND YIELD 1 (PAY1) optimizes plant architecture and increases grain yield in high-yielding rice cultivars by affecting auxin polar transport and distribution (9). Rice PIN PROTEIN 5B (OsPIN5b), an auxin efflux carrier-like gene, affects plant architectural traits, including plant height, shoot and root biomass, panicle length, and yield parameters by changing auxin homeostasis, transport, and distribution (10). Loss-of-function of IPA1 INTERACTING PROTEIN 1 (IPI1), which encodes a RING-finger E3 ligase, causes an increase in rice yield as a result of improved rice plant architectural traits (tiller number and panicle size) (11). In recent years, the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated Cas9) gene-editing system has emerged as a feasible and efficacious method to facilitate precision breeding in plants (12). The system has been used widely to edit disease resistance genes (13–16) and yield-related genes in rice. Individual yield-related rice traits, such as tiller number (17), grain quality (18), grain size (19), and panicle length (20), have been improved by gene editing. Rice plant architecture is a complex of important agronomic traits and is crucial for grain yield. The molecular characterization of genes that control plant architecture will help to promote the breeding of high-yielding super rice cultivars using the CRISPR/Cas9 system.
Programmed cell death (PCD) is an essential process in the life cycle of animals and plants. In plants, PCD plays a critical role in plant development [e.g., aerenchyma formation, senescence (21), and panicle size (22)] and responses to abiotic stresses (e.g., heat, drought, and cold) as well as biotic challenges (23). Human Programmed cell death 5 (PDCD5) participates in apoptosis caused by DNA damage, and decreased PDCD5 expression may be associated with carcinoma formation and malignant progression (24). In Arabidopsis (Arabidopsis thaliana), AtPDCD5 participates in PCD in the responses to UV-B DNA damage (25) and dark-induced senescence (26). Previously, we cloned rice PDCD5 (OsPDCD5) and showed that overexpression of OsPDCD5 causes many PCD symptoms, including inhibited leaf growth, DNA laddering, reduced total protein content, and mitochondrial dysfunction (27). Down-regulation of OsPDCD5 enhances salt stress tolerance by inhibiting the PCD pathways and regulating stress-related genes in rice (28). OsPDCD5 is induced by UV-B radiation (29). Based on the potential functions of PCD to influence multiple phenotypes, we speculated whether OsPDCD5 may affect important agronomic traits in rice.
In this study, we generated strong evidence that negative regulation of OsPDCD5 may enhance grain yield and improve plant architecture. We show that OsPDCD5 affects rice yield by negatively regulating several genes involved in the biosynthesis and signaling pathways of gibberellin (GA), cytokinin, and auxin. Our results lead us to hypothesize that manipulation of genes involved in PCD, such as OsPDCD5, shows potential utility for improvement of yield in future precision breeding of rice.
Results
OsPDCD5 Knockout Lines Show Greatly Improved Plant Architecture and Grain Yield.
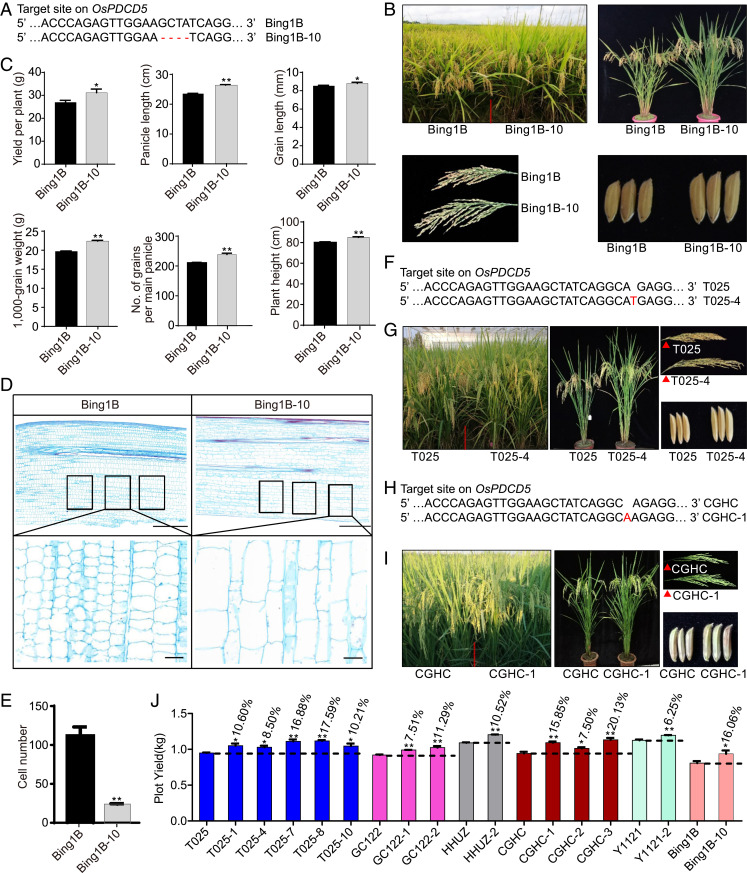
To determine the phenotypic effects of OsPDCD5 on grain yield and yield-related traits, we generated a CRISPR/Cas9 construct targeting the second exon of OsPDCD5 and transformed the construct into Bing1B, a recently released Xian (indica) maintainer line that is currently being promoted for adoption. Compared with Bing1B (the wild-type), the knockout line Bing1B-10 (harboring a 4-bp deletion in OsPDCD5) showed a significant (P < 0.05) increase in grain yield per plant by 4.2 g (15.7%), which was largely attributable to development of larger panicles (by 12.3%), longer grains (by 3.3%), increased 1,000-grain weight (by 13.8%), and increased number of grains per main panicle (by 13.3%), accompanied by a significant increase in plant height (by 5.1 cm or 6.4%) (Fig. 1 A–C). The RNAi-mediated suppression of OsPDCD5 in Bing1B resulted in plants that showed significantly increased plant height and larger panicles (SI Appendix, Fig. S1 A and B). The increase in plant height over that of Bing1B was mainly due to significant reduction in cell number and increase in cell size of the culm (Fig. 1 D and E) and reduced cell density mm−2 (SI Appendix, Fig. S1 C and D). These changes resulted in increased lengths of the basal to uppermost internodes (SI Appendix, Fig. S1E). In addition, we overexpressed OsPDCD5 in Nipponbare; the transgenic plants showed reductions in plant height and panicle length (SI Appendix, Fig. S1 F and G).
Fig. 1.
Phenotypes of OsPDCD5 knockout lines. (A) Target site in the OsPDCD5 sequence in the OsPDCD5 knockout line Bing1B-10. (B) Plant architecture, panicle, and grain phenotypes of the wild-type (Bing1B) and Bing1B-10. (C) Pleiotropic traits of Bing1B and Bing1B-10. (D) Stem longitudinal sections and a magnified view of the boxed areas. (Scale bars, 500 μm for the longitudinal sections and 50 μm for the magnified regions.) (E) Number of cells in the boxed sections in D. (F) Target site in the OsPDCD5 sequence in the OsPDCD5 knockout line T025-4. (G) Plant architecture, panicle, and grain phenotypes of the wild-type T025 and T025-4. (H) Target site in the OsPDCD5 sequence in the OsPDCD5 knockout line CGHC-1. (I) Plant architecture, panicle, and grain phenotypes of the wild-type CGHC and CGHC-1. (J) Plot yield of OsPDCD5 knockout lines (three replicates). The data are the mean ± SEM. *P < 0.05, **P < 0.01. “- - -” indicates the percentage increase in plot yield for the knockout lines (Student’s t test).
