Abstract
Background: Malaria is a disease caused by hemoparasites of the Plasmodium genus. Non-human primates (NHP) are hosts of Plasmodium sp. around the world. Several studies have demonstrated that Plasmodium sp. emerged from Africa. However, little information is currently available about Plasmodium falciparum in the neotropical NHP and even less in Ecuador. Indeed, the objective of our study was to identify by molecular phylogenetic analyses the Plasmodium species associated with NHP from the Western Amazon region of Ecuador, and to design a molecular taxonomy protocol to use in the NHP disease ecology. Methods: We extracted DNA from faecal samples (n = 26) from nine species of captive (n = 19) and free-ranging (n = 7) NHP, collected from 2011 to 2019 in the Western Amazon region of Ecuador. Results: Using a pan-Plasmodium PCR, we obtained one positive sample from an adult female Leontocebus lagonotus. A maximum likelihood phylogenetic analysis showed that this sequence unequivocally clustered with Plasmodium falciparum. Conclusions: The identification of Plasmodium sp. in NHP of the Ecuadorian Amazon would be essential to identify their role as potential zoonotic reservoirs, and it is also important to identify their origin in wildlife and their transmission in captive NHP.
Keywords: Leontocebus lagonotus, malaria, faecal samples
1. Introduction
Malaria is a disease transmitted by mosquitoes and caused by a hemoparasite of the genus Plasmodium. The protozoan Plasmodium spp. belongs to the Apicomplexa groups; they have the ability to parasite cells thanks to a group of organelles called the apical complex. Plasmodium inhabits red blood cells and hepatocytes [1]. It causes high fever, anemia, headaches, and diarrhea, among others. This mosquito-borne disease is found in humans and in several vertebrates such as birds [2,3], bats [4], antelopes and reptiles [5], and non-human primates (NHP) [6,7,8,9]. The latter are an important source of infections for Plasmodium sp. in humans. In OW (Old World) monkeys, Plasmodium sp. has been detected in chimpanzees (Pan troglodytes) [10], baboons (Papio anubis), Tantalus monkeys (Chlorocebus aethiops), red patas monkeys (Erythrocebus patas) [6], long-tailed and pig-tailed macaques (Macaca fascicularis and Macaca nemestrina) [7], and recently in a lemur species (Propithecus verreauxi) and Macaca radiata [11], to name a few. In the neotropics, there is evidence of natural infection in humans with Plasmodium brasilianum in Venezuela [12] and Plasmodium simium in Brazil [13]. These two species of Plasmodium naturally infect NHP. Plasmodium brasilianum has been identified in at least 49 species of New World (NW) NHP [14,15,16,17,18,19,20] (Table 1). Plasmodium simium has been found in less species, around 4 NW NHP species [18,21,22,23]. There are 25 known species of Plasmodium and some of those (P. vivax, P. malariae, P. falciparum, P. ovale, P. knowlesi) are responsible for human malaria [24,25,26]. Plasmodium falciparum is one of the most malignant species of malaria and it originated from human migration into the NW [10,27,28].
Table 1.
Plasmodium species found in neotropical non-human primates.
| Host | Location | Plasmodium Species | Sampling (Invasive Non-Invasive) |
Detection Methods | References |
|---|---|---|---|---|---|
| Alouatta seniculus. | Brazil | Plasmodium sp. | Invasive | Conventional microscopy (GIEMSA) PCR |
[17] |
|
Alouatta caraya
Alouatta guariba clamitans Alouatta seniculus macconnelli Sapajus apella |
Brazil French Guiana |
Plasmodium vivax | Invasive | Microscopy Enzyme-linked Immunosorbent assay IFA ELISA PCR Real-time PCR |
[22,45,46] |
|
Alouatta sp. Alouatta seniculus Alouatta seniculus straminea Alouatta caraya Alouatta guariba clamitans Alouatta guariba guariba Aotus nigriceps Alouatta g. clamitans Ateles sp. Ateles belzebuth Ateles chamek Ateles paniscus Aotus nigriceps Bracytheles arachnoides Cacajao calvus Cacajao rubicundus Callicebus bruneus Callicebus dubuis Callicebus moloch Callicebus personatus Callicebus torquatus Callithrix geoffroyi Cebus sp. Chiropotes albinasus Chiropotes chiropotes Chiropotus sp. Chiropotes satanas Lagothrix cana cana Lagothrix lagotricha lagotricha Lagothrix lagotricha poeppigii Leontopithecus chrysomelas Leontopithecus rosalia Mico humeralifer Pithecia monachus Pithecia irrorata Pithecia pithecia Saguinus martinsi martinsi Saguinus martinsi ochraceous Saguinus midas niger Saguinus midas Saimiri sp. Saimiri sciureus Saimiri sciureus sciureus Saimiri sciureus boliviensis Saimiri ustus Sapajus apella apella Sapajus apella macrocephalus Sapajus robustus Sapajus xanthosternos |
French Guyana Brazil Venezuela |
Plasmodium brasilianum | Invasive | Blood smears Conventional microscopy (GIEMSA) PCR ELISA |
[14,15,16,17,18,19,20] |
|
Alouatta guariba clamitans Cebus sp. Sapajus robustus Sapajus xanthosternos |
Brazil | Plasmodium simium | Invasive Non-Invasive |
Blood smears PCR PCR from faecal samples Nested-PCR |
[18,21,22,23] |
|
Alouatta caraya
Alouatta guariba Alouatta puruensis Alouatta seniculus macconnelli Ateles chamek Callicebus bruneus Lagothrix cana cana Sapajus apella |
Brazil French Guyana |
Plasmodium falciparum | Invasive | ELISA IFA PCR |
[14,45] |
Several genes are used for molecular identification and phylogenetic studies of Plasmodium species [29,30]. However, the small subunit ribosomal RNA gene is widely used for molecular characterization and phylogenetic studies [31,32,33]. Indeed, it has both highly conserved and very variable domains. This gene was used to study the phylogenetic relationships [34,35,36] and host specificity of Plasmodium sp. [19]. More than 49 species of NW monkeys are known to be infected with Plasmodium sp. [37,38]. In Ecuador, only a few studies on Plasmodium sp. were realized [39]. Avian Plasmodia were studied in the Galapagos [40,41]. In humans, several studies with molecular markers have identified population origins of P. falciparum in the Northwest of Ecuador [42,43], but only one study yielded sequence information [44]. The aim of this study was to monitor Plasmodium sp. in the Amazon region of Ecuador, to identify potential zoonotic reservoirs, and to identify the origins of malaria parasites in wildlife and potential human–monkey transmission with captive NHP.
