Significance
The large cytosolic GTPase dynamin-related protein 1 (Drp1) self-assembles around mitochondria to mechanoenzymatically constrict and divide the double-membrane–bound organelle. Although Drp1 recruitment to the mitochondrial surface is contingent upon its binding to protein adaptors, subsequent or coincident interactions with the outer membrane–localized, mitochondria-specific phospholipid cardiolipin (CL) is critical for stress-induced mitochondrial fission. Here, using a combination of multimodal NMR spectroscopy and various biophysical, biochemical, and cellular approaches, we show the intrinsically disordered variable domain of Drp1 undergoes an ordered helical structural transition to penetrate the membrane bilayer and selectively bind CL. The mutation of key residues within an identified CL-binding motif (CBM) results in defective mitochondrial division coupled to the formation of morphologically distinct “donut” mitochondria.
Keywords: dynamin, cardiolipin, NMR, intrinsically disordered, mitochondria
Abstract
Mitochondria form tubular networks that undergo coordinated cycles of fission and fusion. Emerging evidence suggests that a direct yet unresolved interaction of the mechanoenzymatic GTPase dynamin-related protein 1 (Drp1) with mitochondrial outer membrane–localized cardiolipin (CL), externalized under stress conditions including mitophagy, catalyzes essential mitochondrial hyperfragmentation. Here, using a comprehensive set of structural, biophysical, and cell biological tools, we have uncovered a CL-binding motif (CBM) conserved between the Drp1 variable domain (VD) and the unrelated ADP/ATP carrier (AAC/ANT) that intercalates into the membrane core to effect specific CL interactions. CBM mutations that weaken VD–CL interactions manifestly impair Drp1-dependent fission under stress conditions and induce “donut” mitochondria formation. Importantly, VD membrane insertion and GTP-dependent conformational rearrangements mediate only transient CL nonbilayer topological forays and high local membrane constriction, indicating that Drp1–CL interactions alone are insufficient for fission. Our studies establish the structural and mechanistic bases of Drp1–CL interactions in stress-induced mitochondrial fission.
Mitochondria are dynamic organelles remodeled continuously via regulated cycles of fission and fusion (1, 2). Mitochondrial dynamics and morphology are inextricably linked to the organelle’s essential functions in ATP production, calcium homeostasis, and apoptosis (3), the dysregulation of which precipitates various human diseases including neurodegeneration (4). Therefore, an accurate description of the molecular mechanisms underlying mitochondrial fission and fusion is critical for targeted therapeutic intervention.
Distinct dynamin superfamily proteins (DSPs) catalyze mitochondrial fission and fusion (1, 5). Drp1 mediates fission and is recruited from the cytosol onto the mitochondrial surface via interactions with various membrane-integrated protein adaptors, chiefly Mff and MiD49/51 (6). Subsequent Drp1 helical self-assembly around predisposed mitochondrial division sites coupled to GTP-dependent mechanoenzymatic membrane constriction culminates in fission (7, 8). By contrast, fusion is mediated by two separate membrane-embedded DSPs, mitofusins (Mfn1/2) and optic atrophy 1 (OPA1), that catalyze the respective merger of the mitochondrial outer (MOM) and inner membranes (MIM) (1, 5).
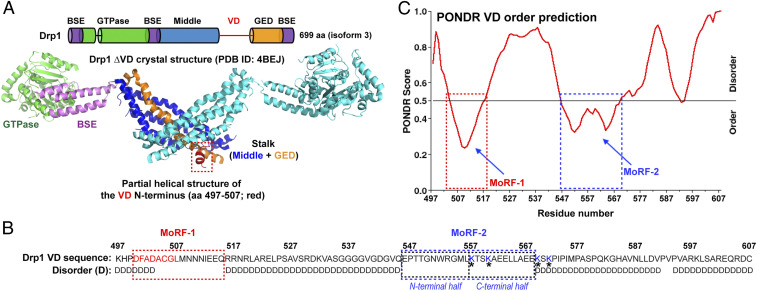
Drp1 is a multidomain protein composed of four distinct regions: the GTPase (G) domain, bundle signaling element (BSE), stalk, and VD (1) (Fig. 1A). The G, BSE, and stalk domains comprise the structured mechanoenzymatic core involved in Drp1 helical self-assembly and assembly-dependent cooperative GTPase activity (5). The VD, located at the base of the molecule (Fig. 1A), is, however, intrinsically disordered (9). Nonetheless, the VD engages in functionally essential interactions with phospholipids and/or protein adaptors at the target membrane (9–11). The VD is absent from, or insufficiently resolved in, any available Drp1 structure until date (12–14).
Fig. 1.
Prediction of Drp1 VD MoRFs as potential CL-binding motifs (CBMs). (A) Domain organization of Drp1 (Top) and a corresponding color-coded ribbon representation of a monomer in the Drp1 dimer (Δ514 to 602) three-dimensional structure (12) (Protein Data Bank identification: 4BEJ; Bottom). The numbering corresponds to the 699 aa ubiquitous Drp1 isoform 3 studied here. The partial helical structure of the VD N terminus is highlighted by a box. (B) Drp1 VD sequence (aa 497 to 607 of isoform 3). Disorder (D) as predicted by the PONDR (Predictor of Naturally Disordered Regions) algorithm in C is indicated below. The residues comprising MoRFs 1 and 2 are boxed. The structurally resolved helical region of MoRF-1 is highlighted in red. MoRF-2 is separated into N- and C-terminal halves with the four K residues previously implicated in CL binding (11) shaded in blue and denoted by *. (C) PONDR VL-XT (Variously characterized Long disordered regions and X-ray characterized Terminal disordered regions) analysis of VD. Segments with low PONDR scores (below 0.5 and approaching zero) are predicted to acquire order (or a stable fold) upon ligand interactions. High scores (above 0.5) represent disorder. The two boxed regions predicted to acquire order correspond to MoRFs 1 and 2.
A preponderance of recent evidence indicates that a direct interaction between the VD and the mitochondria-specific phospholipid CL plays a critical role in mitochondrial fission, especially under stress conditions such as apoptosis and mitophagy (11, 15–17). CL, abundant in the MIM, is nevertheless found in relatively minor but significant quantities (3 to 6%) in the MOM (18, 19). Locally, CL content can approach 25%, for example, at MIM–MOM contact sites (20). Upon the induction of apoptosis or mitophagy, CL is actively translocated from the MIM to the MOM, where it binds and localizes proapoptotic (e.g., tBid, Bax) and mitophagy-related (e.g., LC3) proteins at the mitochondrial surface (21–26). How CL externalization under these conditions is coupled to mitochondrial hyperfragmentation essential for the engulfment of damaged mitochondria by autophagosomes during mitophagy, or for the synchronous release of cytochrome c during apoptosis, still remains unresolved (26–28).
CL promotes Drp1 helical self-assembly on model membranes in vitro and robustly stimulates Drp1 GTPase activity (15). GTP-regulated Drp1–CL interactions alter membrane phase behavior and catalyze the formation of local, narrow membrane tube constrictions primed for fission (29). Yet, how Drp1 specifically recognizes and interacts with CL remain unknown, and the current paradigm for mitochondrial fission does not invoke a role for direct Drp1–phospholipid interactions (8, 10).
The Drp1 VD is necessary and sufficient for specific CL interactions in vitro (9, 29). Yet, none of the four previously implicated VD Lys (K) residues were found to be directly responsible for VD binding to anionic CL (9, 11). No high-resolution structures of the VD–CL complex currently exist, and the nature, residue identity, and functional consequences of specific VD–CL interactions in mitochondrial fission remain unknown. Here, using a comprehensive toolkit of structural, biophysical, and cell biological approaches, we demonstrate that the intrinsically disordered VD undergoes a disorder-to-order structural transition on CL-containing membranes and preferentially binds CL via specific hydrophobic acyl chain interactions. While VD interactions selectively alter CL motion within the membrane, Drp1 GTP hydrolysis-driven conformational rearrangements only transiently modulate the bilayer lipid topology and are thus incapable of fission. We identify a WRG motif conserved between the Drp1 VD and the unrelated ADP/ATP carrier (AAC) that is directly and specifically involved in CL binding, the perturbation of which impairs Drp1–CL interactions in vitro and mitophagy-induced mitochondrial hyperfragmentation in vivo. Thus, Drp1–CL interactions are essential for stress-induced mitochondrial fission.
