Abstract
Human pluripotent stem cells (hPSCs) have generated significant interest in the scientific community based on their potential applications in regenerative medicine. However, numerous research groups have reported a propensity for genomic alterations during hPSC culture that poses concerns for basic research and clinical applications. Work from our laboratory and others has demonstrated that amplification of chromosomal regions is correlated with increased gene expression. To date, the phenotypic association of common genomic alterations remains unclear and is a cause for concern during clinical use. In this study, we focus on trisomy 17 and a list of candidate genes with increased gene expression to hypothesize that overexpressing 17q25 located ARHGDIA will confer selective advantage to hPSCs. HPSC lines overexpressing ARHGDIA exhibited culture dominance in co-cultures of overexpression lines with nonoverexpression lines. Furthermore, during low-density seeding, we demonstrate increased clonality of our ARHGDIA lines against matched controls. A striking observation is that we could reduce this selective advantage by varying the hPSC culture conditions with the addition of ROCK inhibitor (ROCKi). This work is unique in (1) demonstrating a novel gene that confers selective advantage to hPSCs when overexpressed and may help explain a common trisomy dominance, (2) providing a selection model for studying culture conditions that reduce the appearance of genomically altered hPSCs, and (3) aiding in elucidation of a mechanism that may act as a molecular switch during culture adaptation.
Keywords: pluripotent, stem cells, genome instability, self renewal, ARHGDIA, karyotype
Introduction
Human pluripotent stem cells (hPSCs) are defined by their ability to self-renew, undergo tri-lineage differentiation, and maintain a normal karyotype [1]. However, earlier results from our laboratory and others demonstrate a propensity for hPSCs to acquire abnormal genomic signatures upon prolonged propagation [2–5]. Moreover, the presence of genomic alterations can be increased by specific hPSC passaging methodologies [4,6]. Two general approaches for passaging hPSCs involve enzymatic dissociation into single cells or separation into small clumps. The poor survival of hPSCs under single-cell dissociation is well documented [7,8].
Passaging hPSCs as single cells has the following advantages: (1) increased numbers for scale-up, (2) standardization of differentiation protocols, and (3) clonal genetic manipulation [8–11]. Unfortunately, increased genomic instability during single-cell passaging is a primary concern and poor single-cell viability adversely affects protocols and impacts large-scale production [4,12]. To increase hPSC single-cell survival at passaging, many researchers have incorporated small molecules in the culture medium based on inhibiting RHOA-ROCK-pMLC pathway to improve single-cell plating efficiencies and reduce apoptosis [13]. However, use of ROCK inhibitor (ROCKi) in culture medium is contentious, since apoptosis is proposed to purge cultures of cells with genomic damage and chromosomal alterations [14–16].
Strong selective pressure may exist for genomic alterations that increase clonal survival and are antiapoptotic [17,18]. Indeed, the high recurrence of specific genomic species in hPSC culture, such as trisomy 12, 17, and 20, is consistent with strong selective pressure suggesting that in hPSC culture adaptation, comprising of mutation followed by selection, selection is a particularly strong force in the emergence of genomic variants [3]. To date, the phenotype behind common genomic alterations has been unclear, and more specifically, the functionally relevant genes located on these genomic loci causative for culture selection have remained elusive [4].
We take the approach of passaging hPSCs as single cells to (1) reproducibly generate genomic abnormalities for further study, (2) better understand single-cell passaging's influence on genomic instability, and (3) determine conditions that may reduce genomic instability during single-cell passaging. We hypothesize that increasing cell viability will reduce selection of genomic variants and promote propagation of genomically normal hPSCs. This work is unique in demonstrating that increasing expression of ARHGDIA, a gene located on chromosome 17q25, confers selective advantage to hPSCs, and by providing a culture selection model for studying conditions potentially reducing the appearance of genomically altered hPSCs.
Materials and Methods
Cell culture, stem cell characterization, and karyotype analysis
HPSC lines BG01, H1, H9, iPSC (IMR-90) (WiCell Research Institute, Madison, WI), and BG01(v) were maintained on inactivated mouse embryonic fibroblasts (iMEFs) or BD Matrigel in DMEM/F-12, 20% knockout serum replacement, 2 mM l-glutamine, 1% nonessential amino acids, 50 U/mL penicillin, 50 μg/mL streptomycin (all from Gibco/Invitrogen), 0.1 mM beta mercaptoethanol (Sigma), and 4 ng/mL bFGF (Sigma). For BD Matrigel, the medium was conditioned on iMEFs. Cells were enzymatically passaged by sequential dissociation using 1 mg/mL type IV collagenase (Gibco) and 0.05% trypsin-ethylene-diamine tetra-acetic acid (trypsin-EDTA; Invitrogen) or manually passaged by fire-pulled Pasteur pipette.
G-banding, stem cell characterization, and embryoid body (EB) differentiation protocols are previously described [19] and further details can be found in Supplementary Data S1. Before the single-cell dissociation and genomic instability experiments, normal diploid karyotype for BG01, H1, H9, and iPSC (IMR-90) was directly assessed in this study or previously reported by our laboratory as follows: BG01 [3], H1 (Supplementary Table S1), H9 [19], and iPSC (IMR-90) [19]. In accordance with federal regulations regarding the protection of human research subjects (32 CFR 219.101(b)(4)), and due to the fact that the cell lines used were from sources part of the NIH stem cell registry, the VCU Office of Research Compliance determined that the project was exempt from Institutional Review Board (IRB) oversight and human research subjects protection regulations.
