Abstract
Three-dimensional (3D) cell culture systems, such as tumor organoids and multicellular tumor spheroids, have been developed in part as a result of major advances in tissue engineering and biofabrication techniques. 3D cell culture offers great capabilities in drug development, screening, testing, and precision medicine owing to its physiological accuracy. However, since the inception of 3D systems, few methods have been reported to successfully analyze cell viability quantitatively within hydrogel constructs. In this study, we describe and compare commercially available viability assays developed for two-dimensional (2D) applications for use in 3D constructs composed of organic, synthetic, or hybrid hydrogel formulations. We utilized Promega's CellTiter-Glo®, CellTiter-Glo 3D, and CellTiter 96® MTS Assay along with Thermo Fisher's PrestoBlue™ assay to determine if these assays can be used accurately in 3D systems. Compared with direct cell viability commonly used in 2D cell culture, our results show cellular health output inaccuracies among each assay in differing hydrogel formulations. Our results should inform researchers of potential errors when using cell viability measurements in 3D cultures and conclude that microscopic imaging should be used, in combination, for validation.
Impact statement
Three-dimensional (3D) tissue organoids models are a valuable tool not only for studying drug toxicity but also for understanding human embryonic development, intra-tissue morphogenesis, and mechanisms of disease. In cancer organoids, such 3D models may be used for preclinical chemotherapy screening and for understanding cell death and viability mechanisms under physiologically relevant conditions. Cell viability assays are necessary for assessing the effect of biological reagents on cellular health and have been used on in vitro cell cultures for many years. With the increase of 3D systems in cellular biology research to determine therapeutic efficacy, two-dimensional assays that measure cell viability are being used outside their intended use on 3D constructs. In this study, we assess the accuracy of using various commercially available cell viability assays on different 3D hydrogel constructs to help researchers understand expected variability in their experimentation along microscopic imaging validation.
Keywords: viability assay, hydrogel, organoid, 3D culture
Introduction
Cell viability assays are essential for assessing the effect of biological reagents on the health of cells and have been used on in vitro, two-dimensional (2D) monolayer cultures for many years. These assays are common among researchers to evaluate compound toxicity and cell growth effects in the early phases of drug development and discovery. Compared with direct cell counting or automated systems such as the Incuyte® cytotoxicity assay, flow cytometry, or immunohistochemistry (IHC), there are different classifications for indirect viability assays: (1) luminometric assays; (2) fluorometric assays; and (3) colorimetric assays.1
Determining the best method among these assay types is important for obtaining accurate and consistent results. When selecting the cell viability assays to be used, different parameters have to be considered such as detection mechanism, test compounds, desired output data, specificity, cost, and sensitivity. Each cell viability type provides a measurable output to calculate cell health at various time points to gather data such as IC50 values, dose–response curves, growth rates, drug response, and so on.
Recently, advances in cellular biology research have identified limitations in traditional 2D monolayer cell cultures for studying disease and functionality.2 Three-dimensional (3D) cell culture systems such as organoids and multicellular spheroids have been recently developed in biomedical research. Organoids and other 3D constructs consist of cells that are encapsulated in an exogenous extracellular matrix (ECM) or synthetic hydrogel to allow for the preservation of the cell–cell and cell–ECM interactions in multiple dimensions that are necessary for typical cell function.3,4 These 3D models have been used for experimenting on both healthy and cancerous tissues to study targeted therapies, specific cell population interactions, and modeling poorly understood cellular mechanisms. Biomaterial technologies have also led to 3D bioprinting, an automated technique that deposits cells and hydrogels simultaneously, creating 3D organ-like structures for either tissue models for testing or human transplantation.5
Hydrogels are a network of physically or chemically cross-linked polymer molecules that can be designed with a broad range of biological and synthetic materials to recapitulate many features of native ECM.6 A protein-based hydrogel material such as collagen (Col1), the most abundant ECM component in the body, is commonly used in 3D culture systems owing to its biophysical and cell-adhesive properties.7–9 Synthetic material such as polyacrylamide (PA) or polyethylene glycol (PEG) can be incorporated into hydrogels in addition to hyaluronic acid (HA) or polypeptides that have useful biophysical properties, and be chemically modified for cell-adhesion capabilities.10,11 In comparison with naturally derived collagen based, synthetic gels are amenable to chemical modifications to provide a more tunable and stable environment and can even be combined with organic components to form hybrid hydrogels.
With the paradigm shift of cell culture techniques moving from 2D to 3D, cell viability assays need to be assessed for their effectiveness in different hydrogel formulations.12–15 In this study, we tested different types of viability assays, which were originally developed for 2D monocultures, however, being used to determine cellular health in 3D cultures containing different hydrogel formulations.
Methods
Cell culture
Human metastatic colorectal carcinoma cells, HCT-116 (No. CCL-247; ATCC, Manasses, VA), were maintained in Dulbecco's modified Eagle medium (DMEM; Lonza) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were cultured in conditions of 37°C and 5% CO2 and maintained in plastic 15 cm tissue-treated dishes. Cells were cultured to 80–90% confluence before being harvested for use or passaged. Cells were detached from the plates with trypsin/EDTA (Lonza) and resuspended in media at known concentration using a cell counter before further use in studies. Drug treatment was performed by exposing constructs to 1 mM of 5-fluorouracil (5-FU) in dimethyl sulfoxide (DMSO) for 72 h before fixation.
Human patient tumor specimens were collected according to institutional guidelines under RB00040474: Organoid Technology: a patient-specific tumor-on-a-chip platform for determining chemotherapy response prior to the initiation of treatment for abdominal malignancies.
