Abstract
The rapid development of efficacious and safe vaccines against coronavirus disease 2019 (COVID-19) has been instrumental in mitigating the transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Moreover, the emergence of SARS-CoV-2 variants raised concerns on the efficacy of these vaccines. Herein, we report two cases of breakthrough infections with the P1 variant in patients vaccinated with CoronaVac, which is one of the two vaccines authorized for emergency use in the Brazilian immunization program. Our observations suggest that the vaccine reduced the severity of the disease and highlight the potential risk of illness following vaccination and subsequent infection with the P1 variant as well as for continued efforts to prevent and diagnose infection in vaccinated persons.
Keywords: SARS-CoV-2, COVID-19, vaccine
1. Introduction
Over the past 12 months of the unprecedented COVID-19 pandemic characterized by high morbidity and mortality rates, the development of vaccines capable of containing COVID-19 has become a global priority. In Brazil alone, as of 25 April 2021, 14,237,078 individuals were infected by SARS-CoV-2 with 386,416 deaths. Globally, there are 146,067,511 recorded infections with 3,092,497 deaths [1]. According to the World Health Organization (WHO), as of 9 April 2021, 96 vaccines were in various stages of clinical development [2]. Currently, the CoronaVac (Butantan Institute, São Paulo, Brazil and Sinovac Life Sciences, Beijing, China) is one of the vaccines that has received emergency use authorization (EUA) by Agência Nacional de Vigilância Sanitária (ANVISA) for use in Brazil, with efficacy rates against symptomatic COVID-19 of 50.7% and symptomatic disease requiring assistance at 50.7% and 83.7%, respectively [3].
However, throughout the duration of CoronaVac’s clinical trial, the unrestrained SARS-CoV-2 transmission and spread has allowed the emergence of SARS-CoV-2 variants, which are classified as Variants of High Consequence (VOHC), Variants of Interest (VOI), and Variants of Concern (VOC). Variants of Concern are considered those with increased transmissibility and severity as well as significant reduction in the neutralization capacity by antibodies generated in the course of previous infection or vaccination [4]. Three VOCs have been currently circulating in Brazil: British B.1.1.7, South African B.1.351, and the Brazilian B.1.1.28 or P1 [5], whose emergence has raised concerns of the efficacy of the COVID-19 vaccines. In the United States, as of 30 April 2021, 10,262 SARS-CoV-2 vaccine breakthrough infections had been reported out of nearly 101 million fully vaccinated persons (0.0001%), of which 2725 (27%) were asymptomatic, 995 (10%) required hospitalization, and 160 (2%) were fatal [6]. Herein, we describe the clinical outcomes of breakthrough infections with the P1 variant in two patients vaccinated with CoronaVac.
2. Materials and Methods
This study describes the two cases of patients vaccinated against SARS-CoV-2 with CoronaVac, who required ventilation support during their hospitalization following SARS-CoV-2 infection. Both patients were enrolled among 651 participants in the stage 3 clinical trial of CoronaVac, which was carried out by the Butantan Institute in 16 Brazilian centers with 12,396 participants enrolled between 21 July and 16 December 2020 (ClinicalTrials.gov, accessed on 25 June 2021; Identifier: NCT0445659). Unblinding of the vaccine trial’s results took place approximately one month prior to their hospital admission, confirming the enrollment of both patients in the vaccine group.
Epidemiological and clinical data (symptoms and radiologic observations) were obtained from electronic records. Naso/oropharyngeal samples were submitted for further diagnostic tests at the Laboratório de Pesquisas em Virologia (LPV), which is located within the Faculdade de Medicina de São José do Rio Preto (FAMERP). This study was submitted and approved by the Ethical Review Board of the Faculdade de Medicina de São José do Rio Preto (FAMERP), São Paulo, Brazil (protocol 34634620.1.2003.5415, 14 July 2020).