Microarray analysis
Statistical analysis was performed in the R environment. Arrays were normalized by cyclic loess and signal intensities summarized by GCRMA. P values were adjusted by Benjamini and Hochberg correction with significance determined at an FDR <0.05. See Supplementary Data S1 for further detail on the arrays, chromosomal distribution, and ontology analysis.
Generation of ARHGDIA overexpression lines
LentiORF ARHGDIA w/Stop Codon (Open Biosystems) was used for the expression construct. The plasmid was purified using Qiagen Maxi Prep. Lentivirus was generated using HEK293 cells with psPAX2 and pMD.2 plasmids. The viral supernatant was concentrated using the Lenti-X concentrator. Lentivirus was added to hPSCs in the presence of polybrene. See Supplementary Data S1 for further detail.
Competition assay
HPSCs (Arg) and hPSC (WT) were maintained independently by manual passaging until use in specific experiments. Upon initial enzymatic passage, cells were 40 μm filtered and seeded as mixed cultures onto Matrigel™ or iMEFs. At each passage, cultures were enzymatically dissociated into single cells, 40 μm filtered, and the hPSC (Arg) percentage quantitated by flow cytometry using the Accuri C6 instrumentation. The percentage of GFP-positive hPSCs was gated against control plots of hPSC (WT) cells. See Supplementary Data S1 for further detail.
Results
Genomic alterations are observed under single-cell passaging
For all experiments conducted, g-banding confirmed a normal karyotype for hPSC lines: BG01, H1, H9, and iPSC (IMR-90), as shown by our laboratory [3,19]. Euploid hPSC lines were manually dissociated as aggregates for replating before initiating our genomic instability experiments. For genomic instability studies each line, H1, H9, iPSC (IMR-90), and BG01(v), was enzymatically (E) dissociated into single cells and seeded onto iMEFs. Prior observations from our laboratory suggest 25 passages are sufficient to observe genomic abnormalities [3], so we initiated g-band karyotyping after this enzymatic passage window (Fig. 1 and Supplementary Table S1).
FIG. 1.
Karyogram of four hPSC lines serially passaged as single cells. H9 line at passage 71 corresponding to E-26 exhibiting trisomy 17 (A). BG01(v) line exhibiting multiple trisomies on 12, 14, 17, 20, and X and a +der(16) (B). hiPSC exhibiting trisomy 17 and gain of X (C). H1 line with a segmental duplication on 15p11.2 (D). hPSC, human pluripotent stem cell.
Karyotyping the BG01(v) line validated the existence of previously reported genomic alterations on chromosomes 12, 14, 17, and X. Under continuous culture, the H9 line was karyotyped at two passages, E-26 and E-105. At E-26, 67% of the karyotyped H9 cell line exhibited trisomy 17, and at E-105, trisomy 17 and 12 were identified, suggesting an accumulation of genomic alterations upon extended culture. The H9 associated trisomy 17 at passage E-105, contained a deletion of p11.2. Trisomy 17 mosaicism of the H9 line at E-26 suggests capture of the initial culture dominance of this genomic species. The iPSC line, at passage E-31, was trisomy for 17 and harbored a gain of X. The H1 line was karyotyped at two enzymatic passage points, E-29 and E-58. While the initial H1, E-29 sample was euploid, at E-58, we found a previously unreported structural gain on chromosome 15 at p11.2.
Candidate gene identification for positive selection of trisomy 17
Culture dominance by variant hPSCs arises from phenotypic selection with genotypic association [17]. Therefore, we performed transcriptome analysis using Affymetrix HGU-133a microarrays to identify transcriptional patterns and candidate genes conferring selective advantage. Differential gene analysis of the trisomy BG01(v) line against the previously reported euploid H1 line [20,21] demonstrated an enrichment of significantly increased genes on the amplified chromosomes compared to the diploid chromosomes, P value = 0.0046, confirming that transcription levels correlate with genomic copy number (Supplementary Fig. S1).
We further sought to determine potential biomarkers. Utilizing four databases Gene Ontology [22], PluriNet [23], Cancer Genome Project [24], and Genomic Instability [25] for a priori information, we collected a list of annotated genes to intersect with our differential gene expression analysis. Approximately 2,031 genes were included, in which we focused on those genes with an FDR <0.05, located on autosomal chromosomes 12, 14, or 17 (Table 1).
Table 1.