3D constructs and hydrogel formulations
We utilized three hydrogel formulations to test viability assay accuracy on encapsulated HCT-116 cells at a cellular volume of 20,000, 80,000, and 160,000 cells per 10, 25, 50, and 100 μL constructs. HCT-116 cells were trypsinized from 2D culture dishes and counted, then suspended in desired hydrogel formulations described hereunder on polydimethylsiloxane-coated plates (PDMS, DOW Sylgard 184). Forty-eight-well plates were precoated with PDMS by placing ∼100 μL PDMS at the bottom of each well to prevent cell adherence at differing volumes and to create a hydrophobic surface to ensure uniform gel distribution and shape between hydrogels. Cellular shape, although not analyzed in detail, remained round and in small aggregates evenly throughout the hydrogel after the overnight incubation. Each hydrogel at every volume was carefully dispensed to exhibit a semisphere shape. After polymerization or crosslinking, DMEM was added to each well and incubated at 37°C overnight to allow the cells to adhere within the construct.
The biological Type I Rat Tail Collagen (Col1; No. 354236; Corning) hydrogel was prepared per manufacturer protocol at a concentration of 2 mg/mL on ice. When ready, the neutralized Col1 mixture was added to the cells followed by gelation at 37°C for 30 min. The synthetic thiol-modified hyaluronan and gelatin hydrogel (HyStem®; Advanced Biomatrix, San Diego, CA) was prepared per manufacturer protocol with the addition of a photoinitiator. In brief, heprasil (thiolated HA with conjugated heparin groups) and gelin-S (thiolated gelatin) were dissolved in water containing 0.05% (w/v) 2-hydroxy-4′-(2-hydroxyethoxy)-2-thylpropiophenone photoinitiator (Sigma, St. Louis, MO) to make 1% (w/v) solutions. Extralink, a PEGDA crosslinker, was dissolved in water containing the photoinitiator to make a 2% (w/v) solution. Heprasil, gelin-S, and extralink were then mixed with cells in a 2:2:1 ratio by volume, mixed, and irradiated with ultraviolet (UV) light (365 nm, 18 W/cm2) for 2 s at a distance of 3 cm to initiate a thiolene stepwise cross-linking reaction.
Finally, the organic/synthetic hybrid hydrogel, hyaluronic acid:methacrylated Col1 (HA:Col1), was prepared as previously published.16 Methacrylated Col1 (Advanced Biomatrix) was prepared at 6 mg/mL per manufacturer's instructions excluding the provided photoinitiator. Before use with HA, collagen was neutralized using manufacturer provided neutralization solution at 85 μL of solution per 1 mL of collagen. HA was prepared at 2 mg/mL by resuspending Heprasil (heparinized and thiolated HA; ESI BIO, Alameda, CA) in 1 mL deionized water with 0.1% w/v photoinitiator. The neutralized methacrylated Col1 and Heprasil were then added with cells at a 3:1 ratio by volume, mixed, and irradiated with UV light for 2 s at a distance of 3 cm to cross-link.
Viability assays
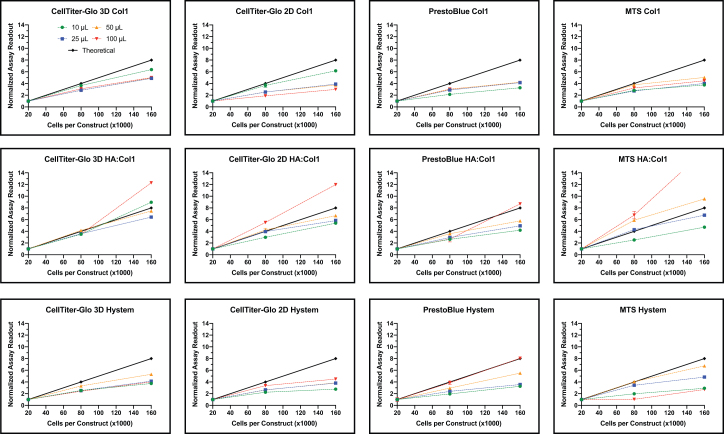
We determined the readout accuracy of four indirect viability assays on the above hydrogel formulations. First, we validated each assay using a serial dilution of HCT-116 cell suspension to identify the standard curve for each test (Supplementary Fig. S1). These results established a theoretical linear slope within the recommended readout range of each assay as a slope of 5.0E-5 when normalized based on the first reading, shown in black (Fig. 1).
FIG. 1.
Cell viability assays across different hydrogel formulation and construct size. Each viability assay (CT3D, left; CT2D, center left; PB, center right; MTS, right) was used to assess viability of HCT-116 cells encapsulated in different hydrogel formulations (Col1, top; HA:Col1, middle; HyStem®, bottom) at various cell numbers and sizes (10 μL, green; 25 μL, blue; 50 μL, orange; 100 μL, red). HCT-116 cells were encapsulated and allowed to equilibrate in culture media before the assays were run per manufacture instructions. The data were normalized based on initial reading for each construct size. A theoretical slope of 5E-05 (black) was calculated based on a cell number-dependent linear output. Experiments were run in duplicates (n = 6, ±SEM). Graphs and calculations were generated with GraphPad Prism. 2D, two-dimensional; 3D, three-dimensional; HA:Col1, hyaluronic acid:methacrylated Col1; PB, PrestoBlue™. Color images are available online.
We decided to use Promega's CellTiter-Glo® (CT2D) and CellTiter-Glo 3D (CT3D) assay that measures ATP as an indicator of viability and generates a luminescent readout that claims to be more sensitive than other methods.17,18 These assays work by lysing the cells with their proprietary lytic component and differ solely by their lytic capabilities. Promega has demonstrated that CT3D assay has a higher ability to lyse within small spheroids and therefore is hypothesized to work best in our studies. CT2D and CT3D assays were used per manufacturer protocol. Reagents and plates were allowed to equilibrate at room temperature before adding 100 μL of reagent to each 48-well plate containing one construct and 100 μL of DMEM. The plates were mixed vigorously for 5 min to induce cell lysis before incubating at room temperature for 25 min to stabilize the luminescent signal. Finally, 100 μL of the solution was transferred to a white opaque-walled 96-well plate to be read on a luminometer (Varioskan™ LUX; Thermo Fisher) as relative luminescence units (RLU) with an integration time of 1 s.
