Abstract
Protecting food crops from viral pathogens is a significant challenge for agriculture. An integral approach to genome-editing, known as CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and CRISPR associated protein 9), is used to produce virus-resistant cultivars. The CRISPR/Cas9 tool is an essential part of modern plant breeding due to its attractive features. Advances in plant breeding programs due to the incorporation of Cas9 have enabled the development of cultivars with heritable resistance to plant viruses. The resistance to viral DNA and RNA is generally provided using the Cas9 endonuclease and sgRNAs (single-guide RNAs) complex, targeting particular virus and host plant genomes by interrupting the viral cleavage or altering the plant host genome, thus reducing the replication ability of the virus. In this review, the CRISPR/Cas9 system and its application to staple food crops resistance against several destructive plant viruses are briefly described. We outline the key findings of recent Cas9 applications, including enhanced virus resistance, genetic mechanisms, research strategies, and challenges in economically important and globally cultivated food crop species. The research outcome of this emerging molecular technology can extend the development of agriculture and food security. We also describe the information gaps and address the unanswered concerns relating to plant viral resistance mediated by CRISPR/Cas9.
Keywords: CRISPR/Cas9, food crops, plant viral diseases, resistance mechanisms and strategies, challenges
1. Introduction
Food crops are vulnerable to various diseases including bacteria, fungi and viruses, resulting in major economic losses. Thus, crop resistance can be improved against various pathogens [1,2]. Disease control approaches rely on resistant cultivars and agrochemicals that are typically highly effective once deployed. Protecting crops from emerging pests and diseases and developing resistant cultivars from the perspective of higher production are significant challenges [3]. Because pesticides used in the field are usually not highly selective, they can also affect other beneficial organisms when killing pathogens, and thus disrupt the ecological balance. The development of disease-resistant crop varieties is an efficient and environmentally responsible integrated agriculture policy [4]. Various genome editing and sophisticated molecular technologies for different transgenic crop plants are combined in modern plant breeding. Thus, improved crop varieties can be obtained with enhanced disease resistance, which is known as resistance plant breeding. This transgenic technology enables plant breeders to crossbreed crop species and insert desired genes from non-related species and/or organisms into crop plants [5].
Genetic diversity with enhanced virus protection is an indispensable part of virus resistance [6]. New breeding techniques (NBTs) comprise the latest and most effective biological methods for the accurate genetic manipulation of single or several target genes. They use a site-driven nuclease to add double-stranded breaks in DNA at specified regions. These breaks are usually repaired through various host-genome cell repair mechanisms, thus resulting in either small insertions or deletions via non-homologous end-joining (NHEJ) or a modified gene carrying the predetermined nucleotide changes copied from a repair matrix via homologous recombination (HR) [7]. CRISPR/Cas9 system highlight the reality that this technique requires fewer skills and financial resources and has a higher accuracy rate for gene editing performance, as in the Cas9 system, crops require off-target cleavage and different goal choices relative to the other nucleases available [8].
CRISPR/Cas9 is considered a highly promising genome-editing method in crops due to its unique features, such as reliable precision, multiple-gene editing, limited off-target impact, greater output, and simplicity [9]. This particular mechanism invades foreign DNA fragments of virus particles and enables them to detect and degrade the DNA or RNA sequences for further invasion [10]. CRISPR/Cas9 technology manipulates the defense mechanism against plant viruses by identifying and destroying pathogenic genes that invade them. It can also be deployed to develop crop cultivars with enhanced resistance against various plant viruses. This approach has revolutionized research in virus resistance due to its sequence-specific nuclease capability [11]. The use of association genetics focused on single nucleotide polymorphisms (SNPs) and other common molecular markers has also increased in plant breeding, generating important high throughput data for quantitative trait loci (QTL) recognition. The major QTL has been used in crop varieties to provide the quantitative resistance to plant viruses, combined with the usage of primary resistance (R) genes incorporated into cultivars with superior agronomic characteristics [12]. In this review, we address the key features of the CRISPR/Cas9 genome-editing technique and its implications for new food crop cultivar evolutions with improved plant virus resistance.
2. Virus-Resistance Mechanism of CRISPR/Cas9 in Crops
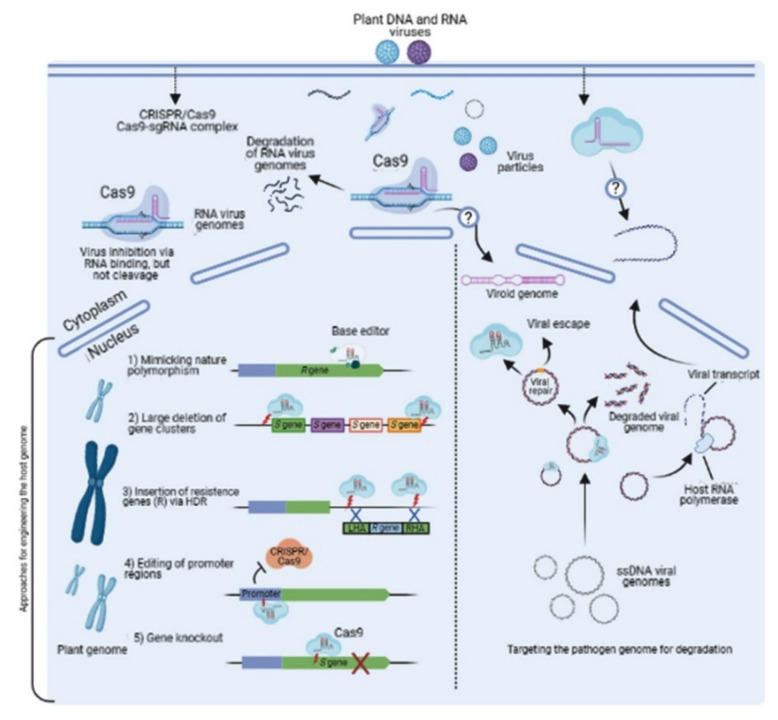
The CRISPR/Cas9 method identifies and targets a pathogen’s genetic material via a three-step procedure: (i) acquisition, (ii) expression and (iii) interference [13]. The first step, acquisition, involves the invasive foreign DNA as a spacer from viruses or plasmids, which are divided into short fragments, and recognized and incorporated into the CRISPR locus. CRISPR loci have been copied and used to produce short RNA (crRNA), directing the effector endonuclease genes through simple complementarity to target the virus components. In general, the protospacer adjacent motif (PAM) includes a short stretch (2–5 bp) of retained nucleotides that obtain the DNA fragment (spacer) for identification. Mutations in viral genomes and PAM overcome the CRISPR-mediated immunity from pathogen attacks (Figure 1) [14].
Figure 1.
Virus resistance via CRISPR/Cas9 (illustration is adapted and modified from Zaidi et al. [14]).
CRISPR/Cas9 expression includes effectively transcribing the large pre-CRISPR RNA (pre-crRNA) attained from the CRISPR locus, thus converting it into several crRNAs with the aid of the Cas9 protein and the tracrRNA molecule [15]. The tracrRNA combines with the crRNA repeat area through base complementarity and facilitates pre-crRNA processing in crRNA (Figure 2) [16]. The activated crRNAs join the CRISPR-associated antiviral protection complex (CASCADE) and help to identify and base-pair a particular target area of foreign DNA [17].
Figure 2.
Mechanism of CRISPR/Cas9 system (illustration is adapted and modified from Arora et al. [16]).
