Significance
The “spiny fin,” comprising the anterior part of the dorsal and anal fins, is an evolutionary novelty that contributed to the success of the spiny-rayed fishes. This domain contains heavily ossified spines that serve as defense mechanism and differ from the posterior flexible soft rays. We show that the partitioning of the median fins into spines and soft rays is established through canonical developmental mechanisms responsible for the anterior–posterior patterning of appendages. Furthermore, the coloration of the anal fin in males appears to be genetically linked to soft-ray identity. Comparative analysis including nonacanthomorph fins indicates that the convergent evolution of fin spines across fishes likely involved the repeated exaptation of a deeply conserved developmental program from the anterior fin.
Keywords: fin spine, acanthomorph, evolutionary key innovation, evo-devo, exaptation
Abstract
With over 18,000 species, the Acanthomorpha, or spiny-rayed fishes, form the largest and arguably most diverse radiation of vertebrates. One of the key novelties that contributed to their evolutionary success are the spiny rays in their fins that serve as a defense mechanism. We investigated the patterning mechanisms underlying the differentiation of median fin Anlagen into discrete spiny and soft-rayed domains during the ontogeny of the direct-developing cichlid fish Astatotilapia burtoni. Distinct transcription factor signatures characterize these two fin domains, whereby mutually exclusive expression of hoxa13a/b with alx4a/b and tbx2b marks the spine to soft-ray boundary. The soft-ray domain is established by BMP inhibition via gremlin1b, which synergizes in the posterior fin with shh secreted from a zone of polarizing activity. Modulation of BMP signaling by chemical inhibition or gremlin1b CRISPR/Cas9 knockout induces homeotic transformations of spines into soft rays and vice versa. The expression of spine and soft-ray genes in nonacanthomorph fins indicates that a combination of exaptation and posterior expansion of an ancestral developmental program for the anterior fin margin allowed the evolution of robustly individuated spiny and soft-rayed domains. We propose that a repeated exaptation of such pattern might underly the convergent evolution of anterior spiny-fin elements across fishes.
Teleost fishes comprise ∼50% of extant vertebrate species and display an astonishing diversity in body plans (1–4). Among the ∼30,000 species of teleosts, the spiny-rayed fish—or Acanthomorpha—are evolutionarily the most successful lineage with over 18,000 species, representing approximately one third of all living vertebrates (1, 2, 5). Spiny-rayed fishes evolved relatively recently, during the Early Cretaceous (133 to 150 Mya) (6), and underwent their primary radiation after the Cretaceous–Paleocene mass extinction (ca. 66 Mya) when their lineage came to dominate many aquatic ecosystems (4, 6–10). One of the characteristics that has strongly contributed to the ecological and evolutionary success of the spiny-rayed fishes is fin spines in dorsal and anal median fins (2, 3, 11). Acanthomorph fin spines are mostly present on the anterior part of the dorsal, anal, and sometimes pectoral and pelvic fins and differ from soft rays by increased ossification, lack of segmentation, fusion of lateral half-segments (hemitrichia), and ending in a sharp point instead of bifurcating (11) (Fig. 1A). The main function of fin spines is to serve as a defense mechanism against gape-limited predators (2, 3, 11, 12), and as such, they strongly suggest a causal link to the success of the Acanthomorpha. Interestingly, anterior spines have evolved independently in other successful lineages of teleosts (2, 11–13), such as the Ostariophysians, in particular catfish and carps, underscoring their adaptive significance. However, in none of these lineages has this resulted in such persistent and pronounced individualization and modularization of separate median fin domains as present in acanthomorph fishes (Discussion).
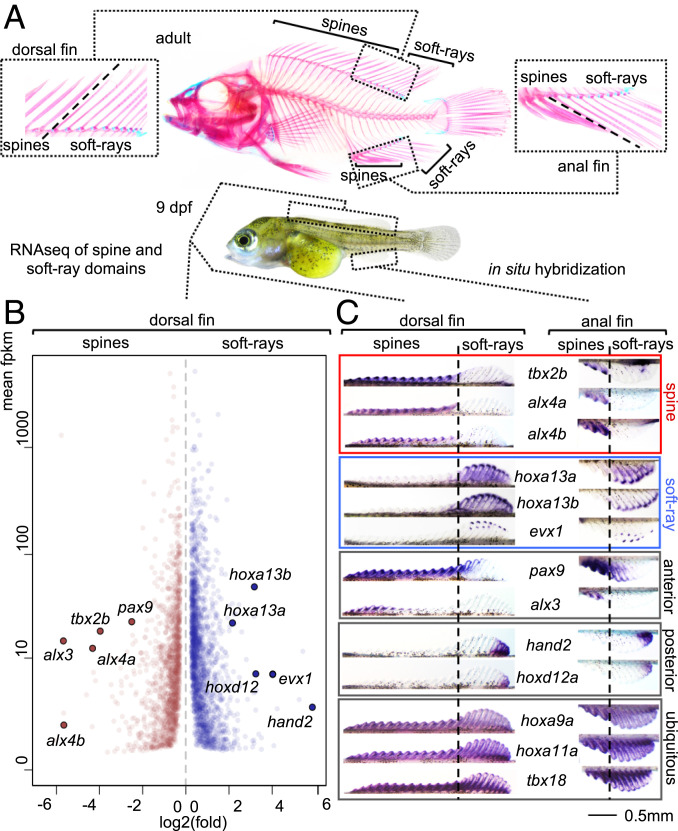
Fig. 1.
(A) Skeleton of adult A. burtoni shows division of the dorsal and anal fins into spine and soft-ray domains. Left Inset shows transition of spiny and soft-ray domains for the dorsal fin, Right Inset for the anal fin. We investigated the developmental basis for the individuation of the spiny and soft-rayed fin domains using RNA-seq analysis (B) and in situ hybridization (C) using 8 to 9 dpf embryos. Tbx2b and alx4a/b define the fin spine territory while hoxa13a/b and evx1 are expressed exclusively in soft rays. Additional transcription factor genes identified in the RNA-seq show an anterior (alx3/pax9) or posterior (hand2/hoxd12) bias but do not segregate with the spine to soft-ray transition. Expression of hoxa9a, hoxa11a, and tbx18 is ubiquitous throughout the fins and underscores shared origins of spines and soft rays. Anterior is to the left. The skeleton and embryo in A are from ref. 25.
In the acanthomorph dorsal and anal fins, the spiny and soft-rayed parts form distinct morphological and developmental units that behave as separate evolutionary modules (14). Examples of extreme morphological specialization of the spiny fin as compared to the soft rays are the Remora’s suction disk (15, 16), the Frogfishes’ ilicium/esca complex (17) and the dorsal part of the Triggerfishes’ “locking mechanism” (18). Species such as the Asian leaf fishes (e.g., Nandus oxyrhynchus) (19) further exemplify the divergence between spiny and soft-rayed fins. As many ambush-hunting fish, they have translucent soft-rayed fins and heavily pigmented spiny fins, whereby the transparency of the unpigmented soft rays enhances camouflage as slight undulations of those fin parts serve to keep the fish stationary. Altogether, this suggests a modularization that is distinct beyond the mere morphological difference between spines and soft rays and also determines pigmentation as well as function and further underscores the adaptive significance of individuated spine and soft-ray fin modules.