Next, we picked Thermo Fisher's PrestoBlue™ (PB) cell viability reagent as our fluorometric assay. PB works by indirectly determining cell viability through the use of a nontoxic, cell-permeable, and resazurin-based solution that is reduced by metabolically active cells to a fluorescent resorufin.19,20 PB assay was used per manufacturer protocol. In brief, PB reagents were allowed to equilibrate to room temperature before adding 20 μL of cell viability reagent to the 48-well plates containing 180 μL of DMEM and one construct. The plate was then incubated at 37°C for a predetermined 1.5 h protected from light. This time was determined by incubating cells at the highest concentration and determining the upper time limit. Finally, the content of each well was gently mixed and 100 μL was transferred to a black 96-well plate and read for fluorescence at 560 nm excitation and 590 nm emission (Varioskan LUX; Thermo Fisher).
Finally, we used Promega's CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) to identify metabolically active cells through tetrazolium reduction to a colorimetric formazan.21,22 MTS assay was used per manufacturer protocol. The reagent was warmed to room temperature before adding 40 μL to each well containing 200 μL of media and one construct. After 1.5 h of incubation at 37°C, the contents were mixed and 100 μL was placed in a 96-well plate for absorbance reading at 490 nm (Varioskan LUX; Thermo Fisher).
Hydrogel dissociation
Col1 hydrogels were dissociated using an enzymatic cocktail of collagenases to determine if breaking down the hydrogel would increase the viability assay readout. Stock solutions of collagenases type I, III, and IV (Worthington, Lakewood, NJ) were prepared fresh in Hank's balanced salt solution (HBSS; Sigma). The final digestion cocktail was prepared in serum-free DMEM at 25 U/mL of type I collagenase, 100 U/mL of type III collagenase, and 200 U/mL of type IV collagenase. Individual constructs were washed twice in serum-free DMEM before being placed in 100 μL of digestion cocktail in microcentrifuge tubes. The tubes were incubated at 37°C on a gentle shaker for 1 h. After digestion, 100 μL of media with serum was added to the cocktail to inhibit further enzymatic activity along with PB or MTS reagents and continued with incubation. For the CT3D and CT2D assays, reagents were added directly to cells once at room temperature and continued with standard protocol. Assays were determined unaffected by the digestion cocktail by using this method on cell suspensions (not shown).
Pore size measurements
Scanning electron microscope (SEM) was used to image cell-free hydrogels. Methods above were used to create cell-free hydrogels that were then placed into individual 1.5 mL Eppendorf tubes and frozen in −80°C overnight. The hydrogels were lyophilized for 48 h, broken in half, placed onto SEM pucks (now exposed inner hydrogel facing up), and gold sputter coated. Images were taken using SEM (FlexSEM 1000; Hitachi, Tokyo, Japan) at various magnifications. Images were taken of four representative areas within each hydrogel. Images were exported and ImageJ FIJI suite was used to find pore area for each hydrogel.
Rheological measurements and analysis
Hydrogel stiffness was determined using a Discovery HR2 Rheometer (TA Instruments) by applying a sinusoidal strain on the cell-free material. The elastic moduli of the 100 μL constructs were determined through generation of a force–displacement curve through compression testing with a flat, 12 mm, round geometry that was set to compress the organoid and collect force and gap distance measurements every 0.25 s. Samples were discarded after compression. Stress values were generated by dividing force measurements by sample area, determined through digital imaging for each sample individually. Strain values were generated by subtracting the gap distance from the sample height and dividing the total sum by the height. Stress (y-axis) and strain (x-axis) were then plotted to yield a stress–strain curve consisting of two phases: an initial amorphous phase and a subsequent crystalline phase occurring after a curve elbow. Elastic modulus was calculated using the slope of the amorphous phase.
Immunohistochemistry
IHC analysis was performed on construct sections to detect direct cell viability. Constructs were fixed in 4% paraformaldehyde overnight at 4°C, then washed with phosphate-buffered saline (PBS), and stored in 70% ethanol before paraffin processing. Paraffin processing was carried out by slowly dehydrating the constructs in increasing concentrations of isopropyl alcohol for 5-min intervals from 60% to 100%, then xylene for two washes at 10 min each, and finally warm paraffin for three washes for 10 min each before embedding in paraffin. After paraffin processing and embedding, 5 μm sections were cut using a microtome (Leica Microsystems, Inc., Buffalo Grove, IL) and mounted to slides. For all stains, slides were baked for 2 h at 60°C followed by deparaffinization and rehydration. Hematoxylin and eosin (H&E) staining was performed by core facilities at the Wake Forest Institute for Regenerative Medicine (WFIRM).
For IHC, all incubations were performed at room temperature. Antigen retrieval was performed using Proteinase K (DAKO, Carpinteria, CA). Samples were permeabilized with 0.05% Triton-X in PBS for 5 min. Nonspecific antigen blocking was performed using protein block solution (No. ab156024; Abcam, Cambridge, MA) incubation for 30 min. Slides were then incubated with the appropriate primary antibody against Cleaved Caspase-3 (No. 9661; Cell Signaling Technology) and Ki-67 (No. ab16667; Abcam) at 1:400 dilutions in a humidified chamber overnight at 4°C. Slides were then washed and incubated for 1 h with the appropriate secondary antibody (Nos. 20015 and 20111; Biotium, Fremont, CA). Slides were exposed to DAPI for 5 min and mounted with Prolong Gold (Invitrogen) before imaging. Relevant control slides were prepared for each condition and each antibody combination by excluding the primary antibody incubation. Stained slides were imaged utilizing laser excitation and were captured with an Olympus BX63 microscope (Olympus, Center Valley, PA) with an Olympus DP80 camera (Olympus).