In the CRISPR/Cas9 method, DNA interference requires a single Cas9 protein [18]. The crRNA guides the Cas9 protein into the target site of the foreign DNA to break down during the interference step and provides immunity against pathogen attacks [13]. Cas9 is an immense protein with several domains (the amino terminus RuvC domain and the centrally-located HNH nuclease domain) and two short RNA segments, namely, crRNA and trans-activating crRNA (tracrRNA). The Cas9 protein promotes adaptation, engages in pre-crRNA processing toward crRNA, and implements specified DNA double-strand breaks (DSBs) led by the tracrRNA and RNAse III-specific double-stranded RNA [19]. The creation and development of CRISPR/Cas9 structures are relatively simple, affordable, and without intellectual property obstacles. The CRISPR/Cas9 tool, crRNA, and tracrRNA components can be fused together into the sgRNA that guides Cas9 to implement the targeting of specific DSBs (Figure 2) [16]. The design of sgRNAs is also straightforward, thus favouring genome-editing. The CRISPR/Cas9 method was initially designed to cause cleavage in DNA in vitro at different sites [19,20]. This method has recently been implemented to edit genomes in bacteria, fungi, viruses, yeast and many other organisms for successful selective mutagenesis [20,21,22,23].
3. Plant Virus-Resistance Strategies Using Cas9 Endonuclease
The CRISPR/Cas9 technology allows for the development of a broader range of CRISPR variants useful for different applications and has been successfully demonstrated to engineer virus-resistant crop cultivars. However, gene disruption is one of the most common applications of the CRISPR/Cas9 tool (Figure 3) [24] and helps to overcome the error-prone behaviour of cellular NHEJ (DNA-repair machinery). The insertion/deletion (InDel) of nucleotides at sgRNA-targeted sites introduces a frameshift mutation and disrupts gene function [25]. In the context of virus resistance, this strategy has been employed to engineer resistance by disrupting the susceptible (S) gene(s) function, which alters the plant–virus interaction, resulting in reduced viral fitness in the host plant. Another approach can also be utilized to introduce InDels in the promoter region rather than the coding region of a gene [24]. CRISPR-mediated promoter disruption blocks the entire gene expression and the effector-binding site of susceptible plants by preventing a virus effector from binding to the promoter [26]. Moreover, due to the gene clusters, it is feasible to use this particular approach to engineer virus resistance in plants. Deleting the chromosomal fragments adjacent to S gene clusters may produce durable virus resistance in different hosts. In addition to virus resistance, CRISPR-mediated gene insertion opens an avenue to study important S genes [27]. The functional analysis of susceptibility genes enables us to understand the regulation of gene expression. The host proteins play key roles in the pathogenicity of the genome, resulting in their expression and localization during viral infection. Recent studies have recognized several resistance (R) gene(s) in wild species and proved the successful transfer of resistance in cultivated crop species [25]. This tool can replace the improper and poorly performing R genes from cultivated crop species with a functional R gene from a virus-resistant cultivar via multiplexed homology directed repair (HDR) methodology. The biomimicry mechanism introduces the CRISPR-mediated mutations, which convert the target gene as the sequence of a virus-resistant cultivar [26]. This approach is beneficial to introduce only specific mutations associated with virus resistance traits instead of replacing the whole gene (Figure 3) [24]. In this manner, it can be assumed that the differences in the nucleotide sequences of the target genes among the cultivated species and wild varieties are not sufficiently significant to confer durable resistance to various plant viruses. In this case, the Cas9 tool can be utilized to modify a particular gene for stable plant resistance. The durability of resistance primarily relies on the appropriate molecular strategies within Cas9 mechanisms that are applied.
Figure 3.
Plant virus-resistance strategies using Cas9 (illustration is adapted and modified from Zaidi et al. [24]).
4. Deletion and Insertion of the Target Gene
Multiple sgRNA sequences can be utilized to drive the numerous double-strand breaks (DSBs) through CRISPR/Cas9 at the appropriate site of the target gene. Two sgRNA bindings will generate two DSBs at the respective sites before the start codon and after the stop codon of the gene of interest. These DSBs then remove the DNA fragments carrying the gene of interest before repairing the cellular non-homologous end-joining (NHEJ) machinery (Figure 4) [28]. The sgRNA bindings can be designed for any gene region carrying a precise trinucleotide protospacer adjacent motif (PAM). This approach deletes the expanded chromosomal fragments, including the individual genes [29,30].
Figure 4.
The deletion and insertion of the target gene (illustration is adapted and modified from Ghosh et al. [28].
The CRISPR/Cas9 tool can be employed to enhance disease resistance via alteration of the susceptible (S) genes. These S genes disrupt the plant proteins, which are essential and primarily multifunctional. Thus, plant health and productivity are also retarded. An alternate approach, such as cis-regulatory component and promoter editing, can alter gene expression instead of gene disruption. It is sometimes crucial to utilize resistance (R) genes against virulent pathogens. In contrast, the host–pathogen interaction has not been fitted well, and the S genes have not been explored extensively [31]. In such cases, the Cas9 technique can be utilized for the insertion of the R genes. Insertion of the CRISPR-mediated gene operates through an alternative pathway that works when Cas9 generates the sgRNA-directed DSBs; this pathway uses cellular homology-directed repair (HDR) instead of non-homologous end-joining (NHEJ) machinery (Figure 4) [28]. The delivery fragment carrying an R gene, which is surrounded by the homologous sequence of DSB ends, is adjunct with the Cas9-gRNA complex. This cassette directs the insertion of the HDR-mediated R gene between the two DSB ends. This approach was practiced to introduce many genes at the distinct genomic regions [32].
5. Off-Target Mutations in CRISPR/Cas9
The CRISPR/Cas9 technology evaluates the high specificity in targeting the genomic regions, such as irradiation-induced mutagenesis. This tool raises questions about how a sgRNA can target the entirely complementary genomic DNA sequences and other genomic sites (off-target regions) and the extent of targeting and potentially provoke unexpected damage. There are two types of expected off-target effects in genomic sites: higher sequence similarities to the target and the unexpected off-target in irrelevant genomic sites. Gene sequence information is essential to predict the expected off-target outcomes [25]. Random off-target mutations have become more frequent when mismatches occurred from distant genomic regions [33]. Recent studies have screened polyploidy progenies via CRISPR/Cas9 knockout to clarify the off-target issue in crops. This may bind the lower effective sequences with the mismatches (usually 1 to 3). The appropriate design of the CRISPR/Cas9 tool can avoid the expected off-target mutations, but unexpected off-target mutations cannot be avoided because of their spontaneous lower frequency mutations of plants [27].
6. Overcoming Off-Target Effects
To enhance the specificity and effectiveness of the CRISPR/Cas9 system, it is vital to analyze and address its possible off-target mutations. Off-target effects generated by the CRISPR/Cas9 system can be identified in plants via qRT-PCR resequencing [34]. The off-target effects have become a potential concern for the CRISPR/Cas9 system. However, few studies have addressed this issue in various organisms. Recently, the high-resolution structure of the SpCas9–sgRNA complex has removed the mismatch tolerance mechanism between the sgRNAs and the targeted DNA sequences [35]. This finding can be considered a significant advance towards understanding the specificity and molecular mechanism of the CRISPR/Cas9 system. To overcome the off-target effects, a mutant Cas9 endonuclease was used to induce the distinct cleavage only at one DNA strand of the targeted site and activate the homologous recombination to enhance the specificity of the target region [27]. The Cas9-sgRNA complex reduced off-target mutations by 50- to 1500-fold by introducing DSBs. The significant numbers of Cas9/sgRNA delivered, and the ratio of Cas9:sgRNA, have also been found to minimize off-target effects. A higher concentration of the Cas9:sgRNA ratio results in higher off-target rates than a lower Cas9/sgRNA concentration [25].