Results
Prediction of VD MoRFs as Potential CBMs.
Size-exclusion chromatography-coupled multiangle light scattering (SEC-MALS) analysis indicated that the isolated VD (amino acid [aa] 497 to 607 of Drp1 isoform 3; Fig. 1B) is predominantly monomeric in solution (SI Appendix, Fig. S1A). Circular dichroism (CD) spectroscopy revealed that the disordered VD has α-helical propensity that is stabilized in presence of either CL-containing membranes or the electronegative amphiphile trifluoroethanol (TFE) (SI Appendix, Fig. S1 B–E). The sequence analysis algorithm PONDR (Predictor of Naturally Disordered Regions) discerned molecular recognition features (MoRFs) within the VD with a predicted propensity to gain secondary structure upon interactions (30) (Fig. 1C). Two regions were identified and named “MoRF-1” and “MoRF-2” (Fig. 1 B and C). MoRF-1 (aa 500 to 518; Fig. 1B) was predicted to form an amphipathic α-helix (SI Appendix, Fig. S2A) that could adapt to the membrane–water interface but lacked an overall positive charge expected for binding negatively charged CL (31). Moreover, a part of MoRF-1 (Fig. 1A, red) is visible in the Drp1 crystal structure (12) as a helical segment (Fig. 1A, colored red) that makes extensive contacts with the adjacent GTPase effector domain (GED) region. Consistent with its primary role in protein–protein interactions, Ala (A) substitutions of its bulky nonpolar residues dramatically affected Drp1 oligomerization in solution but not CL-stimulated GTPase activity (SI Appendix, Fig. S2 B and C). MoRF-2 (aa 546 to 568; Fig. 1B), together with adjacent K569 and K571, featured more positively charged residues but would not constitute an amphipathic helix (SI Appendix, Fig. S2A). In MoRF-2, bulky nonpolar residues are prevalent in the N-terminal half, whereas charged residues predominate in the C-terminal half (Fig. 1B). Notably, this includes the four aforementioned K residues previously considered to be essential for CL binding (9, 11).
Solution NMR Reveals the Drp1 CBMs.
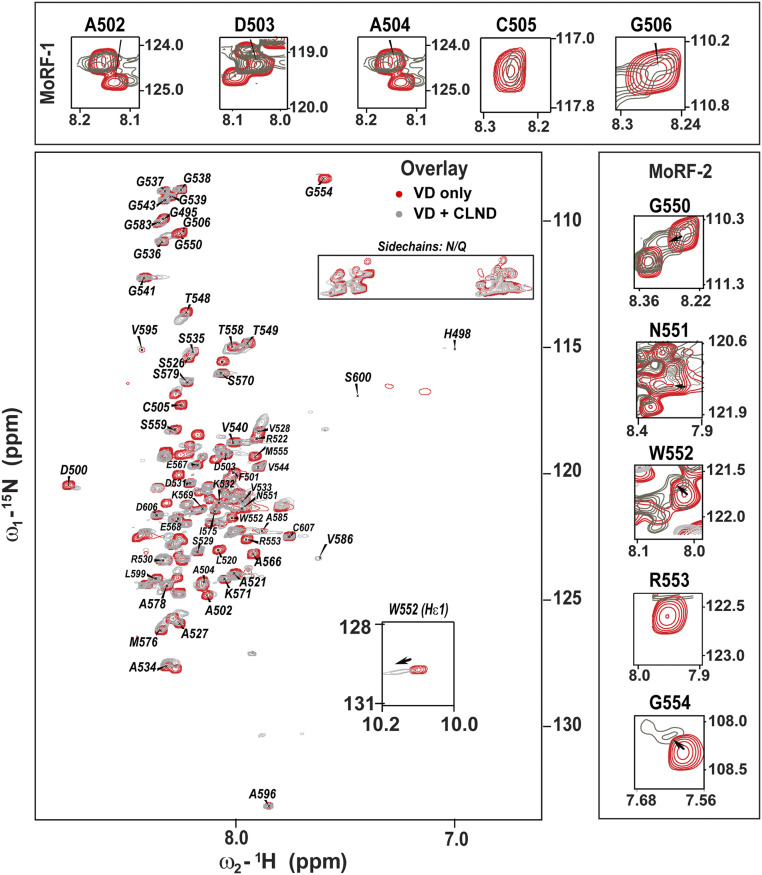
Solution NMR measurements in the absence and presence of CL-containing lipid nanodiscs (CLND) (32, 33) revealed the identity of Drp1 VD residues involved in direct CL interactions. The spectral dispersion from ∼7.6 to 8.8 ppm (>1.1 ppm) showed that the isolated VD is partly folded in solution (Fig. 2, red spectrum, and SI Appendix, Fig. S3A). Despite peak overlap, we unambiguously assigned 59 of the 111 total VD residues (SI Appendix, Table S1). The residue-specific Cα chemical shifts (SI Appendix, Fig. S4 A–D) revealed two short α-helical segments comprising residues 500 to 506 and 550 to 555, remarkably consistent with the predicted MoRF-1 and MoRF-2 regions (Fig. 1B).
Fig. 2.
Solution NMR spectroscopy maps the Drp1 VD–CL binding interface. An overlay of 1H-15N heteronuclear single quantum coherence spectra of Drp1 VD in the absence (red) and presence of CLND (gray) is shown. The spectra are also shown separately in SI Appendix, Fig. S3. CLND interactions lead to extensive peak broadening and a few significant CSPs. (Insets) CSPs and line broadening in helix-forming MoRF-1 and MoRF-2 regions are shown.
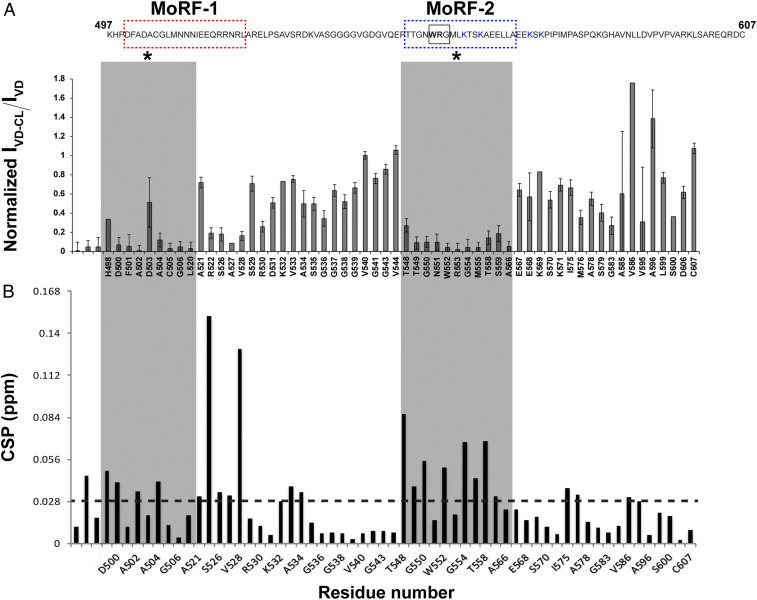
NMR chemical shifts are highly sensitive to environmental changes, resulting in chemical shift perturbations (CSPs) for, for example, lipid-interacting residues (32, 33). We observed that parts of both MoRF-1 and MoRF-2 displayed significant CSPs and extensive line broadening upon CLND interactions (Fig. 3 A and B). Upon CLND binding (Fig. 2, gray spectrum, and SI Appendix, Fig. S3B), ∼48 peaks (∼50% of signals) were exchange broadened, with many becoming effectively “invisible” in these solution NMR experiments. The data indicated an exchange process with a binding affinity (KD) in the low micromolar range. Notably, the CLND-induced spectral changes were centered primarily on MoRF-1 and MoRF-2 (Fig. 3 A and B). Focusing on the MoRF-1 and MoRF-2 spectral regions (Fig. 2, Insets) revealed that their residues show both exchange broadening and CSPs, indicating direct involvement in CLND interactions (Fig. 3 A and B).
Fig. 3.