IHC images were imported as uncompressed files into VisioPharm software (Broomfield, CO) for analysis and quantification. An application was developed and modified using the VisioPharm software. In brief, a script was written to deconvolve each immunofluorescence or H&E signal, and then isolate the nuclei using the DAPI or hematoxylin stain. After each cell was segmented, a second script was written to deconvolve the fluorescence signal and quantify the cells expressing Caspase-3 and/or Ki-67 markers. A third and final script was written to identify nuclear fragmentation. These results were imported in Microsoft Excel and calculated for significance.
Statistical analysis
All experiments were performed in duplicate or greater. Quantitative results are presented as mean and standard deviation. We plotted each normalized data point along with the corresponding standard error of the mean after subtracting blank (Fig. 1). The slope and coefficient of determination (R2) of the linear regression line was calculated using Microsoft Excel and GraphPad Prism.
Experimental Results
Cell viability assays remain a vital resource in drug discovery and efficacy studies in cellular biology research; however, the evolving field of hydrogels and organoids requires a better assessment of cellular health. Unfortunately, researchers use viability assays developed and optimized for 2D cultures for testing the health of cells in 3D constructs. In this study, we tested different hydrogel formulations (Col1, HA:Col1, HyStem) to determine the readout accuracy of four viability assays (CT3D, CT2D, PB, and MTS) in 3D constructs at various volumes (10, 25, 50, and 100 μL) and differing cell numbers (20,000, 80,000, and 160,000 cells) (Fig. 1).
We plotted each normalized data point along with the corresponding standard error of the mean (Fig. 1). We define “output” as the assay readout that has been normalized based on the lowest cell volume, luminescence for CT3D and CT2D, fluorescence for PB, and absorbance for MTS. For example, a luminescence of 4.5 × 107 RLU (after subtracting the blank) for a 10 μL construct containing 80,000 cells was normalized based on the smallest cellular volume of that construct size (10 μL at 20,000 cells in this case), which was valued at “1” (originally 1.5 × 107 RLU). Therefore, the normalized assay readout for the 80,000 value is now 3.0 (originally 4.5 × 107 RLU). The linear line for each assay and construct size was graphed (Fig. 1). For Col1, we saw CT3D had close association with the theoretical slope, for lower volume constructs where the other three assays exhibited slopes much further from the theoretical. All the assays tested in HA:Col1 hydrogels exhibited slopes in seemingly close proximation to the theoretical line. For HyStem, however, the ATP assays CT3D and CT2D had slopes further from the theoretical in comparison with MTS and PB.
The slope and coefficient of determination (R2) of the linear regression line was calculated and is given in Table 1. An R2 value will determine how close the data are to the fitted regression line and indicate response variable variation. Slope values within 50% of the theoretical slope, ranging from 2.5E-05 to 7.5E-05, are highlighted in italics. The R2 values that are ≥0.99 are highlighted in bold. These highlighted values indicate a slope that is comparable with the theoretical value with low variance and gives a visual representation of the data. The results show that all assay readouts for cells encapsulated in Col1 were lower than the theoretical values; however, all CT3D slopes were within close range of the theoretical slope (Table 1). Similarly, the R2 values followed the same trend with all but the 100 μL constructs tested with CT3D were >0.99 in Col1. The three other viability assays had a much lower output than the theoretical when tested on Col1 constructs.
Table 1.
Output Accuracy of the Different Viability Assays
| CellTiter-3D |
CellTiter-2D |
Presto Blue |
MTS |
|||||
|---|---|---|---|---|---|---|---|---|
| Slope | R2 | Slope | R2 | Slope | R2 | Slope | R2 | |
| Collagen | ||||||||
| 10 μL | 3.813E-05 | 0.9940 | 3.661E-05 | 0.9898 | 1.613E-05 | 0.9930 | 1.922E-05 | 0.9301 |
| 25 μL | 2.758E-05 | 0.9967 | 2.048E-05 | 0.9862 | 2.211E-05 | 0.9617 | 2.120E-05 | 0.9730 |
| 50 μL | 2.798E-05 | 0.9979 | 1.934E-05 | 0.9758 | 2.259E-05 | 0.9512 | 2.813E-05 | 0.9157 |
| 100 μL | 2.836E-05 | 0.9877 | 1.418E-05 | 0.9995 | 2.215E-05 | 0.9404 | 2.421E-05 | 0.9381 |
| HA:collagen | ||||||||
| 10 μL | 5.752E-05 | 0.9833 | 3.137E-05 | 0.9994 | 3.137E-05 | 0.9910 | 2.655E-05 | 0.9997 |
| 25 μL | 3.863E-05 | 0.9970 | 3.391E-05 | 0.9561 | 3.391E-05 | 0.9945 | 4.057E-05 | 0.9736 |
| 50 μL | 4.598E-05 | 0.9954 | 3.999E-05 | 0.9735 | 3.999E-05 | 0.9825 | 6.027E-05 | 0.9725 |
| 100 μL | 8.225E-05 | 0.9486 | 7.851E-05 | 0.9996 | 7.851E-05 | 0.9468 | 1.208E-04 | 0.9908 |
| HyStem® | ||||||||
| 10 μL | 1.927E-05 | 0.9743 | 1.236E-05 | 0.9046 | 1.619E-05 | 0.9999 | 1.364E-05 | 0.9910 |
| 25 μL | 2.226E-05 | 0.9954 | 1.978E-05 | 0.9662 | 1.813E-05 | 0.9827 | 2.683E-05 | 0.9443 |
| 50 μL | 3.049E-05 | 0.9843 | 2.012E-05 | 0.9728 | 3.220E-05 | 0.9999 | 4.066E-05 | 0.9866 |
| 100 μL | 2.141E-05 | 0.9976 | 2.435E-05 | 0.9189 | 5.052E-05 | 0.9992 | 1.269E-05 | 0.8287 |
The slope and R2 values of the linear regression for each viability assay was calculated in Microsoft Excel in different of hydrogel formulations, as indicated. Slope values within 50% of the theoretical slope, ranging from 2.5E-05 to 7.5E-05, are highlighted in italics. The R2 values that are ≥0.99 are highlighted in bold. These values were picked to help visually interpret the data.