Viral RNA was extracted from 140 ul of nasal swabs using QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions. One-step real-time polymerase chain reaction (RT-qPCR) was performed using primers and probes targeting the envelope (E) and nucleocapsid (N) of the SARS-CoV-2 genome and human RNAse P designed GeneFinder COVID-19 PLUS RealAmp Kit (OSANG Healthcare Co., Ltd, Gyeonggi-do, Korea). Variant tracking was achieved by the amplification of a 798 bp fragment from the Spike genomic region that includes the Receptor Binding Domain (RBD) by conventional PCR using a set of specific primers (Forward SARS-CoV2_S1_PF: 5′-GAGTCCAACCAACAGAATC-3′ and Reverse SARS-CoV-2_S1_PR: 5′-GAATCTCAAGTGTCTGTGG-3′) and Sanger sequencing using BigDye Terminator v.3.1 (Applied Biosystems, Waltham, MA, USA). Readings were performed in a 3130xl Genetic Analyzer (Applied Biosystems, Waltham, MA, USA), contigs were assembled using the Electropherogram Quality Analysis tool (http://asparagin.cenargen.embrapa.br/phph/, accessed on 17 April 2021), and sequences were aligned using Clustal W [7] available in the BioEdit package [8]. Complete genomic sequences of the variants were obtained by next-generation sequencing. Briefly, library preparations were carried out following the instructions provided by an Illumina CovidSeq Test (Illumina Inc, San Diego, CA, USA). The quality and size of the libraries were verified by Agilent 4150 TapeStation (Agilent Technologies Inc, Santa Clara, CA, USA). Libraries were pooled in equimolar concentrations, and the sequencing was implemented on an Illumina MiSeq system, using a MiSeq Reagent Kit v2 (2 × 150 cycles) (Illumina Inc, San Diego, CA, USA). The quality of FASTQ sequencing data was checked using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc, accessed on 25 April 2021), and trimming was performed in Geneious Prime v. 2021.1, using the software BBDuk v. 37.25, in order to remove adapters and low-quality bases. A minimum Phred score of Q30 and minimal read length of 75 base pairs (bp) were used. The cleaned paired-end reads were mapped against the hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124) genome, which is available in the EpiCov database in GISAID (https://www.gisaid.org/, accessed on 7 May 2021), considering a minimum 50 bp overlap, minimum identity of 90%, and maximum mismatches of 10% per read. SNPs were identified using default settings and minimum coverage of 5 reads per site. The genomes of the both samples under study were aligned against the previous sequences obtained by Sanger sequencing in order to confirm the mutations sites and then submitted to Pangolin COVID-19 Lineage Assigner Tool to confirm the variant classification [9]. Finally, the phylogenetic analysis was performed to support the results of lineage assignment. Consensus sequences (hCoV-19/Brazil/SP-SJRP14/2021, GISAID access EPI_ISL_1754186 and hCoV-19/Brazil/SP-SJRP15/2021, GISAID access EPI_ISL_1941583) were aligned with 103 SARS-CoV-2 complete genomes from Brazil, retrieved from GISAID (Table S1) and the reference genome hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124). Sequencing alignment was carried out in MAFFT multiple alignment software version 7.271 [10] with default parameters. A maximum-likelihood tree was built in IQ-TREE (v. 2.1.3), using GTR+F+I as the nucleotide substitution model and 1000 bootstrap samples.
3. Results
3.1. Case 1
A 60-year-old man with a history of diabetes mellitus type 2, high blood pressure, and obesity degree I (BMI: 32.3 kg/m2), who was a resident of São José do Rio Preto, São Paulo State, Brazil, and no recent travel history, reported on 10 February 2021 with anosmia, malaise, and myalgia. He had been vaccinated with two doses of CoronaVac through enrolment in a clinical trial developed by the Butantan Institute in October 2020 (first dose: 8 October, second dose: 27 October; diagnosis of breakthrough infection 106 days following administration of the 2nd vaccine dose). Unblinding of the trial was in January 2021, which confirmed his enrollment in the vaccine group.