NMR identification of the Drp1 CBMs. (A) Differential line broadening plot of 15N-labeled Drp1 VD in the presence of CLND. The data were referenced to the V540 signal. The residues corresponding to MoRF-1 and MoRF-2 experienced extensive peak attenuation relative to other residues upon CLND interactions (shaded in gray and marked by *). The VD sequence is shown above for easy reference, and the NMR-ascertained MoRF regions are marked by color-coded boxes as in Fig. 1B. The WRG motif is boxed. Error bars were calculated using S/N obtained from the Sparky software. Error bars for overlapped peaks are not plotted. (B) CSP plot shows that the greatest density of CLND-induced perturbations occurs in MoRF-1 and MoRF-2 and correlates well with the extensive line broadening observed for these regions upon CL interactions. The dashed line at 0.028 ppm represents the average CSP, the values above which are considered to be significant.
Given its potential involvement in specific CL interactions (see above), we focused on MoRF-2 that includes two (K557, K560) of the four K residues previously implicated in Drp1–CL interactions (11). The greatest density of CSPs and line broadening was found in the relatively hydrophobic N-terminal half of MoRF-2, whereas the two assigned K residues (K569, K571) immediately adjacent to the charged C-terminal half did not show any significant CSPs (Fig. 3 A and B). We conclude that these K residues, consistent with our previous biophysical studies (9), are likely not engaged in direct VD–CL interactions. In striking contrast, a specific cluster of residues in the N-terminal half of MoRF-2 underwent extensive line broadening coupled to significant, detectable CSPs: the WRG motif of residues 552 to 554 featuring the sole tryptophan (W552) in VD (Figs. 2 and 3 A and B). Whereas membrane-interacting MoRF-2 residues became more ordered, residues flanking MoRF-2 showed increased peak intensities and reduced linewidths (SI Appendix, Table S2) indicating an increase in their mobility, which likely facilitates MoRF-2 binding through entropic contributions. Interestingly, the WRG motif itself is partially ordered in the solution state of the VD, as 1H-1H homonuclear NOESY measurements showed an interaction between the R553 side chain and the G554 backbone amide (SI Appendix, Fig. S4E).
A Conserved WRG Sequence in MoRF-2 Likely Constitutes the Specific Drp1 CBM.
A consensus CBM has not yet been identified in CL-interacting proteins, suggesting that CL recognition elements may not be readily identifiable by sequence homology alone (31). We used published three-dimensional structures of membrane proteins cocrystallized with CL to survey for structural elements also found in VD.
Remarkably, the abovementioned “WRG” sequence was identified to be a conserved motif between the Drp1 VD and the CL-bound AAC (also known as adenine nucleotide translocase or ANT) (34, 35) (SI Appendix, Fig. S5A). The AAC WRG segment is also α-helical and resides close to the lipid–water interface in direct contact with CL, mimicking our findings on VD MoRF-2 (SI Appendix, Fig. S5B). Surprisingly, the central “R” of the AAC WRG motif does not engage in direct electrostatic interactions with the CL phosphates but “snorkels” upward to form part of a positively charged surface at the lipid–water boundary (SI Appendix, Fig. S5 B and C) (34). Instead, the two CL headgroup phosphates form hydrogen bonds with the exposed backbone amide groups of two G residues, including the G from the WRG motif. Moreover, the “W” of the AAC WRG motif engages in direct hydrophobic interactions with one of the four acyl chains of CL. Thus, against expectations, CL-specific binding appeared to be derived from hydrophobic and backbone H-bonding interactions instead of electrostatics (31, 36).
MoRF Mutations Impair VD-CL Binding and CL-Stimulated Drp1 Activity.
An evaluation by mutagenesis of the involvement of the two MoRFs in various in vitro Drp1 activities including CL-dependent membrane binding, self-assembly, and stimulation of GTPase activity revealed the cooperative, yet disparate, nature of their membrane association.
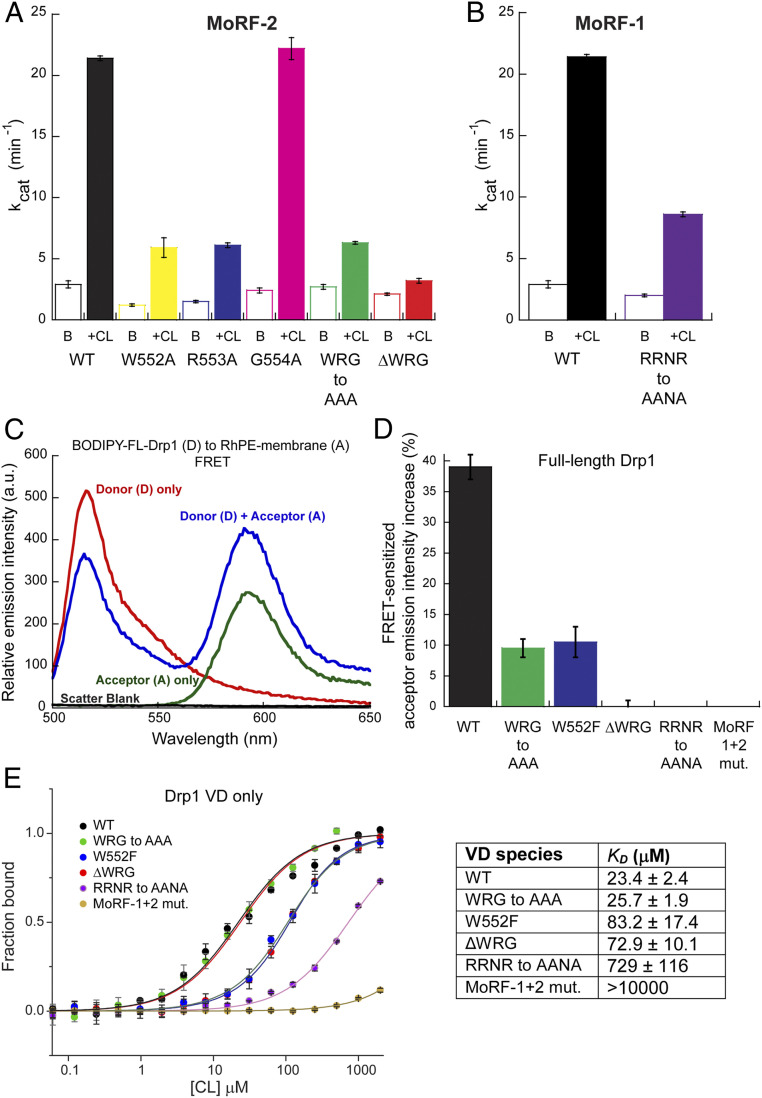
In MoRF-2, Ala (A) substitution of either W552 or R553, or substitution of the entire motif (WRG-to-AAA Drp1), resulted in a pronounced loss of CL-stimulated GTPase activity (Fig. 4A). Deletion of the motif (ΔWRG Drp1) had the greatest impact, essentially abrogating the activity (Fig. 4A). The critical role of W552 was revealed by the very conservative W552F substitution that still significantly impaired CL-stimulated GTPase activity (SI Appendix, Fig. S6A). Consistent with the involvement of the G554 backbone amide, a G554A substitution did not affect CL-stimulated GTPase activity (Fig. 4A). Importantly, in solution, these MoRF-2 mutations affected neither Drp1 oligomerization (SI Appendix, Fig. S6 B and A) nor the characteristic capacity of Drp1 to form rings and helices in the presence of the nonhydrolyzable GTP analog, GMP-PCP (SI Appendix, Fig. S7C). Yet, none of the WRG mutants were capable of self-assembling on, and tubulating, CL-containing liposomes (SI Appendix, Fig. S8).
Fig. 4.
MoRF mutations that impair CL interactions. (A and B) Basal (B) and CL-stimulated (+CL) GTPase activities of WT Drp1 in comparison to various MoRF-2 (A) and MoRF-1 (B) mutants. (C) Representative emission spectra of BODIPY-labeled WT Drp1 (donor) in the absence and presence of 1 mol% RhPE (acceptor) in DOPC/DOPE/CL liposomes. FRET was detected by a decrease in donor emission intensity in the presence of acceptor accompanied by a FRET-sensitized increase in acceptor emission upon donor excitation. The A-only trace shows direct excitation of the acceptor at the donor excitation wavelength, which represents the background. Scatter from liposomes was negligible. (D) FRET-sensitized rhodamine emission intensity increase upon incubation of BODIPY-labeled WT Drp1 or mutants with RhPE-labeled, 25 mol% CL-containing liposomes is plotted as a percentage. Data shown are averages ± SEM. (E) MST measurements of WT VD or mutants (0.35 μM final) binding to pure CL (100 mol%) liposomes. Note the logarithmic scale of the x-axis. (Right) The apparent equilibrium dissociation constant or binding affinity (KD) is tabulated. Data shown are averages ± SEM.