Significantly Increased Genes Located on Trisomy Chromosomes in BG01(v) Line
| Symbol | Cytoband | Ontology | Phenotype |
|---|---|---|---|
| KRAS | 12p12.1 | Ras-mediated signal transduction | Cancer |
| SOCS2 | 12q | Antiapoptosis | |
| DYRK2 | 12q15 | DNA damage response, induction of apoptosis | |
| RFC5 | 12q24.2-q24.3 | DNA repair, DNA replication, checkpoint sensor | Genomic instability, self-renewal |
| PSMD9 | 12q24.31-q24.32 | Mitotic cell cycle | |
| POLE2 | 14q21-q22 | DNA repair | |
| GPHN | 14q23.3 | establishment of synaptic specificity at neuromuscular junction | Cancer |
| ALKBH1 | 14q24.3 | DNA dealkylation | |
| DYNC1H1 | 14q32.3-qter | Mitotic spindle organization and biogenesis | |
| USP22 | 17p11.2 | Positive regulation of mitotic cell cycle | |
| RNMTL1 | 17p13.3 | RNA processing | Self-renewal |
| PAFAH1B1 | 17p13.3 | Establishment of mitotic spindle orientation | |
| RPA1 | 17p13.3 | DNA repair, checkpoint signaling | Cancer predisposition, genomic instability, self-renewal |
| NF1 | 17q11.2 | Positive regulation apoptosis, negative regulation of cell proliferation | Cancer |
| TIAF1 | 17q11.2 | Antiapoptosis | |
| PSMD11 | 17q11.2 | Mitotic cell cycle | Self-renewal |
| COL1A1 | 17q21.33 | extracellular matrix | Cancer |
| RAD51C | 17q22-q23 | DNA repair, HR | Genomic instability |
| SOX9 | 17q24.3-q25.1 | Apoptosis, Cell proliferation | |
| SEP9 | 17q25 | Cell cycle, GCPR signal transduction | Cancer |
| EXOC7 | 17q25.1 | Centriolar satellite | |
| ARHGDIA | 17q25.3 | Antiapoptosis |
Significant genes located on chromosomes 12, 14, or 17 with an FDR <0.05. A gene's ontology is bold if part of genomic instability [22] or an annotation of interest from Gene Ontology. Self- renewal genes are from the PluriNet gene set and phenotype cancer from the Cancer Genome Project.
HR, homologous recombination.
Chromosome 17 is one of the most common genomic abnormalities in hPSCs, occurring in vitro during hPSC culture and in vivo in human embryonic carcinoma cells [26]. In our experiments, three hPSC lines [BG01(v), H9, and iPSC] exhibited trisomy 17 during long-term propagation. In addition, published reports suggest that 17q25 may be a minimal amplicon for genomically altered hPSCs [17]. Interestingly, 17q25 is reported to be the only species conserved genomic amplification between homo sapiens and syntenic locus of mus musculus and rhesus macaque [27]. Therefore, we focused on the significant genes located on 17q25.
Of the three overexpressed genes, SEPTIN 9, EXO7, and ARHGDIA, ARHGDIA caught our attention for its established role in the RHO-ROCK pathway [12,13,28]. ROCKi is commonly used to reduce dissociation-induced cell death resulting from loss of e-cadherin-mediated cell-cell contact [8]. ARHGDIA inhibits the activation of RHOA by preventing the GDP exchange for GTP [29]. Since RHOA activation is necessary for ROCK activation, we hypothesized that overexpression of ARHGDIA would reduce activation of RHOA and therefore lead to increased single-cell survival conferring selective advantage to hPSCs (Supplementary Fig. S2).
ARHGDIA-transduced cell lines maintain pluripotency
To test our selective advantage hypothesis, we generated, by lentiviral transduction, two hPSC lines, H9 (Arg) and BG01 (Arg), which constitutively overexpress ARHGDIA, and similarly generated matched controls for GFP reporter only, H9 (GFP) and BG01 (GFP). Both hPSC (Arg) lines demonstrate increased ARHGDIA expression by quantitative polymerase chain reaction (qPCR) and Western blot, validating our experimental system (Supplementary Figs. S8 and S9). By densitometry, fold increase on ARHGDIA protein abundance was 3.74 for BG01 (Arg) and 10.52 for H9 (Arg). In the variant lines, the comparative increase in ARHGDIA levels for the BG01(v) and H9 (v) lines is 2.68 and 2.12, respectively.