To evaluate the application potential of OsPDCD5 for optimization of rice plant architecture and increase in grain yield, we knocked out OsPDCD5 in 10 commercially grown or newly developed elite Xian and Geng (japonica) rice cultivars (T025, CGHC, GC122, Y1121, X1122, HHUZ, WSSM, R8117, YG4227, and WYG27) using the CRISPR/Cas9 system. Compared with the corresponding wild-types, all OsPDCD5 knockout lines showed significantly improved grain yield and plant architecture. The plot grain yield exceeded that of the corresponding wild-types by 10% for T025-1, T025-10, GC122-2, and HHUZ-2; 15% for Bing1B-10, T025-7, T025-8, and CGHC-1; 20.1% for CGHC-3; and 6 to 10% for T025-4, GC122-1, CGHC-2, and Y1121-2, thus ranging from 6.3 to 20.1% (Fig. 1 F–J and SI Appendix, Fig. S1 H and I). These results indicated that knockout of OsPDCD5 by the CRISPR/Cas9 system resulted in a generally increased yield with relatively small genetic background effects. We also recorded grain quality traits (chalkiness, brown rice rate, alkali spreading value, gel consistency, milled rice rate, head-milled rice rate, and amylose content) of the wild-type T025 and OsPDCD5 knockout lines T025-4 and T025-7 using 500-g mature seeds of each line. Compared with the wild-type, T025-4 and T025-7 showed increases in milled rice rates and gel consistency, and a decrease in amylose content (Table 1). Taken together, these results showed that OsPDCD5 is a potentially valuable gene resource for improvement of grain yield and plant architecture of rice.
Table 1.
Grain quality of two rice OsPDCD5 knockout lines and the wild-type T025
| Quality trait | Units | Cultivar name | OsPDCD5 knockout lines | |
| T025 | T025-4 | T025-7 | ||
| Chalkiness | % | 4.21 ± 0.44 | 3.35 ± 0.10 | 4.88 ± 0.13* |
| Brown rice rate | % | 78.42 ± 0.13 | 78.12 ± 0.66 | 79.25 ± 0.30** |
| Milled rice rate | % | 64.78 ± 1.77 | 68.95 ± 0.23** | 69.66 ± 0.07** |
| Head-milled rice rate | % | 56.64 ± 2.19 | 56.53 ± 0.16 | 64.20 ± 0.10** |
| Alkali spreading value | grade | 6.89 ± 0.19 | 4.18 ± 0.22** | 4.72 ± 0.18** |
| Gel consistency | mm | 65.79 ± 2.89 | 78.40 ± 0.75** | 90.26 ± 2.64** |
| Amylose | % | 21.94 ± 0.57 | 18.47 ± 0.08** | 18.04 ± 0.96** |
T025-4 and T025-7 are OsPDCD5 knockout lines of the wild-type T025. Data represent the mean ± SEM of three replicates. *P < 0.05, **P < 0.01 (Student’s t test).
To understand the molecular mechanisms of the phenotypic effects of OsPDCD5 on rice yield-related traits, we performed a series of experiments as described in the following sections.
Expression and Characterization of OsPDCD5 in Rice.
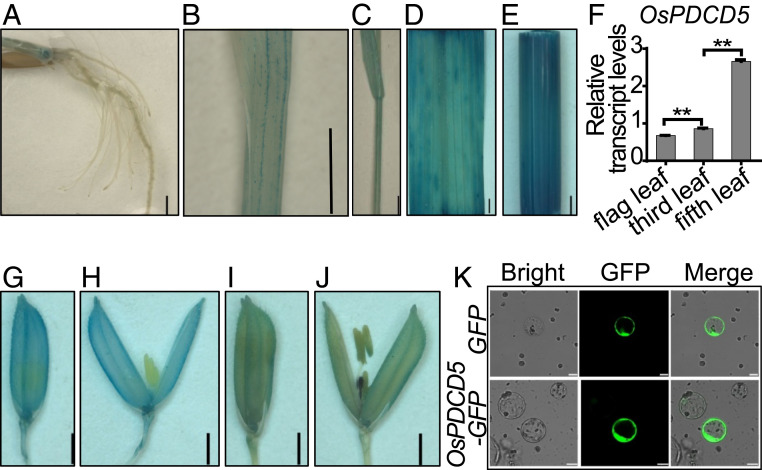
To understand the tissue expression pattern of OsPDCD5 in rice, we generated a transgenic β-glucuronidase (GUS) reporter line (OsPDCD5 promoter::GUS) by transforming the reconstructed vector pCAMBIA1301/OsPDCD5 promoter into the Xian cultivar Bing1B. At the seedling stage, GUS expression was observed in the roots (Fig. 2A), leaf vascular bundles (Fig. 2B), and internodes (Fig. 2C) of the transgenic plants. The expression levels in the culms and leaves were higher at the heading stage (Fig. 2 D and E) than at the seedling stage (Fig. 2 B and C). In the flag, third, and fifth leaves at the heading stage, OsPDCD5 expression levels were higher in older leaves than in young leaves (Fig. 2F). These results were consistent with previous findings (29) and indicated that OsPDCD5 was expressed strongly in mature tissues. OsPDCD5 expression was also detected in the glume at the early heading stage (Fig. 2 G and H) and in the anther at the late heading stage (Fig. 2 I and J). In addition, transient expression assays in rice protoplasts showed that the OsPDCD5-GFP (green fluorescent protein) fusion protein was localized in the nucleus and cytoplasm (Fig. 2K), which was consistent with the subcellular localization of Arabidopsis AtPDCD5 (25). Overall, our findings suggested that OsPDCD5 was a broadly functioning PCD-associated gene in rice.
Fig. 2.
Expression pattern and subcellular localization of OsPDCD5. (A–E and G–J) Root (A), leaf (B), and internode (C) of 14-d-old seedlings, leaf (D) and culm (E) at the heading stage, glume at the early heading stage (G and H), and glume at the late heading stage (I and J) in a rice transgenic GUS reporter line (OsPDCD5 promoter::GUS). (Scale bars, 2 mm.) (F) Expression levels of OsPDCD5 in leaves at the mature stage in the wild-type Bing1B (three replicates). Tubulin was the loading control. The expression levels are the mean ± SEM, **P < 0.01 (Student’s t test). (K) Subcellular localization of OsPDCD5 in rice protoplasts. (Scale bars, 10 μm.)
Transcriptome Analysis of OsPDCD5 Knockout Plants.