This individuation that affects a range of phenotypic traits is reminiscent of anatomical modules determined by master control genes that specify different ontogenetic outcomes for serially homologous elements. Examples of such systems are for instance the hox codes in the axial skeleton (20–22) or the hindbrain (23). Thus, selector genes act upstream in the hierarchy of differentiation to initiate alternative downstream developmental trajectories for meristic structures (24).
In this work, we set out to unravel the developmental basis underlying the patterning of discrete spiny and soft-ray domains using the direct-developing cichlid fish Astatotilapia burtoni (25). Cichlids belong to the Acanthomorpha and possess a spiny fin. The established model-system zebrafish and medaka are not suited to address this question because zebrafish is not an acanthomorph, and medaka has secondarily lost the spiny fin. A. burtoni has the typical division of spine and soft-ray territories in dorsal and anal fins, as well as soft-ray–specific pigment pattern in males (egg spots). An understanding of the genetic basis for the specification of spine and soft-ray domains will help to elucidate the evolutionary origin of these modules at the base of the acanthomorph radiation as well as provide insight into how spines repeatedly emerged across fish clades as a diversity promoting trait.
Results
Mutually Exclusive alx4/tbx2b and hoxa13a/hoxa13b Expression Marks the Spine to Soft-Ray Boundary.
We previously described the ontogeny of the spiny and soft-rayed domains in the dorsal and anal fins of A. burtoni. Spine and soft-ray territories differentiate simultaneously between 4 to 10 dpf (days postfertilization) from continuous Anlagen located along the dorsal and ventral midline (25). The development of fin elements as either soft rays or spines reflects a binary developmental trajectory since intermediate forms do not occur. The partitioning of the fins into two morphologically discrete domains therefore suggests the existence of a code of “master control” genes that direct a developmental choice for the differentiation into soft rays or spines. We performed RNA-sequencing (RNA-seq) on prospective spiny and soft-rayed parts of the dorsal fin of 9 dpf embryos to identify differentially expressed transcription factor genes (Fig. 1 A and B). In the soft-rayed posterior part of the fin, hoxa13a, hoxa13b, hoxd12, hand2, and evx1 are strongly up-regulated, while the spiny part of the fin shows strong expression of alx4a, alx4b, alx3, tbx2b, and pax9. To determine their specificity for spiny or soft-rayed fin domains, the expression of these genes was analyzed using whole mount in situ hybridization (Fig. 1C). In both dorsal and anal fins, we find a strong association of hoxa13a/b and evx1, and their anterior limit of expression marks the spine to soft-ray boundary. In line with its function in zebrafish (26), evx1 is expressed in the forming segment boundaries of the soft rays. Hoxd12 and hand2, however, associate with a more posterior part of the fin, away from the spine to soft-ray boundary. Alx4a, alx4b, and tbx2b associate with the spiny part of the fin and posteriorly demarcate the spine to soft-ray boundary. Pax9 is expressed with an anterior bias but clearly overlaps the soft-ray territory while alx3 is expressed in the anterior-most part of the spiny domain. Additional fin patterning genes hoxa9a, hoxa11a, and tbx18 show ubiquitous fin expression and indicate a largely shared developmental program of the two fin domains, consistent with the spiny fin being a relatively young evolutionary modification. Analysis in a time series from 4 to 7 dpf shows that from 5 dpf onwards alx4a and hoxa13a/hoxa13b stably delineate spine and soft-ray domains whereas this is the case for tbx2b from 6 dpf onwards (SI Appendix, Fig. S1).
BMP Inhibition through gremlin1b Establishes the Soft-Ray Territory in Synergy with shh.
The division of fins into spiny and soft-ray domains reflects an anterior–posterior organization of the median fins. Therefore, we set out to investigate the role of canonical signaling mechanisms used to pattern the anterior–posterior axis of the appendages in the establishment of this division. In limbs and fins, sonic hedgehog (shh) secreted from a ZPA (zone of polarizing activity) is essential for correct anterior–posterior patterning (27–32). Shh expression in a posterior ZPA is an ancestral feature of gnathostome paired and median fins (27, 28, 33, 34). In A. burtoni dorsal and anal fins, we observe first shh expression in a ZPA starting at 5 dpf, becoming strongly expressed at 6 dpf, after which shh disappears from the ZPA and becomes expressed in the distal tips of the forming soft-ray and spine elements (Fig. 2B and SI Appendix, Fig. S2). Treatment during 4 to 6 dpf with the shh agonist SAG induces an anterior expansion of hoxa13b in the dorsal and anal fins while the expression of alx4a and tbx2b becomes more anteriorly restricted—indicating an anterior shift of the spine to soft-ray boundary (Fig. 2C and SI Appendix, Fig. S3). Analysis of the expression of gli1, which is a downstream target of shh and provides a read out for the range of shh signaling (27, 28), suggests that in untreated embryos at 6 dpf shh signaling extends anterior of the ZPA for the length of about two to three somites (Fig. 2B). That is, less than half the extent of the forming soft-ray domain, which develops over the width of 6 to 7 somites (SI Appendix, Fig. S1). Furthermore, inhibition of shh through treatment with the shh antagonist cyclopamine from 4 to 6 dpf fully abolishes gli1 expression but does not lead to a strong displacement of the anterior–posterior position of the spine to soft-ray boundary as indicated by alx4a/tbx2b and hoxa13b expression (Fig. 2C and SI Appendix, Fig. S3) (although the expression levels of hoxa13b are decreased within the prospective soft-ray domain). Thus, this suggests that while shh appears capable of expanding the soft-ray territory, the normal specification of the soft-ray domain occurs at least in part through another, shh-independent, mechanism.
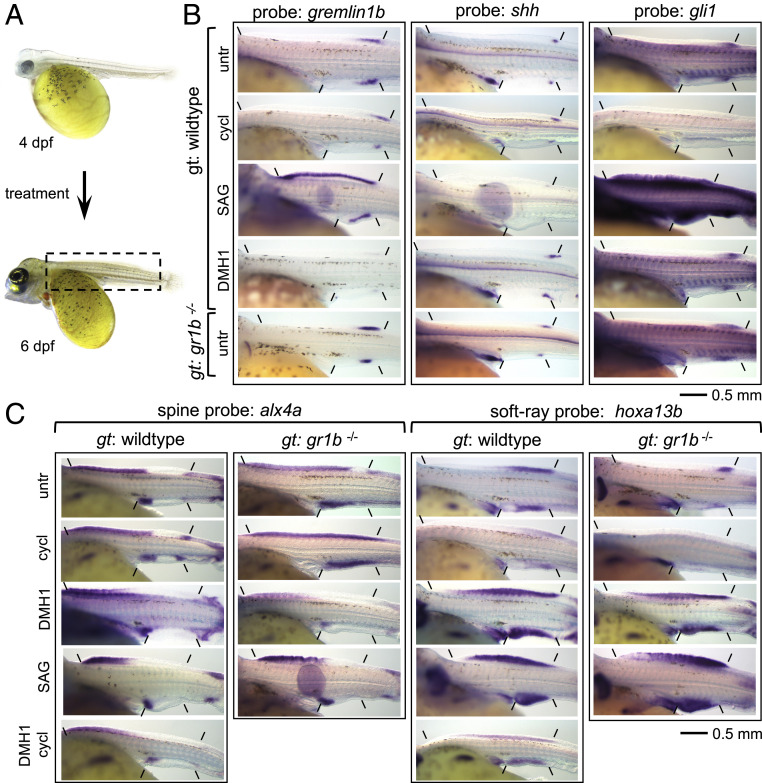
Fig. 2.