2. Results
A total of 26 faecal DNA samples were analysed from captive and free-ranging NHP, in the Western Amazon region of Ecuador. After DNA extraction and using a pan-Plasmodium PCR, one positive sample was obtained from an adult female Leontocebus lagonotus (representing 3.85%). This animal came from a rescue centre in Pastaza. As with the other rescue centres in Ecuador, most NHP from this rescue centre have been donated by families or confiscated by the police during roadside checks, with this individual’s information being uncertain.
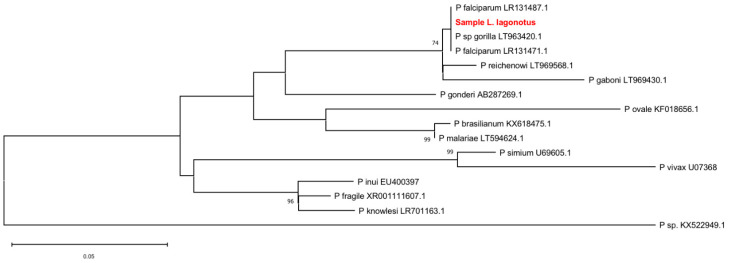
We sequenced the amplicon and then aligned it with sequences from other Plasmodium sp. species available in the GenBank. The maximum-likelihood phylogeny (ML) tree (Figure 1) yielded a topology with the different Plasmodium species clades and an internal (derivated) monophyletic clade comprised by P. falciparum including the sequence obtained from the Leontocebus lagonotus plus P. reichenowi sequences and P. gaboni. The ML recovered a clear reciprocal monophyly between falciparum and reichenowi as sister groups both with a strong support group by bootstrapping values of 74%. The arrangements of other sister clades were recovered: P. simium + P. vivax; P. inui + (P. fragile + P. knowlesi); P. ovale + (P. brasilianum + P. malariae). All sister groups were strongly supported by bootstrapping values of above 96%. In the analysis performed, the sequence of the amplified Plasmodium from the NHP L. lagonotus’ faeces is located within the P. falciparum clade, supporting its molecular identification with this species in the phylogenetic species sense.
Figure 1.
Evolutionary relationships of the Leontocebus lagonotus isolate described in this study (bolded and highlighted in red) compared to representative members of the Plasmodium genus. The tree is based on the maximum-likelihood phylogeny of the partial small subunit ribosomal RNA gene. The phylogenetic analysis was performed using the Tamura 3-parameter substitution model implemented in MEGA X. Bootstrap percentages > 70% (1000 resamplings) are indicated at the nodes. GenBank accession numbers are indicated for each strain. The scale bar indicates nucleotide substitutions per site.
3. Discussion
DNA from only 26 of the 109 faecal samples collected from 109 NHP was analysed because the quality of the DNA from the other 83 samples was insufficient for the detection of Plasmodium, possibly due to the presence of some inhibitors that prevent the amplification of DNA. We detected Plasmodium falciparum in a faecal sample from Leontocebus lagonotus (3.85%), contributing to the parasite ecology of NW NHP. The percentage of detection is not high, but it is in line with previous published results in scientific literature in Latin America (e.g., [23]).
Malaria parasites display host specificity [47], such as avian malaria in birds [48,49], and malaria in apes [50]. However, it is estimated that the diversification of Plasmodium was rapid 16–24 million years ago (MYA) or 26–38 MYA, which differs from the divergence times of the hosts (reptiles to mammals) 75–310 MYA. This difference in the divergence times between parasites and hosts suggests that there has been no co-divergence with the hosts [51]. In primates, two groups of Plasmodium have been identified. The first includes the species P. malariae/P. ovale/P. hylobatid, which infect Old World primates. The second group includes the species P. falciparum/P. reichenowi, which infect humans and NHP. In NW monkeys, P. brasilianum and Plasmodium simium are known to be the cause of primate malaria. Plasmodium falciparum is known to be a cause of human malaria, a result of a recent cross-species transmission of a parasite between gorillas and humans [52]. However, P. falciparum is also known to naturally infect at least eight species of NW NHP [14,45]. Our sample is located in the clade of P. falciparum, and close to P. recheinowi. This relationship was already observed in previous studies [51,53,54]. It has been reported that Plasmodium falciparum and P. reichenowi are part of a monophyletic clade [55].
Non-human primates in captivity from this study are in close contact with care takers and tourists, which may increase the possibility of parasite transmission [56]. However, the origin of these captive animals is unknown. This assumption makes us wonder whether the infection occurred in a sylvatic environment or whether it occurred during the captive period of this NHP. If it occurred during the sylvatic environment, the transmission could be the result of a natural infection via mosquitoes. Leontocebus lagonotus is known to be widely distributed in Western Amazonia in Ecuador, i.e., in Pastaza and Morona Santiago provinces [57], which according to the Center for Disease Control and Prevention (CDC) [58], are areas of malaria (Plasmodium vivax and Plasmodium falciparum). In 2019, Pastaza and Morona Santiago where the two provinces with the highest number of malaria (Plasmodium falciparum and Plasmodium vivax) cases in humans [59]. If it occurred during the captive period, further studies should be carried out to determine if the vector responsible for the human–NHP transmission is present in this region because there are no reports of the vector in the area.