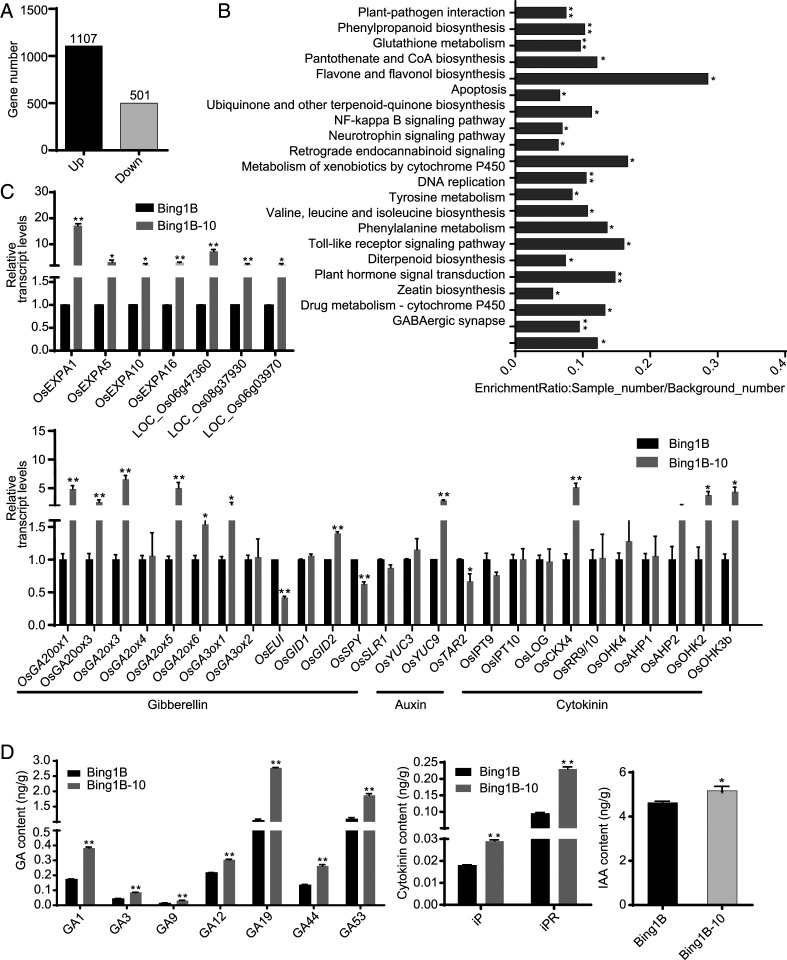
RNA-sequencing (RNA-seq) analysis of the leaf transcriptome of wild-type Bing1B and Bing1B-10 plants at the tillering stage was conducted (Fig. 3). A total of 1,608 differentially expressed genes (DEGs) were detected (false-discovery rate P < 0.05), of which 68.8% (1,107 genes) were up-regulated and 31.2% (501 genes) were down-regulated in the Bing1B-10 plants compared with the expression levels in Bing1B plants (Fig. 3A). Gene ontology (GO) analysis indicated that these DEGs were significantly (P < 0.05) enriched for cellular component, molecular function, and biological process terms. Among biological processes, most DEGs were associated with metabolic and cellular processes. With regard to cell composition, the majority of DEGs were associated with cell components and organelles. Most DEGs in the molecular function category were associated with binding function and catalytic activity (SI Appendix, Fig. S2). A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that the DEGs were mainly enriched in apoptosis, plant hormone signal transduction, and zeatin biosynthesis (Fig. 3B). Comparison between Bing1B-10 and Bing1B of the transcript levels of expansin-related genes and those involved in the GA, cytokinin, and auxin biosynthetic and signaling pathways revealed that the expression levels of these genes (e.g., OsGA20ox1, OsGA3ox1, OsEXPA1, OsEXPA5, and OsEXPA10) differed significantly between Bing1B-10 and Bing1B leaves (Fig. 3C). As a result, we detected significant differences in the endogenous GA, cytokinin, and indole-3-acetic acid (IAA) contents between Bing1B-10 and Bing1B. Specifically, the GA1, GA3, GA9, GA12, GA19, GA44, GA53, isopentenyladenosine, isopentenyladenosine riboside, and IAA contents in Bing1B-10 leaves were higher than those in Bing1B leaves (Fig. 3D). These results indicated that the biosynthesis of auxin, GA, and cytokinin was promoted in the OsPDCD5-knockout Bing1B-10 plants, as were the associated signaling pathways, including expansin-related genes.
Fig. 3.
Transcriptome of the rice OsPDCD5 knockout line Bing1B-10 and the wild-type Bing1B. (A) Number of up- and down-regulated genes (false-discovery rate P < 0.05) in leaves at the tillering stage of Bing1B-10 compared with Bing1B. (B) KEGG pathway enrichment analysis of DEGs. (C) Transcript levels of expansin-related genes and genes involved in the GA, auxin, and cytokinin biosynthesis and signaling pathways at the tillering stage. Actin was used as the loading control. (D) GA contents at the tillering stage, cytokinins (isopentenyladenosine and isopentenyladenosine riboside) content at the seedling stage, and IAA content at the tillering stage in Bing1B-10 and Bing1B. The data are the mean ± SEM (three replicates in C and D): *P < 0.05, **P < 0.01 (Student’s t test).
OsPDCD5 Interacts with OsAGAP and OsAGAP Positively Regulates Plant Architecture.