The effect of interference with shh and BMP signaling on the establishment of spine and soft-ray territories. (A) All expression domains were analyzed at 6 dpf, all chemical treatments were initiated at 4 dpf. Inset in 6 dpf embryo shows the approximate extent of the part of the embryos shown in B and C. (B) Analysis of the expression of gremlin1b, shh, and gli1 (indicated “probe”) in WT embryos treated with cyclopamine, SAG, DMH1, and in gremlin1b−/− embryos. (C) Analysis of the spine to soft-ray transition using the spine marker alx4a and the soft-ray marker hoxa13b in treatment with cyclopamine, SAG, DMH1, and a combination of DMH1 and cyclopamine. Experiments were performed on WT and gremlin1b−/− embryos (except the combination of DMH1 + cyclopamine) and observed on a minimum of 6/6 embryos per probe per treatment/genotype. Anterior and posterior limits of dorsal and anal fin Anlagen are indicated. gt: genotype, untr: untreated, cycl: cyclopamine. Anterior is to the left. The embryos in A are from ref. 25.
In limbs, shh activates the secreted BMP antagonist gremlin1 (35), which together with BMP4 provides a mechanism downstream of shh to regulate digit identity (36). In the dorsal and anal fins of A. burtoni, gremlin1b becomes expressed at 4 dpf. In the dorsal fin, its expression initially extends anterior of the vent but becomes subsequently restricted to approximately the soft-ray territory at 5 dpf and continues to regress more posteriorly during 6 and 7 dpf (Fig. 2B and SI Appendix, Fig. S2). No expression of the other gremlin homologs gremlin1a, gremlin2, or any other BMP antagonists investigated (noggin1, noggin2, chordin, chordin-like-2) was detected during early fin development (SI Appendix, Fig. S4). BMP4 is expressed throughout the fin, although there may be a bias toward higher expression in the gremlin1b territory (SI Appendix, Fig. S2). We investigated the function of gremlin1b and BMP signaling during fin patterning by generating A. burtoni CRISPR/Cas9 knockout lines (Materials and Methods) and through gain of function by mimicking the inhibitory effect of gremlin1b with the small molecule BMP-receptor inhibitor DMH1. Gremlin1b knockout leads to a more posteriorly restricted hoxa13b domain of approximately the size of the shh signaling zone as inferred from gli1 expression (Fig. 2C). At the same time, alx4a and tbx2b domains become expanded posteriorly, altogether indicating a posterior shift of the spine to soft-ray boundary. The gain of function approach induced the opposite effect with an anteriorly expanded hoxa13b domain and anteriorly shifted alx4a and tbx2b domains (Fig. 2C and SI Appendix, Fig. S3) These anterior shifts are also induced in the gremlin1b knockout treated with DMH1, which therefore rescues the posterior shifts observed in untreated gremlin1b−/− embryos (Fig. 2C). The posteriorized and anteriorized spine to soft-ray boundaries observed in gremlin1b−/− and DMH1-treated embryos are maintained during development (hoxa13b-, hoxa13a-, alx4a-, and tbx2b-stained embryos shown at 9 dpf in SI Appendix, Fig. S3). Therefore, BMP inhibition in the posterior fin by gremlin1b is required for the delimitation of the alx4a/tbx2b and hoxa13b domains and influences the anterior–posterior position of the spine to soft-ray boundary.
In tetrapod limbs, gremlin1b is activated by, and acts downstream of, shh signaling (35). In A. burtoni, SAG treatment leads to widespread up-regulation of gremlin1b expression (Fig. 2B). Cyclopamine treatment however, reduces but does not eliminate posterior gremlin1b domain (Fig. 2B). This observation of shh-independent gremlin1b expression is consistent with its early activation at 4 dpf before shh in the ZPA becomes detectable (SI Appendix, Fig. S2). BMP inhibition by DMH1 strongly down-regulates gremlin1b expression, whereas it appears locally up-regulated in the gremlin1b−/− embryos (Fig. 2B). This suggests that, as in limbs, BMP and shh are upstream of gremlin1b (36) but that in median fins these signaling pathways act in part redundantly. In the context of auto- and cross-regulatory interactions of these pathways, we observe that shh in the ZPA is strongly down-regulated with SAG treatment and up-regulated with cyclopamine treatment, suggesting the presence of an autoregulatory negative feedback loop (Fig. 2B) as has also been observed during limb development (37). Furthermore, DMH1 treatment slightly enhances shh expression in the ZPA but does not increase signaling (as judged by gli1 expression) to an extent that it explains the far anterior shift of the soft-ray to spine boundary (Fig. 2B). Altogether, these experiments suggest that shh and gremlin1b are acting independently upstream of the specification of the soft-ray domain.
We further tested this hypothesis by combining shh activation and inhibition conditions with gremlin1b knockout and BMP inhibition. Embryos treated with a combination of cyclopamine and DMH1 display a similar expansion of hoxa13b and reduction of alx4a and tbx2b domains as treatment with DMH1 alone (Fig. 2C and SI Appendix, Fig. S3), showing that BMP inhibition can posteriorize the fin independently of shh. In gremlin1b−/− embryos treated with cyclopamine, the posterior residual patch of hoxa13b expression disappears completely and alx4a and tbx2b domains now extend throughout the length of the dorsal and anal fin, indicating a complete absence of a soft-ray domain (Fig. 2C and SI Appendix, Fig. S3). Gremlin1b knockout embryos treated with SAG resemble wild-type (WT) embryos treated with SAG (Fig. 2C and SI Appendix, Fig. S3), confirming that hoxa13b expansion and alx4a/tbx2b reduction can occur independent of BMP inhibition by gremlin1b. Therefore, the posterior soft-ray territory is synergistically patterned by shh and gremlin1b, whereby gremlin1b determines the position of the spine to soft-ray boundary in WT fish.
Interference with BMP Signaling Induces Homeotic Transformations of Soft Rays into Spines and Vice Versa.