4. Materials and Methods
4.1. Study Sites and Sampling
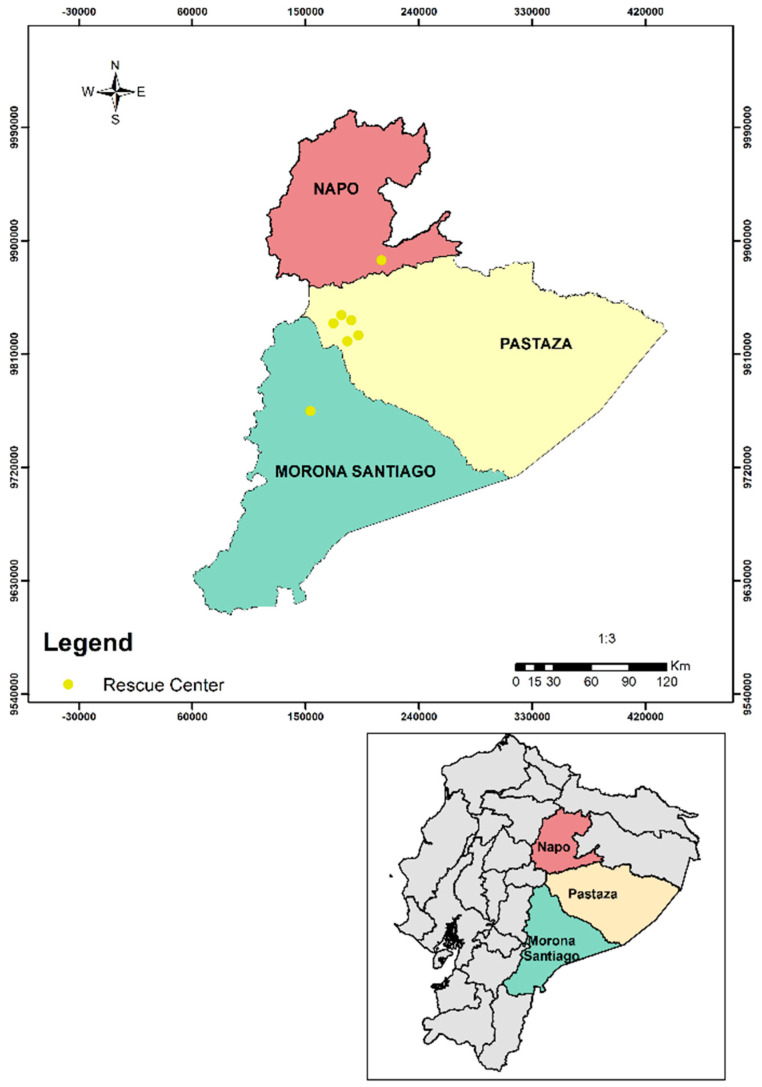
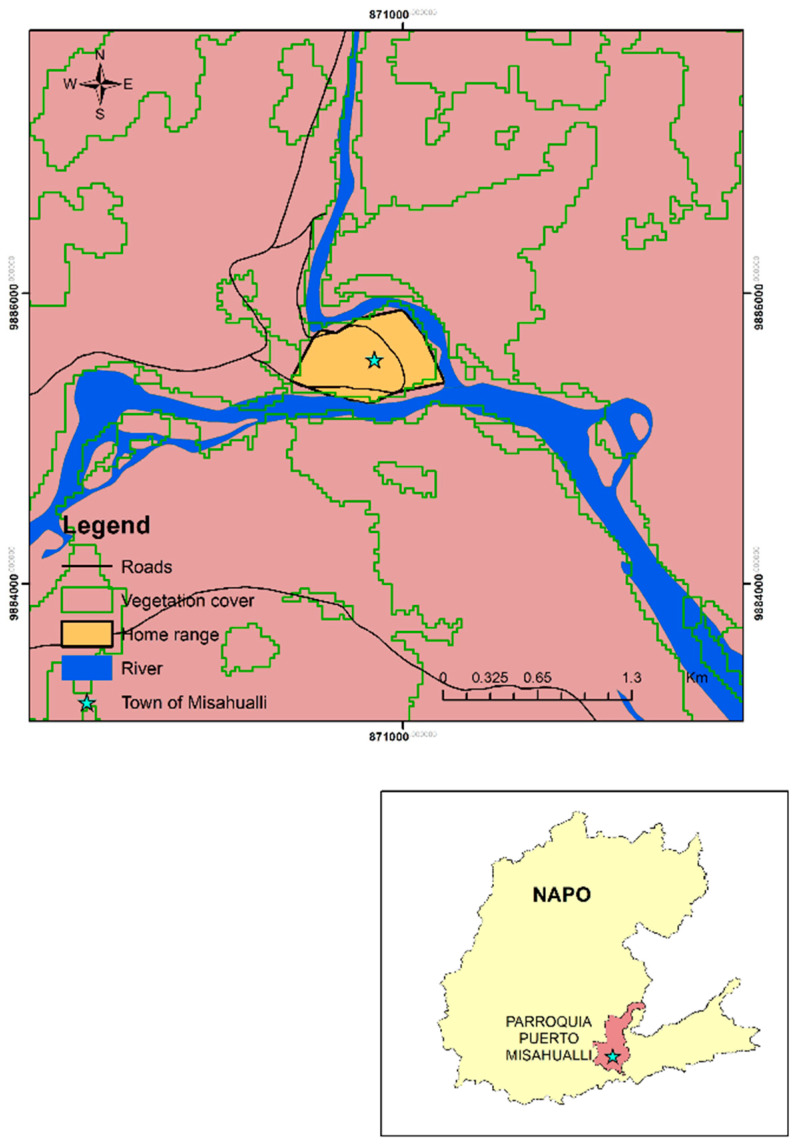
This study was performed in Puyo (Pastaza), Tena (Napo), and Macas (Morona Santiago), three cities in the Western Amazon region (Figure 2). We examined Plasmodium spp. in two populations, one captive and one free-ranging. The captive population was studied in wildlife refuges. Most NHP from wildlife refuges had been donated by families or confiscated by the police during roadside checks. The free-ranging population lived in the small town of Misahualli (Tena, Napo) (1°2′7.0″ S, 77°39′59.4″ W) (Figure 3).
Figure 2.
Rescue centres surveyed in the Amazon region of Ecuador.
Figure 3.
Location of free-ranging population of Cebus yuracus surveyed in the Amazon region of Ecuador.
Faecal samples are an important source of information about pathogens (viruses, prokaryotes, or eukaryotes) that infect primate species. The analysis of molecular faecal samples offers a non-invasive option that becomes a valid alternative to the traditional sampling methods (blood and tissue samples) of primates [60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77]. Studies in conservation genetics have used faecal samples for DNA extraction prioritizing species that are in some category of protection or threat [78,79]. In this study, we collected 109 faecal samples from 109 NHP. However, the DNA quality was not good enough on 83 samples to detect Plasmodium. Indeed, we analysed faecal samples (n = 26) from nine species of captive (n = 19) and free-ranging (n = 7) NHPs (Table 2), between 2011 and 2019, in the Western Amazon region of Ecuador from Proyecto Primates Ecuador. Individuals were followed daily from 8 a.m. to 6 p.m. and we collected the samples immediately after defecation to avoid getting confused with samples from other species. All animals were individually identified to facilitate the results analysis. For the molecular analyses, samples were stored in 50 mL Falcon tubes in 99% alcohol at −20 °C to prevent the degradation of DNA. In addition, 600 µL of faeces suspension (1:3; 1 part of faecal sample and 3 parts of ethanol 96–100%) was centrifuged for 2 min at 239 g and the pellet was washed with 1 mL of PBS Buffer (Oxoid, Hampshire, England). This solution (pellet +PBS) was centrifugated for 5 min and the supernatant was discarded. This washing step was repeated three times. Next, the pellet was re-suspended in 600 µL of 2% PVPP (polyvinylpolypyrolidone—Sigma), and frozen overnight at −20 °C to facilitate the capture of phenols in the sample. DNA extraction was performed twice on different days using the QIAamp Stool FAST Mini Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer’s instructions. To prevent cross-contamination, sample preparation, DNA extraction, and the PCR were performed in completely different and separated rooms. Furthermore, the master mix was assembled in a DNA-free room. Ultraviolet light sterilization was performed before and after each procedure. The PCR was carried out using primers targeting the small subunit of 18S ribosomal RNA. The primers used were described by dos Santos et al. [80] (Table 3).