In MoRF-1, on the other hand, Ala (A) substitutions of its bulky hydrophobic residues did not appreciably affect CL-stimulated GTPase activity (SI Appendix, Fig. S2C), reinforcing the idea that MoRF-1 instead acts through electrostatics. Remarkably, comparing the Drp1 VD sequence to its counterpart in the ortholog, CmDnm1, from the primitive red alga Cyanidioschyzon merolae revealed a conserved “RRNR” motif located at the Drp1 MoRF-1 C terminus (SI Appendix, Fig. S5D), despite very little overall VD homology (37). Consistent with our notion, complete Ala (A) substitution of the three positively charged R residues (RRNR-to-AANA Drp1) resulted in a substantial loss of CL-stimulated GTPase activity (Fig. 4B) while affecting neither Drp1 oligomerization (SI Appendix, Fig. S7B) nor the capacity to form rings and helices in solution (SI Appendix, Fig. S7C) (15). Yet, like the MoRF-2 WRG mutants, RRNR-to-AANA Drp1 was incapable of remodeling CL-containing liposomes (SI Appendix, Fig. S8).
Förster resonance energy transfer (FRET) measurements between BODIPY-labeled wild type (WT)Drp1 or mutants (donor) and rhodamine-PE (RhPE)-labeled, 25 mol% CL-containing liposomes (acceptor) indicated very little to no binding of the MoRF-1 or MoRF-2 mutants with membranes (Fig. 4 C and D). Interestingly, MoRF-2 substitution mutants (WRG-to-AAA and W552F) retained partial membrane association, whereas the MoRF-1 RRNR-to-AANA mutant and the combined MoRF-1 + MoRF-2 mutant (RRNR-to-AANA + WRG-to-AAA mutant, termed “MoRF-1+2 mutant”) did not stably or detectably bind membranes. Thus, MoRF-1–mediated electrostatic interactions are key to stable VD–CL association. Microscale thermophoresis (MST) measurements of pure CL binding (KD) to monomeric Drp1 VD (38) also showed that electrostatic interactions of the MoRF-1 RRNR motif dominate VD-CL binding (Fig. 4E), while the combined MoRF-1+2 mutant essentially abolished CL binding. CD measurements showed no disorder-to-order transition for MoRF-1 and MoRF-2 mutants in the presence of CL (SI Appendix, Fig. S9). Together, these data indicated positive cooperativity between MoRF-1 and MoRF-2 in CL binding and suggested that essential, nonspecific electrostatic interactions of the MoRF-1 RRNR motif with CL precede and facilitate specific, short-range hydrophobic CL interactions of the MoRF-2 WRG motif.
MoRF Mutations Impair Fission and Induce Donut Mitochondria Formation.
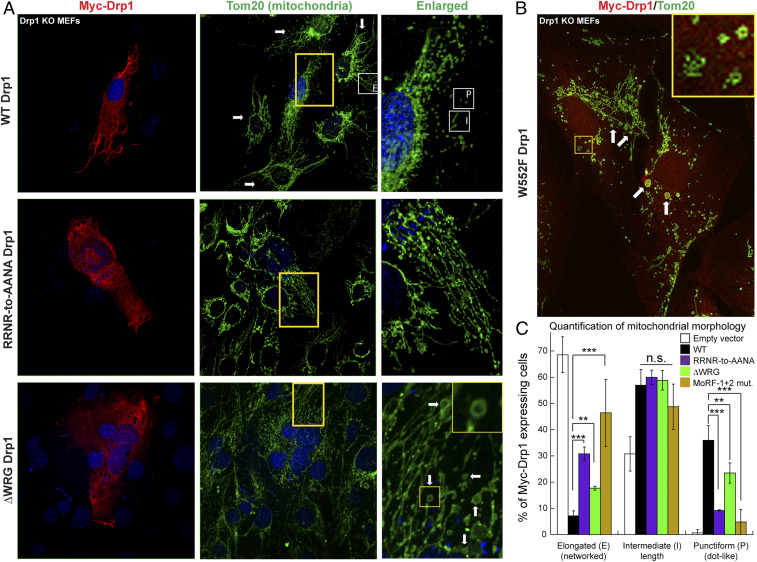
In vivo, the overexpression of MoRF-1 or MoRF-2 mutants in endogenous Drp1 knockout mouse embryonic fibroblasts (Drp1 KO MEFs) resulted in the impairment of mitochondrial hyperfragmentation but to different extents (Fig. 5). In the case of ΔWRG and W552F Drp1, it manifestly gave rise to a mitochondrial morphology previously unknown for any other Drp1 mutant (15, 39). WT Drp1 expression, as expected, restored mitochondrial fission and fragmentation in Drp1 KO MEFs, which otherwise displayed extended and hyperfused mitochondria (Fig. 5 A, Top and C). However, the equivalent expression of MoRF-1 RRNR-to-AANA Drp1 resulted in a significant retention of hyperfused mitochondria accompanied by the lack of punctiform (rounded) or hyperfragmented mitochondria (Fig. 5 A, Middle and C). The MoRF-2 WRG-to-AAA Drp1 mutant produced a similar phenotype, although the impairment of fission was less pronounced (SI Appendix, Fig. S10). In comparison, mitochondrial hyperfragmentation was almost entirely ablated in MoRF-1+2 Drp1–expressing cells, consistent with the complete loss of CL-stimulated GTPase activity (Fig. 5C and SI Appendix, Fig. S11 A and B, Top). Relative to MoRF-1 RRNR-to-AANA– and MoRF-1+2 Drp1–expressing cells, cells expressing ΔWRG Drp1 produced more punctiform mitochondria, albeit significantly less than WT Drp1, suggestive of partial or basal mitochondrial fission activity (Fig. 5C). Remarkably, ΔWRG- or W552F-Drp1 expression resulted in the conspicuous appearance of ring-like or donut-shaped toroidal mitochondria alongside elongated mitochondrial fragments (Fig. 5A, Bottom and B). This morphology was previously ascribed to excessive Mfn1/2-catalyzed mitochondrial fusion coupled to impaired Drp1-mediated fission (40–42). This altered morphology sharply contrasted with the perinuclear clustering of hyperfused mitochondria produced by other fission-inhibiting Drp1 mutants such as K38A Drp1 (39) and 4KA Drp1 (29). Together, these data firmly established that productive mitochondrial fission is significantly impaired in the absence of specific VD–CL interactions. Consistent with a role for Mfn1/2 in the donut mitochondria formation (42), no such morphology was observed in W552F Drp1-expressing Mfn1/2 KO MEFs, which characteristically displayed hyperfragmented mitochondria (SI Appendix, Fig. S12).
Fig. 5.
MoRF mutations impair fission and induce donut mitochondria formation. (A and B) Confocal immunofluorescence images of Drp1 KO MEFs expressing either Myc-WT Drp1 or mutants (red) and stained for mitochondria (green). Arrows in A, Top Middle point to hyperfused mitochondrial networks in untransfected Drp1 KO MEFs. Regions demarcated by yellow boxes are enlarged and shown to the right. Examples of elongated “E,” intermediate “I,” and punctiform “P” mitochondria are marked by white boxes in Top, Middle, and Right. (Insets) Donut mitochondria of various shapes and sizes prominently found in ΔWRG and W552F Drp1-expressing cells are pointed by arrows and are shown enlarged. (C) Classification of Myc-Drp1-expressing cells based on the predominant mitochondrial morphology displayed. Mean ± SD is plotted. Statistical significance was determined using two-way ANOVA with post hoc Tukey’s test for comparison of multiple groups. n.s. not significant, **P ≤ 0.01, ***P ≤ 0.001.