Next, we strived to assess the phenotypic consequences of interference with the shh and BMP pathways. Morphological differentiation between spine and soft-ray elements, as indicated by the presence of fin segments and the development of spine tips, first occurs in A. burtoni around 10 dpf (25). Cyclopamine and SAG treatments induced widespread pleiotropic effects outside of the fins and severely compromised embryonic viability beyond 8 dpf, that is, before the morphological differences between spines and soft rays are established and therefore preclude such morphological analyses. DMH1 treatment or loss of gremlin1b is, however, well tolerated with phenotypic consequences that appear primarily in the fins and thus allow further morphological analyses of the extent of spine and soft-ray territories. In the dorsal fins of gremlin1b mutants, we observe a posterior shift of the spine to soft-ray boundary caused by a homeotic transformation of the anterior soft rays into spines as indicated by the presence of a spiny tip, the absence of segmentation, and the anterior fusion of the hemitrichia (Fig. 3A and SI Appendix, Fig. S5) (WT/heterozygous (n = 21): 13 to 14 spines, 9 to 10 soft ray; gremlin1b−/− (n = 16): 15 to 20 spines, 3 to 6 soft rays). In the anal fin, a similar posterior expansion of the spine domain is observed whereby only 3 to 4 soft rays are maintained (Fig. 3C and SI Appendix, Fig. S5) (WT/heterozygous (n = 21): 3 spines, 8 to 10 soft ray; gremlin1b−/− (n = 16): 4 to 7 spines, 1 to 6 soft rays). The preservation of soft-ray identity in the posterior fin is consistent with the presence of a posterior patch of hoxa13b expression that arises in a shh-dependent manner in gremlin1b−/− embryos (Fig. 2C). The inhibition of BMP signaling through DMH1 treatment for a 24-h window during 4 to 5 dpf results in the opposite phenotype in the dorsal fin with an anterior transformation of spines into soft rays (Fig. 3B and SI Appendix, Fig. S5) (n = 7, spines 3 to 10, soft rays 14 to 21). This treatment induces the same soft-ray expansion in a gremlin1b−/− background (SI Appendix, Fig. S6) (n = 5/5). In the anal fin, no significant shift in number of soft rays and spines is observed (P = 0.06, two-sided t test, n = 6, spines 2 to 3, soft rays 9 to 11) (SI Appendix, Fig. S5), suggesting that additional genetic factors besides BMP signaling determine the presence of the 3 anterior fin spines in the anal fin.
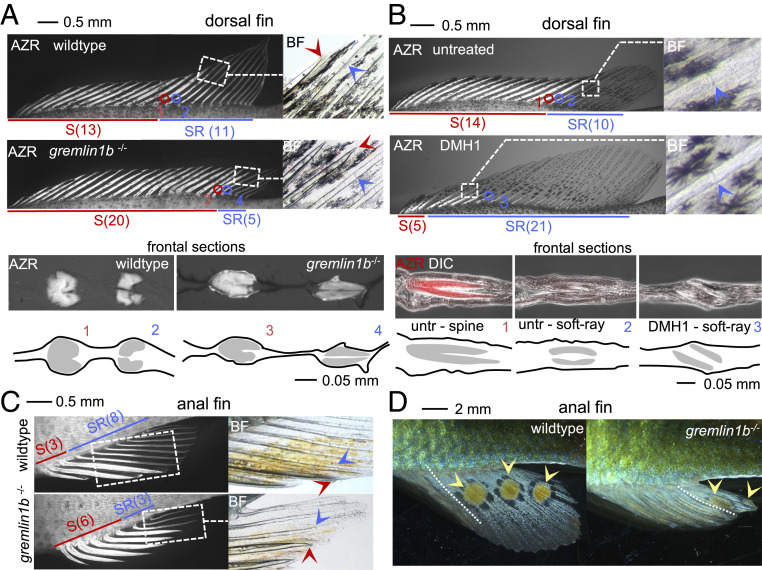
Fig. 3.
Interference with BMP signaling induces homeotic transformations in dorsal and anal fins. (A–C) Fin morphology of dorsal (A and B) and anal fins (C) in WT (A–C), gremlin1b−/− (A and C), and DMH1-treated (B) fish at approximately 1 mo postfertilization. Bony structures were visualized using Alizarin red (“AZR”), and fins were imaged using fluorescence microscopy. Insets shown (dashed boxes) were taken using brightfield microscopy (“BF”). In A and B, transversal sections at the level of the spine to soft-ray boundary are shown, which in A was imaged using fluorescence microscopy (Alizarin red fluorescence in white) and in B using fluorescence microscopy (Alizarin red fluorescence in false color red) and differential interference contrast microscopy (“DIC”) (in white). The dorsal fins of gremlin1b−/− fish in A show an expanded spine domain (indicated by red line) and reduced soft-ray domain (indicated by blue line) indicating soft-ray to spine homeotic transformations. Alizarin red staining visualizes the heavier ossification of spines as compared to soft rays. Insets show spine (red arrowhead spine tip) and soft-ray (blue arrowhead segment boundary) characters, at the spine to soft-ray transition. Transversal sections through the spine to soft-ray boundary confirm the presence of fused and unfused hemisegments in spines and soft rays, respectively (section position is indicated with circles and numbers). The DMH1-treated fish shown in B show the opposite transformation displaying spine to soft-ray transformations. The Inset shows segments in the most anterior soft ray (blue arrowhead). Transversal sections confirm the presence of unfused soft-ray–like elements in the anterior fin. C shows a comparison of gremlin1b−/− and WT anal fins showing soft-ray to spine transformations. Insets indicate spine and soft-ray characters at the spine to soft-ray boundary. A quantitative analysis of spine and soft-ray counts is provided in SI Appendix, Fig. S5A. (D) Egg spots are present on the soft-ray part of the anal fins of male A. burtoni. In gremlin1b−/−, the egg spots have shifted posterior together with the soft-ray domain. A quantitative analysis of egg spots distribution is provided in SI Appendix, Fig. S5B. AZR: Alizarin red, BF: brightfield, S: spine, SR: soft ray. Anterior is to the left.
Altogether, the observed homeotic transformations of spines to soft rays and vice versa underpin that BMP inhibition by gremlin1b is a primary determinant of soft-ray identity as also suggested by the analysis of developmental marker genes.
Gremlin1b Mutants Display Homeotic Transformations in Anal Fin Coloration.
The individuation of the soft-rayed and spiny domains of the male anal fin in A. burtoni is also reflected in its coloration. The mouth-brooding African cichlids evolved egg spots, or “egg dummies,” apparently to increase the chances of fertilization during courtship (25, 38). The distribution of egg spots in the anal fin typically shows a bias toward the posterior side of the fin overlapping with the soft rays while being absent from the spiny part. To investigate whether egg spots are in fact part of the same genetic modules that determine soft-ray and spine development, we analyzed the presence of egg spots at 3 mo of age in WT/heterozygous and gremlin1b−/− males derived from two gremlin1+/− crosses. Comparison of mutant with WT or heterozygous fish (which are WT in appearance with respect to spine and soft-ray distribution) shows an altered distribution of egg spots on the fin. Concomitant with the posterior shift of the soft-ray domain, the egg spots in these fish are present more posteriorly, and egg spots were never observed to overlap with the spiny-fin domain. In the cross analyzed, WT and heterozygous fish have an average of 3.5 egg spots whereas gremlin1b homozygous mutant fish have an average of 2 egg spots (WT/heterozygous n = 8; gremlin1b−/− n = 9, P = 0.0002, two-sided t test) (Fig. 3D and SI Appendix, Fig. S5). In the same cohort, WT and heterozygous male egg spots are present over 57% of the length of the fin, whereas this is reduced to 28% in homozygous gremlin1b mutant fish (P = 9.6 × 10−6, two-sided t test) (SI Appendix, Fig. S5). Therefore, the distribution of egg spots in the anal fin appears to be determined by the same upstream patterning mechanism as that inducing the soft-ray and spiny-fin domains, whereby the posterior reduction of the soft-ray domain results in a concomitant posterior shift in the presence of egg spots.