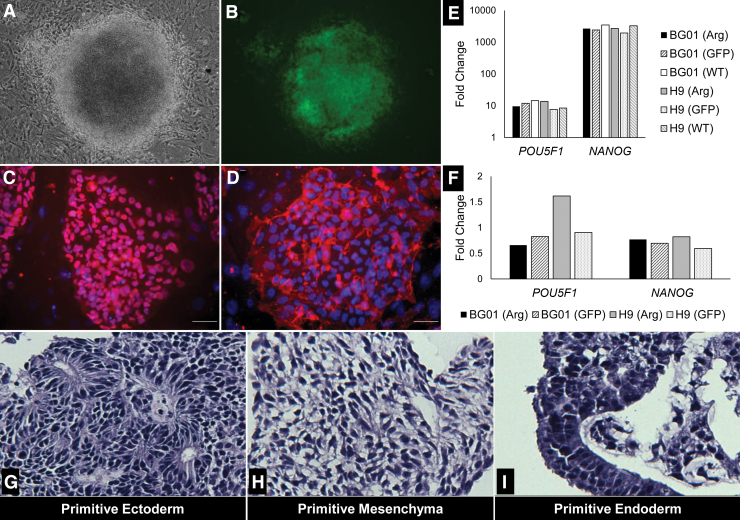
To confirm that ARHGDIA overexpression did not adversely influence self-renewal and differentiation, we characterized our hPSC (Arg) lines for pluripotency. Our transduced lines exhibit typical hPSC cobblestone, colony morphology on iMEFs (Fig. 2A, B) and Matrigel (data not shown). Interestingly, the H9 (Arg) colonies on iMEFs appeared to have increased cell-cell contact, demonstrated by greater multilayer colony density (Supplementary Fig. S3). After 30 passages, the hPSC (Arg) lines exhibited by immunocytochemistry positive protein expression for characteristic stem cell markers, OCT4 and SSEA4 (Fig. 2C, D and Supplementary Figs. S4 and S5). When comparing gene expression by qPCR of OCT4 and NANOG in hPSC (Arg) lines against human dermal fibroblasts (hDFs), we observed fold changes of one and two orders of magnitude, respectively. Across hPSC lines, the fold change patterns against the hDFs were similar. Quantitative PCR of NANOG and the OCT4 gene POU5F1 in H9 (Arg) and BG01 (Arg) relative to hPSC (GFP), hPSC (WT), and hDF controls confirms self-renewal of these lines (Fig. 2E, F). Through EB formation, we validated by histopathology that H9 (Arg) EBs stained for all three primitive germ layers, indicating hPSC (Arg) lines retained their differentiation capacity (Fig. 2G–I). Taken together, these results indicate ARHGDIA overexpression does not adversely affect pluripotency.
FIG. 2.
HPSCs overexpressing ARHGDIA demonstrate hallmark pluripotency. H9 (Arg) exhibits dense and tightly packed standard pluripotent stem cell colony formation (A) and concomitant GFP signal indicating positive ARHGDIA overexpression (B); magnification is 4 × . Positive nuclear expression of OCT4 and cell surface marker SSEA4, (C, D), respectively; DAPI-blue (C, D), OCT4-red (C), SSEA4-red (D); Scale bar = 50 μm. HPSC transcription factor gene expression relative to human dermal fibroblast (E). hPSC (Arg) experimental and HPSC (GFP) controls were validated for hPSC transcription factor gene expression against WT lines (F). For each cell line, n = 3 (E, F). Hematoxylin and eosin-stained histologic sections of EBs from H9 (Arg) lines. The trilineage differentiation is indicative of pluripotency (G–I). Magnification is 20 × . Arg, ARHGDIA; EBs, embryoid bodies; GFP, green fluorescent protein; WT, wild type. Color images available.
ARHGDIA confers selective advantage and increases clonality
For the H9 (Arg) line, we used two assays to test our hypothesis that ARHGDIA overexpression will improve hPSCs cultured as single cells: the first, a competition-based assay, consisted of co-cultures of cells overexpressing ARHGDIA versus nonoverexpressing cells in which the transduced ARHGDIA cells co-express GFP. The second assay tested clonality of single cells seeded at low density.
Competition co-culture experiments were performed on two independent H9 (Arg) sublines, H9 (Arg) s.1 and H9 (Arg) s.2. For each subline, the competition-based assay was initiated as a mixture of H9 [Arg(+)] and H9 [Arg(−)] co-cultures, discriminated by GFP co-expression. At each passage, the percentage of GFP-positive cells was measured by flow cytometry. Both H9 (Arg) s.1 and H9 (Arg) s.2 sublines exhibited strong competitive advantage when passaged as single cells. The initial subpopulations of H9 (Arg+) s.1 and H9 (Arg+) s.2 were 53.2% and 42.0%, respectively. After serial, single-cell passaging, the total H9 [Arg(+)] percentages for the H9 (Arg) s.1 and H9 (Arg) s.2 lines reached 90.6% and 93.5%, respectively (Supplementary Figs. S6 and S7).
Next, we determined whether H9 (Arg) cells exhibited increased clonality. H9 (Arg), H9 (GFP), and H9 (WT) were manually passaged until clonal analysis. Clonality was tested on the first enzymatic passage to control for selective pressure. On day 7, colonies were alkaline phosphatase (AP) stained to aid visualization and validate pluripotency [30]. The observed average colony numbers for H9 (Arg), H9 (GFP), and H9 (WT) are 125.66, 31.44, and 42.83, respectively (Fig. 3). Therefore, clonality of H9 (Arg) cells increased 4.0- and 2.93-fold relative to H9 (GFP) and H9 (WT) with P values of 0.0023 and 0.0004, respectively. Together, the competition and clonality experiments support the conclusion that H9 (Arg) cells have competitive advantage over H9 (WT) cells.
FIG. 3.
ARHGDIA overexpression increases clonality. H9 control and experimental lines were continually propagated by manual dissection before clonal survival analysis. H9s were plated at 100 cells/cm2 under initial enzymatic dissociation and 40 μm filtering. Colonies were AP stained and counted on day 7. H9 (Arg) line overexpressing ARHGDIA has significantly increased number of colonies in low-density single-cell seeding. *Indicates significance (P < 0.01) for H9 (Arg) compared to H9 (WT) and H9 (GFP): N = 10, N = 8, and N = 9, respectively. AP, alkaline phosphatase.