7. CRISPR/Cas9 Toolkit in Crop Improvement
Due to its potentiality, low cost, and simplicity, Cas9 has become a widely adopted genome-editing tool in numerous living organisms, including plant species. Editing genes allow the contemporaneous alteration of multiple genetic loci, thus accelerating various commercial crop improvements and enhancing food safety globally [36]. A number of recent studies using this technique for genome-editing have focused on different economically important food crops necessary for modern agriculture. It has been found that this technique could be utilized for improvement of specific traits, such as crop yield, grain quality and disease resistance [37]. CRISPR/Cas9 has been practiced in the genome-editing of rice (Oryzae sativa). Several Cas9-sgRNAs were designed to effectively delete the small fragments of the dense and erect panicle1 (DEP1) gene in the Indica rice line. These improvements in traits that contribute to yield, such as reduced plant height and dense and upright panicles, have been executed in the production of mutant plants [38]. This method was utilized to introduce mutations into the GmFT2a gene, an essential integrator in photoperiodism of the flowering pathway of soybean plants. The newly developed soybean cultivars exhibited late flowering, which results in the increased size of the vegetative growth. This mutation was also firmly inherited among the following generations [39]. CRISPR/Cas9 was used to generate mutations in the flowering suppressor gene SELF-PRUNING5G (SP5G) in tomato plants to manipulate the photoperiod response. These mutations were achieved using Cas9 and caused rapid flowering and augmented the dense growth habit of tomato plants, resulting in an immediate increase in early yield [40]. The development of resistance against Xanthomonas citri subsp. citri, which causes bacterial citrus canker, was undertaken using Cas9 technology. A promoter gene CsLOB1 was targeted, which promotes the development of canker in citrus plants. The developed transgenic lines exhibited improved resistance against canker in citrus compared to the wild types [41]. Bread wheat cultivars mutated using the CRISPR/Cas9 toolkit exhibited improved resistance to infection with Blumeria graminis f. sp. tritici. This finding also showed the development of powdery mildew resistance in wheat [42].
8. Genetics of Plant Virus Resistance
Although viruses are relatively simple genetic entities, many of their mechanisms by which diseases are generated, and by which plants can resist these effects by natural resistance, remain unknown [43]. The most effective and sustainable approach to protect plants from virulent viruses is the development of genetic resistance against these viruses. The rigorous study of plant resistance (R) genes, in which genetic variability occurs that alters the plant’s suitability, raises numerous fundamental questions regarding the molecular, biochemical, cellular and physiological mechanisms involved in the plant–virus interaction [5]. Recently, significant advances have been made in molecular mechanisms associated with natural virus resistance genes. Both the dominant and recessive resistance genes have been characterized at the molecular level to understand new principles of innate immunity to viruses associated with gene silencing. Thus, it has been possible to undertake genome sequencing of many crop species, and this technology is now being utilized on a larger scale. These advances also provide new opportunities to tackle the barriers to virus resistance [3]. Resistance breeding should not depend on extreme molecular characterization of resistance gene alleles and the target a virulence (avr) determinant of a virus. In a practical sense, the successful deployment of a potential resistance gene into a crop depends more upon the identification of a positive phenotype, on the dissection of the phenotype, leading to the identification of genetic markers for marker-assisted selective breeding (MAS) and on an understanding of how the novel resistance will behave in different genetic backgrounds and under pathogen pressure in the field [44]. However, the focus is mainly on monogenic dominant resistance to bacterial and fungal pathogens, and the common mechanisms can also be employed in virus resistance. Low resistance durability is a barrier to the appearance and increased frequency of resistance-breaking (RB) variants among virus populations. The genetic changes required for a virus to overcome plant-resistance mechanisms and the effects of these changes on the fitness of the virus are key determinants of resistance durability.
9. Economic Importance of Plant Viral Diseases in Food Crops
Plant viruses have immense importance in agriculture because many infect different food crops such as vegetables, fruits and staple grains, causing a deterioration in productivity and quality. One virus may infect several crop species, and different kinds of viruses usually attack each species of crop. Viruses cause disease by consuming cells or killing them with toxins and utilizing cellular substances during multiplication, taking up space in cells and disrupting cellular processes. Almost all viral diseases appear to cause dwarfing or stunting of the entire crop plant and a reduction in the total yield. These effects may be severe and cause striking symptoms on the stem, fruit and roots; alternatively, they may or may not cause any symptom development. Plants may show acute severe symptoms soon after inoculation that may lead to the death of young shoots or the entire host plant. If the host survives the initial shock phase, the symptoms tend to become milder (chronic symptoms) in the subsequently developing parts of the plant, leading to partial or even total recovery.
However, in some diseases, symptoms may increase progressively in severity and may result in a gradual or quick decline in the health of the plant. The most common types of plant symptoms produced by systemic virus infections are mosaics and ring spots. Mosaics are characterized by light-green, yellow or white areas with the typical green color of the leaves, flowers and fruits. Depending on the intensity or pattern of discolorations, mosaic symptoms may be described as mottling, streaks, a ring pattern, a line pattern, vein clearing, vein banding or chlorotic spotting. Ring spots are characterized by chlorotic or necrotic rings on the leaves and sometimes also on the fruits and stems. In many ring spot diseases, the symptoms tend to disappear later, but the virus remains. The extent of economic yield loss due to viral infection in many crop varieties is summarized in Table 1 and Table 2 present specific examples of key resistance techniques by CRISPR/Cas9 used in several food crops from 2015 to 2021.
Table 1.
A list of viruses causing diseases in food crops via insect vectors and estimated yield loss in different regions of the world; results summarized from G. N. Agrios (2005) [45].
| Virus | Virus Family | Damaging Food Crop | Distribution | Vector | Crop Yield Loss |
|---|---|---|---|---|---|
| African cassava mosaic virus (ACMV) | Geminiviridae | Cassava | Africa and all cassava growing countries | Whitefly | 20–90% |
| Banana bunchy top virus (BBTV) | Nanoviridae | Banana | All banana growing countries | Aphid | Up to 100% |
| Banana streak virus (BSV) | Caulimoviridae | Banana | All banana growing countries | Mealybug | 6–15% |
| Barley yellow dwarf virus (BYDV) | Luteoviridae | Barley, oats, rye and wheat | Global | Aphid | 30–50%, |
| Bean yellow dwarf virus (BeYDV) | Potyviridae | Bean, peas and other legumes and non-legumes | Global | Aphid and mechanical infection | 30–70% |