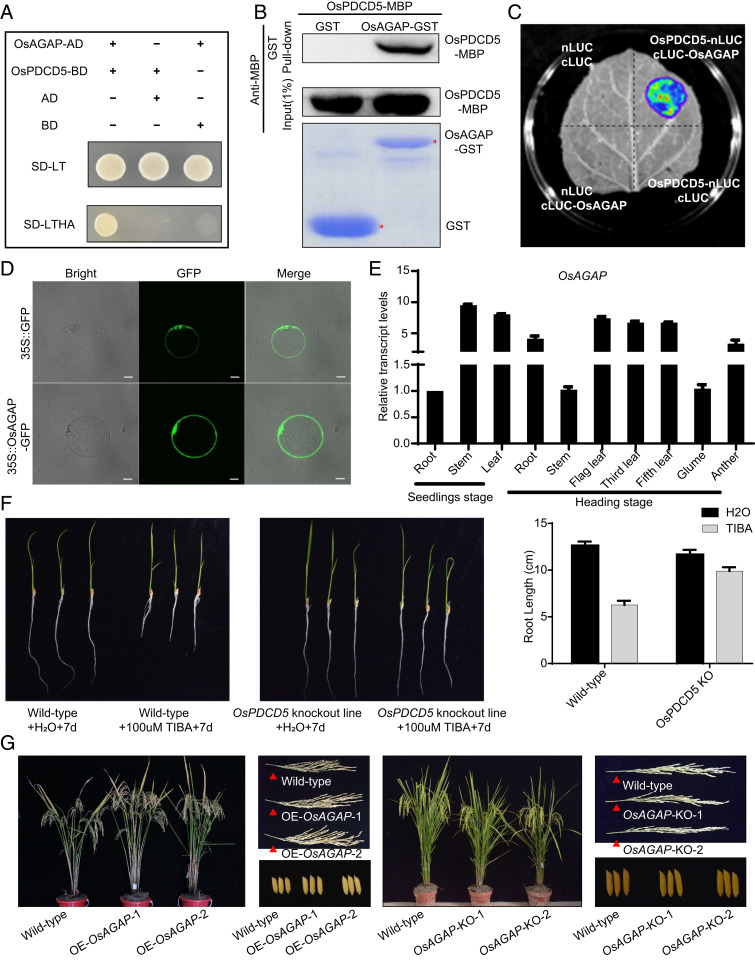
To identify candidate OsPDCD5-interacting proteins that participate in rice growth and development, we performed yeast two-hybrid screening of a normalized rice cDNA prey library prepared from calli, leaves at the seedling and booting stages, young panicles, and grains. This assay led to identification of OsAGAP as the most likely protein that interacts with OsPDCD5. To confirm this hypothesis, we cloned the full-length cDNA of OsPDCD5 and confirmed that OsPDCD5 and OsAGAP were capable of interacting in yeast cells (Fig. 4A). To validate this result, we performed in vitro and in vivo protein interaction assays. We produced the glutathione S-transferase (GST) and maltose‐binding protein (MBP) fusion proteins OsAGAP-GST and OsPDCD5-MBP using Escherichia coli. Pull-down assays showed that OsAGAP-GST, but not GST alone, retained OsPDCD5-MBP (Fig. 4B). The interaction was validated using a split-luciferase assay, which showed that luciferase (LUC) activity was reconstituted when OsPDCD5-nLUC and cLUC-OsAGAP were coexpressed in tobacco (Nicotiana benthamiana) leaves. No luciferin signal was observed in the coexpression of OsPDCD5-nLUC (LUC N terminus) and cLUC (LUC C terminus) or in the coexpression of nLUC and OsAGAP-cLUC (Fig. 4C). In addition, the localization of OsAGAP in the nucleus and cytoplasm (Fig. 4D) was consistent with that of OsPDCD5. Consistent with OsPDCD5, OsAGAP was expressed from the seedling to the heading stages in all examined tissues (Fig. 4E). We compared the OsAGAP transcript level in OsPDCD5 overexpression and RNAi-suppressed lines (OE-OsPDCD5 and RNAi-OsPDCD5, respectively) with that in the corresponding wild-types. The transcript level of OsAGAP in RNAi-OsPDCD5 lines was higher than that in the wild-type Bing1B, whereas the transcript level in OE-OsPDCD5 lines was lower than that in the wild-type Nipponbare (SI Appendix, Fig. S3).
Fig. 4.
Interaction of OsPDCD5 and OsAGAP. (A) Yeast two-hybrid assay of OsPDCD5 with OsAGAP. Full-length coding sequences of OsAGAP and OsPDCD5 were cloned into AD (the prey plasmid pGADT7) or BD (the bait plasmid pGBKT7). Yeast cells transformed with the plasmids were grown on control SD-LT medium and selective SD-LTHA medium. (B) Pull-down assay. OsAGAP was fused with the GST tag, and OsPDCD5 was fused with the MBP tag. After coincubating the two proteins, they were immunoprecipitated with glutathione-Superflow resin and detected using anti-MBP antibodies. (C) Split-luciferase assay. OsPDCD5 and OsAGAP were fused separately with the N-terminal (nLUC) and C-terminal (cLUC) portions of firefly luciferase (LUC). Different combinations of constructs were agro-infiltrated into tobacco leaves, and chemiluminescence was imaged after adding the substrate luciferin. (D) Subcellular localization of OsAGAP in rice protoplasts. (Scale bars, 10 μm.) (E) Expression analysis of OsAGAP in Bing1B as revealed by quantitative real-time PCR (three replicates). Tubulin was used as the loading control. (F) Root elongation in response to TIBA treatment. (G) Phenotypes of transgenic lines overexpressing OsAGAP (OE-OsAGAP-1 and OE-OsAGAP-2) and the wild-type Bing1B, and of OsAGAP knockout lines (OsAGAP KO-1 and OsAGAP KO-2) and the wild-type CGHC.
OsAGAP plays an important role in regulating the auxin influx pathway of rice by mediating root polar auxin transport (PAT) (30), and indirectly regulates many aspects of plant growth and development (31). Overexpression of OsAGAP impairs PAT by regulating vesicle trafficking pathways and affects the localization of the Arabidopsis auxin-influx carrier AUX1 (31). To determine if this was also the case in rice, we treated the OsPDCD5 knockout line (Bing1B-10) with 100 µM 2,3,5-triiodobenzoic acid (TIBA), a PAT inhibitor, for 7 d. The TIBA-induced inhibition of root elongation was much stronger in the wild-type than in the OsPDCD5 knockout line. In the OsPDCD5 knockout line, root growth showed decreased sensitivity to TIBA. Thus, the increase in resistance to TIBA suggested that the OsPDCD5 knockout line was defective in PAT (Fig. 4F). Importantly, compared with the wild-type, the OsAGAP overexpression lines showed significant increases in plant height, panicle length, grain length, 1,000-grain weight, number of grains per main panicle, and yield per plant (Fig. 4G and SI Appendix, Fig. S4). The OsAGAP knockout lines showed decreases in plant height, panicle length, and number of grains per main panicle (Fig. 4G and SI Appendix, Fig. S4). Taken together, these results indicated that OsPDCD5 regulated rice yield and plant architecture at least partially through interacting with OsAGAP.
Evolutionary Relationships of OsPDCD5.
OsPDCD5 was predicted to encode a protein of 129 amino acids that contained a double-stranded DNA-binding domain (SI Appendix, Fig. S5A). Homologous proteins were detected by BLAST searches of the UniProt database (https://www.uniprot.org/). A multiple sequence alignment of OsPDCD5 and the detected homologs was used to construct a phylogenetic tree using the neighbor-joining method. Homologs of OsPDCD5 were identified in many diverse plant species, including rice, Arabidopsis, white poplar (Populus alba), Hall’s panicum (Panicum hallii), wormwood (Artemisia annua), eelgrass (Zostera marina), cotton (Gossypium raimondii and Gossypium hirsutum), and Sitka spruce (Picea sitchensis) (SI Appendix, Fig. S5B), and in major grain crops, such as sorghum (Sorghum bicolor), foxtail millet (Setaria italica), wheat (Triticum aestivum), barley (Hordeum vulgare), and maize (Zea mays) (SI Appendix, Fig. S5C). These homologs all shared more than 77% sequence identity with OsPDCD5; the highest identities observed were with OsPDCD5 sequences from the wild rice species Oryza glaberrima (100%) and Oryza brachyantha (96.9%) (SI Appendix, Table S1). The wide-ranging presence and high similarity of OsPDCD5 homologs indicated that OsPDCD5 was highly conserved throughout the plant kingdom, implying that it performs fundamentally important biological functions.