WRG–CL Interactions Catalyze Mitophagy-Induced Mitochondrial Hyperfragmentation.
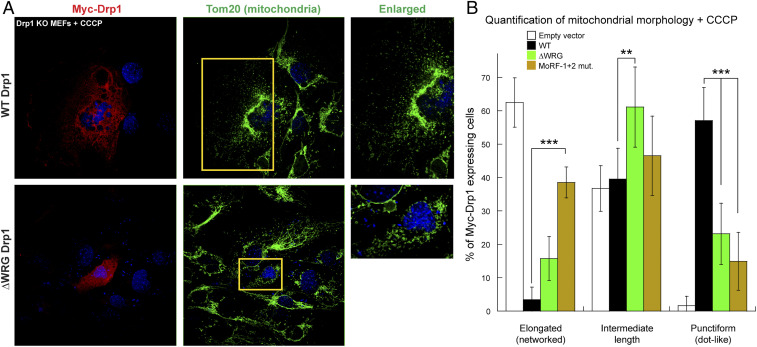
As mitochondrial fission in ΔWRG Drp1-expressing cells was only partially impaired under physiological conditions (Fig. 5C), we tested the mutant’s efficacy in stress-induced excessive mitochondrial hyperfragmentation as occurs during mitophagy (24). To this end, we treated Drp1 KO MEFs with the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) that depolarizes mitochondria (43) and induces mitophagy by actively promoting CL externalization to the MOM (24) (Fig. 6A). Remarkably, under these stress conditions, ΔWRG Drp1-expressing cells were more severely impaired in mitochondrial hyperfragmentation and were comparable to MoRF-1+2 Drp1–expressing cells in the production of significantly less punctiform mitochondria (Fig. 6B and SI Appendix, Fig. S11 B, Bottom). Interestingly, no significant differences in mitochondrial morphology were observed for MoRF-1+2 Drp1–expressing cells in the absence and presence of CCCP (Figs. 5C and 6B), indicating that MoRF-1 RRNR–lipid interactions are essential for fission regardless of environmental conditions. On the other hand, our data indicate that specific MoRF-2 WRG–CL interactions are particularly critical for stress-induced mitochondrial hyperfragmentation.
Fig. 6.
ΔWRG and MoRF-1+2 mutant Drp1-expressing cells resist mitophagy-induced mitochondrial hyperfragmentation. (A) Same as Fig. 5A but in the presence of the mitophagy-inducer CCCP. (B) Same as Fig. 5C but in the presence of CCCP. Statistically significant differences are observed for all three mitochondrial phenotypes between WT and mutant Drp1-expressing cells. Mean ± SD is plotted. Statistical significance was determined using two-way ANOVA with post hoc Tukey’s test for comparison of multiple groups. **P ≤ 0.01, ***P ≤ 0.001.
Lipid-Packing Defects Enable Drp1–CL Recognition but Do Not Facilitate Hemifission or Fission.
A recent study inferred that Drp1–CL interactions alone are sufficient for membrane fission (44). Solid-state NMR (ssNMR), together with electron microscopy (EM), was used to examine how Drp1 binds to, and affects the integrity of, CL-containing model membranes in vitro (9, 29). Drp1 was self-assembled on CL-containing membranes (at 1:200 protein/lipid ratio) and allowed to hydrolyze GTP. Using negative-stain transmission EM as earlier (29, 45), we observed Drp1-decorated membrane tubes with intermittent narrow diameters (constrictions) with no evidence of membrane fission (vesiculation) (Fig. 7A). Using 31P ssNMR techniques sensitive to such phases, we sought direct evidence of nonbilayer structures that are a prerequisite for membrane fission. The obtained 31P ssNMR spectra were characteristic of fluid lipid bilayers under all conditions studied (Fig. 7B) with no evidence of stable nonbilayer structures. The binding of isolated VD yielded analogous ssNMR results, again with no sign of stable nonbilayer phases, even when using higher amounts of protein (1:50 P/L ratio) (SI Appendix, Fig. S13A). Thus, 31P ssNMR detected no significant signals from stable nonbilayer phases (requisite hemifission stalk intermediates), which are easily identified by their qualitatively different spectral features (46) (as illustrated in SI Appendix, Fig. S13 B–D).
Fig. 7.
Drp1 VD intercalates into the membrane core to bind CL but does not induce stable nonbilayer phases. (A) Negative-stain transmission EM of the Drp1 ssNMR sample shows Drp1-induced membrane tubulation and GTP-dependent local membrane constriction (yellow boxes; arrow points to a constricted region). (B) Static 31P ssNMR of DOPC/PE/CL liposomes in the presence (blue) and absence (dashed) of bound Drp1 shows the characteristic line shape of a liquid crystalline lipid bilayer. Binding by Drp1 in presence of GTP (Bottom) leads to a narrowing of the overall 31P line shape, indicating an increased lipid headgroup wobble. (C) 31P MAS ssNMR of DOPC/CL liposomes without (Top) and with (Bottom) bound VD shows two peaks corresponding to the DOPC and CL lipid species. (D) T2 relaxation measurements reveal different amounts of dephasing for VD-bound CL versus PC. (E) Upon VD binding, the T2 relaxation curve for DOPC is unchanged (Bottom), but the T2 time for CL is significantly shortened (Top), indicating a slowing down of CL lateral diffusion by bound VD. (F–H) GTPase activity of Drp1 incubated with liposomes containing: (F) CL species of differing acyl chain lengths and saturation in a 85/15 DOPC/CL species context, (G) an increasing mole fraction of DOPE in the DOPC/CL background, and (H) an increasing concentration of CL in the presence of 20% DOG. Data are averages ± SD.
Complementary 31P magic-angle spinning (MAS) ssNMR measurements of the mixed lipid bilayers revealed the isotropic signals of the individual lipid types (Fig. 7C). In a 31P T2 relaxation measurement, we observed the relaxation-induced signal losses (Fig. 7D), which are sensitive to the rate of lipid diffusion (47). In protein-free liposomes, the decay curves for PC and CL were indistinguishable (Fig. 7E, dashed curves), showing that the two lipids experience the same diffusion rates. Upon addition of 2 mol% VD, the PC relaxation remained unchanged, but the CL relaxation was significantly increased (Fig. 7E, solid curves). Thus, VD binding selectively decreases the in-plane diffusion of CL.
Also, in the static 31P spectra (Fig. 7B and SI Appendix, Fig. S13A), the increase in ssNMR linewidth (due to the faster T2 relaxation) is outweighed by a concomitant reduction in the 31P chemical shift anisotropy. This change is indicative of an increased wobble of the lipid phosphates upon VD binding (47, 48), which can be due to CL clustering, an intercalation of VD into the subinterface region, or a combination of both. Hydrophobic VD membrane interactions, supported by the ssNMR data above, would predict that membrane-surface defects facilitate VD–CL interactions and, thus, Drp1 activity (49, 50). Assessing CL-stimulated GTPase activity as a measure of this expected connection between membrane structure and Drp1 function revealed that, whereas no significant difference in activity was observed between CL species of differing acyl chain lengths and unsaturation (Fig. 7F), the incorporation of differently sized lipid headgroups had a significant impact. Membrane-surface defects enhanced by replacing cylindrical PC with cone-shaped (but electrically neutral) PE, or by incorporating diacylglycerol (DAG) lipid species, caused linear increases in CL-stimulated GTPase activity (Fig. 7 G and H). DAG, which facilitates mitochondrial fission in vivo (51), also reduced the effective CL concentration needed for the stimulation of Drp1 GTPase activity to less than 10% (Fig. 7H), matching physiologically relevant MOM CL levels (<6 mol%) (Fig. 7G). No CL-stimulated GTPase activity was previously observed for Drp1 at CL concentrations ≤10 mol% in the absence of DAG (15). Thus, effective VD intercalation into CL-containing membranes facilitated by membrane-surface defects determines Drp1 activity.