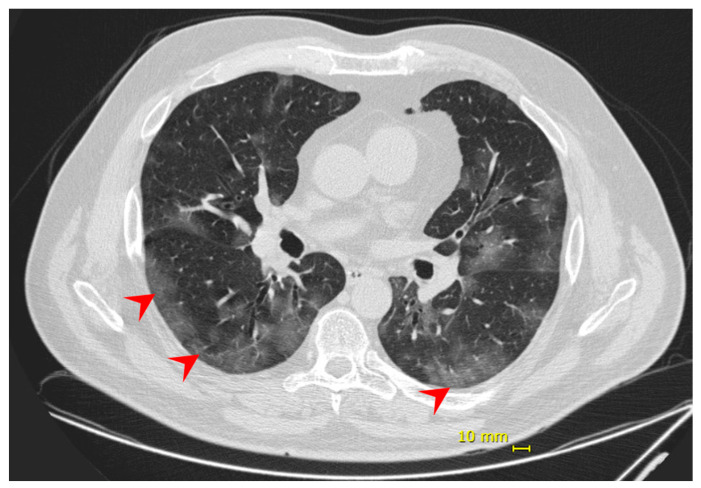
Eight days following the onset of symptoms, the patient complained of dyspnea and decreased peripheral saturation of oxygen (SpO2). He was admitted at the hospital with blood pressure 125/85 mmHg; heart rate 98 bpm; respiratory rate 18 ipm; temperature 35.1 °C; and SpO2 88% without supplemental oxygen. No additional alterations were observed in the physical exam. Chest computed tomography (chest CT) showed evidence of bilateral multifocal ground-glass opacities (25–50%) predominantly peripheral and subsegmental atelectasis (Figure 1), and COVID-19 diagnosis was confirmed by a positive RT-PCR on the submitted nasal/oropharyngeal swab sample. The patient was monitored in the intensive care unit (ICU) and received supplemental oxygen by high flow nasal catheter until one day before the discharge. No intubation or other ventilation device was needed. During hospitalization, the patient also received dexamethasone 6 mg, once a day, for 10 days, and enoxaparin 40 mg, as a prophylaxis regimen. He was discharged 12 days from hospital admission and 20 days from symptoms onset, having fully recovered.
Figure 1.
Patient 1—chest computed tomography evidenced bilateral multifocal ground-glass opacities (25–50%) predominantly peripheral (red arrow).
3.2. Case 2
On 26 February 2021, a 55-year-old male resident of São José do Rio Preto, São Paulo State, Brazil, without comorbidities, BMI: 25.9 kg/m2, was admitted at our hospital complaining of sore throat, headache, malaise, chills, coryza, and sneezing. A COVID-19 diagnosis was confirmed by a positive RT-PCR on the submitted nasal/oropharyngeal swab sample. He had a recent trip to Espírito Santo State, Brazil, returning 4 days before the onset of symptoms. He had been vaccinated with two doses of CoronaVac in October 2020 (first dose: 9 October, second dose: 27 October; diagnosis of breakthrough infection 122 days following administration of the 2nd vaccine dose) through enrolment in a clinical trial developed by the Butantan Institute. Unblinding of the trial in January 2021 confirmed his enrollment in the vaccine group.
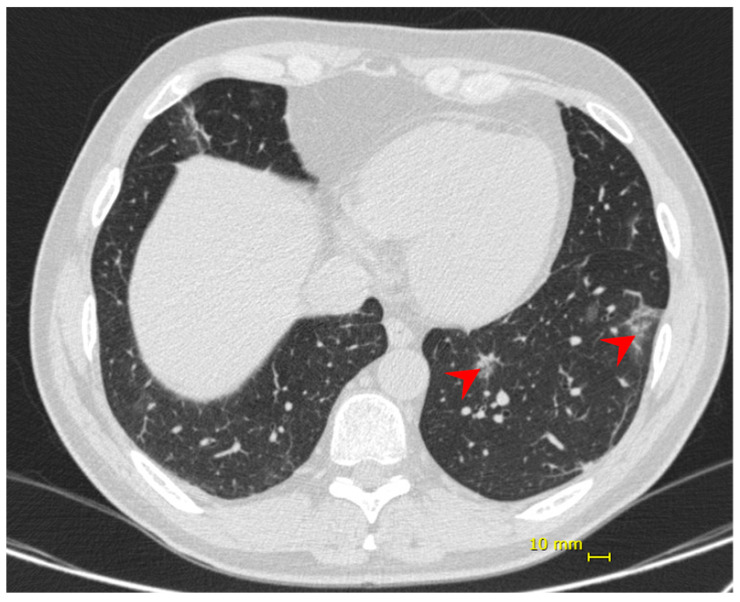
Nine days following the onset of symptoms, the patient presented dyspnea and hypoxia (SpO2 84% without supplemental oxygen), blood pressure 97/54 mmHg, heart rate 86 bpm, respiratory rate 31 ipm, temperature 36.9 °C. No additional alterations were observed in the physical exam. Chest CT showed bilateral multifocal ground-glass opacities (<25%) predominantly peripheral and subsegmental atelectasis (Figure 2). He received supplemental oxygen by high-flow nasal catheter, dexamethasone 6 mg, once a day, for 10 days, and enoxaparin 40 mg as prophylactic dose. His discharge took place 6 days from his hospital admission and 14 days from the onset of symptoms.