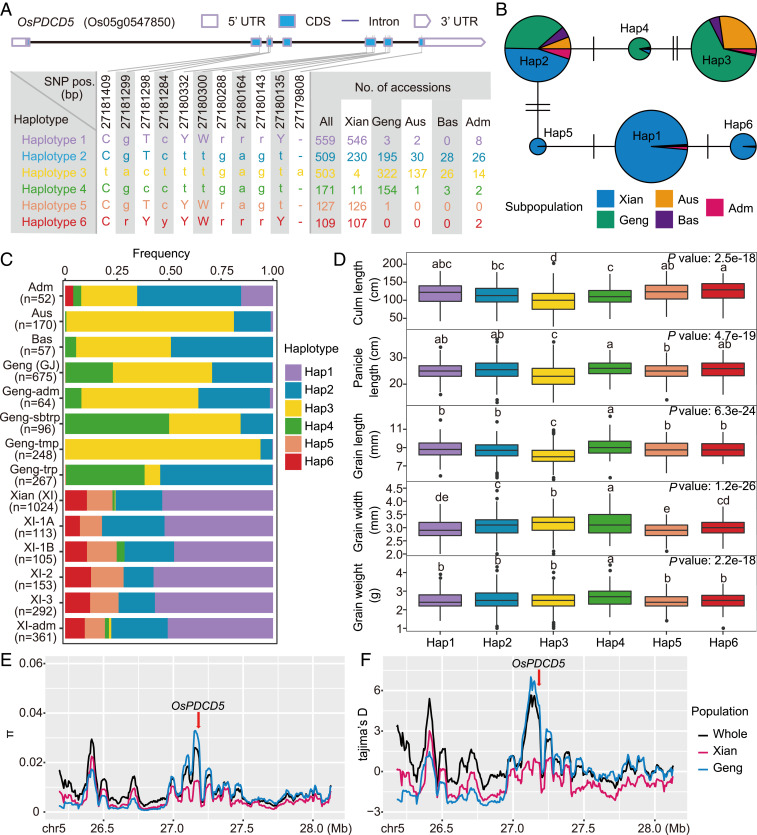
To understand the evolutionary relationships of OsPDCD5 among the subspecies of rice, we analyzed the polymorphism of OsPDCD5 in the 3K RG dataset (32). We detected six major haplotypes of OsPDCD5 (Fig. 5A) using 11 single-nucleotide polymorphisms (SNPs) in the coding sequence (CDS) region in 1,978 accessions of the 3K RG dataset (rare haplotypes of <100 accessions are not shown). The frequencies of the major haplotypes differed significantly between the subspecies Xian and Geng except for Hap2 (Fig. 5 B and C), suggesting that OsPDCD5 may have contributed to the differentiation of Xian and Geng. ANOVA revealed significant differences in five yield-related traits (culm length, panicle length, grain length, grain width, and grain weight) among the six haplotypes (Hap). Interestingly, for the culm, panicle, and grain lengths, Hap1, Hap2, Hap4, Hap5, and Hap6 showed significantly higher mean trait values than those of Hap3 (Fig. 5D). These phenotypic improvements of Hap1, Hap2, Hap4, Hap5, and Hap6 were apparently caused by the deletion of base A at the 27,179,808-bp position in OsPDCD5 because the deletion caused 4 amino acid changes in the tail end of the gene, and delayed the stop codon with an additional 16 amino acids (Fig. 5A and SI Appendix, Fig. S5 D and E). With regard to the nucleotide diversity (π) and Tajima’s D statistic in the ∼2-Mb region flanking OsPDCD5 in the Xian and Geng subpopulations, we observed that the Geng population had a significantly higher π value than the Xian population (Fig. 5E). The Geng population also showed a positive peak in Tajima’s D in the 2-Mb genomic region around OsPDCD5 compared with that of the Xian population (Fig. 5F), indicating that OsPDCD5 had undergone strong balancing selection in the Geng subpopulation.
Fig. 5.
Genetic diversity of OsPDCD5 in the 3K RG dataset. (A) Haplotypes of OsPDCD5 (Os05g0547850) in 1,978 accessions of 3K RG (rare haplotypes of <100 accessions are not shown) using 11 SNPs in the CDS region. Lowercase letters represent synonymous mutations, whereas uppercase letters indicate nonsynonymous mutations. (B) Haplotype network of OsPDCD5 in 3K RG. (C) Haplotype frequency of OsPDCD5 in subpopulations of 3K RG. (D) Performance distribution of different haplotypes of OsPDCD5 on culm length, panicle length, grain length, grain width, and grain weight in 3K RG. Different letters on the boxplots indicate statistically significant differences (P < 0.01, Duncan’s new multiple range test). (E and F) Nucleotide diversity (π) and Tajima’s D for ∼2-Mb genomic region flanking OsPDCD5 in 3K RG. The red arrow indicates the position of OsPDCD5.
Discussion
PCD is a complex multifaceted process involved in many aspects of the growth and development of animals and plants. As an essential process leading to cell apoptosis during the normal life cycle of plants (33–35), PCD is also an important response to external abiotic and biotic stresses (36). We have previously shown that overexpression of OsPDCD5 naturally induces plant PCD and that constitutive expression of antisense-OsPDCD5 results in increased salt tolerance (28). In the present study, we demonstrated that knockout of OsPDCD5 significantly increased grain yield in diverse genetic backgrounds. The increase in yield of the OsPDCD5 knockout lines may be attributable to their increased source (improved plant architecture and increased net photosynthetic rate) and sink capacity (increased number of grains per plant and heavier grains) (SI Appendix, Figs. S1 H and I, and S6). The increased source and sink capacity of the OsPDCD5 knockout lines was not associated with any change in harvest index (SI Appendix, Fig. S7). These results were consistent with our current knowledge of the yield potential of cereal crops (37).
Taken together, these results suggest the potential for improvement of rice productivity by manipulation of OsPDCD5-regulated PCD in rice. Sequence comparison among the six major haplotypes at OsPDCD5 revealed that the deletion of an adenine residue at position 27,179,808 bp was responsible for a functional change of OsPDCD5 and significant differences in yield-related traits were observed between Hap3 and Haplotypes 1, 2, 4, 5, or 6 in the 3K RG dataset (Fig. 5D). However, no base deletion was detected in the OsPDCD5-edited cultivars (SI Appendix, Fig. S5D), which also showed improved yields compared with those of the corresponding wild-types. Our results show that Hap3 has undergone strong balancing selection in temperate Geng cultivars. Hap4 showed different functions to those of Hap3, and was present at a high frequency among the subtropical and tropical Geng cultivars, especially the former cultivars. However, a subspecies-level differentiation of haplotypes was not observed among the five subspecies of the Xian population (Fig. 5C). This is probably the main reason why the Geng population showed a significantly higher π and Tajima's D values than the Xian population. Considering that Hap3 and Hap4 have different functions and are rare in the Xian population, rice cultivars can be improved with Hap3 and Hap4 (without deletions) according to the different objectives in future molecular breeding. Taken together, these results suggest there is substantial potential for improvement of rice productivity by accurate manipulation of OsPDCD5-regulated PCD using CRISPR technology.