Analysis of the Dorsal Fin Pattern in Nonacanthomorph Spiny and Nonspiny Catfish.
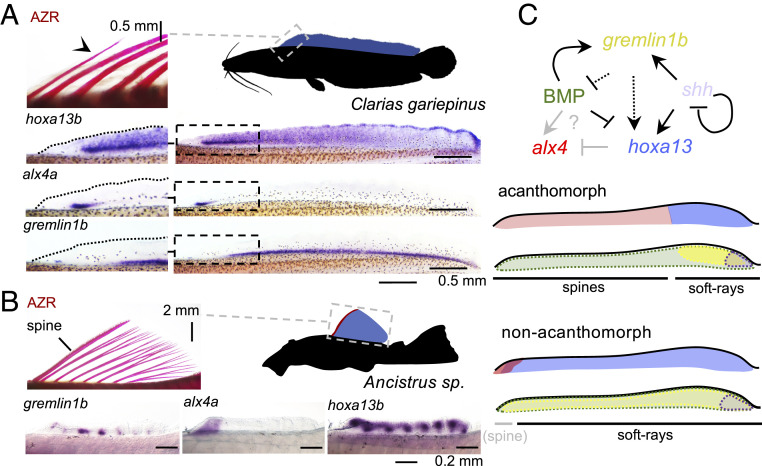
Anterior spines have convergently evolved in several clades of nonacanthomorph teleosts such as catfish and carps. We wanted to further understand the relationship between dorsal fin patterns and the repeated emergence of fin spines. Furthermore, the dorsal fin pattern of nonacanthomorphs could provide information concerning the evolutionary origin of the acanthomorph fin pattern. We thus compared the anterior–posterior patterning observed in A. burtoni with that in nonacanthomorph species with median fins consisting of soft rays only or in those with convergently evolved fin spines. The nonacanthomorph zebrafish possesses soft rays only, and alx4a is expressed in the anterior-most fin rays of the dorsal and anal fins (39), tentatively suggesting that the spine pattern derives from a domain originally confined to the anterior fin margin. Zebrafish, however, has a narrow dorsal fin that is restricted to the posterior part of the body and that is about the size of the A. burtoni soft-ray domain. This leaves open the possibility that wider and further anteriorly extending nonacanthomorph fins show a similar extended alx4 domain as A. burtoni. We investigated the expression of alx4a, hoxa13b, and gremlin1b expression in embryos of the African catfish (Clarias gariepinus), which has an extended dorsal fin (Fig. 4A) comprised of soft rays only and lacks the typical anterior spine found in many catfish species. Consistent with its soft-ray identity, hoxa13b and gremlin1b expression extends anterior throughout most of the dorsal fin. As in zebrafish, alx4a expression is confined to the anterior fin margin. Analysis in South American Ancistrus catfish whose anterior-most dorsal fin element has convergently evolved into a spine, for hoxa13b, gremlin1b, and alx4a, shows a similar pattern (Fig. 4B). In this species, the expression of all three genes overlaps in the first dorsal fin element, which will develop into a spine. Therefore, anteriorly limited expression of alx4a is also apparent in nonacanthomorph fish. This domain is, however, restricted much more anteriorly, and the gremlin1b/hoxa13 domain extends along the anterior–posterior fin axis. Intriguingly, the anterior domain can coincide with the development of either a soft ray (as in zebrafish and Clarias) or a spine (as in Ancistrus) (Discussion).
Fig. 4.
Anterior–posterior dorsal fin patterning in acanthomorphs and nonacanthomorphs. (A) African catfish (C. gariepinus) have an extended dorsal fin (blue) comprised of soft rays only. The Alizarin red bone staining on the Left shows the anterior-most fin elements, and arrowheads indicate segment boundaries. In line with its soft-ray identity, hoxa13b and gremlin1b are expressed throughout most of the anterior–posterior fin axis. Alx4a expression is detected in a domain in the anterior fin similar to that in zebrafish (39). (B) Ancistrus catfish have a dorsal fin that is restricted to the anterior part of the trunk. This fin consists of posterior soft rays and a single anterior spine. Hoxa13b is expressed throughout the anterior–posterior extent of the fin, including the first elements, as is gremlin1b. Alx4a expression is confined to the spine and therefore may be involved in the individualization of this element compared to the posterior domain. (C) Model for the signaling network establishing the soft-ray domain in acanthomorph and nonacanthomorph teleosts. In acanthomorphs, the soft-ray domain is established via gremlin1b, which acts posteriorly in synergy with shh to activate hoxa13 through the inhibition of BMP signaling. The absence of these posterior signals results in posterior expansion of alx4 expression and the spine domain either through direct activation by BMP or loss of repression by hoxa13 proteins. In nonacanthomorphs, the soft-ray signature extends throughout the anterior–posterior fin axis, and alx4 is only expressed in the anterior fin margin, possibly related to the convergent evolution of spiny elements in nonacanthomorphs such as catfish. AZR: Alizarin red. Anterior is to the left.
Discussion
Spiny fins can be considered an evolutionary key innovation that arose as a novel module in the spiny-rayed fishes and added to the evolvability and thereby evolutionary success of the teleost body plan. Here, we show that the specification of spine and soft-ray domains during embryonic development is the result of a canonical signaling network involved in the patterning of the anterior–posterior fin axis, whereby posterior expression of gremlin1b and shh specify the soft-ray domains (Fig. 4C). In WT A. burtoni, the primary determinant of the spine to soft-ray boundary is BMP inhibition by gremlin1b, and alterations in BMP signaling induce homeotic transformations in fin identity. Interestingly, modulation of BMP levels is capable of inducing homeotic transformations in digits (40) and tooth identity (41). Therefore, spines and soft rays form another example of a deeply homologous function of BMP signaling in “specifying discrete identities amongst meristic structures” (quotation from ref. 40).
During tetrapod limb development, shh and BMP inhibition via gremlin1 are part of a regulatory loop including FGFs expressed in the distal ectoderm, which are required for ZPA survival (27–31, 35). We therefore investigated the potential role of FGFs in the establishment of soft-ray and spiny-fin domains. Fgf16 is expressed along the anterior–posterior extent of the distal edge of the dorsal and anal fins and is slightly up-regulated by DMH1 and SAG treatment whereas it is somewhat down-regulated by cyclopamine treatment and in gremlin1b−/− embryos (SI Appendix, Fig. S7). Altogether, this potentially indicates a conserved position of ectodermal FGF signaling downstream of shh and gremlin1. Treatment with the FGF antagonist BGJ398 from 4 to 7 dpf results in complete abortion of fin outgrowth, equally affecting spine and soft-ray domains and consistent with the relatively homogenous expression along the fin anterior–posterior axis. (SI Appendix, Fig. S7). Therefore, while important for fin outgrowth, ectodermal FGF signaling is not a major factor determining the anterior–posterior division of the dorsal and anal fins into spine and soft-ray territories.