| Bean common mosaic virus (BCMV) and bean yellow mosaic virus (BYMV) | Potyviridae | BCMV infects the only bean, BYMV also infects peas and yellow summer squash | Global | Aphid and seeds from infected plants | Up to 100% |
| Bean golden mosaic virus (BGMV) | Geminviridae | Bean, other legume crops and Malvaceous weeds | Global | Whitefly | Up to 100% |
| Beet severe curly top virus (BSCTV) | Geminiviridae | Sugar beet, bean, melons, spinach, sugar beet and tomato | Global | Leafhopper | Up to 100% |
| Beet yellows virus (BYV) | Closteroviridae | Sugar beets, spinach and table beets | All sugar beet growing countries | Aphid | Up to 50% |
| Cassava brown streak virus (CBSV) | Potyviridae | Cassava | East Africa including Kenya, Mozambique and Tanzania | Whitefly | Up to 30%, reduced market price up to 90% |
| Cauliflower mosaic virus (CaMV) | Caulimoviridae | Vegetables | Many parts of the world | Aphid | 20–50% |
| Cotton leaf curl kokhran virus (CLCuKoV) | Geminiviridae | Okra | China, India, Pakistan and Philippines | Whitefly | Up to 55% |
| Cucumber mosaic virus (CMV) | Bromoviridae | Bananas, beans, beets, celery, crucifers, cucumbers, melons, peppers, spinach, squash, tomatoes and vegetables | Global | Aphid | 80% and above |
| Cucumber vein yellowing virus (CVYV) | Potyviridae | Cucumber, melon or watermelon and other crops of Cucurbitaceae family | Global | Whitefly | Up to 70% |
| Grapevine fan leaf virus (GFLV) | Secoviridae | Grape | Global | Nematodes | Up to 80% |
| Lettuce infectious yellows virus (LIYV) | Closteroviridae | Lettuce, cantaloupe, carrot, melon, squash, sugar beet, squash | Global | Whitefly | 30–100% |
| Lettuce mosaic virus (LMV) | Potyviridae | Lettuce, marigold, pea and sweet pea | Global | Aphid and infected seeds | 55–85% |
| Maize dwarf mosaic virus (MDMV) | Potyviridae | Maize | Global | Aphid | Up to 70% |
| Maize streak virus (MSV) | Geminiviridae | Maize, rice, wheat, millet and sugarcane | India and southern part of Africa | Leafhopper | Up to 100% |
| Merremia mosaic virus (MeMV) | Geminiviridae | Hot pepper, sweet pepper and so on. | Where irrigated crop production practiced worldwide | Whitefly | 70–80% |
| Okra yellow vein mosaic virus (OYVMV) | Geminiviridae | Okra | Global | Whitefly | More than 90% |
| Papaya ring spot virus-W (PRSV-W) | Potyviridae | Papaya and cucurbits | Global | Aphid | Up to 100% |
| Plum pox virus (PPV) | Potyviridae | Apricot, nectarine, peach and plum | Global including Asia, Europe and North America | Aphid, budding and grafting | 80–100% |
| Potato leafroll virus (PLRV) | Luteoviridae | Potato | Global | Aphid | Up to 90% |
| Potato virus X (PVX) | Alphaflexiviridae | Potato, pepper, tobacco and tomato | Global | Handling of the plant materials | 30–40% |
| Potato virus Y (PVY) | Potyviridae | Potato, pepper, tobacco and tomato | Global | Aphid | Up to 70% |
| Rice tungro bacilliform virus (RTBV) and Rice tungro spherical virus (RTSV) | Caulimoviridae (RTBV), Secoviridae (RTSV) | Rice | South and Southeast Asia | Leafhopper | Up to 100% |
| Soybean mosaic virus (SMV) | Potyviridae | Soybean and various crops of Fabaceae and Leguminosae family | Global | Aphid | 35–90% |
| Squash leaf curl virus (SLCV) | Geminiviridae | All cucurbits such as squash, cucumber, watermelon and cantaloupe | Southern region of California | Whitefly | 70–80% |
| Sugarcane mosaic virus (SCMV) | Potyviridae | Corn, sorghum, sugarcane and other crops of Gramineae family | Global | Aphid | Up to 40% |
| Tomato mosaic virus (ToMV) | Virgaviridae | Tomato and other Solanaceous crops | Global | Infected seeds | 20–90% |
| Tomato mottle virus (TMoV) | Geminiviridae | Common bean, tobacco and tomato | Global | Whitefly | Up to 95% |
| Tomato ring spot virus (TomRSV) | Secoviridae | Tomato, strawberries, raspberries, grapes and apple | Global | Nematodes | 50–80% |
| Tomato spotted wilt virus (TSWV) | Bunyaviridae | Lettuce, papaya, peanut, pineapple, tomato and various fruits and vegetables | Tropics and subtropics and a few temperate regions of the world | Thrip | 50–90% |
| Tomato yellow leaf curl Sardinia virus (TYLCSV) | Geminiviridae | Watermelon, tomato, squash, potato, pepper, melon, cotton, cassava and bean | Global particularly prevalent in tropics and subtropics regions | Whitefly | Up to 100% |
| Tomato yellow leaf curl virus (TYLCV) | Geminiviridae | Tomato and many food crops | Global | Whitefly | Up to 100% |
| Turnip mosaic virus (TuMV) | Potyviridae | Brussels, sprouts, cabbage and cauliflower | Global | Aphid | Up to 70% |
| Watermelon mosaic virus (WMV) | Potyviridae | Peas and Leguminous crops | Global | Aphid | Up to 100% |
| Wheat dwarf virus (WDV) | Geminiviridae | Wheat and barley | Throughout Europe | Leafhopper | Up to 75% |
| Zucchini yellow mosaic virus (ZYMV) | Potyviridae | Cucumber, muskmelon, watermelon, zucchini squash. | Global | Aphid | Up to 70% |
Table 2.
A meta-analysis of CRISPR/Cas9 strategies used for virus resistance in selected food crops; data summarized from 2015–2021 published studies.
| Virus in Food Crops | Experimental/Model Host | Targeted Gene Region(s) | Key Strategies of CRISPR/Cas9 | Reference |
|---|---|---|---|---|
| Banana streak virus (BSV) | Arabidopsis thaliana | BSOLV and eBSOLV | The gRNAs were designed to target the sequences of BSOLV and eBSOLV via the CRISPR/Cas9 technology. Three gRNAs based on their specificity to their target site (targeting sequence S1, S2 and S3 from ORF1, ORF2 and ORF3, respectively) and minimal potential off-targets were introduced into the triploid Musa genome (Gonja Manjaya, AAB). The gRNA cassette, containing OsU6 promoter followed by two BbsI restriction sites and tracer RNA scaffold, was amplified from pZKOsU6-gRNA plasmid and cloned into pENTR-D/Topo. One gRNA from each ORF of the BSV genomic sequence was synthesized and cloned into pMR185 to generate the gRNA modules. The Cas9 endonuclease was employed in the plasmid of Arabidopsis codon-optimized and regulated by parsley ubiquitin promoter (PcUbi). | [46] |
| Bean yellow dwarf virus (BeYDV) |
A. thaliana; Nicotiana benthamiana |
IR, CP, and Rep protein | CRISPR/Cas9 technology was successfully utilized in engineering resistance to the bean yellow dwarf virus (BeYDF). Baltes et al. [46] established a transient assay to detect the activity of Cas9 and sgRNA in N. benthamiana using BeYDV. They used double 35S promoter and AtU6/At7SL RNA polymerase III promoter to express Cas9 and sgRNAs, respectively. The enhanced green fluorescent protein (eGFP) gene replaced the coat protein genes and movement protein for Cas9 and sgRNAs activity assessment against BeYDV in N. benthamiana. | [47] |
| Beet severe curly top virus (BSCTV) | N. bethamiana | CP and Rep protein | The CRISPR/Cas9 tool constructed two vectors (pV86-401 and pC86-401) in which the Cas9 protein is driven by one or two BSCTV promoters and sgRNA complex is driven by AtU6 promoter. The expression of Cas9 under both pV86 and pC86 promoters was significantly induced after BSCTV accumulation in N. benthamiana and pC86 promoter appeared to enable higher-level induction than the pV86 promoter. Previously employed pV86-sgRNA and pC86-sgRNA vectors using four highly active sgRNAs were then constructed to determine the system efficiency against BSCTV. | [48] |
| Cassava brown streak virus (CBSV) | Morchella esculenta | eIF4E, nCBP-1 and nCBP-2 | Cassava brown streak virus (CBSV) is a significant threat to cassava production in Africa. For the disease development in the host, CBSV requires the interaction between “viral genome-linked protein (VPg)” and “eukaryotic translation initiation factor 4E (eIF4E) isoforms” of the host. The nCBP clade was consistently associated with VPg protein and given priority due to its functional characterization. The CRISPR/Cas9 system was employed to produce mutant alleles of nCBP isoforms in cassava. Five constructs were combined, simultaneously targeting the different locations within nCBP-1 and nCBP-2 clades. | [49] |