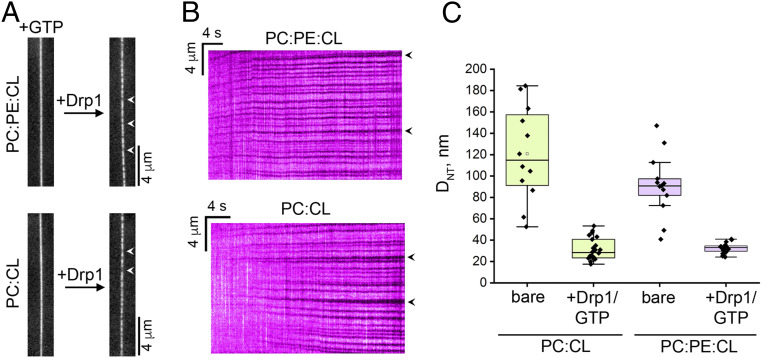
The impact of conical lipids was also observed in Drp1 capacity to constrict freely suspended biomimetic membrane nanotubes (NTs) (52, 53) composed of PC and CL in the absence and presence of PE (Fig. 8). In either case, GTP-regulated Drp1 self-assembly on NTs monitored using fluorescence microscopy revealed the formation of locally constricted membrane regions (Fig. 8 A and B), whose diameters matched previous reports (54) (Fig. 8C). Importantly, the presence of PE facilitated more consistent constrictions (Fig. 8C), supporting a catalytic and/or a regulatory role for conical lipids and associated membrane-surface defects in Drp1-mediated membrane remodeling.
Fig. 8.
Drp1–CL interactions are not sufficient for membrane fission in vitro. (A) Fluorescence micrographs showing a lipid NT before (Left) and after (Right) addition of unlabeled WT Drp1 (1 μM final) in the presence of GTP (1 mM final). NTs were composed of either PC:PE:CL:RhPE 39:35:25:1 mol% (Top) or PC:CL:RhPE 75:24:1 mol% (Bottom). RhPE fluorescence is monitored. Spots with decreased RhPE fluorescence along the NTs (arrowheads) upon Drp1 addition correspond to localized NT constriction upon GTP-regulated self-assembly of a Drp1 helical scaffold. (B) Representative kymographs showing the growth in time of the Drp1 helical scaffolds (arrowheads). Data correspond to the initial frames from Movies S1 (with PE) and S2 (without PE) upon addition of Drp1. RhPE fluorescence is monitored. Note that the NTs did not undergo fission in either condition 7 min after detecting the first Drp1 scaffold on the NT membrane (36 NTs with PE and 181 NTs without PE from three independent experiments). (C) Box plots show the NT diameters (DNT) (SI Appendix) before (bare) and after Drp1 addition in the presence of GTP as in A. Error bars show SD.
Critically, regardless of condition, the final diameters of the constricted NT regions remained well above the bilayer thickness, therefore precluding membrane fission (55) (Fig. 8C and Movies S1 and S2). These results are consistent with EM observations of Drp1-decorated membrane tubes upon GTP addition here (Fig. 7A and SI Appendix, Fig. S14) and in previous studies (29, 45). Instead, we observed NT membrane buckling (Movies S1 and S2) likely because of restricted lipid (CL) diffusion upon VD interactions as indicated by the ssNMR data (Fig. 7E). Based on these collective data, we conclude that Drp1–CL interactions alone do not suffice for either membrane hemifission or fission.
Discussion
The fission of damaged, dysfunctional mitochondrial segments into short, punctiform units is essential for their engulfment by autophagosomes during mitophagy (26, 27). We demonstrate that specific interactions of Drp1 with MOM-localized CL, externalized under stress conditions, catalyzes such mitochondrial hyperfragmentation (Fig. 9, Top). Our current working model (Fig. 9, Middle) suggests that stable Drp1–CL interactions accompany a local disorder-to-order transition of the VD affected by the electrostatic association of the MoRF-1 RRNR motif with negatively charged CL headgroups coupled with the hydrophobic membrane intercalation and direct acyl chain interactions of the CL-specific MoRF-2 WRG motif. This CL-induced structural reorganization and membrane anchoring of the VD presumably relieves VD autoinhibition of Drp1 self-assembly (9), promotes GTP hydrolysis-dependent Drp1 membrane constriction, and potentiates CL nonbilayer forays underneath the dynamic Drp1 helical scaffold toward the creation of the requisite membrane hemifission intermediate (Fig. 9, Middle) (29). Stable membrane hemifission is, however, likely achieved through cooperative Drp1 interactions with additional cofactors [conceivably with stress-activated Drp1 receptors such as Fis1 (56) or MiD49/51 (57)] that, in concert with membrane tension, enable leak-free mitochondrial fission and pole separation (Fig. 9, Middle) (1). Weakened or unstable VD–CL interactions as observed with select MoRF-1 and MoRF-2 mutants in this study (Fig. 9, Bottom), on the other hand, likely cause CL degradation to profusogenic phosphatidic acid (PA) by Drp1-associated mitochondrial phospholipase D (17). Hyperactivation of Mfn1/2 by excess PA presumably catalyzes mitochondrial hyperfusion (58) and, under yet-unresolved conditions (40–42), promotes donut mitochondria formation (Fig. 9, Bottom). Based on our collective results, we further postulate that while housekeeping mitochondrial fission (biogenesis) and maintenance of the basic fission–fusion balance by Drp1 are mostly CL independent, the increased rates and efficacy of fission observed during stress conditions, including mitophagy (24) and apoptosis (28), strongly rely on specific Drp1–CL interactions.
Fig. 9.
Model for CL-dependent Drp1-catalyzed stress-induced mitochondrial fission. (Top) Proposed pathway for CL externalization-induced Drp1 self-assembly and mitochondrial hyperfragmentation toward mitophagy. (Middle and Bottom) Specific interactions of the VD MoRF-1 and MoRF-2 motifs with CL elicit a disorder-to-order transition that enables VD membrane insertion and stabilization of Drp1 self-assembly toward hemifission and fission. Certain MoRF-1 and MoRF-2 mutants are impaired in this transition, leading to weakened or entirely abrogated Drp1–membrane interactions. Unbound MOM CL is converted to PA by the action of mitochondrial phospholipase D. Profusogenic PA stimulates Mfn1/2 activity, leading to mitochondrial hyperfusion and, in the case of W552 mutants, the formation of mitophagy-resistant donut mitochondria.
Signaling lipids such as phosphatidylinositol-4,5-bisphosphate (PIP2) and DAG utilize cognate stereospecific binding pockets in the structured globular domains (e.g., PH and C1 domains) of diverse protein partners to achieve interaction specificity (59). However, no such structures nor consensus binding elements have been uncovered yet for CL-interacting proteins, including Drp1 (31, 60). We here demonstrate that the conserved WRG sequence of the VD constitutes the canonical Drp1 CBM. VD–CL recognition is accomplished via an apparent disorder-to-order structural transition involving the MoRF-2 helix that facilitates direct hydrophobic contact of the conserved Trp residue (W552) with the CL acyl chain moiety as observed in AAC/ANT (34, 35). We further postulate that the preferential binding of Drp1 to other charged but cone-shaped lipids such as PA and phosphatidylglycerol (PG), albeit weakly compared to CL (15), likely involves analogous hydrophobic VD interactions. This may explain why in CL-deficient cells, the noted accumulation of PG, a CL precursor and a minor anionic lipid under normal conditions, compensates for the lack of CL and restores mitochondrial fission (61–64).
Our data highlight the central importance of W552 of the WRG motif, reminiscent of the common involvement of aromatic amino acids in lipid-binding motifs, including CBMs (31, 65–68). Nonetheless, another key finding is that the Drp1 VD relies on a complex and combinatorial set of molecular interactions to achieve specific CL binding. These include surprising backbone hydrogen bonding and hydrophobic membrane contact, alongside more expected charge-based electrostatic interactions. Positively charged Lys and Arg have been previously shown to utilize their long side chains as “snorkels” to regulate protein membrane immersion depth, while nearby W residues act as membrane interfacial anchors (69) to regulate helix orientation at the lipid–water interface (70–72). Noting that Trp–lipid interactions in model peptides also facilitate the induction of nonbilayer lipid phases (69, 72), we propose that the WRG motif of the Drp1 VD functions similarly and may play a direct role in catalyzing downstream membrane hemifission and fission events. Its combinatorial lipid interactions, coupled to potential GTP-dependent VD conformational rearrangements akin to those of the dynamin PH domain (73, 74), may induce the lipid tilt necessary for the creation of the requisite hemifission stalk intermediate (75). It is thus conceivable that the multivalent membrane interactions of a helical Drp1 polymer function cooperatively to transiently tilt the CL molecules within the bilayer plane [by tugging on their acyl chains as previously demonstrated through a GTP-dependent increase in membrane nonbilayer propensity (9, 29)] to achieve narrow membrane constrictions for fission.