In A. burtoni, the anterior–posterior pattern in dorsal and anal fins differs from that in their pectoral fins. In the latter, hoxa13a/b are expressed throughout the anterior–posterior extent of the fin (42, 43) and alx4a/b remain restricted to the anterior-most fin domain (39, 44). This appears to be a deeply conserved pattern that is for instance also present in shark pectoral fins (32, 45, 46). Also, in A. burtoni pectoral fins gremlin1b is expressed throughout most of the anterior–posterior axis of the pectoral fin Anlage (SI Appendix, Fig. S8) and does not show the posterior bias observed in dorsal and anal fins. Overall, the patterning of the median fins in nonacanthomorph, Clarias, Ancistrus, and zebrafish (39) therefore resembles a pectoral fin pattern (although the median fin expression pattern of gremlin1b in zebrafish remains to be determined) and may therefore represent a shared ancestral pattern among median and paired fins that became modified in the median fins of spiny-rayed fish. This would have involved an expansion of the anterior pattern and a concomitant reduction of the soft-ray domain (Fig. 4C). Whether in the ancestral fin pattern gremlin1b acts to establish the posterior domain remains to be investigated by loss of function approaches in nonacanthomorphs. It is however suggestive that in A. burtoni gremlin1b loss does not lead to reduction of hoxa13b expression or expansion of alx4a expression in pectoral fins (SI Appendix, Fig. S8). This therefore might hint at a newly evolved posteriorizing role of gremlin1b in acanthomorphs median fins.
It is noteworthy that nonacanthomorphs frequently have a modified first fin element. For instance, in zebrafish and goldfish the first soft ray does not branch distally, and in many catfish species and carps a “spine” develops at this position. This suggests that an individuation of the anterior-most fin exists in nonacanthomorphs, which is consistent with the anterior domain of alx4 expression in the fin margin of zebrafish (39), Clarias, and Ancistrus catfish. The tendency for more robustly ossified or spiny anterior fin ray elements is a trend present throughout fishes in both paired and median fins. Additional examples are the anterior fin spine in catfish (47) and sturgeon pectoral fins (48), robustly ossified anterior fin rays in tetrapodomorphs (49), the anterior fin spine that evolved convergently in chimaeras (50), and acanthodians (spiny sharks) and stem sharks (e.g., hybodonts) (51). It is plausible that convergently evolved spines all rely on the same deeply homologous anterior fin individualization. Importantly however, this module appears restricted to the first few anterior-most fin elements only in all lineages except for the Acanthomorpha, which show a strong posterior expansion. Furthermore, spines in nonacanthomorph teleosts are different from those in acanthomorphs because the former initially develop as segmented elements that are indistinguishable from soft rays (47) (developing Ancistrus catfish dorsal fin shown in SI Appendix, Fig. S9). Therefore, in addition to the expansion of the anterior fin identity, a change in the downstream interpretation of this pattern (in the form of exaptation) was needed for the evolution of true fin spines and the consolidation of a robustly individualized anterior spiny-fin module in the acanthomorphs. Altogether, such changes in fin architecture allowed the emergence of the spiny-rayed fishes and initiated one of the most successful and diverse of vertebrate radiations.
Materials and Methods
In Situ Hybridization.
In situ hybridization was carried out according to Woltering et al., 2009 (21), 2014 (52), 2015 (20), and 2020 (44). The reported shifts in expression domains in the inhibitor experiments and gremlin1b−/− embryos were observed with complete penetrance.
Cloning of Probes.
Probes were cloned in pGEMT (Promega A3600) vector using PCR from A. burtoni. C. gariepinus, or Ancistrus sp. embryonic copyDNA. A primer table is provided in SI Appendix, Table S1. The A. burtoni hoxa11a, hoxa13a, hoxa13b, hoxd12, and alx4b probes were described before (42, 44). Catfish sequences were identified by BLAST (basic local alignment search tool) against C. gariepinus and Acistrus sp. embryonal/larval RNA-seq libraries, and messengerRNA sequences for alx4a, hoxa13a, hoxa13b, gremlin1a, and gremlin1b were deposited in GenBank under accession nos. MW846856 to MW846866. Correct identification of “a” and “b” ohnologs was confirmed by generation of maximum likely hood gene trees and microsynteny analysis (also reference SI Appendix, Figs. S10 and S11 for gremlin1 and alx4).
Small Molecule Treatment Experiments.
Embryos were treated using the following concentrations: 1 μM SAG (Selleckchem S7779) (dissolved at 10 mM in DMSO), 5 μM cyclopamine (Selleckchem S1146) (dissolved at 50 mM in ethanol), 1 μM DMH1 (Selleckchem S7146, dissolved at 20 mM in DMSO), and 1 μM BGJ398 (Selleckchem S2183, dissolved at 10 mM in DMSO). Embryos were cultured in 30 mL equilibrated tap water (approximately pH 8, 9°dH) with addition of 0.01μg/mL Methylene blue and penicillin/streptomycin (Sigma P4333) diluted 1:1,000 in Ø8.5 cm plastic Petri dishes on an orbital shaker at 33 rpm at 28 °C in a heating incubator. Embryos were cultured at a maximum density of 20 embryos per dish (but usually less) and treated from 4 to 6 dpf for ∼48 h (with the exception of BGJ398). Chemicals were added to the dish upon start of treatment, and embryos were kept in the same medium until the point of fixation (with 4% PFA buffered with 1× PBS (phosphate buffered saline) overnight at 4 °C, afterward storage in 100% ethanol at −20 °C). For the FGF inhibition experiment, embryos were cultured from 4 to 7 dpf for ∼60 h in 1 μM BGJ398, which was added at 4 dpf, and the embryos were kept in the same medium until the point of analysis. For the phenotypic analysis of DMH1 treatments, embryos were treated in 1 μM DMH1 from mid 4 to mid 5 dpf for ∼24 h and subsequently transferred to normal culturing medium and raised under standard conditions until the point of analysis. Mock treatments were performed using DMSO and ethanol, which do not result in phenotypic alterations.
RNA-Seq Analysis.
RNA-seq was performed in triplicate using dissected soft-ray and spine territories of 9 dpf embryos using 10 individuals per sample. RNA was extracted using the ReliaPrep RNA Tissue Miniprep System (Promega Z6111) using the fibrous tissue protocol, and sequencing libraries were generated using TruSeq RNA Library Preparation Kit v2 (Illumina RS-122-2001). Samples were sequenced on an Illumina HiSeq2500 125 bp (base pairs) paired ends, and reads were demultiplexed and trimmed using Trimmomatic (version [v.] 0.36). The tuxedo pipeline for transcriptome assembly and quantification was used (53). Briefly, TopHat and Bowtie2 were used to map reads to the A. burtoni genome (v. 1.0). Cufflinks was used to assemble transcripts, to assemble a merged transcriptome, and to conduct differential gene expression analysis. Data (29,293 transcripts) were then imported into R (v. 3.6.3), and transcripts that showed no expression in at least one out of three replicates in at least one of the two groups (ray or spine) were excluded. Additionally, genes with extremely low expression (average FPKM [fragments per kilobase of transcript per million mapped reads] >0.5) were also excluded (20,592 transcripts, 17,733 of which were annotated and 17,597 were unique). Raw P values obtained from Cuffdiff were corrected for multiple testing using the false discovery method for transcripts. Raw sequence data have been deposited in National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA718487 (54) with accession nos. SAMN18537261–SAMN18537266 (55–60).
Phenotype Analysis.