Table 2.
Non-human primate species sampled in the Ecuadorian Amazon.
| Habitat Settings | Non-Human Primate Species | n | Sex | Age | |||
|---|---|---|---|---|---|---|---|
| Male | Female | Adult | Subadult | Juvenile | |||
| Captive | Alouatta seniculus | 4 | 0 | 4 | 1 | 2 | 1 |
| Ateles belzebuth | 1 | 1 | 0 | 1 | 0 | 0 | |
| Callicebus lucifer | 1 | 1 | 0 | 1 | 0 | 0 | |
| Cebuella pygmaea | 1 | 1 | 0 | 1 | 0 | 0 | |
| Cebus yuracus | 2 | 1 | 1 | 1 | 1 | 0 | |
| Lagothrix lagotricha | 2 | 0 | 2 | 1 | 0 | 1 | |
| Leontocebus fuscicollis | 3 | 2 | 1 | 2 | 1 | 0 | |
| Saimiri sciureus | 3 | 2 | 1 | 1 | 1 | 1 | |
| Sapajus apella | 2 | 1 | 1 | 1 | 0 | 1 | |
| Free ranging | Cebus yuracus | 7 | 5 | 2 | 4 | 1 | 2 |
Table 3.
Sequences of the primers.
| Reaction | Primer | Oligonucleotide Sequence | |
|---|---|---|---|
| First reaction | rPLU1 | 5′-TCAAAGATTAAGCCATGCAAGTGA 3′ | forward |
| rPLU6R | 5′-CGTTTTAACTGCAACAATTTTAA-3′ | reverse | |
| Second Reaction | rPLU3 | 5′-TTTTTATAAGGATAACTACGGAAAAGCTGT-3′ | forward |
| rPLU4 | 5′-TACCCGTCATAGCCATGTTAGGCCAATACC-3′ | reverse |
The molecular identification was performed in two reactions (nested PCR) as described by [80], with adaptations. The amplification in the first reaction consisted of 95 °C for 5 min, 95 °C for 30 seg; 50 cycles of 55 °C for 30 seg, 72 °C for 1 min; and a final extension step at 72 °C for 5 min, with a product of 600 bp. In addition, the amplification in the second reaction consisted of an initial denaturation at 95 °C for 5 min, 95 °C for 30 seg; 50 cycles of 58 °C for 30 seg and 72 °C for 1 min; and a final extension at 72 °C for 5 min. The products of the second reaction (240 bp) were observed using the electrophoresis of an agarose gel under UV light. Amplicons were cut, extracted using NucleoSpin gel and the PCR clean-up kit (Macherey-Nagel, Düren, Germany) and sequenced (Sanger sequencing) by Eurofins (Hamburg, Germany). Every PCR reaction contained a negative and a positive control. Sterile filtered pipette tips were used in all stages of the methodology to prevent contamination. We changed the pipette tip after each sample to avoid false-positive reactions/cross-contamination and all laboratory consumables were not reused.
A positive control for Plasmodium spp. was obtained using DNA extracted from the spleen of a Belgian blackbird (Turdus merula) collected in 2018 [81,82].
4.2. Molecular Identification
The sequence was uploaded to GenBank under the accession number submission MZ156589. Sequence reconstruction was performed using Assembler by MacVector software 17.5.5. The first sequence identity was confirmed by BLAST in NCBI resources. A total of 15 sequences from 14 species of Plasmodium were retrieved from GenBank and included as sister groups in order to obtain a wide geographic diversity and taxonomic representation [83] (Table 4). DNA sequences were aligned using MacVector 17.5.5 [84] by the ClustalW algorithm with high gap creation and extension penalties by 30.0 and 10.0, respectively, searching for a strong positional homology. The evolutionary history was inferred by using the maximum likelihood method and the Tamura 3-parameter model. The tree with the highest log likelihood (−932.64) is shown. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 0.3352)). The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 16 nucleotide sequences. There was a total of 241 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.
Table 4.
GenBank accession numbers of Plasmodium species sequences.
| Plasmodium Species | ID Genbank | Host | Country |
|---|---|---|---|
| Plasmodium sp. | LT963420.1 | Gorilla sp. | Unknown |
| P. falciparum | LR131487.1 | Unknown (Genome assembly) | Unknown (Genome assembly) |
| P. falciparum | MZ156589 | Leontocebus lagonotus | Ecuador |
| P. falciparum | LR131471.1 | Unknown (Genome assembly) | Unknown (Genome assembly) |
| P. reichenowi | LT969568.1 | Unknown (Genome assembly) | Unknown (Genome assembly) |
| P. gaboni | LT969430.1 | Unknown (Genome assembly) | Unknown (Genome assembly) |
| P. gonderi | AB287269.1 | Cercocebus atys | Central Africa |
| P. ovale | KF018656.1 | Homo sapiens | China |
| P. brasillianum | KX618475.1 | Sapajus flavius | Brazil |
| P. malariae | LT594624.1 | Unknown (Genome assembly) | Unknown (Genome assembly) |
| P. fragile | XR001111607.1 | Unknown | Unknown |
| P. inui | EU400397 | Macaca fascicularis | Thailand |
| P. simium | U69605.1 | Saimiris sciureus | Colombia |
| P. vivax | U07368 | Unknown | CDC Strain |
| Plamodium sp. | KX522949.1 | Anopheles nuneztovari | Brazil |
| P. knowlesi | LR701163.1 | Unknown (Genome assembly) | Unknown (Genome assembly) |
The robustness for all the analyses was estimated using bootstrapping with 1000 pseudoreplicates and shown in percentage.
5. Conclusions
The results of this study provide evidence of Plasmodium falciparum in a species of NW NHP, and the potential risk of zoonotic malaria transmission. The present study, by identifying the presence of the parasite in NHP, suggests the need to promote continuous and systematic diagnoses and monitoring of malaria in these animals. In addition, wildlife trafficking and management should be incorporated into public health policies for the prevention of malaria as an emerging zoonotic disease.
Acknowledgments
We would like to thank Ministerio del Ambiente for its support.
Author Contributions
Conceptualization, C.S. and G.A.C.B.; methodology, G.A.C.B., M.-M.G., J.-C.N., E.M.; software, M.-M.G., J.-C.N.; validation, C.S., M.-M.G., S.M.-S., G.A.C.B., W.B.-O.; formal analysis, G.A.C.B., M.-M.G., J.-C.N., S.M.-S.; investigation, G.A.C.B., C.S.; resources, G.A.C.B., S.M.-S.; data curation, M.-M.G., S.M.-S.; writing—original draft preparation, G.A.C.B.; writing—review and editing, C.S., M.-M.G., S.M.-S., J.-C.N.; visualization, G.A.C.B., S.M.-S.; supervision, C.S.; project administration, G.A.C.B.; funding acquisition, W.B-O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded the Academy of Research and Higher Education (ARES) through an institutional support program entitled “Hemoparasites and arboviruses in non-human primates of the Ecuadorian Amazon using non-invasive techniques”, which involves the Universidad Central del Ecuador and the University of Liège in Belgium. We also had a grant from UISEK number: DII-UISEK-P011617-2 (JCN).