Figure 2.
Patient 2—chest computed tomography showing bilateral multifocal ground-glass opacities (<25%) predominantly peripheral (red arrow).
The clinical and laboratory results of both cases are presented below in Table 1. As in other studies with severe cases (reviewed by [11,12]), lymphocytopenia, increased lactate dehydrogenase, and C-reactive protein were observed.
Table 1.
Clinical and laboratory findings during the progressing of COVID-19 in two patients vaccinated with CoronaVac. In bold: out of range results from reference values.
| Patient 1 | Patient 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Signs and symptoms | Anosmia, malaise, and myalgia | Sore throat, headache, coryza, malaise, chills, sneezing | ||||||
| Worsening day | D8-dyspnea and SpO2 88% | D9-dyspnea and SpO2 84% | ||||||
| Clinical management | Supplemental oxygen by high flow nasal catheter. Dexamethasone and enoxaparin as prophylactic dose | Supplemental oxygen by high flow nasal catheter. Dexamethasone and enoxaparin as prophylactic dose | ||||||
| Chest CT | Bilateral multifocal ground-glass opacities (25–50%) predominantly peripheral and subsegmental atelectasis | Bilateral multifocal ground-glass opacities (<25%) predominantly peripheral, and subsegmental atelectasis | ||||||
| Discharge | Recovery, 20 days from symptoms onset | Recovery, 14 days from symptoms onset | ||||||
| Laboratory findings | Admission | Recovery | Discharge | Admission | Clinical Worsening | Recovery | Discharge | Reference values |
| Hemoglobin (g/dL) | 17.7 | 15.6 | 15.3 | 13.3 | 13.8 | 13.2 | 13.2 | 12–16 |
| Hematocrit (%) | 51.5 | 44.2 | 43.9 | 39.1 | 39.8 | 39.8 | 44.3 | 36–47 |
| Leukocytes (cells/mm3) | 5880 | 7400 | 7020 | 5150 | 7160 | 8990 | 11,200 | 4–11 × 103 |
| Neutrophils (cells/mm3) | 4720 | 5790 | 4090 | 4630 | 5580 | 6837 | 7470 | 1.6–7.7 × 103 |
| Lymphocytes (cells/mm3) | 730 | 760 | 1840 | 780 | 860 | 899 | 1650 | 0.6–3.9 × 103 |
| Platelets (×103) | 120 | 322 | 327 | 123 | 125 | 168 | 214 | 150–450 × 103 |
| CRP (mg/dL) | 5.14 | 1.42 | 0.16 | 1.96 | 9.8 | 2.27 | 1.99 | up to 0.50 |
| LDH (U/L) | 393 | 393 | 317 | 178 | 324 | 288 | 287 | 1–250 |
| Dimer D (ug/mL) | 0.42 | 0.34 | 0.33 | 0.34 | 1.26 | 0.97 | 0.7 | up to 0.500 |
| AST (U/L) | 46 | 18 | 26 | 106 | 15–24 | |||
| ALT (U/L) | 36 | 28 | 16 | 185 | 15–24 | |||
| GGT (U/L) | 67 | 29 | 34 | 173 | up to 123 | |||