| Cauliflower mosaic virus (CaMV) | A. thaliana | CP | The target sites in the CaMV CP gene were selected using standard Cas9 protein. Linear arrays of Arabidopsis U6 promoter: sgRNA units were designed and subsequently synthesized. When controlling viruses using the CRISPR-Cas9 system, both Cas9 and sgRNAs are consistently expressed in the cells. Recruiting Cas9 to viral DNA depends on the presence and abundance of sgRNAs. However, due to the existence of folded dsRNA domains in sgRNAs, siRNAs can be formed to contain the alien RNAs. | [50] |
| Cucumber mosaic virus (CMV) | N. benthamiana | PAMs | CMV was artificially injected into A. thaliana and N. benthamiana through vector pCR01. The pCR01 vector contained F. novicida Cas9 (FnCas9) a codon-optimized protein that is driven by an enhanced 35S promoter and a short-range RNA (sgRNA) guide that is driven by an AtU6 promoter. Complementary oligonucleotides were synthesized based on target gene sequences and were inserted inside the pCR01 vector efficiently to construct 23 corresponding vectors of the pCR01-sgRNA complex. | [51] |
| Cucumber Vein Yellowing Virus (CVYV), Papaya ring spot virus-W (PRSV-W) and Zucchini yellow mosaic virus (ZYMV) | Crocus sativus | eIF4E | CMV was artificially injected into A. thaliana and N. benthamiana through vector pCR01. The Cas9/sgRNAs complex was constructed to target the eIF4E gene in cucumber plants. The sgRNA1 sequence was expected to destroy the intact eIF4E gene, and the sgRNA2 sequence to allow the translation of two-thirds portions of the total protein products. One diploid genome with another single eIF4E gene and homozygous mutant plants were propagated to knock out the expression of the eIF4E gene. The Cas9/sgRNA complex was employed to disrupt the function of the recessive eIF4E gene and thus enhance virus resistance in cucumber. | [52] |
| Rice tungro spherical virus (RTSV) | Oryza sativa | eIF4G | Natural resistance to RTSV is a recessive trait that is controlled by a gene, namely, translation initiation factor 4 gamma gene (eIF4G). Mutations that occurred within eIF4G genes were generated utilizing the CRISPR/Cas9 technology to develop new resistance sources in the RTSV-susceptible variety IR64. The final products containing RTSV resistance were found to no longer have the Cas9 sequence under greenhouse conditions. | [53] |
| Turnip mosaic virus (TuMV) | A. thaliana | eIF4E and eIF(iso) 4E | The CRISPR/Cas9 technology was applied to generate the significant genetic resistance against TuMV in A. thaliana plants by deletion of a known host factor (eIF(iso)4E), which was strictly needed for viral existence. Transgenic delivery of the Cas9-sgRNA complex through CRISPR/Cas9 technology has proved its feasibility for the segregation of the transgene originated by induced mutation at the targeted eIF(iso)4E location, initially to generate stable and heritable mutations except for any persistent transgene. This approach is hypothesized as the reason why the recessive gene allele eIF(iso)4E exhibits more durable resistance against TuMV infection; the possible presence of VPg polymorphisms performing via eIF(iso)4E is an independent pathway. | [54] |
| Tomato yellow leaf curl Sardinia virus (TYLCSV) | N. benthamiana | CP and IR | The virus highly conserved a non-nucleotide sequence, which forms a stem-loop-like structure inside the IR sites. This conserved structure was directly involved in the Rep binding site in virus replication and contained some illegal bidirectional gene promoters. The IRs were also strain-specific and associated only with targeted Rep proteins. Authentic TYLCSV-IR-sgRNA was utilized to target the virus TYLCSV. Infectious clones of TYLCSV were injected in N. benthamiana plants via agro-infection overexpressing the CRISPR/Cas9 method. The IR target sequences were examined by loss assay assessment of the SspI enzyme, which confirmed the complete absence of recognized indels. | [55] |
| Tomato yellow leaf curl virus (TYLCV) | N. benthamiana; Solanum lycopersicum | CP, IR and Rep sequences | Agrobacterium-mediated transfer DNA (T-DNA) modification was used to express the sgRNAs cassettes by U6-26s promoter and Cas9 protein by CaMV-35S promoter in N. benthamiana plants. The U6-sgRNA cassette and Cas9 protein were cloned within a binary vector controlling by the CaMV-35S promoter and transferred in the N. benthamiana leaf discs utilizing Agrobacterium tumefaciens. The results proved the ability of CRISPR/Cas9 technology to target the infectious strains of TYLCV in the CP, IR and Rep sequences in transgenic N. benthamiana plants. | [27,55,56,57] |
| Wheat dwarf virus (WDV) | Hordeum vulgare | CP, MP, LIR and Rep | To identify the multiple target regions, the WDV genome was mapped for efficient CRISPR/Cas9 target sequences encircling the PAMs motif. Four different target sites were selected that showed no off-target effects and were capable of attacking several viral DNA sequences. The sgRNA WDV1 exhibits the complementarity overlapping in the CP and MP coding regions, sgRNA WDV2 targets the Rep/Rep A coding regions remaining in the N-terminus of the proteins, sgRNA WDV3 targets the LIR region and sgRNA WDV4 targets the Rep protein region and encodes the C-terminus of the protein. | [58] |
10. Challenges
The CRISPR/Cas9 platform is a groundbreaking advancement that allows the engineering of interference against plant pathogens. In-depth research is required to completely develop the effectiveness of this method to globally combat hunger due to crop losses caused by viral diseases. One of the concerns of the CRISPR/Cas9 tool is the risk of newly emerging recombinant viruses that are resistant to the Cas9 endonuclease [59]. These recombinant viruses are capable of escaping CRISPR/Cas9 targeting due to few InDels target of the sgRNA sites. These mutants are not lethal to viral replication, but they impede the sgRNA sites from identifying the targeted genomic sequence encompassing the Cas9 endonuclease [25]. The mutation and recombination of endogenous genes may lead to their functional inactivation, whereas the variations of virus sequences produced by CRISPR/Cas9 can facilitate viral evolution. It is well known that mutation and recombination are the major driving forces of plant viruses [60].
CRISPR/Cas9 targeting viral open reading frames (ORFs) can generate more viral variants, resulting from the viral escape events at different levels. Cas9 may disseminate these mutant viruses from plants and with multiple sgRNAs [26]. In addition to targeting or interfering with the viral genome to inhibit its infection, CRISPR/Cas9 can generate new viral variants as genome-editing by-products that speed up the evolution of the virus, causing the produced transgenic crops to lose their resistance capacity against these viruses [44]. These limitations are shortcomings of the CRISPR/Cas9 tool against some plant viruses. Several molecular approaches can be used to combat these recombinant viral escapes; for instance, utilizing multiplex genome engineering to target the different sites of the virus genome can significantly inhibit the virus replication [58].
A number of reports were found regarding resistance genes in many economically important food crops, including the 20 viruses listed in Table 3. Cas9 technology could be a potential genomic tool to determine the host–virus interaction mechanisms with the listed genes (Table 3). Currently, these viruses are controlled using chemical management, which has little or no effectiveness. In addition, synthetic inorganic chemicals are toxic to soil and water and are harmful to health. Alarmingly, hazardous chemical molecules continue to recycle within the agroecosystems.
Table 3.