Institutional Review Board Statement
This study was approved by the Ministerio del Ambiente Ecuador under the permit number MAE-DNB-CM-2015-0028-M-002.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References and Note
- 1.Goater T.M., Goater C.P., Esch G.W. Parasitism: The Diversity and Ecology of Animal Parasites. Cambridge University Press; Cambridge, UK: 2014. p. 524. [Google Scholar]
- 2.Böhme U., Otto T.D., Cotton J.A., Steinbiss S., Sanders M., Oyola S.O., Nicot A., Gandon S., Patra K.P., Herd C., et al. Complete avian malaria parasite genomes reveal features associated with lineage-specific evolution in birds and mammals. Genome Res. 2018;28:547–560. doi: 10.1101/gr.218123.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensch S., Stjernman M., Hasselquist D., Ostman O., Hansson B., Westerdahl H., Pinheiro R.T. Host specificity in avian blood parasites: A study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc. R. Soc. B. 2000;267:1583–1589. doi: 10.1098/rspb.2000.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boundenga L., Ngoubangoye B., Mombo I.M., Tsoubmou T.A., Renaud F., Rougeron V., Prugnolle F. Extensive diversity of malaria parasites circulating in Central African bats and monkeys. Ecol. Evol. 2018;8:10578–10586. doi: 10.1002/ece3.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boundenga L., Makanga B., Ollomo B., Gilabert A., Rougeron V., Mve-Ondo B., Arnathau C., Durand P., Moukodoum N.D., Okouga A.-P., et al. Haemosporidian parasites of antelopes and other vertebrates from Gabon, Central Africa. PLoS ONE. 2016;11:e0148958. doi: 10.1371/journal.pone.0148958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mbaya A.W., Aliyu M.M., Nwosu C.O., Ibrahim U.I. Captive wild animals as potential reservoirs of haemo and ectoparasitic infections of man and domestic animals in the arid- region of Northeastern Nigeria. Vet. Arhiv. 2008;78:429–440. [Google Scholar]
- 7.Lee K.S., Divis P.C.S., Zakaria S.K., Matusop A., Julin R.A., Conway D.J., Cox-Singh J., Singh B. Plasmodium knowlesi: Reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springer A., Fichtel C., Calvignac-Spencer S., Leendertz F.H., Kappeler P.M. Hemoparasites in a wild primate: Infection patterns suggest interaction of Plasmodium and Babesia in a lemur species. Int. J. Parasitol. Parasites Wildl. 2015;4:385–395. doi: 10.1016/j.ijppaw.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueiredo M.A.P., Santi S.M.D., Manrique W.G., André M.R., Machado R.Z. Serological and molecular techniques applied for identification of Plasmodium spp. in blood samples from nonhuman primates. Rev. Bras. Parasitol. Vet. 2018;27:363–376. doi: 10.1590/s1984-296120180043. [DOI] [PubMed] [Google Scholar]
- 10.Escalante A.A., Freeland D.E., Collins W.E., Lal A.A. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixit J., Zachariah A., Sajesh P.K., Chandramohan B., Shanmuganatham V., Karanth K.P. Reinvestigating the status of malaria parasite (Plasmodium sp.) in Indian non-human primates. PLoS Negl. Trop. Dis. 2018;12:e0006801. doi: 10.1371/journal.pntd.0006801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalremruata A., Magris M., Vivas-Martínez S., Koehler M., Esen M., Kempaiah P., Jeyaraj S., Perkins D.J., Mordmüller B., Metzger W.G. Natural infection of Plasmodium brasilianum in humans: Man and monkey share quartan malaria parasites in the Venezuelan Amazon. EBioMedicine. 2015;2:1186–1192. doi: 10.1016/j.ebiom.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasil P., Zalis M.G., Pina-Costa A.D., Siqueira A.M., Junior C.B., Silva S., Areas A.L.L., Pelajo-Machado M., Alvarenga D.A.M.D., Santelli A.C.F.D.S., et al. Plasmodium simium causing human malaria: A zoonosis with outbreak potential in the Rio de Janeiro Brazilian Atlantic forest. bioRxiv. 2017 doi: 10.1101/122127. [DOI] [Google Scholar]
- 14.Araújo M.S., Messias M.R., Figueiró M.R., Gil L.H.S., Probst C.M., Vidal N.M., Katsuragawa T.H., Krieger M.A., Silva L.H.P.D., Ozaki L.S. Natural Plasmodium infection in monkeys in the state of Rondônia (Brazilian Western Amazon) Malar. J. 2013;12:180. doi: 10.1186/1475-2875-12-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thoisy B.D., Michel J.-C., Vogel I., Vié J.-C. A survey of hemoparasite infections in free-ranging mammals and reptiles in french Guiana. J. Parasitol. 2000;86:1035–1040. doi: 10.1645/0022-3395(2000)086[1035:ASOHII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 16.Deane L. Simian malaria in Brazil. Mem. Inst. Oswaldo Cruz. 1992;87:1–20. doi: 10.1590/S0074-02761992000700001. [DOI] [PubMed] [Google Scholar]
- 17.Fandeur T., Volney B., Peneau C., Thoisy B. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology. 2000;120 doi: 10.1017/S0031182099005168. [DOI] [PubMed] [Google Scholar]
- 18.Alvarenga D.A.M.D., Pina-Costa A.D., Sousa T.N.D., Pissinatti A., Zalis M.G., Suaréz-Mutis M.C., Lourenço-de-Oliveira R., Brasil P., Daniel-Ribeiro C.T., Brito C.F.A.D. Simian malaria in the Brazilian Atlantic forest: First description of natural infection of capuchin monkeys (Cebinae subfamily) by Plasmodium simium. Malar. J. 2015;14:81. doi: 10.1186/s12936-015-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarenga D.A.M., Pina-Costa A., Bianco C., Moreira S.B., Brasil P., Pissinatti A., Daniel-Ribeiro C.T., Brito C.F.A. New potential Plasmodium brasilianum hosts: Tamarin and marmoset monkeys (family Callitrichidae) Malar. J. 2017;16:71. doi: 10.1186/s12936-017-1724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arruda M.D., Nardin E.H., Nussenzweig R.S., Cochrane A.H. Sero-epidemiological studies of malaria in Indian tribes and monkeys of the Amazon basin of Brazil. Am. J. Trop. Med. Hyg. 1989;41:379–385. doi: 10.4269/ajtmh.1989.41.379. [DOI] [PubMed] [Google Scholar]