OsPDCD5 encodes a predicted protein with a double-stranded DNA-binding domain (SI Appendix, Fig. S5A) similar to its human homologs. Up-regulation of OsPDCD5 may induce PCD, including DNA damage, reactive oxygen species production, and mitochondrial swelling (38). At present, there are few reports of PCD-associated genes affecting yield. For example, the paab1-1 mutant harbors a mutation in OsALMT7 and the apical spikelet in the paab1-1 mutant undergoes PCD. OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport (22). The early senescence 2 (es2) mutant exhibits cell death and elevated reactive oxygen species accumulation in the leaves, and the es2 mutation causes premature leaf senescence and influences yield-related traits in rice (39). Thus, a natural question concerns the reason that negative regulation of OsPDCD5 causes simultaneous increase in salt tolerance, improvement in plant architecture, and increase in yield. The present transcriptome analysis indicated that OsPDCD5 regulated expression level of large numbers of genes involved in the biosynthesis of auxin, GA, and cytokinin, as well as their signaling pathways, and thus interfered with plant growth and reproduction, and responses to external stimuli (Fig. 3C).
We further show that the pleiotropic effects of OsPDCD5 were at least partially achieved through its direct interaction with OsAGAP (Fig. 4 A–C) because OsAGAP overexpression lines showed similar increases in plant height, panicle length, and number of grains per main panicle (Fig. 4G). OsAGAP affects PAT by regulating vesicle trafficking pathways and the localization of the presumptive auxin-influx carrier AUX1 (31). Interestingly, we observed that OsAGAP knockout lines exhibited the opposite phenotypes to the overexpression lines (Fig. 4G). In addition, the OsPDCD5 knockout line showed decreased sensitivity to the PAT inhibitor TIBA (Fig. 4F). These results suggest that altered polar transport-mediated auxin distribution was at least partially responsible for the observed phenotypic effects on yield-related traits and plant architecture of the OsPDCD5 knockout lines (40, 41). In this respect, OsPDCD5 was indicated to function as a hormone facilitator of PCD in rice, but it remains unclear how OsPDCD5 influenced the biosynthesis of auxin, GA, and cytokinin, which requires elucidation in future. The present results suggest there is considerable potential for simultaneous improvement of the productivity and stress tolerance of rice, and probably other cereals, by fine-tuning PCD through accurate manipulation of important PCD-associated genes, such as OsPDCD5. Further investigation of PCD in plants is crucial to understand the regulatory mechanisms of development- and defense-associated processes.
Accession Codes.
Sequence data from this article are available at the Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/) under accession numbers Os05g0547850 (OsPDCD5) and Os02g0198300 (OsAGAP); both cDNAs were cloned from Bing1B.
Materials and Methods
Plant Materials and Growth Conditions.
OsPDCD5 knockout lines and the corresponding wild-types were germinated and transplanted in an experimental field (Taicang, China; 31°33′40″N, 121°09′18″E) in summer and in a different experimental field (Hainan, China; 18°18′52″–18°28′29″N, 109°03′05″–115°15′58″E) in winter. All the planting was done according to well-established standard practices, with intra- and interrow spacing of 13.3 cm and 26.4 cm, respectively. Field management followed standard rice production practices. Knockout homozygous T1 mutants were selected to backcross with the receptor parent and the F2 population was generated by single-seed descent with self-pollination. The F2 population was used for the genotypic and phenotypic analysis. Knockout mutants were confirmed by PCR with the primers listed in SI Appendix, Table S2. The rice (O. sativa) cultivars used in this study are listed in SI Appendix, Table S3. Student’s t test was used to evaluate the significance of differences between two samples.
Transgene Constructs and Targeted Gene Editing.
For OsPDCD5 overexpression, the full-length OsPDCD5 protein-coding sequence (CDS: 387 bp) was amplified from Bing1B and cloned into the vector pMD19-T, then introduced into the plant binary vector pCAMBIA1304 to generate the expression vector pCaMV35S::OsPDCD5. For OsPDCD5 down-regulation, the OsPDCD5 CDS (+1 to +387) was placed in both the sense and antisense orientations separated by an intron sequence in the vector pTCK303 to generate an RNAi vector. To generation knockout plants using CRISPR/Cas9 technology, single-guide RNA targeting 5′-GCCCAGAGTTGGAAGCTATC-3′ was cloned downstream of the OsU6 promoter in the CRISPR/Cas9 binary vector BGK032 (Biogle Technology). These constructs were introduced into the corresponding rice cultivars by Agrobacterium-mediated transformation using standard protocols. The transgenic rice plants were confirmed by quantitative real-time PCR (qPCR) or PCR detection and direct sequencing.
Plant Photosynthesis Measurements.
The photosynthetic parameters were measured on attached, fully developed leaves of Bing1B-10 and Bing1B at the tillering stage using a portable system (GFS-3000; Heinz Walz). The following parameters were measured: transpiration, vapor pressure deficit, internal CO2 concentration, stomatal conductance, and net photosynthetic rate. The GFS-3000 instrument was preheated 60 min before the measurements were recorded. Airflow was maintained at 750 μM s−1. In the leaf chamber of the GFS-3000, the air temperature was kept constant (20 °C), the CO2 mass was 0.05%, and the relative humidity was 50%. Data were recorded after the parameters of each group had stabilized.
Rice Quality Measurements.
Chalkiness, brown rice rate, milled rice rate, head-milled rice rate, amylose content, gel consistency, and alkali spreading value were determined as described previously (42). Briefly, about 500 g of grains harvested from T025 (wild-type) and OsPDCD5 knockouts T025-4 and T025-7 were dried for quality analysis. A 150-g sample of the dried grains was passed twice through a dehusker, polished, then separated into broken and unbroken grains. The brown rice rate, milled rice rate, and head rice rate were expressed as percentages of the total (150 g) grains. Chalkiness was evaluated visually on 100 milled grains. Grain samples comprising at least 20% white-belly, white-core, and white-back grains or a combination of these categories were considered to be chalky. Gel consistency, alkali spreading value, and amylose content were measured according to the Rice Quality Measurement Standards (Ministry of Agriculture, People’s Republic of China, 1988).
Primers.
The sequences of all primers used to construct vectors in this study are listed in SI Appendix, Table S4.
Yeast Two-Hybrid Assays.
Yeast two-hybrid screening was conducted in accordance with the protocol provided by Clontech. Briefly, cDNA encoding OsPDCD5 was inserted into the bait plasmid PGBKT7 and a cDNA library of all tissues was incorporated into the prey plasmid PGADT7 (Clontech). Yeast cells containing the bait and prey plasmids were grown on SD−Trp/−Leu/−His/−Ade medium. Colony PCR and sequencing were performed to verify the prey plasmid sequences. Interaction of OsPDCD5 and OsAGAP was assayed. The cDNA encoding OsPDCD5 was inserted into the bait plasmid PGBKT7, and the cDNA encoding OsAGAP was cloned into the pGADT7 vector. Both vectors were cotransformed into the yeast strain AH109 and by coating the transformant on solid −Trp/−Leu medium. After 3 d at 30 °C, randomly selected cells were transferred to −Trp/−Leu and −Trp/−Leu/−His/−Ade plates to test the OsPDCD5–OsAGAP interaction.