A list of identified virus resistance genes in 20 food crops, is expected to apply CRISPR/Cas9 to control viral diseases.
| SL. No. | Virus in Food Crop | Identified Virus Resistant Gene | Reference |
|---|---|---|---|
| 01 | African cassava mosaic virus (ACMV) | CMD1 (recessive resistance gene), CMD2 (major dominant gene) and CMD3 (conferring resistance) | [61,62] |
| 02 | Banana bunchy top virus (BBTV) | BBTV DNA-R and BBTV DNA-S1 | [63] |
| 03 | Barley yellow dwarf virus (BYDV) | Bdv1, Bdv2, Bdv3, Bdv4 and Ryd2 | [64,65] |
| 04 | Bean common mosaic virus (BCMV) | I (dominant resistance gene), bc-u, bc-1, bc-12, bc-2, bc-22, and bc-3 (recessive resistance gene) | [66] |
| 05 | Bean yellow mosaic virus (BYMV) | rym4/5 | [67] |
| 06 | Bean golden mosaic virus (BGMV) | bgm-1 and bgm-2 | [68,69] |
| 07 | Beet yellows virus (BYV) | III, V and VI QTLs | [70] |
| 08 | Grapevine fanleaf virus (GFLV) | F13, EcoRI and StyI | [71] |
| 09 | Lettuce mosaic virus (LMV) | eIF(iso)4G1,mo11 and mo12 (recessive resistance gene), Mo2 (dominant resistance gene) | [72] |
| 10 | Maize dwarf mosaic virus (MDMV) | Mdm1 | [73] |
| 11 | Maize streak virus (MSV) | msv1 | [74] |
| 12 | Plum pox virus (PPV) | eIF(iso)4G1, eIF(iso)4E, eIFiso4G11, PpDDXL ParP-1 to Par-P-6, ParPMC1 and ParPMC2 | [75] |
| 13 | Potato leafroll virus (PLRV) | Rladg | [76] |
| 14 | Potato virus X (PVX) | Rx1 and Rx2 | [77] |
| 15 | Potato virus Y (PVY) | Ryadg, Rysto , Y-1, pvr1, pvr21, pvr22 + pvr6, pot-1 | [78] |
| 16 | Rice tungro bacilliform virus (RTBV) | RTBV ORF IV and RTBV-CP | [79] |
| 17 | Squash leaf curl virus (SLCV) | slc-2 | [80] |
| 19 | Sugarcane mosaic virus (SCMV) | Scmv1 and Scmv2 | [81] |
| 19 | Soybean mosaic virus (SMV) | Rsv1, Rsv3, Rsv4, Rsv5, Rsv7, Rsv8, Rsv15 and Rsv20 | [82] |
| 20 | Watermelon mosaic virus (WMV) | Wmv1551 | [83] |
11. Research Opportunities
This review highlighted that CRISPR/Cas9 technology has been used to develop crop varieties resistant to numerous significant viral pathogens. Furthermore, this technique may also provide an important opportunity to research these viruses, resulting in benefits to humans. Viruses cause numerous diseases in the most economically significant crop plants, and CRISPR-mediated virus resistance has been used to target the virus DNA and RNA. This approach has some limitations; notably, viruses have the capacity to escape and generate resistance-blocking strains. Specific utilization of the host susceptibility factors involved in plant–virus interactions is a possible solution for this shortcoming. Moreover, the well-distinguished susceptible (S) gene(s) available for other pathogens are usually absent for viruses. Considerable research has been conducted to identify and understand the mechanisms of virus infection, and fewer potentially relevant plant genes have also been recognized. These comprehensive reviews show that the host susceptibility factors may potentially target the S genes to develop resistance against the crop virus, which is a promising future approach for agriculture. The mutation among the plants generated using the CRISPR/Cas9 method is usually stable and heritable in nature. This mutation can be easily separated from the Cas9/sgRNA complex to avoid further alterations by Cas9/sgRNA, thus helping to develop transgene-free progeny in only one generation. The integration of Cas9/sgRNA in the CRISPR/Cas9 toolkit for virus resistance assists in the development of virus-resistant transgenic plants. The plasmid-free delivery of pre-assembled complexes is another essential technique for generating virus-resistant plants that contain no foreign DNA in the respective virus genome. Identifying potential genes or DNA sequences that can be utilized as useful targets for gene editing is also essential. For example, the fission of preserved non-coding regions with the virus genome contributes to the same taxonomic community’s resistance to specific viruses. Using the prokaryotic CRISPR/Cas9 framework means that the resistance produced would be more effective because, unlike the RNA interference method, viruses lack the machinery to prevent their inhibitory effects. The Cas9 protein and short-range in vitro transcribed RNA guide can be incorporated directly into the cells as a ribonucleo protein complex (RNP), which does not require the integration of transgenes into the genome. These approaches apply to RNA or DNA genome viruses. In addition, the rate of recombination and mutation of viral DNA also remains to be explored immediately after the cleavage virus genome enters the host cell. Among other questions, it is also unknown whether the multiplexing of sgRNAs in transgenic plants may promote resistance to diverse strains of plant viruses, because it is naturally covered with the crRNA gene within the prokaryotic CRISPR arrays.
The presence of off-target results continues to be investigated using CRISPR/Cas9 in vivo conditions among the individual hosts. In the CRISPR/Cas9 method, the target RNA binds the DNA, and pre-designed sequences remaining inside the RNA lead the Cas9 protein to break DNA strands at the correct positions. DNA cutting is achieved by extracting and adding the appropriate sequences of the target DNA. Homologous recombination of DNA restores the DNA damage and causes mutations. However, it also often contributes to unintended mutations because it is not appropriate to complement a total of 20 base pairs in the short-range RNA guide sequence to bind the target DNA. This may also result in the attachment of lead RNA in an inappropriate location and cutting of the non-targeted DNA, resulting in improper mutations and perhaps causing the loss of the essential segment of DNA sequences with necessary details. However, research is being undertaken to allow more practical application of CRISPR/Cas9 because the Cas9 protein can be managed to target specific DNA strands by modifying the sequences of the RNA guide.
12. Conclusions
The CRISPR/Cas9 method shows promise as a useful genome-editing technology for producing different virus-resistant crop cultivars with improved yield, quality, and abiotic/biotic stress tolerance. The off-target effects are among the limitations for large-scale field application of the CRISPR/Cas9 tool in virus-resistant crop development. This challenge may be overcome by applying more effort to the machine learning (ML) process. At present, the impact of the CRISPR/Cas9 methodology on plant physiological traits and potential virus mutants is inadequately documented. Global collaboration using open field trial data (from multiple climates and locations) on Cas9-treated crop plant–virus–environment interactions, and the subsequent growth and yield performance of staple food crops, warrants further attention.
Acknowledgments
Authors thank Pankaj Bhowmik, Laboratory of Cell Technologies, Genome-Editing and Trait Development, National Research Council, Saskatoon, Canada for proofreading this manuscript.