- 21.Abreu F.V.S.D., Santos E.D., Mello A.R.L., Gomes L.R., Alvarenga D.A.M.D., Gomes M.Q., Vargas W.P., Bianco-Júnior C., Pina-Costa A.D., Teixeira D.S., et al. Howler monkeys are the reservoir of malarial parasites causing zoonotic infections in the Atlantic forest of Rio de Janeiro. PLoS Negl. Trop. Dis. 2019;13:e0007906. doi: 10.1371/journal.pntd.0007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa D.C., Cunha V.P.D., Assis G.M.P.D., Souza Junior J.C.D., Hirano Z.M.B., Arruda M.E.D., Kano F.S., Carvalho L.H., Brito C.F.A.D. Plasmodium simium/Plasmodium vivax infections in southern brown howler monkeys from the Atlantic Forest. Mem. Inst. Oswaldo Cruz. 2014;109:641–653. doi: 10.1590/0074-0276130578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assis G.M.P.D., Alvarenga D.A.M.D., Costa D.C., Souza Junior J.C.D., Hirano Z.M.B., Kano F.S., Sousa T.N.D., Brito C.F.A.D. Detection of Plasmodium in faeces of the New World primate Alouatta clamitans. Mem. Inst. Oswaldo Cruz. 2016;111:570–576. doi: 10.1590/0074-02760160222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.William T., Rahman H.A., Jelip J., Ibrahim M.Y., Menon J., Grigg M.J. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax Malaria in Sabah, Malaysia. PLoS Negl. Trop. Dis. 2013;7:e2026. doi: 10.1371/journal.pntd.0002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lalremruata A., Jeyaraj S., Engleitner T., Joanny F., Lang A., Belard S., Mombo-Ngoma G., Ramharter M., Kremsner P.G., Mordmuller B., et al. Species and genotype diversity of Plasmodium in malaria patients from Gabon analysed by next generation sequencing. Malar. J. 2017;16:398. doi: 10.1186/s12936-017-2044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miguel-Oteo M., Jiram A.I., Ta-Tang T.H., Lanza M., Hisam S., Rubio J.M. Nested multiplex PCR for identification and detection of human Plasmodium species including Plasmodium knowlesi. Asian Pac. J. Trop. Dis. 2017;10:299–304. doi: 10.1016/j.apjtm.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Prugnolle F., Durand P., Ollomo B., Duval L., Ariey F., Arnathau C., Gonzalez J.-P., Leroy E., Renaud F. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathog. 2011;7:e1001283. doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues P.T., Valdivia H.O., Oliveira T.C.D., Alves J.M.P., Duarte A.M.R.C., Cerutti-Junior C., Buery J.C., Brito C.F.A., Souza J.C.D., Hirano Z.M.B., et al. Human migration and the spread of malaria parasites to the New World. Sci. Rep. 2018;8:1993. doi: 10.1038/s41598-018-19554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buitrago S.P., Garzón-Ospina D., Patarroyo M.A. Size polymorphism and low sequence diversity in the locus encoding the Plasmodium vivax rhoptry neck protein 4 (PvRON4) in Colombian isolates. Malar. J. 2016;15:501. doi: 10.1186/s12936-016-1563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pacheco M.A., Cepeda A.S., Bernotienė R., Lotta I.A., Matta N.E., Valkiūnas G., Escalante A.A. Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int. J. Parasitol. 2018;48:657–670. doi: 10.1016/j.ijpara.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato H., Leo N., Katakai Y., Takano J.-I., Akari H., Nakamura S.-i., Une Y. Prevalence and molecular phylogenetic characterization of Trypanosoma (megatrypanum) minasense in the peripheral blood of small neotropical primates after a quarantine period. J. Parasitol. 2008;94:112–1138. doi: 10.1645/GE-1513.1. [DOI] [PubMed] [Google Scholar]
- 32.Khan S.M., Debnath C., Pramanik A.K., Xiao L., Nozaki T., Ganguly S. Molecular characterization and assessment of zoonotic transmission of Cryptosporidium from dairy cattle in West Bengal, India. Vet. Parasitol. 2010;171:41–47. doi: 10.1016/j.vetpar.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Bertelsen M.F., Meyland-Smith F., Willesen J.L., Jefferies R., Morgan E.R., Monrad J. Diversity and prevalence of metastrongyloid nematodes infecting the red panda (Ailurus fulgens) in European zoos. Vet. Parasitol. 2010;172:299–304. doi: 10.1016/j.vetpar.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 34.Duval L., Fourment M., Nerrienet E., Rousset D., Sadeuh S.A., Goodman S.M., Andriaholinirina N.V., Randrianarivelojosia M., Paul R.E., Robert V., et al. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc. Natl. Acad. Sci. USA. 2010;107:10561. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leclerc M.C., Hugot J.P., Durand P., Renaud F. Evolutionary relationships between 15 Plasmodium species from New and Old World primates (including humans): A 18S rDNA cladistic analysis. Parasitology. 2004;129:677–684. doi: 10.1017/S0031182004006146. [DOI] [PubMed] [Google Scholar]
- 36.Chua T.H., Manin B.O., Daim S., Vythilingam I., Drakeley C. Phylogenetic analysis of simian Plasmodium spp. infecting Anopheles balabacensis Baisas in Sabah, Malaysia. PLoS Negl. Trop. Dis. 2017;11:e0005991. doi: 10.1371/journal.pntd.0005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuart M.D., Pendergast V., Rumfelt S., Pierberg S., Greenspan L.L., Glander K.E., Clarke M.R. Parasites of wild howlers (Alouatta spp.) Int. J. Primatol. 1998;19:493–512. doi: 10.1023/A:1020312506375. [DOI] [Google Scholar]