Although we contend that the WRG motif of MoRF-2 is the primary site that mediates specific Drp1 VD–CL interactions, our data also identify a key role for MoRF-1. While engaged in autoinhibitory protein–protein interactions in the solution conformation of Drp1 (9, 76), it could rearrange upon target membrane binding to engage with negatively charged lipids, including CL, via its conserved RRNR motif. Multivalent CL binding may facilitate the formation of “CL nanodomains” underneath the helical Drp1 scaffold as suggested by previous kinetic data that indicated that each VD binds more than one CL molecule (29). The reorientation of the MoRF-1 α-helix could elicit larger structural rearrangements in the self-assembled Drp1 polymer to allosterically achieve the observed CL stimulation of Drp1 GTPase activity.
For a CL-binding domain, a remarkable feature of the Drp1 VD is that it lacks an overall positive charge. There is nearly an equal representation of positively (K + R = 17) and negatively charged (D + E = 15) residues, whereas hydrophobic residues (55 out of 111 total) predominate the structure overall. This feature supports the idea that hydrophobic acyl chain interactions are important to achieve specific CL binding. This contrasts with other CL-binding proteins like cytochrome c that depend on high levels of positive charges (77). This may be necessitated by the required control over CL topological transitions and to avoid inopportune disruptions of membrane bilayer integrity. Based on its similarity to AAC/ANT-CL binding, we propose that the MoRF-2 helix, featuring the WRG motif, balances flexibility (via G554) and structural rigidity (via W552 and R553) as needed for these CL interactions. We further propose a two-step model of CL binding, wherein nonspecific electrostatic interactions between the numerous K and R residues of the Drp1 VD with a local negatively charged membrane surface (with or without CL) initiate membrane binding, albeit weakly and nonspecifically. CL recognition and sequestration underneath the growing Drp1 helical scaffold, as previously demonstrated (29), concentrate the packing defects among cone-shaped CL, therefore facilitating hydrophobic VD membrane insertion and consolidating high-affinity Drp1-CL binding.
While our results regarding the lack of fission by Drp1 alone contradict the conclusions of ref. 44, they are otherwise consistent with multiple previous investigations that employed either tagless or N-terminally His6-tagged Drp1 to report robust GTP hydrolysis-dependent local high membrane curvature generation (or stabilization) without any accompanying fission or membrane vesiculation (12, 29, 45, 78, 79). We surmise that the altered conformational and assembly–disassembly dynamics of the C-terminally tagged Drp1 construct employed in ref. 44 that exhibits diminished cooperative GTPase activity on CL-containing membranes (44), coupled to the potential restriction of GTP hydrolysis-induced oligomer disassembly as previously observed with GFP-tagged Drp1 (79), are likely contributors to the observed abrupt NT membrane destabilization and rupture. The influence of affinity tags appended to the conformationally sensitive BSE region on Drp1 oligomerization equilibria, assembly–disassembly dynamics, and associated membrane remodeling activities is currently under thorough investigation. Other experimental aspects, including a disparate response of the freely suspended NTs used in this study (and ref. 54) versus polymer-supported NTs used in the discrepant study (44) may also contribute to the observed differences in vitro. Regardless, the overwhelming majority of data from past studies corroborate our conclusion that Drp1–CL interactions alone are insufficient for membrane fission. In the current work, our ssNMR and EM studies provide independent and orthogonal support. Based on these results and previous observations from other groups, we conclude that additional force factors, including contributions from cytoskeletal elements that regulate Drp1 activity, for example, F-actin or microtubules (80–83), are likely required for physiologically relevant membrane fission.
In summary, our comprehensive studies firmly establish an essential role for Drp1 VD–CL interactions in stress-induced mitochondrial fission in vivo and provide insights into the molecular mechanisms underlying CL recognition and binding.
Materials and Methods
A detailed description of the methodologies used in this study including protein purification and labeling, the preparation of lipid templates, far ultraviolet CD spectroscopy, computational analyses, solid-state and solution NMR, fluorescence and negative-stain transmission EM, GTPase assay, FRET, MST, SEC-MALS, and cell biology experiments is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank David Chan (Caltech) for the provision of Mfn1/2 KO MEFs, Emily Wang (Case Western Reserve University) for initial CD measurements, Jason Mears (Case Western Reserve University) for EM of Drp1 polymers, Michael Delk (University of Pittsburgh) for assistance with the ssNMR, James Conway and Alexander Makhov (both University of Pittsburgh) for the use of the EM facility, and Alessia Lasorsa (University of Groningen) for control ssNMR measurements. The research in this study was supported by NIH R01 Grant GM121583 awarded to R.R. P.C.A.v.d.W. was supported by NIH R01 Grant GM112678, and J.C.B. was supported by NIH T32 Grant GM088119 and the Achievement Rewards for College Scientists Foundation. NIH R01 Grant GM112491 to M.B. supported his and M.M.’s contributions, while NIH R01 Grant NS088192 supported X.Q., Y.S., and R.W. A.S. was supported by Spain Ministry of Science and Innovation/National Research Agency/European Regional Development Fund Grant PGC2018-099971-B-I00 and EUR2019-103830, and Basque Government Grant IT1270-19. J.M.M.G. acknowledges the predoctoral fellowship from the University of the Basque Country. The 750-MHz ssNMR instrument was funded by NIH Grant S10RR033477. We thank Blake Hill (Medical College of Wisconsin) for critical comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023079118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Ramachandran R., Mitochondrial dynamics: The dynamin superfamily and execution by collusion. Semin. Cell Dev. Biol. 76, 201–212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labbé K., Murley A., Nunnari J., Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 30, 357–391 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Roy M., Reddy P. H., Iijima M., Sesaki H., Mitochondrial division and fusion in metabolism. Curr. Opin. Cell Biol. 33, 111–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh K., Nakamura K., Iijima M., Sesaki H., Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 23, 64–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramachandran R., Schmid S. L., The dynamin superfamily. Curr. Biol. 28, R411–R416 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Richter V., Singh A. P., Kvansakul M., Ryan M. T., Osellame L. D., Splitting up the powerhouse: Structural insights into the mechanism of mitochondrial fission. Cell. Mol. Life Sci. 72, 3695–3707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman J. R., et al., ER tubules mark sites of mitochondrial division. Science 334, 358–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBride H. M., Frost A., Cell biology: Double agents for mitochondrial division. Nature 540, 43–44 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Lu B., et al., Steric interference from intrinsically disordered regions controls dynamin-related protein 1 self-assembly during mitochondrial fission. Sci. Rep. 8, 10879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bui H. T., Shaw J. M., Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr. Biol. 23, R891–R899 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bustillo-Zabalbeitia I., et al., Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS One 9, e102738 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fröhlich C., et al., Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 32, 1280–1292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francy C. A., Clinton R. W., Fröhlich C., Murphy C., Mears J. A., Cryo-EM studies of Drp1 reveal cardiolipin interactions that activate the helical oligomer. Sci. Rep. 7, 10744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalia R., et al., Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 558, 401–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macdonald P. J., et al., A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol. Biol. Cell 25, 1905–1915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ugarte-Uribe B., Müller H. M., Otsuki M., Nickel W., García-Sáez A. J., Dynamin-related protein 1 (Drp1) promotes structural intermediates of membrane division. J. Biol. Chem. 289, 30645–30656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adachi Y., et al., Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol. Cell 63, 1034–1043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daum G., Lipids of mitochondria. Biochim. Biophys. Acta 822, 1–42 (1985). [DOI] [PubMed] [Google Scholar]
- 19.Ha E. E., Frohman M. A., Regulation of mitochondrial morphology by lipids. Biofactors 40, 419–424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ardail D., et al., Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265, 18797–18802 (1990). [PubMed] [Google Scholar]