2D, two-dimensional; 3D, three-dimensional; HA, hyaluronic acid.
Cells encapsulated in HA:Col1 experienced a more accurate readout to the slope in all assays tested compared with Col1 with all slope values being within range of the theoretically calculated slope except for the 100 μL constructs (Table 1). The calculated R2 of the regression were all >0.94; however, values >0.99 were obtained with different assays and construct sizes. Finally, cells encapsulated in HyStem exhibited lower signal as seen in Col1 constructs (Table 1). Surprisingly, CT2D and CT3D performed poorly with one slope value within 50% of the theoretical.
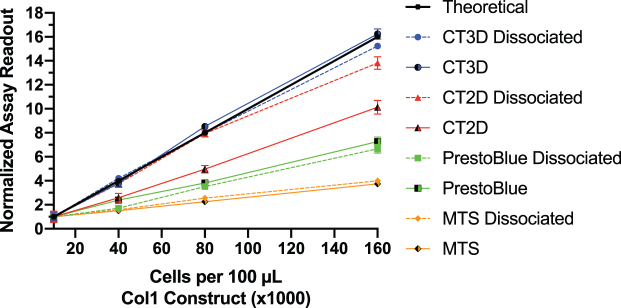
Next, we explored potential explanations for the differential accuracy of the different viability assays in different 3D hydrogels. To do this, we compared the viability of cells inside the 3D hydrogels to cells “freed” from the hydrogel through digestion of the 3D constructs. We chose to use Col1 for these studies, with HCT-116 cells at various cell quantities of 10,000, 40,000, 80,000, and 160,000 in 100 μL constructs (Fig. 2). The cell:hydrogel mixture at the desired concentrations were briefly cultured and then either dissociated gently using collagenase for 1 h or left as a whole construct. To exclude a possibility of an interference owing to the presence of digestion enzymes, HCT-116 cell suspensions were treated with digestion solution and showed no effect on the viability results (not shown).
FIG. 2.
Comparison between assays performed on cells encapsulated in 3D constructs versus cells released through hydrogel digestion. Each cell viability assay was tested on increasing quantities of HCT-116 cells (10,000, 40,000, 80,000, and 160,000) in 100 μL Col1 constructs. The constructs were either kept whole (solid line) or digested (dashed line) with a digestion cocktail of type I collagenase (25 U/mL), type III collagenase (100 U/mL), and type IV collagenase (200 U/mL) for 1 h in serum-free DMEM, immediately before running each assay. The data were normalized based on initial reading at 10,000 cells. A theoretical slope (black) was calculated based on a cell number-dependent linear output. Experiments were carried out in duplicates (n = 6, ±SEM). Graphs and calculations were generated with GraphPad Prism. DMEM, Dulbecco's modified Eagle medium. Color images are available online.
The results show that the CT3D showed near theoretical readings in the 3D constructs and in dissociated cells. CT2D increased accuracy when used on dissociated cells compared with 3D construct. The PB and MTS assays, however, did not improve in accuracy when used on dissociated cells and remained unchanged in readout and slope compared with 3D construct. These results show inaccuracies when using the resazurin and tetrazolium-based assays compared with the ATP-based assays in Col1 construct even after dissociation.
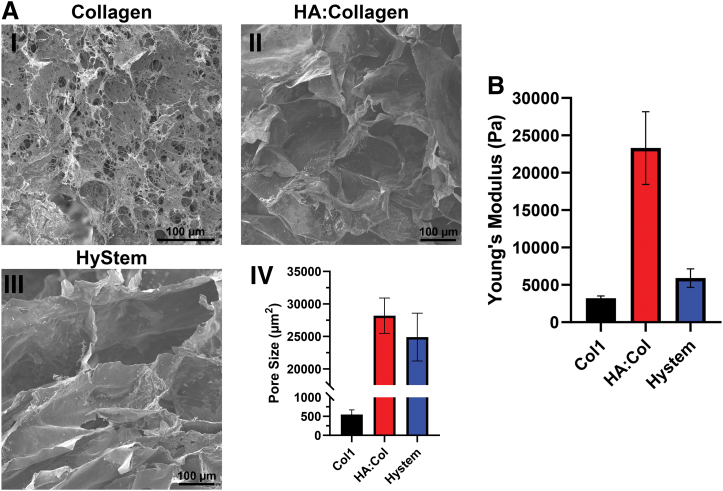
Cell-free hydrogel constructs were analyzed for porosity and stiffness. SEM images of frozen, lyophilized, and broken cell-free constructs were used to determine average pore size and quantified using ImageJ (Fig. 3A.IV). The data show that Col1 constructs had very small pores (∼500 μm2; Fig. 3A.I) in comparison with pores in the crosslinked HA:Col (∼33,000 μm2; Fig. 3A.II) and HyStem (∼25,000 μm2; Fig. 3A.II) constructs. Rheological measurements, however, show that Col1 constructs were the least stiff (∼3,000 Pa), whereas HA:Col has a much higher Young's modulus (∼23,000 Pa) versus HyStem (∼6,000 Pa) (Fig. 3B). Altogether, these results indicate that the smaller pore size in Col1 could be the major factor that is limiting reagent diffusion of the PB and MTS assays, causing them to have decreased detection.