- 38.Figueiredo M.A.P., Santi S.M.D., Manrique W.G., André M.R., Machado R.Z. Identification of Plasmodium spp. in Neotropical primates of Maranhense Amazon in Northeast Brazil. PLoS ONE. 2017;12:e0182905. doi: 10.1371/journal.pone.0182905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pérez F., Martin-Solano S., Sáenz F., Minda-Aluisa E., Navarro J.C., Carillo-Bilbao G.-A. Detection of Plasmodium sp. from Fecal Samples of Non-Human Primates from the Cities of Tena, Puyo and Macas Using Nested-PCR; Proceedings of the RED Santo Domingo Investiga; Santo Domingo, Dominican Republic. 25–29 September 2018. [Google Scholar]
- 40.Levin I.I., Colborn R.E., Kim D., Perlut N.G., Renfrew R.B., Parker P.G. Local parasite lineage sharing in temperate grassland birds provides clues about potential origins of Galapagos avian Plasmodium. Ecol. Evol. 2016;6:716–726. doi: 10.1002/ece3.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perlut N.G., Parker P.G., Renfrew R.B., Jaramillo M. Haemosporidian parasite community in migrating bobolinks on the Galapagos Islands. Int. J. Parasitol. Parasites Wildl. 2018;7:204–206. doi: 10.1016/j.ijppaw.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vera-Arias C.A., Castro L.E., Gómez-Obando J., Sáenz F.E. Diverse origin of Plasmodium falciparum in northwest Ecuador. Malar. J. 2019;18:251. doi: 10.1186/s12936-019-2891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sáenz F.E., Morton L.C., Okoth S.A., Valenzuela G., Vera-Arias C.A., Vélez-Álvarez E., Lucchi N.W., Castro L.E., Udhayakumar V. Clonal population expansion in an outbreak of Plasmodium falciparum on the northwest coast of Ecuador. Malar. J. 2015;14:497. doi: 10.1186/s12936-015-1019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker J., McCarthy J., Gatton M., Kyle D.E., Belizario V., Luchavez J., Bell D., Cheng Q. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J. Infect. Dis. 2005;192:870–877. doi: 10.1086/432010. [DOI] [PubMed] [Google Scholar]
- 45.Duarte A.M.R.D.C., Porto M.A.L., Curado I., Malafronte R.S., Hoffmann E.H.E., Oliveira S.G., Silva A.M.J., Kloetzel J.K., Gomes A.D.C. Widespread occurrence of antibodies against circumsporozoite protein and against blood forms of Plasmodium vivax, P. falciparum and P. malariae in Brazilian wild monkeys. J. Med. Primatol. 2006;35:87–96. doi: 10.1111/j.1600-0684.2006.00148.x. [DOI] [PubMed] [Google Scholar]
- 46.Volney B., Pouliquen J.F., Thoisy B., Fandeur T. A sero-epidemiological study of malaria in human and monkey populations in French Guiana. Acta Trop. 2002;82 doi: 10.1016/S0001-706X(02)00036-0. [DOI] [PubMed] [Google Scholar]
- 47.Siao M.C., Borner J., Perkins S.L., Deitsch K.W., Kirkman L.A. Evolution of host specificity by Malaria parasites through altered mechanisms controlling genome maintenance. mBio. 2020;11:e03272-19. doi: 10.1128/mBio.03272-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loiseau C., Harrigan R.J., Robert A., Bowie R.C., Thomassen H.A., Smith T.B., Sehgal R.N. Host and habitat specialization of avian malaria in Africa. Mol. Ecol. 2012;21:431–441. doi: 10.1111/j.1365-294X.2011.05341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iezhova T.A., Valkiünas G., Bairlein F. Vertebrate host specificity of two avian malaria parasites of the subgenus Novyella: Plasmodium nucleophilum and Plasmodium vaughani. J. Parasitol. 2005;91:472–474. doi: 10.1645/GE-3377RN. [DOI] [PubMed] [Google Scholar]
- 50.Makanga B., Yangari P., Rahola N., Rougeron V., Elguero E., Boundenga L., Moukodoum N.D., Okouga A.P., Arnathau C., Durand P., et al. Ape malaria transmission and potential for ape-to-human transfers in Africa. Proc. Natl. Acad. Sci. USA. 2016;113:5329–5334. doi: 10.1073/pnas.1603008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayakawa T., Culleton R., Otani H., Horii T., Tanabe K. Big bang in the evolution of extant malaria parasites. Mol. Biol. Evol. 2008;25:2233–2239. doi: 10.1093/molbev/msn171. [DOI] [PubMed] [Google Scholar]
- 52.Loy D.E., Liu W., Li Y., Learn G.H., Plenderleith L.J., Sundararaman S.A., Sharp P.M., Hahn B.H. Out of Africa: Origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int. J. Parasitol. 2017;47:87–97. doi: 10.1016/j.ijpara.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rich S.M., Leendertz F.H., Xu G., LeBreton M., Djoko C.F., Aminake M.N., Takang E.E., Diffo J.L.D., Pike B.L., Rosenthal B.M., et al. The origin of malignant malaria. Proc. Natl. Acad. Sci. USA. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otto T.D., Rayner J.C., Böhme U., Pain A., Spottiswoode N., Sanders M., Quail M., Ollomo B., Renaud F., Thomas A.W., et al. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nat. Commun. 2014;5:4754. doi: 10.1038/ncomms5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W., Li Y., Learn G.H., Rudicell R.S., Robertson J.D., Keele B.F., Ndjango J.-B.N., Sanz C.M., Morgan D.B., Locatelli S., et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chapman C.A., Gillespie T.R., Goldberg T.L. Primates and the ecology of their infectious diseases: How will anthropogenic change affect host-parasite interactions? Evol. Anthropol. 2005;14:134–144. doi: 10.1002/evan.20068. [DOI] [Google Scholar]
- 57.Álvarez-Solas S., Torre S.D.L., Tirira D. Tamarín Ensillado de Dorso Rojo Leontocebus lagonotus (Jiménez de la Espada, 1870) In: Tirira D., Torre S.D.L., Ríos G.Z., editors. Estado de Conservación de los Primates del Ecuador. 1st ed. Grupo de Estudio de Primates del Ecuador/Asociación Ecuatoriana de Mastozoología; Quito, Ecuador: 2018. [Google Scholar]
- 58.Gershman M.D., Jentes E.S., Stoney R.J., Tan K.R., Arguin P.M. Yellow Fever Vaccine & Malaria Prophylaxis Information, by Country. In: Brunette G.W., Nemhauser J.B., editors. Yellow Book. Oxford University Press; New York, NY, USA: 2020. p. 720. [Google Scholar]
- 59.Gaceta Epidemiológica—SIVE. Gaceta vectores SE Alert n°52. 2019 (report).