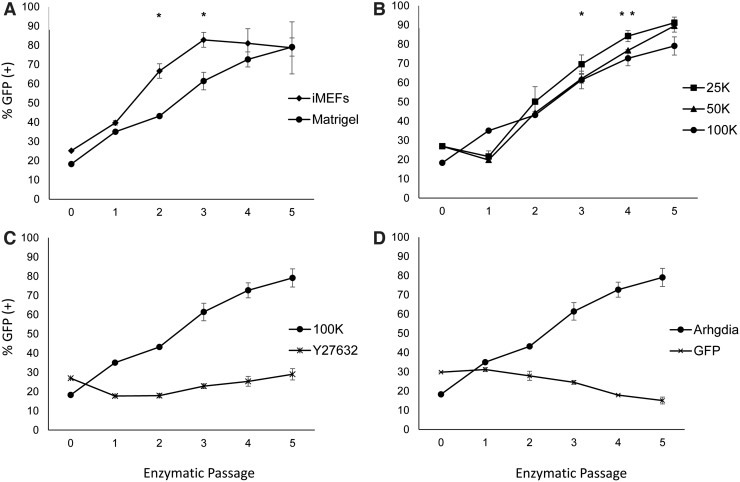
We repeated our competition-based assay on BG01 (Arg) cells to test variable culture conditions, including substrates (i-MEFS and BD Matrigel, seeding density (25K, 50K, and 100K per 35 mm), and ROCKi exposure. Competitive advantage of BG01 (Arg) cells in mixed co-cultures with BG01 (WT) is strikingly observed. In contrast, co-culture experiments of BG01 (GFP) against BG01 (WT) did not exhibit competitive advantage. Each co-culture experimental condition was carried out in parallel biological triplicates.
In their feeder-free cultures, Rosler and colleagues report trisomy 20 as the most frequently observed aberration [31], while trisomy 17 is prominent in our hPSC cultures propagated on iMEFs. Thus, we sought to determine whether iMEFs or Matrigel influenced competitive advantage of BG01 (Arg) versus BG01 (WT) co-cultures. Both substrates showed strong selection for BG01 (Arg) cells. In serial seedings at 100K, by E-3, the BG01 (Arg) cells dominated to 82.87% on iMEFs and comparatively on Matrigel increased to 79.11% at E-5. For both E-2 and E-3, the percent BG01 (Arg) is significantly higher in the iMEF cultures compared to Matrigel, P < 0.01 (Fig. 4A). These results suggest more broadly that substrates can influence culture selection of hPSC genomic variants.
FIG. 4.
BG01 (Arg) hESCs demonstrate competitive advantage across substrates and seeding densities and selection is inhibited by ROCKi, Y27632. BG01 (Arg) lines co-cultured with BG01 (WT) have competitive advantage across substrates (A) and seeding densities in 35 mm Petri dishes (B). ROCKi, ameliorated competitive advantage (C) and BG01 (GFP) control did not exhibit competitive advantage (D). ROCKi, ROCK inhibitor.
Next, we investigated seeding density on BG01 (Arg) and BG01 (WT) co-cultures plated at 25K, 50K, and 100K per 35 mm on Matrigel. Since, low single-cell seeding decreases survival of hPSCs possibly by reduced paracrine signaling [32] or increased cell migration distance [33,34], relative competitive advantage of the BG01 (Arg) to the BG01 (WT) may be greater in co-cultures as seeding density decreases. BG01 (Arg) competitive advantage was clearly demonstrated within five enzymatic passages for each seeding density. The maximal BG01 (Arg) percentages for 25K, 50K, and 100K, are 91.19%, 89.52%, and 79.11% (Fig. 4B). The higher percent of BG01 (Arg) in the 25K samples relative to 50K is significant for E-4 (P < 0.05) and is significant relative to 100K for E-4 and E-5 (P < 0.05). This supports the existence of subtle selection pressure at lower seeding densities.
Finally, we tested the impact of ROCKi, given its role in promoting single-cell viability during plating. We hypothesized that ROCKi would reduce the selective advantage of BG01 (Arg) cells relative to BG01 (WT) by increasing the survival of dissociated BG01 (WT) cells. BG01 (Arg) and BG01 (WT) co-cultures were seeded at 100K on Matrigel in the presence of 10 μM Y27632 containing growth medium. Strikingly, when ROCKi was added to the cultures during plating and subsequently withdrawn at the first medium exchange, we did not observe competitive advantage of BG01 (Arg) cells (Fig. 4C). Starting with an initial BG01 (Arg) percentage of 26.97%, by E-5, BG01 (Arg) cells remained similar at 29.02% of culture. This is in stark contrast to our other BG01 (Arg) experiments, in which competitive advantage was demonstrative. We checked whether BG01 (GFP) control line subpopulations displayed competitive advantage and indeed this was not the case (Fig. 4D).
Combined across BG01 (Arg) and H9 (Arg) co-culture and clonality experiments, our results present compelling evidence that ARHGDIA overexpression confers selective advantage to hPSCs through increased single-cell survival, and this selective advantage can be ameliorated by the addition of ROCKi.