Author Contributions
Conceptualization, S.A.S., C.N.W.C., M.A.R., N.C.P. and J.U.; investigation, S.A.S., M.A.R., N.C.P. and J.U.; writing—original draft preparation, S.A.S.; writing—final submission, M.N.I., M.A.R., S.S. and S.A.S.; revisions and responses to reviewers, M.N.I., S.S. and S.A.S.; funding acquisition, S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received financial support from Universiti Malaysia Sabah Scheme UMS Great code number (GUG0276-2/2018).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have declared that there is no conflict of interest exists.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Velásquez A.C., Castroverde C.D., He S.Y. Plant–Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018;28:R619–R634. doi: 10.1016/j.cub.2018.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savary S., Willocquet L., Pethybridge S.J., Esker P., McRoberts N., Nelson A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019;3:430–439. doi: 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- 3.Esse H.P., Reuber T.L., Does D. Genetic modification to improve disease resistance in crops. New Phytol. 2020;225:70–86. doi: 10.1111/nph.15967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aktar W., Sengupta D., Chowdhury A. Impact of pesticides use in agriculture: Their benefits and hazards. Interdiscip. Toxicol. 2009;2:1–12. doi: 10.2478/v10102-009-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong O.X., Ronald P.C. Genetic engineering for disease resistance in plants: Recent progress and future perspectives. Plant Physiol. 2019;180:26–38. doi: 10.1104/pp.18.01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Bulk R.W. Application of cell and tissue culture and in vitro selection for disease resistance breeding—A review. Euphytica. 1991;56:269–285. doi: 10.1007/BF00042373. [DOI] [Google Scholar]
- 7.Nelson R., Wiesner-Hanks T., Wisser R., Balint-Kurti P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2017;19:21–33. doi: 10.1038/nrg.2017.82. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande K., Vyas A., Balakrishnan A., Vyas D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Genetic Engineering: Robotic Genetic Surgery. Am. J. Robot. Surg. 2016;2:49–52. doi: 10.1166/ajrs.2015.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar V., Jain M. The CRISPR-Cas system for plant genome editing: Advances and opportunities. J. Exp. Bot. 2015;66:47–57. doi: 10.1093/jxb/eru429. [DOI] [PubMed] [Google Scholar]
- 10.Barakate A., Stephens J. An overview of crispr-based tools and their improvements: New opportunities in understanding plant-pathogen interactions for better crop protection. Front. Plant Sci. 2016;7:765. doi: 10.3389/fpls.2016.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., Qi L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marraffini L.A., Sontheimer E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan-e-Ali Zaidi S., Mansoor S., Ali Z., Tashkandi M., Mahfouz M.M. Engineering Plants for Geminivirus Resistance with CRISPR/Cas9 System. Trends Plant Sci. 2016;21:279–281. doi: 10.1016/j.tplants.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora L., Narula A. Gene editing and crop improvement using CRISPR-cas9 system. Front. Plant Sci. 2017;8:1932. doi: 10.3389/fpls.2017.01932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., Van Der Oost J., Regev A., et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaidi S.S., Mahas A., Vanderschuren H., Mahfouz M.M. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 2020;21:1–19. doi: 10.1186/s13059-020-02204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mushtaq M., Mukhtar S., Sakina A., Dar A.A., Bhat R., Deshmukh R., Molla K., Kundoo A.A., Dar M.S. Tweaking genome-editing approaches for virus interference in crop plants. Plant Physiol. Biochem. 2020;147:242–250. doi: 10.1016/j.plaphy.2019.12.022. [DOI] [PubMed] [Google Scholar]
- 26.Cao Y., Zhou H., Zhou X., Li F. Control of Plant Viruses by CRISPR/Cas System-Mediated Adaptive Immunity. Front. Microbiol. 2020;11:593700. doi: 10.3389/fmicb.2020.593700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pramanik D., Shelake R.M., Park J., Kim M.J., Hwang I., Park Y., Kim J.Y. CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int. J. Mol. Sci. 2021;22:1878. doi: 10.3390/ijms22041878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh D., Venkataramani P., Nandi S., Bhattacharjee S. CRISPR-Cas9 a boon or bane: The bumpy road ahead to cancer therapeutics 06 Biological Sciences 0604 Genetics. Cancer Cell Int. 2019;19:1–10. doi: 10.1186/s12935-019-0726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai Y., Chen L., Sun S., Wu C., Yao W., Jiang B., Han T., Hou W. CRISPR/Cas9-mediated deletion of large genomic fragments in Soybean. Int. J. Mol. Sci. 2018;19:3835. doi: 10.3390/ijms19123835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H., Liu B., Weeks D.P., Spalding M.H., Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M., Rehman S., Tang X., Gu K., Fan Q., Chen D., Ma W. Methodologies for improving HDR efficiency. Front. Genet. 2019;10:691. doi: 10.3389/fgene.2018.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Vu T., Sung Y.W., Kim J., Doan D.T., Tran M.T., Kim J.Y. Challenges and Perspectives in Homology-Directed Gene Targeting in Monocot Plants. Rice. 2019;12:1–29. doi: 10.1186/s12284-019-0355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Ge X., Yang F., Zhang L., Zheng J., Tan X., Jin Z.B., Qu J., Gu F. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalinina N.O., Khromov A., Love A.J., Taliansky M.E. CRISPR Applications in Plant Virology: Virus Resistance and Beyond. Phytopathology. 2020;110:18–28. doi: 10.1094/PHYTO-07-19-0267-IA. [DOI] [PubMed] [Google Scholar]
- 35.Manghwar H., Li B., Ding X., Hussain A., Lindsey K., Zhang X., Jin S. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020;7:1902312. doi: 10.1002/advs.201902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Massel K., Godwin I.D., Gao C. Applications and potential of genome editing in crop improvement 06 Biological Sciences 0604 Genetics 06 Biological Sciences 0607 Plant Biology 07 Agricultural and Veterinary Sciences 0703 Crop and Pasture Production. Genome Biol. 2018;19:210. doi: 10.1186/s13059-018-1586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma X., Zhu Q., Chen Y., Liu Y.G. CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Mol. Plant. 2016;9:961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y., Geng L., Yuan M., Wei J., Jin C., Li M., Yu K., Zhang Y., Jin H., Wang E., et al. Deletion of a target gene in Indica rice via CRISPR/Cas9. Plant Cell Rep. 2017;36:1333–1343. doi: 10.1007/s00299-017-2158-4. [DOI] [PubMed] [Google Scholar]
- 39.Cai Y., Chen L., Liu X., Guo C., Sun S., Wu C., Jiang B., Han T., Hou W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 2018;16:176–185. doi: 10.1111/pbi.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soyk S., Müller N.A., Park S.J., Schmalenbach I., Jiang K., Hayama R., Zhang L., Van Eck J., Jiménez-Gómez J.M., Lippman Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017;49:162–168. doi: 10.1038/ng.3733. [DOI] [PubMed] [Google Scholar]
- 41.Peng A., Chen S., Lei T., Xu L., He Y., Wu L., Yao L., Zou X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017;15:1509–1519. doi: 10.1111/pbi.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 43.Kang B.C., Yeam I., Jahn M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005;43:581–621. doi: 10.1146/annurev.phyto.43.011205.141140. [DOI] [PubMed] [Google Scholar]
- 44.Salgotra R.K., Neal Stewart C. Functional markers for precision plant breeding. Int. J. Mol. Sci. 2020;21:4792. doi: 10.3390/ijms21134792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agrios G.N. Plant Pathology. 5th ed. Elsevier Academic Press; Burlington, MA, USA: 2005. [Google Scholar]