- 60.Bairami A., Rezaei S., Rezaeian M. Synchronous identification of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. in stool samples using a multiplex PCR assay. Iran. J. Parasitol. 2018;13:24–30. [PMC free article] [PubMed] [Google Scholar]
- 61.Bezjian M., Gillespie T.R., Chapman C.A., Greiner E.C. Coprologic evidence of gastrointestinal helminths of Forest baboons, Papio anubis, in Kibale National Park, Uganda. J. Wildl. Dis. 2008;44:878–887. doi: 10.7589/0090-3558-44.4.878. [DOI] [PubMed] [Google Scholar]
- 62.Carozzi F.M., Sani C. Fecal collection and stabilization methods for improved fecal DNA test for colorectal cancer in a screening setting. J. Cancer Res. 2013;2013:818675. doi: 10.1155/2013/818675. [DOI] [Google Scholar]
- 63.Cerda-Molina A.L., Hernández-López L., Páez-Ponce D.L., Rojas-Maya S., Mondragón-Ceballos R. Seasonal variations of fecal progesterone and 17β-estradiol in captive female black-handed spider monkeys (Ateles geoffroyi) Theriogenology. 2006;66:1985–1993. doi: 10.1016/j.theriogenology.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 64.Chinchilla M., Guerrero O.M., Gutierrez-Espeleta G.A., Sánchez R., Valerio Campos I. Parásitos en monos carablanca Cebus capucinus (Primates: Cebidae) de Costa Rica. Parasitol. Latinoam. 2007;62:170–175. doi: 10.4067/S0717-77122007000200011. [DOI] [Google Scholar]
- 65.Conga D.F., Bowler M., Tantalean M., Montes D., Serra-Freire N.M., Mayor P. Intestinal helminths in wild Peruvian red uakari monkeys (Cacajao calvus ucayalii) in the northeastern Peruvian Amazon. J. Med. Primatol. 2014;43:130–133. doi: 10.1111/jmp.12092. [DOI] [PubMed] [Google Scholar]
- 66.Jirků M., Pomajbíková K., Petrželková K.J., Hůzová Z., Modrý D., Lukeš J. Detection of Plasmodium spp. in human feces. Emerg. Infect. Dis. 2012;18:634–636. doi: 10.3201/eid1804.110984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nys H.D., Madinda F., Merkel K., Robbins M., Boesch C., Leendertz F., Calvignac-Spencer S. A cautionary note on fecal sampling and molecular epidemiology in predatory wild great apes: Fecal based epidemiology in predatory apes. Am. J. Primatol. 2015;77:833–840. doi: 10.1002/ajp.22418. [DOI] [PubMed] [Google Scholar]
- 68.Nys H.M.D., Calvignac-Spencer S., Boesch C., Dorny P., Wittig R.M., Mundry R., Leendertz F.H. Malaria parasite detection increases during pregnancy in wild chimpanzees. Malar. J. 2014;13:413. doi: 10.1186/1475-2875-13-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., Van der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strier K.B., Ziegler T.E., Wittwer D.J. Seasonal and social correlates of fecal testosterone and cortisol levels in wild male muriquis (Brachyteles arachnoides) Horm. Behav. 1999;35:125–134. doi: 10.1006/hbeh.1998.1505. [DOI] [PubMed] [Google Scholar]
- 71.Ziegler T.E., Santos C.V., Pissinatti A., Strier K.B. Steroid excretion during the ovarian cycle in captive and wild muriquis, Brachyteles arachnoides. Am. J. Primatol. 1997;42:311–321. doi: 10.1002/(SICI)1098-2345(1997)42:4<311::AID-AJP6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 72.Acharya K.R., Dhand N.K., Whittington R.J., Plain K.M. PCR inhibition of a quantitative PCR for detection of Mycobacterium avium subspecies Paratuberculosis DNA in feces: Diagnostic implications and potential solutions. Front. Microbiol. 2017;8:115. doi: 10.3389/fmicb.2017.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Areeqi M.A., Sady H., Al-Mekhlafi H.M., Anuar T.S., Al-Adhroey A.H., Atroosh W.M., Dawaki S., Elyana F.N., Nasr N.A., Ithoi I., et al. First molecular epidemiology of Entamoeba histolytica, E. dispar and E. moshkovskii infections in Yemen: Different species-specific associated risk factors. Trop. Med. Int. Health. 2017;22:493–504. doi: 10.1111/tmi.12848. [DOI] [PubMed] [Google Scholar]
- 74.Arregui G., Enriquez S., Benítez-Ortiz W., Navarro J.-C. Taxonomía molecular de Anopheles del Ecuador mediante ADN mitocondrial (citocromo c oxidasa I) y optimización por parsimonia máxima. Bol. Mal. Salud. Amb. 2015;55:132–154. [Google Scholar]
- 75.Mathay C., Hamot G., Henry E., Georges L., Bellora C., Lebrun L., Witt B.D., Ammerlaan W., Buschart A., Wilmes P., et al. Method optimization for fecal sample collection and fecal DNA extraction. Biopreserv. Biobank. 2015;13:79–93. doi: 10.1089/bio.2014.0031. [DOI] [PubMed] [Google Scholar]
- 76.Zinner D., Wertheimer J., Liedigk R., Groeneveld L.F., Roos C. Baboon phylogeny as inferred from complete mitochondrial genomes. Am. J. Phys. Anthropol. 2013;150:133–140. doi: 10.1002/ajpa.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yasuda K., Oh K., Ren B., Tickle T., Franzosa E., Wachtman L., Miller A., Westmoreland S., Mansfield K., Vallender E., et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taberlet P., Luikart G. Non-invasive genetic sampling and individual identification. Biol. J. Linn. Soc. 1999;68:41–55. doi: 10.1111/j.1095-8312.1999.tb01157.x. [DOI] [Google Scholar]
- 79.Creel S., Spong G., Sands J.L., Rotella J., Zeigle J., Joe L., Murphy K.M., Smith D. Population size estimation in Yellowstone wolves with error-prone noninvasive microsatellite genotypes. Mol. Ecol. 2003;12:2003–2009. doi: 10.1046/j.1365-294X.2003.01868.x. [DOI] [PubMed] [Google Scholar]
- 80.Santos L.C.D., Curotto S.M.R., Moraes W.D., Cubas Z.S., Costa-Nascimento M.D.J., Filho I.R.D.B., Biondo A.W., Kirchgatter K. Detection of Plasmodium sp. in capybara. Vet. Parasitol. 2009;163:148–151. doi: 10.1016/j.vetpar.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 81.Bentz S., Rigaud T., Barroca M., Martin-Laurent F., Bru D., Moreau J., Faivre B. Sensitive measure of prevalence and parasitaemia of haemosporidia from European blackbird (Turdus merula) populations: Value of PCR-RFLP and quantitative PCR. Parasitology. 2006;133:685–692. doi: 10.1017/S0031182006001090. [DOI] [PubMed] [Google Scholar]
- 82.Rouffaer L.O., Steensels M., Verlinden M., Vervaeke M., Boonyarittichaikij R., Martel A., Lambrecht B. Usutu virus epizootic and plasmodium coinfection in Eurasian blackbirds (Turdus merula) in Flanders, Belgium. J. Wildl. Dis. 2018;54:859–862. doi: 10.7589/2017-07-163. [DOI] [PubMed] [Google Scholar]
- 83.Nixon K.C., Carpenter J.M. On outgroups. Cladistics. 1993;9:413–426. doi: 10.1111/j.1096-0031.1993.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 84.MacVectorInc Sequence Analysis Tools for Molecular Biologists. MacVector, Inc.; Apex, NC, USA: 2020. version 17.5.5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.