RNA-Seq Analysis.
Leaves of Bing1B and Bing1B-10 at the tillering stage were harvested and immediately frozen in liquid nitrogen. Total RNA was extracted with TRIzol Reagent (Invitrogen). RNA-seq analysis was performed by Major Bio Briefly, 2 mg total RNA was used for mRNA purification and cDNA synthesis. Double-stranded cDNA was generated by reverse transcription and DNA-dependent DNA synthesis. The amplified DNA fragments were sequenced with an Illumina HiSeq X Ten System platform. For data analysis, the gene-expression level was calculated and normalized to fragments per kilobase of transcript per million mapped reads. The Cufflinks suite (http://cole-trapnell-lab.github.io/cufflinks/) was used to splice transcripts and detect DEGs (P < 0.05, fold-change > 2). DEGs were automatically annotated and manually categorized in accordance with the putative or demonstrated function based on GO. A GO enrichment analysis was performed using the Gene Ontology Resource (http://geneontology.org/). A KEGG pathway enrichment analysis was performed using the KEGG database (https://www.genome.jp/kegg/).
Measurement of Free IAA, GA, and Cytokinin Content.
Endogenous IAA and GA contents were determined by the Greensword Creation Technology Company in accordance with a previously reported method with slight modification (43). Free cytokinin was determined by the same company in accordance with a method described previously (44). For these assays, two samples of young leaves (∼2-g fresh weight per sample) were collected from Bing1B-10 and Bing1B at the seedling (30-d-old) and tillering stages. After extraction and purification, the samples were analyzed by LC-MS/MS.
Split-Luciferase Assay.
The CDS of OsPDCD5 and OsAGAP were fused in-frame with nLUC and cLUC, respectively. These recombinant plasmids and empty vector controls were transformed into Agrobacterium strain GV3101. Equal amounts of bacteria carrying the nLUC or cLUC fusion plasmid were combined and infiltrated into tobacco (N. benthamiana) leaves. The luciferase luminescence signals were imaged 3 d after infiltration using an in vivo plant imaging system (NIGHTSHADE LB 985; Berthold).
Pull-Down Assays.
GST alone and the OsAGAP-GST and OsPDCD5-MBP fusion proteins were expressed in E. coli strain Rosetta. Equal volumes of GST or the OsAGAP-GST fusion protein and glutathione Superflow resin (40 µL) were incubated with the OsPDCD5-MBP fusion protein in 1.0 mL total volume of pull-down buffer (20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1‰ [vol/vol] Tween-20, and protease inhibitor) at 4 °C with gentle upside-down rotation overnight. After extensive washing, 2× SDS loading buffer (40 µL) was added to each of the tubes containing the washed beads, and recombinant proteins were separated by 12% SDS/PAGE and detected by Western blotting using either anti-GST or anti-MBP antibodies.
qPCR.
Total RNA was extracted using the RNAprep pure Plant Kit (Tiangen, DP432) in accordance with the manufacturer’s instructions. First-strand cDNA was synthesized using the PrimeScript RT reagent Kit (Takara, RR036A) and qPCR was performed with SYBR Premix Ex Taq (Takara, RR820A). Relative expression levels were normalized to that of ubiquitin, tubulin, or actin. All primers used in the qPCR analyses are listed in SI Appendix, Table S5.
GUS Staining.
The OsPDCD5 promoter (1 to 1,404 bp upstream of the OsPDCD5 CDS) was cloned into the plasmid pCAMBIA1301 to express the OsPDCD5 promoter and GUS reporter fusion protein. Combinations were transformed into the Xian rice Bing1B by Agrobacterium-mediated transformation. The T0-positive plants were self-crossed and used to generate T1 mutants as the experimental material. Different tissues were added to X-Gluc staining solution and incubated at 37 °C for 12 h in the dark. After the staining was completed, the tissues were decolorized with absolute ethanol. The GUS reporter gene expression site was observed and photographed.
Subcellular Localization.
The full-length cDNA of OsPDCD5 was cloned into the pYL322-d1 vector to express the OsPDCD5-GFP fusion protein at the N terminus of GFP. The fusion construct (35S::OsPDCD5-GFP) and control construct (35S::GFP) were transformed separately into rice protoplasts. The fluorescence was detected by confocal microscopy (Leica Microsystems, TCS SP8) after culture for 14 h.
Histological Analysis.
A histological analysis was performed in accordance with a method described previously with minor modifications (45). Briefly, stems at the heading stage were fixed in FAA (70% ethanol, 5% glacial acetic acid, 5% formaldehyde, and 35% water), vacuum-infiltrated for 10 min, kept at room temperature for 16 h, then dehydrated in a gradient ethanol series. The samples were embedded in Paraplast, sectioned using a microtome (RM2235, Leica), stained with 0.5% Fast Green, and photographed under a microscope (Zeiss). The glume inner surface of mature grains was observed with a scanning electron microscope (TM3000, Hitachi) as described previously (45, 46). Visual observations were conducted at 180× magnification. Cell number was measured using ImageJ software.
Trait Measurements.
All yield traits were measured when the plants had attained maturity. Panicle length, grain length, grain number, yield per plant, plot yield, and 1,000-grain weight were recorded. Yield per plant was scored as the total weight of grains from the entire plant. The number of tillers per plant was scored as the number of reproductive tillers for each plant. For plot yield, 100 plants for each line were grown in a paddy field with three replicates, and 40 plants were collected for yield measurement, except that for Bing1B 30 plants were collected. Grain length, grain number, and 1,000-grain weight were measured using an automatic seed counting and analyzing instrument (Model SC-G; Wanshen). Plant height and panicle length were measured and analyzed.
Population Genetic Analysis of OsPDCD5.
The haplotypes of OsPDCD5 in the 3k RG were classified according to all SNPs with minor allele frequency > 0.01 within the CDS region using the RFGB v2.0 database (47). The haplotypes in at least 100 rice accessions were used for comparative analysis of five yield-related traits (culm length, panicle length, grain length, grain width, and grain weight), which were downloaded from the Rice SNP-Seek Database (32). One-way ANOVA followed by Duncan’s new multiple-range test were performed with the agricolae package in R. Haplotype networks were constructed using the pegas package in R. Nucleotide diversity (π) and Tajima’s D for each 50-kb window across the genome, with an overlapping 5-kb step size, were calculated for the 2-Mb region flanking OsPDCD5 with the Variscan software (v2.0.3) (48).
Supplementary Material
Acknowledgments
We thank Robert McKenzie (Liwen Bianji, Edanz) (https://www.liwenbianji.cn/), for editing the English text of a draft of this manuscript. This work was supported by the Genetically Modified Organisms Breeding Major Projects (2016ZX08001004), the Shanghai Science and Technology Innovation Action Plan (19391900400), and the National Natural Science Foundation of China (31671655 and 31971918).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018799118/-/DCSupplemental.
Data Availability
Sequence data from this article are available at the Rice Annotation Project Database, https://rapdb.dna.affrc.go.jp/ (accession nos. Os05g0547850 [OsPDCD5] and Os02g0198300 [OsAGAP]); both cDNAs were cloned from Bing1B. All other study data are included in the article and SI Appendix.