FIG. 3.
Pore size and stiffness of different cell-free hydrogel construct. Scanning electron microscopy was used to capture individual pore sizes of collagen (A.I), HA:collagen (A.II), and HyStem (A.III), quantified by calculating average pore size area using ImageJ and graphed using GraphPad Prism; ±SEM, n = 40 (A.IV). Hydrogel stiffness was determined using a rheometer to calculate the Young's modulus, in Pascal, of each construct formulation and graphed using GraphPad Prism; ±SEM, n = 6 (B). HA, hyaluronic acid. Color images are available online.
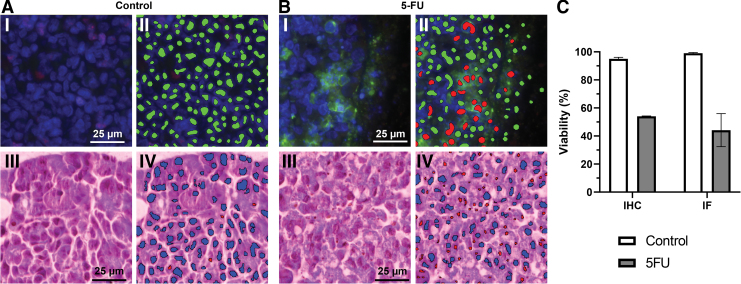
Finally, we performed direct evaluation of cell viability in 3D constructs using histological techniques. Histological assessment not only gives the researcher viability information, but also has the capability of determining where the live and dead cells are spatially in the dense hydrogel. Col1 hydrogel constructs (100 μL) with HCT-116 cells were cultured for 72 h in control media or in the presence of a chemotherapeutic agent, 1 mM 5-FU (Fig. 4). The constructs were fixed, embedded in paraffin, and sections were stained with DAPI (blue), Caspase-3 (green), and Ki-67 (red) (Fig. 4A.I, 4B.I). The stained images were quantified using VisioPharm software that deconvolutes the images (Fig. 4C). The software identified cellular nuclei that are intact and free of caspase-3 (Fig. 4A.II), indicating healthy cells in green, or nuclei that are fragmented and surrounded by caspase-3 (Fig. 4B.II), indicating apoptotic cells in red. These results were validated through quantification of H&E-stained sections of the same Col1 hydrogels (Fig. 4A.III, A.IV, B.III, B.IV). The software successfully detected each intact nucleus as blue and fragmented nucleus as red. This powerful quantification method through histological sections can be useful for determining cellular health in most hydrogel formulation in combination with 3D indirect assays.
FIG. 4.
Direct measurement of cell viability in 3D constructs using histological analyses. HCT-116 cells encapsulated in Col1 hydrogels (100 μL) were cultured for 72 h in standard media (Control, A) or in the presence of 1 mM of 5-5-FU (B). The constructs were embedded in paraffin and 5 μm sections were stained with DAPI (blue), Caspase-3 (green), and Ki-67 (red) as IF (A.I, B.I). Stained images were quantified using VisioPharm software (Broomfield, CO) (A.II, B.II), labeling healthy nuclei (green), and nuclei surrounded by caspase-3 (red). VisioPharm quantification of H&E-stained IHC sections of the same Col1 hydrogels (A.III, A.IV, B.III, B.IV) labeled healthy, intact nuclei (blue) and fragmented nuclei (red). IHC and IF direct viability data were quantified and graphed using VisoPharm analysis as percentage of healthy cells compared with unhealthy cells (Caspase-3 positive or fragmented) (C). Scale bar is the same for all images, 25 μm. Graphed error bars are ±SEM (n = 4). 5-FU, 5-Fluorouracil; H&E, hematoxylin and eosin; IF, immunofluorescent; IHC, immunohistochemistry. Color images are available online.
Discussion
3D cell culture systems have been a focus for culturing tumor cells based on the ability to establish nutrient, waste, and even cellular gradients, among others, for the purpose of creating a more physiologically relevant model of tumor cell growth and chemotherapy treatments.4,23–25 Moreover, when incorporating a hydrogel material, increased cell–cell and cell–ECM interactions will enable embedded cells to act as they would in vivo, thus improving translation to clinical conditions. To establish chemotherapy efficacy in hydrogel systems, multiple indirect cell viability assays have been used and tested as we did in this work, previously establishing large variability between reagents. Many research reports have indicated that ATP-based viability assays are most suitable for 3D spheroid and hydrogel cultures.26–28
In this study, we found varied output inaccuracies among indirect viability assays for different hydrogel formulation (Fig. 1 and Table 1). Promega's CellTiter-Glo 3D, originally designed for small spheroidal models, exhibited the most accurate readouts overall regardless of construct size, cell density, and hydrogel makeup; however, readouts using CT3D were less precise in HyStem hydrogels. Of interest, all four assays fared well in accurately quantifying cell viability in hybrid Col1:HA hydrogel. Furthermore, Promega's MTS assay and Thermo Fisher's PB assay both produced accurate readouts in HyStem hydrogels apart from larger, 100 μL constructs but was very imprecise in Col1 constructs.
Upon further experimentation to determine why the MTS and PB assays performed poorly in the biological Col1 hydrogel, we speculated that the solution either remained “trapped” within the Col1 or the gel interfered with the assay's chemistry even when constructs were digested, and cells freed (Fig. 2). Nevertheless, the Promega's ATP assay designed for 2D monolayers, CT2D, displayed equal viability measurements to CT3D after Col1 digestion indicating the lack of penetration and lytic capabilities of CT2D in hydrogels, which performed the worst of the assays (Fig. 2). The results presented here show major variability between indirect, 2D cell viability assays used in 3D hydrogels and thus we suggest using them with caution when comparing 2D and 3D measurements. However, based on our data, validation using direct methods is essential in cell viability determination embedded in hydrogels, as discussed hereunder.