- 21.Karbowski M., et al., Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 159, 931–938 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucken-Ardjomande S., Montessuit S., Martinou J. C., Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 15, 929–937 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Montessuit S., et al., Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142, 889–901 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu C. T., et al., Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 15, 1197–1205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu C. T., Bayır H., Kagan V. E., LC3 binds externalized cardiolipin on injured mitochondria to signal mitophagy in neurons: Implications for Parkinson disease. Autophagy 10, 376–378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudek J., Role of cardiolipin in mitochondrial signaling pathways. Front. Cell Dev. Biol. 5, 90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twig G., et al., Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank S., et al., The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1, 515–525 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Stepanyants N., et al., Cardiolipin’s propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol. Biol. Cell 26, 3104–3116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohan A., et al., Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Planas-Iglesias J., et al., Cardiolipin interactions with proteins. Biophys. J. 109, 1282–1294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viennet T., et al., Structural insights from lipid-bilayer nanodiscs link α-Synuclein membrane-binding modes to amyloid fibril formation. Commun. Biol. 1, 44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosol S., Contreras-Martos S., Cedeño C., Tompa P., Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules 18, 10802–10828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruprecht J. J., et al., Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, E426–E434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pebay-Peyroula E., et al., Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Cappello A. R., et al., Functional and structural role of amino acid residues in the even-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J. Mol. Biol. 363, 51–62 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Nishida K., et al., Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc. Natl. Acad. Sci. U.S.A. 100, 2146–2151 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bharambe N., Ramachandran R., Microscale thermophoresis (MST ) as a tool for measuring dynamin superfamily protein (DSP)-lipid interactions. Methods Mol. Biol. 2159, 85–92 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M., Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui M., Tang X., Christian W. V., Yoon Y., Tieu K., Perturbations in mitochondrial dynamics induced by human mutant PINK1 can be rescued by the mitochondrial division inhibitor mdivi-1. J. Biol. Chem. 285, 11740–11752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan F. J., et al., CED-9 and mitochondrial homeostasis in C. elegans muscle. J. Cell Sci. 121, 3373–3382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., Hajnóczky G., Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ. 18, 1561–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narendra D., Tanaka A., Suen D. F., Youle R. J., Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamerkar S. C., Kraus F., Sharpe A. J., Pucadyil T. J., Ryan M. T., Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat. Commun. 9, 5239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoppins S., Lackner L. L., Lee J. E., Mears J. A., In vitro and in vivo assays for mitochondrial fission and fusion. Methods Cell Biol. 155, 491–518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cullis P. R., de Kruijff B., Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta 559, 399–420 (1979). [DOI] [PubMed] [Google Scholar]
- 47.Dufourc E. J., Mayer C., Stohrer J., Althoff G., Kothe G., Dynamics of phosphate head groups in biomembranes. Comprehensive analysis using phosphorus-31 nuclear magnetic resonance lineshape and relaxation time measurements. Biophys. J. 61, 42–57 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoop C. L., Sivanandam V. N., Kodali R., Srnec M. N., van der Wel P. C., Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry 51, 90–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L., Rajendram M., Weibel D. B., Yethiraj A., Cui Q., Ionic hydrogen bonds and lipid packing defects determine the binding orientation and insertion depth of RecA on multicomponent lipid bilayers. J. Phys. Chem. B 120, 8424–8437 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Gautier R., et al., PackMem: A versatile tool to compute and visualize interfacial packing defects in lipid bilayers. Biophys. J. 115, 436–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang H., et al., piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev. Cell 20, 376–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velasco-Olmo A., Ormaetxea Gisasola J., Martinez Galvez J. M., Vera Lillo J., Shnyrova A. V., Combining patch-clamping and fluorescence microscopy for quantitative reconstitution of cellular membrane processes with giant suspended bilayers. Sci. Rep. 9, 7255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez Galvez J. M., Garcia-Hernando M., Benito-Lopez F., Basabe-Desmonts L., Shnyrova A. V., Microfluidic chip with pillar arrays for controlled production and observation of lipid membrane nanotubes. Lab Chip 20, 2748–2755 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Ugarte-Uribe B., Prévost C., Das K. K., Bassereau P., García-Sáez A. J., Drp1 polymerization stabilizes curved tubular membranes similar to those of constricted mitochondria. J. Cell Sci. 132, jcs208603 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Frolov V. A., Escalada A., Akimov S. A., Shnyrova A. V., Geometry of membrane fission. Chem. Phys. Lipids 185, 129–140 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Shen Q., et al., Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol. Biol. Cell 25, 145–159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otera H., Miyata N., Kuge O., Mihara K., Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J. Cell Biol. 212, 531–544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frohman M. A., Role of mitochondrial lipids in guiding fission and fusion. J. Mol. Med. (Berl.) 93, 263–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lemmon M. A., Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Palsdottir H., Hunte C., Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Khalifat N., et al., Interplay of packing and flip-flop in local bilayer deformation. How phosphatidylglycerol could rescue mitochondrial function in a cardiolipin-deficient yeast mutant. Biophys. J. 107, 879–890 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pokorná L., et al., Specific degradation of phosphatidylglycerol is necessary for proper mitochondrial morphology and function. Biochim. Biophys. Acta 1857, 34–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raemy E., et al., Cardiolipin or MTCH2 can serve as tBID receptors during apoptosis. Cell Death Differ. 23, 1165–1174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsumura A., et al., Inactivation of cardiolipin synthase triggers changes in mitochondrial morphology. FEBS Lett. 592, 209–218 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Lee S. J., et al., Structural basis of control of inward rectifier Kir2 channel gating by bulk anionic phospholipids. J. Gen. Physiol. 148, 227–237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whorton M. R., MacKinnon R., Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147, 199–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Savinov S. N., Heuck A. P., Interaction of cholesterol with perfringolysin O: What have we learned from functional analysis? Toxins (Basel) 9, E381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polekhina G., Giddings K. S., Tweten R. K., Parker M. W., Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc. Natl. Acad. Sci. U.S.A. 102, 600–605 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Wel P. C., Reed N. D., Greathouse D. V., Koeppe R. E. II, Orientation and motion of tryptophan interfacial anchors in membrane-spanning peptides. Biochemistry 46, 7514–7524 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strandberg E., et al., Lipid dependence of membrane anchoring properties and snorkeling behavior of aromatic and charged residues in transmembrane peptides. Biochemistry 41, 7190–7198 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Bowen M., Brunger A. T., Conformation of the synaptobrevin transmembrane domain. Proc. Natl. Acad. Sci. U.S.A. 103, 8378–8383 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Planque M. R., et al., Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J. Biol. Chem. 274, 20839–20846 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Mehrotra N., Nichols J., Ramachandran R., Alternate pleckstrin homology domain orientations regulate dynamin-catalyzed membrane fission. Mol. Biol. Cell 25, 879–890 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong L., et al., Cryo-EM of the dynamin polymer assembled on lipid membrane. Nature 560, 258–262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marrink S. J., Mark A. E., Molecular view of hexagonal phase formation in phospholipid membranes. Biophys. J. 87, 3894–3900 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Francy C. A., Alvarez F. J., Zhou L., Ramachandran R., Mears J. A., The mechanoenzymatic core of dynamin-related protein 1 comprises the minimal machinery required for membrane constriction. J. Biol. Chem. 290, 11692–11703 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M., et al., Surface-binding to cardiolipin nanodomains triggers cytochrome c pro-apoptotic peroxidase activity via localized dynamics. Structure 27, 806–815.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koirala S., et al., Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. U.S.A. 110, E1342–E1351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montecinos-Franjola F., Bauer B. L., Mears J. A., Ramachandran R., GFP fluorescence tagging alters dynamin-related protein 1 oligomerization dynamics and creates disassembly-refractory puncta to mediate mitochondrial fission. Sci. Rep. 10, 14777 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji W. K., Hatch A. L., Merrill R. A., Strack S., Higgs H. N., Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore A. S., Holzbaur E. L. F., Mitochondrial-cytoskeletal interactions: Dynamic associations that facilitate network function and remodeling. Curr. Opin. Physiol. 3, 94–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hatch A. L., Gurel P. S., Higgs H. N., Novel roles for actin in mitochondrial fission. J. Cell Sci. 127, 4549–4560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mahecic D., et al., Mitochondrial membrane tension governs fission. Cell Rep. 35, 108947 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.