Alizarin red staining was performed according to standard protocols and imaged under fluorescence microscopy. Spine and soft-ray counts given in SI Appendix, Fig. S5 were determined by manual inspection under a dissection binocular.
Animal Husbandry.
A. burtoni were bred and collected at the University of Konstanz as previously described (25). C. gariepinus embryos were kindly provided by Fleuren and Nooyen, Aquaculture ID, Netherlands. Ancistrus sp. embryos were provided by private breeders. Animal experiments were carried out under 35-9185.81/G-18/32, Tierforschungsanlage (University of Konstanz) Aktenzeige T18/07.
Generation of CRISPR/Cas9 gremlin1b Mutant A. burtoni Lines.
Guide RNAs (gRNA) were cloned in pT7gRNA and produced according to ref. 61 using oligos. gRNA-1 FW: TAGGACTCCAGCACTTCGTCGG, gRNA-1 RV:AAACCCGACGAAGTGCTGGAGT, gRNA-2 FW: TAGGTTGCTGCTCCGATTCGTT, gRNA-2 RV: AAACAACGAATCGGAGCAGCAA. A mixture of 1 to 2 nL of two gRNAs at 10 ng/μL each including Cas9 protein (NEB M0646T diluted 1/40) was injected at the one to two cell stage. Embryos were cultured individually in 6-well plates on an orbital skater at 28 °C in the presence of penicillin/streptomycin (Sigma P4333) diluted 1:1,000 and addition of methylene blue. Two independent lines were derived (SI Appendix, Fig. S12): gremlin1b-stopCD38 has an in-frame premature stop codon introduced at codon 38; gremlin1bΔ740 has a 740-bp deletion including the 5` 339 bp and start codon. Both lines gave indistinguishable phenotypes. The gremlin1b-stopCD38 was genotyped using fragment mapping on a capillary sequencer (3130xl Genetic Analyzer, Applied Biosystems) using a 40 cycle PCR with primers M13 tailed FW: CAGGAAACAGCTATGACCACGCATATCTTCTACAGTATGG, RV: GTCTGCGGTTGCTGCTCCGATTC followed by a second one-cycle labeling PCR with a HEX-labeled M13 FW primer: CAGGAAACAGCTATGAC. The gremlin1bΔ740 allele was detected using standard PCR and gel electrophoresis using primers FW: CAGTGCACAGTCGACACCAGTAG, RV: GACGAGCACAATTTCTTGGCTGTG.
Supplementary Material
Acknowledgments
We wish to acknowledge S. Nappe, K. Gergen, J. Gerwin, C. Dickmanns, and M. Holzem for help in the laboratory; C. Kratochwil for use of equipment; members of the A.M. laboratory for useful discussions; S. Boycheva Woltering for critical reading of the manuscript; and A. Pfeifer and M. Schauber for excellent fish care. This project was funded by Deutsche Forschungsgemeinschaft Grant WO-2165/2-1 (to J.M.W.), support from the Young Scholar Fund of the University of Konstanz (to J.M.W.), and European Research Council (ERC) Advance Grant (“GenAdap” 293700) (to A.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101783118/-/DCSupplemental.
Data Availability
RNA-seq dataset of A. burtoni 9 dpf fins and catfish gene sequences data have been deposited in NCBI SRA and NCBI GenBank (SRA: BioProject PRJNA718487 (54), SAMN18537261–SAMN18537266 (55–60); GenBank: MW846856–MW846866) (62–72).
References
- 1.Nelson J. S., Fishes of the World (Wiley, ed. 4, 2006). [Google Scholar]
- 2.Helfman G. S., Colette B. B., Douglas E. F., The Diversity of Fishes (Blackwell Publishing, 1997), pp. 32, 238, 239. [Google Scholar]
- 3.Wainwright P. C., Longo S. J., Functional innovations and the conquest of the oceans by acanthomorph fishes. Curr. Biol. 27, R550–R557 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Alfaro M. E., et al., Explosive diversification of marine fishes at the Cretaceous-Palaeogene boundary. Nat. Ecol. Evol. 2, 688–696 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Smith W. L., Wheeler W. C., Venom evolution widespread in fishes: A phylogenetic road map for the bioprospecting of piscine venoms. J. Hered. 97, 206–217 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Chen W.-J., et al., New insights on early evolution of spiny-rayed fishes (Teleostei: Acanthomorpha). Front. Mar. Sci. 1, 1–17 (2014). [Google Scholar]
- 7.Patterson C., An overview of the early fossil record of acanthomorphs. Bull. Mar. Sci. 52, 29–59 (1993). [Google Scholar]
- 8.Murray M. M., Wilson M. V. H., Four new basal acanthomorph fishes form the late Cretaceous of Morocco. J. Vertebr. Paleontol. 34, 34–48 (2014). [Google Scholar]
- 9.Near T. J., et al., Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. U.S.A. 109, 13698–13703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman M., Explosive morphological diversification of spiny-finned teleost fishes in the aftermath of the end-Cretaceous extinction. Proc. Biol. Sci. 277, 1675–1683 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyle P. B., Cech J. J., Fishes: An Introduction To Ichthyology (Prentice-Hall, 1982), pp. 19, 288. [Google Scholar]
- 12.Price S. A., Friedman S. T., Wainwright P. C., How predation shaped fish: The impact of fin spines on body form evolution across teleosts. Proc. Biol. Sci. 282, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arratia G., “Actinopterygian postcranial skeleton with special reference to the diversity of fin ray elements, and the problem of identifying homologies. ” in Mesozoic Fishes, Volume 4: Homology and Phylogeny, Arratia G., Schultze H.-P., Wilson M. V. H., Eds. (Dr. Friedrich Pfeil, Munich, 2008), pp. 49–101. [Google Scholar]
- 14.Mabee P. M., Crotwell P. L., Bird N. C., Burke A. C., Evolution of median fin modules in the axial skeleton of fishes. J. Exp. Zool. 294, 77–90 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Britz R., Johnson G. D., Ontogeny and homology of the skeletal elements that form the sucking disc of remoras (Teleostei, Echeneoidei, Echeneidae). J. Morphol. 273, 1353–1366 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Friedman M., Johanson Z., Harrington R. C., Near T. J., Graham M. R., An early fossil remora (Echeneoidea) reveals the evolutionary assembly of the adhesion disc. Proc. Biol. Sci. 280, 20131200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietsch T. W., Grobecker D. B., Frogfishes of the World: Systematics, Zoogeography, and Behavioral Ecology (Stanford University Press, 1987). [Google Scholar]
- 18.Matsuura Y., Katsuragawa M., Osteological developments of fins and their supports of larval grey triggerfish, Balistes capriscus. Jpn. J. Ichthyol. 31, 411–421 (1985). [Google Scholar]
- 19.Ng H. H., Vidthayanon C., Ng P. K. L., Nandus oxyrhynchus, a new species of leaf fish (Teleostei: Nandidae) from the Mekong basin. Raffles Bull. Zool. 44, 11–19 (1996). [Google Scholar]
- 20.Woltering J. M., Duboule D., Tetrapod axial evolution and developmental constraints; Empirical underpinning by a mouse model. Mech. Dev. 138, 64–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woltering J. M., et al., Axial patterning in snakes and caecilians: Evidence for an alternative interpretation of the Hox code. Dev. Biol. 332, 82–89 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Mallo M., Wellik D. M., Deschamps J., Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344, 7–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker H. J., Krumlauf R., Segmental arithmetic: Summing up the Hox gene regulatory network for hindbrain development in chordates. Wiley Interdiscip. Rev. Dev. Biol. 6, 1–28 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Wagner G. P., Evolutionary innovations and novelties: Let us get down to business! Zool. Anz. 256, 75–81 (2015). [Google Scholar]
- 25.Woltering J. M., Holzem M., Schneider R. F., Nanos V., Meyer A., The skeletal ontogeny of Astatotilapia burtoni–A direct-developing model system for the evolution and development of the teleost body plan. BMC Dev. Biol. 18, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulte C. J., Allen C., England S. J., Juárez-Morales J. L., Lewis K. E., Evx1 is required for joint formation in zebrafish fin dermoskeleton. Dev. Dyn. 240, 1240–1248 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Tickle C., Towers M., Sonic hedgehog signaling in limb development. Front. Cell Dev. Biol. 5, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McQueen C., Towers M., Establishing the pattern of the vertebrate limb. Development 147, 1–14 (2020). [DOI] [PubMed] [Google Scholar]