Specific properties of different hydrogel types can be the cause of poor translation when adapting 2D cellular assays to 3D constructs, and could alter results owing to autofluorescence, porosity, stiffness, and diffusion specific for each hydrogel type.6 Porosity of a material has major influences on reagents diffusing throughout a hydrogel. With low porosity, assay reagents and metabolites secreted from cells can become trapped in the hydrogel, resulting in skewed and lowered readouts.
From our results, we hypothesize that the low porosity of the natural gelation process of collagen may be trapping the reagents in the hydrogel material, causing lower readouts with PB and MTS, whereas the large pores in HA:Col and HyStem could be a major contributor to better readouts using these assays (Fig. 3A). The issue of diffusion could be remedied by simply incubating the hydrogels in the viability reagents for a longer time to improve accuracy; however, this study has not tested that. Nevertheless, our data indicate that when using collagen hydrogels, ATP-based assays with a lytic component would have less interference with the collagen formulation. The authors would also like to highlight that a single, cancerous and immortalized cell line was used to produce the data of the current. Cell type, origin, and age could potentially lead to different results using these assays and hydrogels.
The indirect viability methods discussed thus far have their limitations and should be used with caution; however, these assays are still used owing to the lack of fast, high-throughput approaches to determine cellular proliferation within hydrogels. Direct methods like BrdU incorporation, fluorescence-activated cell sorting (FACS), and Incucyte® automated system that are primarily performed on cell suspensions should be considered for use in hydrogel constructs that can be digested. Unfortunately, these systems, although direct, still cannot indicate spatially where the viable (or not viable) cell population is within the hydrogel. To gain such information, researchers are using calcein AM with ethidium homodimer-1 staining, for example, Live/Dead®.29–31
The Live/Dead reagents can be used to stain living and dead cells directly in the hydrogel with high penetration and imaged using confocal microscopy to produce Z-stack images of the whole construct and analyzed using numerous software, a method with high-throughput potential. This method is only limited by the microscope's working distance and light penetration/detection depth, which renders a large issue with more complex organoid systems. In addition, standard histological analyses of cellular health using IHC or H&E staining has the capability to determine how cells respond to different insults spatially within a construct (Fig. 4). This method eliminates much of the uncertainty presented using indirect viability assays and Live/Dead assays that has microscope and light penetration limitations. However, the histological analyses lack high-throughput capabilities and are laborious. Several researchers and companies have begun confronting this issue with high-throughput live/dead molecular probes and tissue clearing techniques; however, basic colorimetric, fluorescent, and luminescent viability assays are still predominately used today.30,32
The data presented here are merely an indication that determining viability in 3D constructs is more complex than 2D methodologies and should be used in combination with direct microscopic techniques that are readily available.
Conclusions
The use of 3D tumor models has proven a valuable tool not only for preclinical chemotherapy drug screening but also for understanding cell death and viability mechanisms under physiologically similar conditions. Cell viability assays originally developed for 2D monolayer cultures, are now being used outside their intended use, on 3D constructs. This may result in inaccurate and misleading results leading to unintentional wrong conclusions. In this study, we assessed the accuracy of using multiple commercially available 2D assays in different hydrogel constructs and conclude that careful consideration should be made when approaching which viability methodology to use. Pilot studies should be routine in determining which assay to use with the presented 3D model while considering the hydrogel properties. Nevertheless, we recommend validating any indirect viability assay with a direct, microscope technique like Live/Dead or IHC.
Supplementary Material
Acknowledgments
The authors acknowledge WFIRM core facilities for their assistance in histology and imaging. The authors also acknowledge the WFIRM Regenerative Medicine Clinical Center (RMCC) for providing the PB reagent.
Authors' Contributions
A.J.D. performed experiments, analyzed data, wrote the article and designed the project. M.D., S.D.F., and S.S. conceptualized and designed the project, supervised the work, and contributed to writing the article. All authors reviewed the article.
Disclosure Statement
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding Information
The authors acknowledge funding through the USAMRMC/MTEC/ATI award number W81XWH-15-9-0001. The authors also acknowledge the support of the Wake Forest Baptist Comprehensive Cancer Center Tumor Tissue and Pathology Shared Resource supported by the National Cancer Institute's Cancer Center Support Grant award number P30CA012197 and the support of the United Stated-Israel Binational Science Foundation (BSF) Grant No. 2019124. S.D.F. is supported by a grant from the National Institute of Health (T32CA247819).