| AP (U/L) | 65 | 75 | Up to 104 | |||||
| Creatinine (mg/dL) | 0.7 | 0.6 | 0.7 | 1.3 | 0.9 | 0.9 | 1.1 | 0.7–1.2 |
| pH | 7.44 | 7.49 | 7.46 | 7.46 | 7.43 | 7.42 | 7.35–7.45 | |
| PO2 (mmHg) | 69.7 | 84.6 | 83.8 | 109 | 80.3 | 69.8 | 80–100 | |
| PCO2 (mmHg) | 35.8 | 34.7 | 34.9 | 33.7 | 34.6 | 41.1 | 38–50 | |
| HCO3 (mmol/L) | 23.9 | 26 | 24.5 | 23.8 | 22.6 | 26.4 | 22–26 | |
| EB (mmol/L) | 1.1 | 1.3 | 1.9 | 0.8 | −0.6 | 2.3 | −2.0–2.0 | |
| SatO2 (%) | 91.6 | 95.1 | 93.9 | 98 | 97.6 | 93.9 | 94–100 | |
| Lactate (mmol/L) | 4.1 | 1.7 | 1.6 | 1.3 | 1.5 | 2.3 | 0.5–1.6 | |
3.3. Variant Tracking
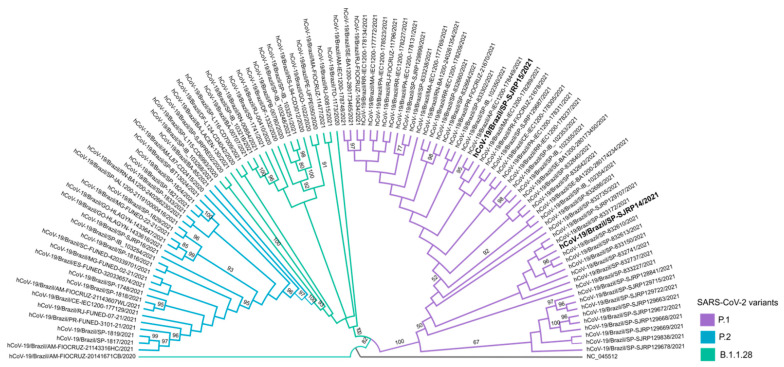
Naso/oropharyngeal samples were submitted for additional tests for variant tracking. The samples of both patients were designated hCoV-19/Brazil/SP-SJRP14/2021 and hCoV-19/Brazil/SP-SJRP15/2021, respectively, and classified as P1 lineage by the Pango lineage assignment tool, as well as by phylogenetic inference with high branch support (Figure 3). Sequence analysis showed that hCoV-19/Brazil/SP-SJRP14/2021 exhibited 13 non-synonymous mutations in the Spike protein. From these changes, ten are defining SNPs of the P1 variant (aa:S:L18F, aa:S:T20N, aa:S:P26S, aa:S:D138Y, aa:S:R190S, aa:S:K417T, aa:S:E484K, aa:S:N501Y, aa:S:H655Y and aa:S:T1027I), and three are amino acid substitutions (S:N196Y, S:D614G, S:V1176F) that have been reported in other lineages, such as B.1.1.28, from which P1 is derived. Additionally, several amino acid changes were also verified in other genome regions (Table 2). The hCoV-19/Brazil/SP-SJRP15/2021 displayed ten non-synonymous substitutions in the Spike protein; from those, nine are well characterized as SNPs from the P1 variant. Moreover, several amino acid mutations were observed in the whole genome, as in hCoV-19/Brazil/SP-SJRP14/2021. The small number of mutations found in hCoV-19/Brazil/SP-SJRP15/2021 compared with hCoV-19/Brazil/SP-SJRP14/2021 is a consequence of less than optimal genome coverage.
Figure 3.
Phylogenetic relationship of hCoV-19/Brazil/SP-SJRP14/2021 and hCoV-19/Brazil/SP-SJRP15/2021 (in bold). Maximum-likelihood tree was built in IQ-TREE (v. 2.1.3), using GTR+F+I as a nucleotide substitution model. Bootstrap: 1000.
Table 2.
Non-synonymous changes identified in the whole-genomes of hCoV-19/Brazil/SP-SJRP14/2021(EPI_ISL_1754186) and hCoV-19/Brazil/SP-SJRP15/2021 (EPI_ISL_1941583). In bold: mutations of concern.