- 29.McGlinn E., Tabin C. J., Mechanistic insight into how Shh patterns the vertebrate limb. Curr. Opin. Genet. Dev. 16, 426–432 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Harfe B. D., et al., Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Rios J., The many lives of SHH in limb development and evolution. Semin. Cell Dev. Biol. 49, 116–124 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Tanaka M., Fins into limbs: Autopod acquisition and anterior elements reduction by modifying gene networks involving 5'Hox, Gli3, and Shh. Dev. Biol. 413, 1–7 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Letelier J., et al., A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet. 50, 504–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahn R. D., Davis M. C., Pappano W. N., Shubin N. H., Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature 445, 311–314 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Panman L., et al., Differential regulation of gene expression in the digit forming area of the mouse limb bud by SHH and gremlin 1/FGF-mediated epithelial-mesenchymal signalling. Development 133, 3419–3428 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Bénazet J. D., et al., A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science 323, 1050–1053 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Butterfield N. C., et al., Patched 1 is a crucial determinant of asymmetry and digit number in the vertebrate limb. Development 136, 3515–3524 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Salzburger W., Braasch I., Meyer A., Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 5, 51 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nachtrab G., Kikuchi K., Tornini V. A., Poss K. D., Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development 140, 3754–3764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahn R. D., Fallon J. F., Interdigital regulation of digit identity and homeotic transformation by modulated BMP signaling. Science 289, 438–441 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Tucker A. S., Matthews K. L., Sharpe P. T., Transformation of tooth type induced by inhibition of BMP signaling. Science 282, 1136–1138 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Woltering J. M., Holzem M., Meyer A., Lissamphibian limbs and the origins of tetrapod hox domains. Dev. Biol. 456, 138–144 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T., Gehrke A. R., Lemberg J., Szymaszek J., Shubin N. H., Digits and fin rays share common developmental histories. Nature 537, 225–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woltering J. M., et al., Sarcopterygian fin ontogeny elucidates the origin of hands with digits. Sci. Adv. 6, eabc3510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onimaru K., et al., A shift in anterior-posterior positional information underlies the fin-to-limb evolution. eLife 4, 1–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamoto K., et al., Heterochronic shift in Hox-mediated activation of sonic hedgehog leads to morphological changes during fin development. PLoS One 4, e5121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubicek K. M., Britz R., Conway K. W., Ontogeny of the catfish pectoral-fin spine (Teleostei: Siluriformes). J. Morphol. 280, 339–359 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Grom K., Pasicka E., Tarnawski K., Comparative anatomy of pectoral girdle and pectoral fin in Russian sturgeon and American paddlefish. Folia Morphol. (Warsz) 75, 173–178 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Stewart T. A., et al., Fin ray patterns at the fin-to-limb transition. Proc. Natl. Acad. Sci. U.S.A. 117, 1612–1620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jerve A., Johanson Z., Ahlberg P., Boisvert C., Embryonic development of fin spines in Callorhinchus milii (Holocephali); implications for chondrichthyan fin spine evolution. Evol. Dev. 16, 339–353 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Janvier P., Early Vertebrates (Clarendon Press, Oxford, 1996). [Google Scholar]
- 52.Woltering J. M., Noordermeer D., Leleu M., Duboule D., Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol. 12, e1001773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C., et al., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Spine and soft-ray differentiation. SRA. https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA718487. Deposited 30 March 2021.
- 55.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., RNAseq of Haplochromis burtoni 9dpf dorsal fin soft-ray domain. SRA. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN18537261. Deposited 30 March 2021.
- 56.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., RNAseq of Haplochromis burtoni 9dpf dorsal fin soft-ray domain. SRA. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN18537262. Deposited 30 March 2021.
- 57.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., RNAseq of Haplochromis burtoni 9dpf dorsal fin soft-ray domain. SRA. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN18537263. Deposited 30 March 2021.
- 58.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., RNAseq of Haplochromis burtoni 9dpf dorsal fin soft-ray domain. SRA. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN18537264. Deposited 30 March 2021.
- 59.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., RNAseq of Haplochromis burtoni 9dpf dorsal fin soft-ray domain. SRA. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN18537265. Deposited 30 March 2021.
- 60.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., RNAseq of Haplochromis burtoni 9dpf dorsal fin soft-ray domain. SRA. https://www.ncbi.nlm.nih.gov/sra/?term=SAMN18537266. Deposited 30 March 2021.
- 61.Jao L. E., Wente S. R., Chen W., Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Ancistrus spec. alx4a mRNA, full CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846856. Deposited 21 March 2021.
- 63.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Ancistrus spec. alx4a mRNA, full CDs. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846857. Deposited 21 March 2021.
- 64.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Ancistrus spec. Hoxa13b mRNA. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846858. Deposited 21 March 2021.
- 65.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Ancistrus spec. Gremln1b mRNA, full CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846859. Deposited 21 March 2021.
- 66.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Ancistrus spec. Gremln1a mRNA, partial CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846860. Deposited 21 March 2021.
- 67.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Clarias gariepinus alx4a mRNA, partial CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846861. Deposited 21 March 2021.
- 68.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Clarias gariepinus gremlin1b mRNA, full CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846862. Deposited 21 March 2021.
- 69.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Clarias gariepinus gremlin1a mRNA, partial CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846863. Deposited 21 March 2021.
- 70.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Clarias gariepinus gremlin1a mRNA, partial CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846864. Deposited 21 March 2021.
- 71.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Clarias gariepinus hoxa13a mRNA, full CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846865. Deposited 21 March 2021.
- 72.Höch R., Schneider R. F., Kickuth A., Meyer A., Woltering J. M., Clarias gariepinus hoxa13b mRNA, full CDS. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/MW846866. Deposited 21 March 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq dataset of A. burtoni 9 dpf fins and catfish gene sequences data have been deposited in NCBI SRA and NCBI GenBank (SRA: BioProject PRJNA718487 (54), SAMN18537261–SAMN18537266 (55–60); GenBank: MW846856–MW846866) (62–72).