References
- 1.Spielmeyer W., Ellis M. H., Chandler P. M., Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U.S.A. 99, 9043–9048 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H., Deng X. W., Development of the, “third-generation” hybrid rice in China. Genomics Proteomics Bioinf. 16, 393–396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang S., Chen W. F., Zhang L. B., Trends in breeding rice for ideotype. Zhongguo Shuidao Kexue 2, 129–135 (1988). [Google Scholar]
- 4.Khush G. S., What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 59, 1–6 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Zhang N., et al., A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell 30, 1461–1475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhardt D., Kuhlemeier C., Plant architecture. EMBO Rep. 3, 846–851 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishiwata A., et al., Two WUSCHEL-related homeobox genes, narrow leaf2 and narrow leaf3, control leaf width in rice. Plant Cell Physiol. 54, 779–792 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Wing R. A., Purugganan M. D., Zhang Q., The rice genome revolution: From an ancient grain to green super rice. Nat. Rev. Genet. 19, 505–517 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Zhao L., et al., PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 83, 528–536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu G., et al., OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83, 913–925 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Wang J., et al., Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29, 697–707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J. F., et al., Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C., et al., A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 18, 313–315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S., et al., Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant Biol. 61, 1201–1205 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Macovei A., et al., Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y. A., Moon H., Park C. J., CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice (N. Y.) 12, 67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Q., et al., Targeted mutagenesis of the rice FW 2.2-like gene family using the CRISPR/Cas9 system reveals OsFWL4 as a regulator of tiller number and plant yield in rice. Int. J. Mol. Sci. 21, 809 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basnet R., Zhang J., Hussain N., Shu Q., Characterization and mutational analysis of a monogalactosyldiacylglycerol synthase gene OsMGD2 in rice. Front. Plant Sci. 10, 992 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji X., et al., The basic helix-loop-helix transcription factor, OsPIL15, regulates grain size via directly targeting a purine permease gene OsPUP7 in rice. Plant Biotechnol. J. 17, 1527–1537 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Y., Wen J., Zhao W., Wang Q., Huang W., Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR-Cas9 system. Front. Plant Sci. 10, 1663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Hautegem T., Waters A. J., Goodrich J., Nowack M. K., Only in dying, life: Programmed cell death during plant development. Trends Plant Sci. 20, 102–113 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Heng Y., et al., OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell 30, 889–906 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams B., Dickman M., Plant programmed cell death: Can’t live with it; can’t live without it. Mol. Plant Pathol. 9, 531–544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L., et al., PDCD5 interacts with Tip60 and functions as a cooperator in acetyltransferase activity and DNA damage-induced apoptosis. Neoplasia 11, 345–354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcone Ferreyra M. L., et al., AtPDCD5 plays a role in programmed cell death after UV-B exposure in Arabidopsis. Plant Physiol. 170, 2444–2460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falcone Ferreyra M. L., Casati P., AtPDCD5 plays a role during dark-senescence in Arabidopsis. Plant Signal. Behav. 11, e1176820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attia K., et al., Overexpression of the OsPDCD5 gene induces programmed cell death in rice. J. Integr. Plant Biol. 47, 1115–1122 (2005). [Google Scholar]
- 28.Yang M. F., et al., Down-regulation of OsPDCD5, a homolog of the mammalian PDCD5, increases rice tolerance to salt stress. Mol. Breed. 31, 333–346 (2013). [Google Scholar]
- 29.Su W., et al., Interaction between programmed cell death 5 and calcineurin B-like interacting protein kinase 23 in Oryza sativa. Plant Sci. 170, 1150–1155 (2006). [Google Scholar]
- 30.Zhuang X., et al., Over-expression of OsAGAP, an ARF-GAP, interferes with auxin influx, vesicle trafficking and root development. Plant J. 48, 581–591 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Du C., Chong K., ARF-GTPase activating protein mediates auxin influx carrier AUX1 early endosome trafficking to regulate auxin dependent plant development. Plant Signal. Behav. 6, 1644–1646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandrov N., et al., SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 43, D1023–D1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koyano T., Kurusu T., Hanamata S., Kuchitsu K., “Regulation of vacuole-mediated programmed cell death during innate immunity and reproductive development in plants.” in Sexual Reproduction in Animals and Plants, Sawada H., Inoue N., Iwano M., Eds. (Springer, 2014), pp. 431–440. [Google Scholar]
- 34.Lord C. E. N., Gunawardena A. H. L. A. N., Programmed cell death in C. elegans, mammals and plants. Eur. J. Cell Biol. 91, 603–613 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Ranganath R. M., Nagashree N. R., Role of programmed cell death in development. Int. Rev. Cytol. 202, 159–242 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Lam E., Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5, 305–315 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Venkateswarlu B., Visperas R. M., Source-sink relationships in crop plants. IRRI Res. Pap. Ser. 125, 1–19 (1987). [Google Scholar]
- 38.Sun F., et al., Investigating the role of OsPDCD5, a homolog of the mammalian PDCD5, in programmed cell death by inducible expression in rice. Plant Mol. Biol. Report. 30, 87–98 (2012). [Google Scholar]
- 39.Yang S., et al., Rice EARLY SENESCENCE 2, encoding an inositol polyphosphate kinase, is involved in leaf senescence. BMC Plant Biol. 20, 393 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blakeslee J. J., Peer W. A., Murphy A. S., Auxin transport. Curr. Opin. Plant Biol. 8, 494–500 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Li R., et al., ADP1 affects plant architecture by regulating local auxin biosynthesis. PLoS Genet. 10, e1003954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., et al., Yield, grain quality and water use efficiency of rice under non-flooded mulching cultivation. Field Crops Res. 108, 71–81 (2008). [Google Scholar]
- 43.Chen M.-L., et al., Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905, 67–74 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Liu Z., et al., Determination of cytokinins in plant samples by polymer monolith microextraction coupled with hydrophilic interaction chromatography-tandem mass spectrometry. Anal. Methods 2, 1676–1685 (2010). [Google Scholar]
- 45.Liu Y., et al., OstMAPKKK5, a truncated mitogen-activated protein kinase kinase kinase 5, positively regulates plant height and yield in rice. Crop J. 5, 707–714 (2019). [Google Scholar]
- 46.Liu Y., Ng P. K., Isolation and characterization of wheat bran starch and endosperm starch of selected soft wheats grown in Michigan and comparison of their physicochemical properties. Food Chem. 176, 137–144 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Wang C.-C., et al., Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0. Plant Biotechnol. J. 18, 14–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilella A. J., Blanco-Garcia A., Hutter S., Rozas J., VariScan: Analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics 21, 2791–2793 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this article are available at the Rice Annotation Project Database, https://rapdb.dna.affrc.go.jp/ (accession nos. Os05g0547850 [OsPDCD5] and Os02g0198300 [OsAGAP]); both cDNAs were cloned from Bing1B. All other study data are included in the article and SI Appendix.