Supplementary Material
References
- 1. Kamiloglu, S., Sari, G., Ozdal, T., and Capanoglu, E.. Guidelines for cell viability assays. Food Front 1, 332, 2020 [Google Scholar]
- 2. Van Zundert, I., Fortuni, B., and Rocha, S.. From 2D to 3D cancer cell models—the enigmas of drug delivery research. Nanomaterials (Basel) 10, 2236, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Devarasetty, M., Mazzocchi, A.R., and Skardal, A.. Applications of bioengineered 3d tissue and tumor organoids in drug development and precision medicine: current and future. BioDrugs 32, 53, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dominijanni, A., Mazzocchi, A., Shelkey, E., et al. Bioengineered tumor organoids. Curr Opin Biomed Eng 13, 168, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knowlton, S., Onal, S., Yu, C.H., Zhao, J.J., and Tasoglu, S.. Bioprinting for cancer research. Trends Biotechnol 33, 504, 2015 [DOI] [PubMed] [Google Scholar]
- 6. Caliari, S.R., and Burdick, J.A.. A practical guide to hydrogels for cell culture. Nat Methods 13, 405, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charoen, K.M., Fallica, B., Colson, Y.L., Zaman, M.H., and Grinstaff, M.W.. Embedded multicellular spheroids as a biomimetic 3D cancer model for evaluating drug and drug-device combinations. Biomaterials 35, 2264, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dominijanni, A., Devarasetty, M., and Soker, S.. Manipulating the tumor microenvironment in tumor organoids induces phenotypic changes and chemoresistance. iScience 23, 101851, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antoine, E.E., Vlachos, P.P., and Rylander, M.N.. Tunable collagen I hydrogels for engineered physiological tissue micro-environments. PLoS One 10, e0122500, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devarasetty, M., Wang, E., Soker, S., and Skardal, A.. Mesenchymal stem cells support growth and organization of host-liver colorectal-tumor organoids and possibly resistance to chemotherapy. Biofabrication 9, 021002, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Erikson, A., Andersen, H.N., Naess, S.N., Sikorski, P., and Davies Cde, L.. Physical and chemical modifications of collagen gels: impact on diffusion. Biopolymers 89, 135, 2008 [DOI] [PubMed] [Google Scholar]
- 12. Idrees, A., Chiono, V., Ciardelli, G., et al. Validation of in vitro assays in three-dimensional human dermal constructs. Int J Artif Organs 41, 779, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li, L., and LaBarbera, D.V. 3D high-content screening of organoids for drug discovery. In: Comprehensive Medicinal Chemistry III. 2017, p. 388 [Google Scholar]
- 14. Walzl, A., Unger, C., Kramer, N., et al. The resazurin reduction assay can distinguish cytotoxic from cytostatic compounds in spheroid screening assays. J Biomol Screen 19, 1047, 2014 [DOI] [PubMed] [Google Scholar]
- 15. Boehnke, K., Iversen, P.W., Schumacher, D., et al. Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. J Biomol Screen 21, 931, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mazzocchi, A., Devarasetty, M., Huntwork, R., Soker, S., and Skardal, A.. Optimization of collagen type I-hyaluronan hybrid bioink for 3D bioprinted liver microenvironments. Biofabrication 11, 015003, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kleijn, A., Kloezeman, J.J., Balvers, R.K., et al. A systematic comparison identifies an ATP-based viability assay as most suitable read-out for drug screening in glioma stem-like cells. Stem Cells Int 2016, 5623235, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crouch, S.P.M., Kozlowski, R., Slater, K.J., and Fletcher, J.. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods 160, 81, 1993 [DOI] [PubMed] [Google Scholar]
- 19. Xu, M., McCanna, D.J., and Sivak, J.G.. Use of the viability reagent PrestoBlue in comparison with alamarBlue and MTT to assess the viability of human corneal epithelial cells. J Pharmacol Toxicol Methods 71, 1, 2015 [DOI] [PubMed] [Google Scholar]
- 20. Lall, N., Henley-Smith, C.J., De Canha, M.N., Oosthuizen, C.B., and Berrington, D.. Viability reagent, PrestoBlue, in comparison with other available reagents, utilized in cytotoxicity and antimicrobial assays. Int J Microbiol 2013, 420601, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barltrop, J.A., Owen, T.C., Cory, A.H., and Cory, J.G.. 5-(3-Carboxymethoxyphenyl)-2-(4,5-dimethylthiazolyl)-3-(4-sulfophenyl)tetrazolium, inner salt (MTS) and related analogs of 3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT) reducing to purple water-soluble formazans as cell-viability indicators. Bioorg Med Chem Lett 1, 611, 1991 [Google Scholar]
- 22. Cory, A.H., Owen, T.C., Barltrop, J.A., and Cory, J.G.. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun 3, 207, 1991 [DOI] [PubMed] [Google Scholar]
- 23. Drost, J., and Clevers, H.. Organoids in cancer research. Nat Rev Cancer 18, 407, 2018 [DOI] [PubMed] [Google Scholar]
- 24. Khawar, I.A., Park, J.K., Jung, E.S., et al. Three dimensional mixed-cell spheroids mimic stroma-mediated chemoresistance and invasive migration in hepatocellular carcinoma. Neoplasia 20, 800, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lv, D., Hu, Z., Lu, L., Lu, H., and Xu, X.. Three-dimensional cell culture: a powerful tool in tumor research and drug discovery. Oncol Lett 14, 6999, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Forsythe, S.D., Devarasetty, M., Shupe, T., et al. Environmental toxin screening using human-derived 3D bioengineered liver and cardiac organoids. Front Public Health 6, 103, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gantenbein-Ritter, B., Potier, E., Zeiter, S., et al. Accuracy of three techniques to determine cell viability in 3D tissues or scaffolds. Tissue Eng Part C Methods 14, 353, 2008 [DOI] [PubMed] [Google Scholar]
- 28. Kijanska, M., and Kelm, J. In vitro 3D spheroids and microtissues: ATP-based cell viability and toxicity assays. In: Markossian S., Sittampalam G.S., Grossman A., et al., eds. Assay Guidance Manual [Internet]. Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004-. Available at: https://www.ncbi.nlm.nih.gov/books/NBK343426/ [PubMed]
- 29. Forsythe, S., Pu, T., and Skardal, A.. Using organoid models to predict chemotherapy efficacy: the future of precision oncology? Expert Rev Precis Med Drug Dev 4, 317, 2019 [Google Scholar]
- 30. Forsythe, S.D., Sasikumar, S., Moaven, O., et al. Personalized identification of optimal HIPEC perfusion protocol in patient-derived tumor organoid platform. Ann Surg Oncol 27, 4950, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Votanopoulos, K.I., Forsythe, S., Sivakumar, H., et al. Model of patient-specific immune-enhanced organoids for immunotherapy screening: feasibility study. Ann Surg Oncol 27, 1956, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dekkers, J.F., Alieva, M., Wellens, L.M., et al. High-resolution 3D imaging of fixed and cleared organoids. Nat Protoc 14, 1756, 2019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.