Discussion
Trisomy 17 is the most common abnormality observed in our hPSC lines and was scored in the H9, BG01v, and iPSC lines. The recurrent trisomy 12 was also observed in the BG01(v) and H9 lines. NANOG is located on chromosome 12 and increased NANOG expression may confer self-renewal benefit to these populations [35]. Of note, trisomies 12 and 17 are hallmarks of germ cell tumors and the presence of these alterations in hPSCs as well as other known variations poses potential clinical concern [26,36,37]. Enzymatic passage may increase genomic instability more than twofold [4]. We observe disproportionate expression of trisomy 17 under single-cell dissociation potentially indicating that a chromosome 17 gain is associated with a clonal phenotype.
Significantly increased genes on recurrent genomic amplifications are candidates for biological relevance in positive selection. In our analysis, ARHGDIA was the most attractive gene for its location, gene expression, and established role in RHOA signaling [12]. Consistent with trisomy, our study is the first to demonstrate that increased expression of a gene located on chromosome 17 confers selective advantage to hPSCs when passaged as single cells.
ARHGDIA overexpression did not seem to adversely affect self-renewal or differentiation, as ARHGDIA-transduced cultures were maintained for several months and differentiated into EBs comprising all three germ layers. This is consistent with the routine use of ROCKi for hPSC culture; however, some laboratories have begun to report using ROCKi in differentiation protocols [38–41].
In H9 cell lines, we tested two independent approaches for ARHGDIA's influence on survival. The H9 [Arg(+)] cells demonstrated competitive advantage against H9 [Arg(−)] cells. In the H9 clonality assay, single-cell survival was drastically increased in H9 (Arg) cells compared to controls strongly supporting clonality as the phenotypic advantage.
The BG01 (Arg) cells dominated co-cultures in as quickly as three passages in the iMEF condition. Across seeding densities on Matrigel, the BG01 (Arg) cells with the lowest seeding density of 2.5 k/cm2 exhibited the fastest culture dominance. Both hPSC migration promoting cell-cell contact [33] and increased trophic paracrine signaling are thought to increase hPSC viability [32,42]. Either of these factors may have reduced the survival of BG01 (WT) at lower seeding densities with a relative increase in selective advantage of the BG01 (Arg) cells. This poses interesting questions regarding the mechanism in which increased levels of ARHGDIA improves survival during single-cell plating, such as through possibly inhibiting ROCK-associated blebbing, facilitating migration, or in an unidentified cell adhesion factor affecting clonal survival.
Ben-David et al. suggest that ROCKi may reduce the rate of genomic adaptation of hPSCs in culture by reducing selective pressure [43]. However, to date there has not been any report directly testing this hypothesis. Of note, Thompson and colleagues did not observe an increase in hPSC mutation frequency when culturing with Y27632 [16]. Our experiments are the first to provide compelling evidence that ROCKis can be used to reduce the selection of competitive subpopulations.
Interestingly, through live-cell imaging, Barbaric and colleagues elucidate the impact of culture adaptation on alleviating the bottlenecks of colony formation with implications for selective advantage of variant hPSCs [44]. Consistent with the observations in our ROCKi co-culture experiments, Y27632 had negligible prosurvival influence on their aneuploid H7 and H14 cultures. Noteworthy, the H7 and H14 lines used in their study are known for trisomy 17, spanning the region containing ARHGDIA. Thus, our two studies may be uncovering similar mechanisms for culture adaptation that we propose is through the RHOA-ROCK-pMLC pathway. While our study emphasizes single-cell survival, it will be of keen interest to investigate, in further detail, ARHGDIA's potential influence on adhesion, migration, and cell cycle kinetics.
The variability of genomic species and time to emergence poses inherent challenges to statistically assess culture conditions influencing hPSC culture genomic adaptation. Several reports have shown that BCL-XL increases the survival of hPSCs and mediates the selective advantage of 20q11.21 amplicon by increasing expression levels [32,45,46]. Together, ARHGDIA and BCL-XL provide biologically relevant candidates for engineering hPSC lines to study culture conditions influencing positive selection of genomic variants. Such genetically defined lines can be used to reproducibly test small molecules, substrates, and three-dimensional culture systems on amenable time scale.
HPSCs are known for high rates of centrosomal instability [47] and genomically mosaic cultures [48,49] indicating an inherent and appreciable mutational background. Thus, for prolonged propagation of genetically normal hPSCs, improving culture conditions to reduce selective advantage of variants is a particularly attractive strategy.
Conclusions
We observe common trisomy 17 in our hPSC cultures and increased gene expression associated with amplified chromosomes. We demonstrate that increasing gene expression of 17q25-located ARHGDIA confers strong selective advantage to hPSCs under enzymatic passage plated as single cells. Our hPSC (Arg) cells exhibit clear increased single-cell survival in our clonality assay and reproducibly dominate our mixed co-cultures. Overexpression of ARHGDIA does not affect pluripotency. We show that subtle culture conditions influence selective advantage of our genetically modified line. Using Y27632 to improve survival of hPSCs ameliorates clonal disadvantage of wild-type hPSCs and reduces selective advantage of our ARHGDIA hPSC lines.
Supplementary Material
Author Disclosure Statement
No competing financial interests exist.
Funding Information
Funding for this work was provided by NSF-CAREER074556 (R.R.R.).