- 46.Tripathi J.N., Ntui V.O., Ron M., Muiruri S.K., Britt A., Tripathi L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019;2:46. doi: 10.1038/s42003-019-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baltes N.J., Hummel A.W., Konecna E., Cegan R., Bruns A.N., Bisaro D.M., Voytas D.F. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants. 2015;1:15145. doi: 10.1038/nplants.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji X., Si X., Zhang Y., Zhang H., Zhang F., Gao C. Conferring DNA virus resistance with high specificity in plants using virus-inducible genome-editing system. Genome Biol. 2018;19:197. doi: 10.1186/s13059-018-1580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez M.A., Lin Z.D., Moll T., Chauhan R.D., Renninger K., Beyene G., Taylor N.J., Carrington J., Staskawicz B., Bart R. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. bioRxiv. 2017:209874. doi: 10.1111/pbi.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H., Soyars C.L., Li J., Fei Q., He G., Peterson B.A., Meyers B.C., Nimchuk Z.L., Wang X. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct. 2018;2:e00047. doi: 10.1002/pld3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang T., Zheng Q., Yi X., An H., Zhao Y., Ma S., Zhou G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018;16:1415–1423. doi: 10.1111/pbi.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandrasekaran J., Brumin M., Wolf D., Leibman D., Klap C., Pearlsman M., Sherman A., Arazi T., Gal-On A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016;17:1140–1153. doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macovei A., Sevilla N.R., Cantos C., Jonson G.B., Slamet-Loedin I., Čermák T., Voytas D.F., Choi I.R., Chadha-Mohanty P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018;16:1918–1927. doi: 10.1111/pbi.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pyott D.E., Sheehan E., Molnar A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 2016;17:1276–1288. doi: 10.1111/mpp.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali Z., Ali S., Tashkandi M., Zaidi S.S., Mahfouz M.M. CRISPR/Cas9-Mediated Immunity to Geminiviruses: Differential Interference and Evasion. Sci. Rep. 2016;6:26912. doi: 10.1038/srep26912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tashkandi M., Ali Z., Aljedaani F., Shami A., Mahfouz M.M. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 2018;13:e1525996. doi: 10.1080/15592324.2018.1525996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ali Z., Abulfaraj A., Idris A., Ali S., Tashkandi M., Mahfouz M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015;16:238. doi: 10.1186/s13059-015-0799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kis A., Hamar É., Tholt G., Bán R., Havelda Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019;17:1004–1006. doi: 10.1111/pbi.13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reddy D.V., Sudarshana M.R., Fuchs M., Rao N.C., Thottappilly G. Genetically engineered virus-resistant plants in developing countries: Current status and future prospects. Adv. Virus Res. 2009;75:185–220. doi: 10.1016/s0065-3527(09)07506-x. [DOI] [PubMed] [Google Scholar]
- 60.Gosavi G., Yan F., Ren B., Kuang Y., Yan D., Zhou X., Zhou H. Applications of CRISPR technology in studying plant-pathogen interactions: Overview and perspective. Phytopathol. Res. 2020;2:1–9. doi: 10.1186/s42483-020-00060-z. [DOI] [Google Scholar]
- 61.Fondong V.N. The search for resistance to cassava mosaic geminiviruses: How much we have accomplished, and what lies ahead. Front. Plant Sci. 2017;8:408. doi: 10.3389/fpls.2017.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houngue J.A., Zandjanakou-Tachin M., Ngalle H.B., Pita J.S., Cacaï G.H., Ngatat S.E., Bell J.M., Ahanhanzo C. Evaluation of resistance to cassava mosaic disease in selected African cassava cultivars using combined molecular and greenhouse grafting tools. Physiol. Mol. Plant Pathol. 2019;105:47–53. doi: 10.1016/j.pmpp.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsao T.T. Ph.D. Thesis. Queensland University of Technology; Brisbane, Australia: 2008. Towards the Development of Transgenic Banana Bunchy Top Virus (BBTV)-Resistant Banana Plants: Interference with Replication. [Google Scholar]
- 64.Jarošová J., Singh K., Chrpová J., Kundu J.K. Analysis of Small RNAs of Barley Genotypes Associated with Resistance to Barley Yellow Dwarf Virus. Plants. 2020;9:60. doi: 10.3390/plants9010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choudhury S., Larkin P., Xu R., Hayden M., Forrest K., Meinke H., Hu H., Zhou M., Fan Y. Genome wide association study reveals novel QTL for barley yellow dwarf virus resistance in wheat. BMC Genom. 2019;20:89. doi: 10.1186/s12864-019-6249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pasev G., Kostova D., Sofkova S. Identification of Genes for Resistance to Bean Common Mosaic Virus and Bean Common Mosaic Necrosis Virus in Snap Bean ( Phaseolus vulgaris L.) Breeding Lines Using Conventional and Molecular Methods. J. Phytopathol. 2014;162:19–25. doi: 10.1111/jph.12149. [DOI] [Google Scholar]
- 67.Zhao Y., Yang X., Zhou G., Zhang T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020;18:328–336. doi: 10.1111/pbi.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Velez J.J., Bassett M.J., Beaver J.S., Molina A. Inheritance of resistance to bean golden mosaic virus in common bean. J. Am. Soc. Hortic. Sci. 1998;123:628–631. doi: 10.21273/JASHS.123.4.628. [DOI] [Google Scholar]
- 69.Blair M.W., Rodriguez L.M., Pedraza F., Morales F., Beebe S. Genetic mapping of the bean golden yellow mosaic geminivirus resistance gene bgm-1 and linkage with potyvirus resistance in common bean (Phaseolus vulgaris L.) Theor. Appl. Genet. 2007;114:261–271. doi: 10.1007/s00122-006-0428-6. [DOI] [PubMed] [Google Scholar]
- 70.Grimmer M.K., Bean K.M., Qi A., Stevens M., Asher M.J. The action of three Beet yellows virus resistance QTLs depends on alleles at a novel genetic locus that controls symptom development. Plant Breed. 2008;127:391–397. doi: 10.1111/j.1439-0523.2008.01515.x. [DOI] [Google Scholar]
- 71.Vigne E., Komar V., Fuchs M. Field safety assessment of recombination in transgenic grapevines expressing the coat protein gene of Grapevine fanleaf virus. Transgenic Res. 2004;13:165–179. doi: 10.1023/B:TRAG.0000026075.79097.c9. [DOI] [PubMed] [Google Scholar]
- 72.German-Retana S., Walter J., Le Gall O. Lettuce mosaic virus: From pathogen diversity to host interactors. Mol. Plant Pathol. 2008;9:127–136. doi: 10.1111/j.1364-3703.2007.00451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones M.W., Redinbaugh M.G., Louie R. The Mdm1 locus and maize resistance to Maize dwarf mosaic virus. Plant Dis. 2007;91:185–190. doi: 10.1094/PDIS-91-2-0185. [DOI] [PubMed] [Google Scholar]
- 74.Nair S.K., Babu R., Magorokosho C., Mahuku G., Semagn K., Beyene Y., Das B., Makumbi D., Lava Kumar P., Olsen M., et al. Fine mapping of Msv1, a major QTL for resistance to Maize Streak Virus leads to development of production markers for breeding pipelines. Theor. Appl. Genet. 2015;128:1839–1854. doi: 10.1007/s00122-015-2551-8. [DOI] [PubMed] [Google Scholar]
- 75.Zuriaga E., Romero C., Blanca J.M., Badenes M.L. Resistance to Plum Pox Virus (PPV) in apricot (Prunus armeniaca L.) is associated with down-regulation of two MATHd genes. BMC Plant Biol. 2018;18:25. doi: 10.1186/s12870-018-1237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carneiro O.L., Ribeiro S.R., Moreira C.M., Guedes M.L., Lyra D.H., Pinto C.A. Introgression of the Rladg allele of resistance to potato leafroll virus in Solanum tuberosum L. Crop. Breed. Appl. Biotechnol. 2017;17:236–243. doi: 10.1590/1984-70332017v17n3a37. [DOI] [Google Scholar]
- 77.Bendahmane A., Köhm B.A., Dedi C., Baulcombe D.C. The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 1995;8:933–941. doi: 10.1046/j.1365-313X.1995.8060933.x. [DOI] [PubMed] [Google Scholar]
- 78.Torrance L., Cowan G.H., McLean K., MacFarlane S., Al-Abedy A.N., Armstrong M., Lim T.Y., Hein I., Bryan G.J. Natural resistance to Potato virus Y in Solanum tuberosum Group Phureja. Theor. Appl. Genet. 2020;133:967–980. doi: 10.1007/s00122-019-03521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ganesan U., Suri S.S., Rajasubramaniam S., Rajam M.V., Dasgupta I. Transgenic expression of coat protein gene of Rice tungro bacilliform virus in rice reduces the accumulation of viral DNA in inoculated plants. Virus Genes. 2009;39:113–119. doi: 10.1007/s11262-009-0359-9. [DOI] [PubMed] [Google Scholar]
- 80.Falk B.W., Nouri S. Special issue: “Plant virus pathogenesis and disease control”. Viruses. 2020;12:1049. doi: 10.3390/v12091049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gustafson T.J., de Leon N., Kaeppler S.M., Tracy W.F. Genetic analysis of sugarcane mosaic virus resistance in the Wisconsin diversity panel of maize. Crop. Sci. 2018;58:1853–1865. doi: 10.2135/cropsci2017.11.0675. [DOI] [Google Scholar]
- 82.Widyasari K., Alazem M., Kim K.H. Soybean resistance to soybean mosaic virus. Plants. 2020;9:219. doi: 10.3390/plants9020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pérez-de-Castro A., Esteras C., Alfaro-Fernández A., Daròs J.A., Monforte A.J., Picó B., Gómez-Guillamón M.L. Fine mapping of wmv 1551, a resistance gene to Watermelon mosaic virus in melon. Mol. Breed. 2019;39:1–15. doi: 10.1007/s11032-019-0998-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.