Supplementary Material
References
- 1. Thomson JA. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147 [DOI] [PubMed] [Google Scholar]
- 2. Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, et al. (2011). Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8:106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mitalipova MM, Rao RR, Hoyer DM, Johnson JA, Meisner LF, Jones KL, Dalton S and Stice SL. (2005). Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol 23:19–20 [DOI] [PubMed] [Google Scholar]
- 4. Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, Baker J, Baker D, Munoz MB, et al. (2011). Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol 29:1132–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Henry MP, Hawkins JR, Boyle J and Bridger JM. (2018). The genomic health of human pluripotent stem cells: genomic instability and the consequences on nuclear organization. Front Genet 9:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imreh MP, Gertow K, Cedervall J, Unger C, Holmberg K, Szoke K, Csoregh L, Fried G, Dilber S, Blennow E and Ahrlund-Richter L. (2006). In vitro culture conditions favoring selection of chromosomal abnormalities in human ES cells. J Cell Biochem 99:508–516 [DOI] [PubMed] [Google Scholar]
- 7. Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, et al. (2010). Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7:225–239 [DOI] [PubMed] [Google Scholar]
- 8. Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A and Ding S. (2010). Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci U S A 107:8129–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ausubel LJ, Lopez PM and Couture LA. (2011). GMP scale-up and banking of pluripotent stem cells for cellular therapy applications. Methods Mol Biol 767:147–159 [DOI] [PubMed] [Google Scholar]
- 10. Hasegawa K, Fujioka T, Nakamura Y, Nakatsuji N and Suemori H. (2006). A method for the selection of human embryonic stem cell sublines with high replating efficiency after single-cell dissociation. Stem Cells 24:2649–2660 [DOI] [PubMed] [Google Scholar]
- 11. Singh AM. (2019). An efficient protocol for single-cell cloning human pluripotent stem cells. Front Cell Dev Biol 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen G, Hou Z, Gulbranson DR and Thomson JA. (2010). Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell 7:240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K and Sasai Y. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 25:681–686 [DOI] [PubMed] [Google Scholar]
- 14. Rocha CR, Lerner LK, Okamoto OK, Marchetto MC and Menck CF. (2013). The role of DNA repair in the pluripotency and differentiation of human stem cells. Mutat Res 752:25–35 [DOI] [PubMed] [Google Scholar]
- 15. Hong Y and Stambrook PJ. (2004). Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci U S A 101:14443–14448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson O, von Meyenn F, Hewitt Z, Alexander J, Wood A, Weightman R, Gregory S, Krueger F, Andrews S, et al. (2020). Low rates of mutation in clinical grade human pluripotent stem cells under different culture conditions. Nat Commun 11:1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H and Andrews PW. (2007). Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25:207–215 [DOI] [PubMed] [Google Scholar]
- 18. Merkle FT, Ghosh S, Kamitaki N, Mitchell J, Avior Y, Mello C, Kashin S, Mekhoubad S, Ilic D, et al. (2017). Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545:229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abraham S, Sheridan SD, Miller B and Rao RR. (2010). Stable propagation of human embryonic and induced pluripotent stem cells on decellularized human substrates. Biotechnol Prog 26:1126–1134 [DOI] [PubMed] [Google Scholar]
- 20. Sato N, Sanjuan IM, Heke M, Uchida M, Naef F and Brivanlou AH. (2003). Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol 260:404–413 [DOI] [PubMed] [Google Scholar]
- 21. Rao RR, Calhoun JD, Qin X, Rekaya R, Clark JK and Stice SL. (2004). Comparative transcriptional profiling of two human embryonic stem cell lines. Biotechnol Bioeng 88:273–286 [DOI] [PubMed] [Google Scholar]
- 22. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, et al. (2008). Regulatory networks define phenotypic classes of human stem cell lines. Nature 455:401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaiser J. (2005). National Institutes of Health. NCI gears up for cancer genome project. Science 307:1182. [DOI] [PubMed] [Google Scholar]
- 25. Aguilera A and Gomez-Gonzalez B. (2008). Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9:204–217 [DOI] [PubMed] [Google Scholar]
- 26. Harrison NJ, Baker D and Andrews PW. (2007). Culture adaptation of embryonic stem cells echoes germ cell malignancy. Int J Androl 30:275–281; discussion 281. [DOI] [PubMed] [Google Scholar]
- 27. Ben-David U and Benvenisty N. (2012). High prevalence of evolutionarily conserved and species-specific genomic aberrations in mouse pluripotent stem cells. Stem Cells 30:612–622 [DOI] [PubMed] [Google Scholar]
- 28. Harb N, Archer TK and Sato N. (2008). The Rho-Rock-Myosin signaling axis determines cell-cell integrity of self-renewing pluripotent stem cells. PLoS One 3:e3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DerMardirossian C and Bokoch GM. (2005). GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol 15:356–363 [DOI] [PubMed] [Google Scholar]
- 30. O'Connor MD, Kardel MD, Iosfina I, Youssef D, Lu M, Li MM, Vercauteren S, Nagy A and Eaves CJ. (2008). Alkaline phosphatase-positive colony formation is a sensitive, specific, and quantitative indicator of undifferentiated human embryonic stem cells. Stem Cells 26:1109–1116 [DOI] [PubMed] [Google Scholar]
- 31. Rosler ES, Fisk GJ, Ares X, Irving J, Miura T, Rao MS and Carpenter MK. (2004). Long-term culture of human embryonic stem cells in feeder-free conditions. Dev Dyn 229:259–274 [DOI] [PubMed] [Google Scholar]
- 32. Bai H, Chen K, Gao YX, Arzigian M, Xie YL, Malcosky C, Yang YG, Wu WS and Wang ZZ. (2012). Bcl-xL enhances single-cell survival and expansion of human embryonic stem cells without affecting self-renewal. Stem Cell Res 8:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li L, Wang BH, Wang S, Moalim-Nour L, Mohib K, Lohnes D and Wang L. (2010). Individual cell movement, asymmetric colony expansion, rho-associated kinase, and E-cadherin impact the clonogenicity of human embryonic stem cells. Biophys J 98:2442–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodin S, Antonsson L, Niaudet C, Simonson OE, Salmela E, Hansson EM, Domogatskaya A, Xiao Z, Damdimopoulou P, et al. (2014). Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat Commun 5:3195. [DOI] [PubMed] [Google Scholar]
- 35. Yoshihara M, Hayashizaki Y and Murakawa Y. (2017). Genomic instability of iPSCs: challenges towards their clinical applications. Stem Cell Rev Rep 13:7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hovatta O, Jaconi M, Tohonen V, Bena F, Gimelli S, Bosman A, Holm F, Wyder S, Zdobnov EM, et al. (2010). A teratocarcinoma-like human embryonic stem cell (hESC) line and four hESC lines reveal potentially oncogenic genomic changes. PLoS One 5:e10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Almstrup K, Hoei-Hansen CE, Wirkner U, Blake J, Schwager C, Ansorge W, Nielsen JE, Skakkebaek NE, Rajpert-De Meyts E and Leffers H. (2004). Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res 64:4736–4743 [DOI] [PubMed] [Google Scholar]
- 38. Cheng YT, Yeih DF, Liang SM, Chien CY, Yu YL, Ko BS, Jan YJ, Kuo CC, Sung LY, et al. (2015). Rho-associated kinase inhibitors promote the cardiac differentiation of embryonic and induced pluripotent stem cells. Int J Cardiol 201:441–448 [DOI] [PubMed] [Google Scholar]
- 39. Maldonado M, Luu RJ, Ramos ME and Nam J. (2016). ROCK inhibitor primes human induced pluripotent stem cells to selectively differentiate towards mesendodermal lineage via epithelial-mesenchymal transition-like modulation. Stem Cell Res 17:222–227 [DOI] [PubMed] [Google Scholar]
- 40. Vernardis SI, Terzoudis K, Panoskaltsis N and Mantalaris A. (2017). Human embryonic and induced pluripotent stem cells maintain phenotype but alter their metabolism after exposure to ROCK inhibitor. Sci Rep 7:42138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aoki H, Yamashita M, Hashita T, Ogami K, Hoshino S, Iwao T and Matsunaga T. (2020). Efficient differentiation and purification of human induced pluripotent stem cell-derived endothelial progenitor cells and expansion with the use of inhibitors of ROCK, TGF-beta, and GSK3beta. Heliyon 6:e03493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Przybyla L and Voldman J. (2012). Probing embryonic stem cell autocrine and paracrine signaling using microfluidics. Annu Rev Anal Chem (Palo Alto Calif) 5:293–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ben-David U, Benvenisty N and Mayshar Y. (2010). Genetic instability in human induced pluripotent stem cells: classification of causes and possible safeguards. Cell Cycle 9:4603–4604 [DOI] [PubMed] [Google Scholar]
- 44. Barbaric I, Biga V, Gokhale PJ, Jones M, Stavish D, Glen A, Coca D and Andrews PW. (2014). Time-lapse analysis of human embryonic stem cells reveals multiple bottlenecks restricting colony formation and their relief upon culture adaptation. Stem Cell Rep 3:142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Avery S, Hirst AJ, Baker D, Lim CY, Alagaratnam S, Skotheim RI, Lothe RA, Pera MF, Colman A, et al. (2013). BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Rep 1:379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nguyen HT, Geens M, Mertzanidou A, Jacobs K, Heirman C, Breckpot K and Spits C. (2014). Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol Hum Reprod 20:168–177 [DOI] [PubMed] [Google Scholar]
- 47. Holubcova Z, Matula P, Sedlackova M, Vinarsky V, Dolezalova D, Barta T, Dvorak P and Hampl A. (2011). Human embryonic stem cells suffer from centrosomal amplification. Stem Cells 29:46–56 [DOI] [PubMed] [Google Scholar]
- 48. Peterson SE, Westra JW, Rehen SK, Young H, Bushman DM, Paczkowski CM, Yung YC, Lynch CL, Tran HT, et al. (2011). Normal human pluripotent stem cell lines exhibit pervasive mosaic aneuploidy. PLoS One 6:e23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baker D, Hirst AJ, Gokhale PJ, Juarez MA, Williams S, Wheeler M, Bean K, Allison TF, Moore HD, Andrews PW and Barbaric I. (2016). Detecting genetic mosaicism in cultures of human pluripotent stem cells. Stem Cell Rep 7:998–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.