| Samples | ORF1a | ORF1b | S | ORF3a | M | ORF8 | ORF9b | N |
|---|---|---|---|---|---|---|---|---|
| hCoV-19/Brazil/SP-SJRP14/2021 | ORF1a:S1188L, ORF1a:K1795Q, ORF1a:S3675-, ORF1a:G3676-, ORF1a:F3677- |
ORF1b:P314L, ORF1b:E1264D |
S:L18F, S:T20N, S:P26S, S:D138Y, S:R190S, S:N196Y, S:K417T, S:E484K, S:N501Y, S:D614G, S:H655Y, S:T1027I, S:V1176F | ORF3a:S253P | − | ORF8:E92K | ORF9b:Q77E | N:P80R, N:R203K, N:G204R |
| hCoV-19/Brazil/SP-SJRP15/2021 | ORF1a:K1795Q | ORF1b:P314L, ORF1b:E1264D | S:L18F, S:T20N, S:D138Y, S:R190S, S:K417T, S:E484K, S:N501Y, S:D614G, S:H655Y, S:T1027I | ORF3a:L94F, ORF3a:S253P | M:T7N, M:I8S | ORF8:E92K | ORF9b:Q77E | N:P80R |
4. Discussion
In December 2019, the first cases of an atypical pneumonia were described in individuals in China, and SARS-CoV-2 was identified as its etiologic agent [13]. The economic and social impact of the new disease, COVID-19, were unimaginable at the time. The virus spread first within China, from where it spread throughout the globe and was declared as a public health emergency of international concern by the World Health Organization (WHO) [14] on 11 March 2020 and right afterwards a pandemic [15]. The first documented case of COVID-19 in Brazil was reported in a Brazilian traveler in February 2020, who had returned from a trip to Lombardy, Italy [16]. Since then, the virus spread uncontrolled throughout the country, reaching 15,003,563 COVID-19-confirmed cases and 416,949 deaths as of 06 May 2021.
Manaus, the capital of Amazonas State, is the largest city in the Amazon, and it was the epicenter of attention for the dire consequences of SARS-CoV-2 in Brazil. The city reported an explosive number of cases at the beginning of the epidemic with high mortality rates [17]. Data showed that up to three-quarters of its population had been infected by SARS-CoV-2 in the first months of 2020, leading epidemiologists to suggest that herd immunity may have contained the epidemic [18,19]. However, the achieved high levels of community exposure were insufficient in preventing a second wave of COVID-19 cases, which was caused by the emergence of a new variant (P1) of SARS-CoV-2 [20], leading to the total collapse of the local health system [21].
Soon, the P1 variant (as well as other variants) spread in many Brazilian states [22], including São Paulo, where São José do Rio Preto is located. P1 is a variant that raised great concern since it is associated with higher transmission and infection rates as well as a reduction in antibody-mediated immunity (reviewed in [23]), which were characteristics attributed to a set of mutations (K417T, E484K, N501Y) in the RBD region of the S protein [24,25] and D614G [26]. The combination of these spike mutations, which is also shared with the B.1.351 variant, has been implicated in the decrease of neutralizing activity, suggesting that neutralizing antibodies are less effective against VOCs with this set of mutations [27,28,29,30]. That led to the hypothesis SARS-CoV-2 VOCs may produce more severe disease and be poorly protected against post-vaccination [31,32]. Herein, we described two cases of breakthrough infections in two CoronaVac-vaccinated individuals that were infected by the P1 SARS-CoV-2 variant and had favorable clinical outcomes.
The concern of vaccine efficacy against variants was raised early in the establishment of the national program of COVID-19 immunization, leading to the development and implementation of step-by-step protocols in diagnosing their circulation and effect on the vaccine efficacy among the population [33]. Such concern has been given more attention as several mutations were detected in spike glycoprotein. Many vaccines against COVID-19 are based on a version of the spike glycoprotein [reviewed in [34]] to inducing neutralizing antibodies, and since that site is the main target of mutations, the immune escape is unclear.
Despite the unprecedented and uncontrolled spread of the virus in Brazil, the rapid approval and vaccine immunization drives among the population offer a growing sense of relief in controlling the epidemic. The safety and immunogenicity of CoronaVac, one of the vaccines administrated in Brazil, had been already presented in a recent phase 1/2 clinical trial in adults [35], and its efficacy data were shown in a randomized, double-blind, placebo-controlled phase 3 clinical trial developed in Brazil [3]. Additional evidence for its efficacy in a high P1 variant transmission setting was recently presented. In this case-control study, vaccination with at least one dose resulted in a 0.50-fold reduction of symptomatic SARS-CoV-2 infection (adjusted vaccine effectiveness, 49.6%; 95% CI, 11.3–71.4), underscoring the need for increase in the vaccination efforts [data not peer reviewed yet [36]]. As of 3 May 2021, 14.05% of the Brazilian population had received at least one vaccine dose, and 6.63% were fully vaccinated [37].
Our study reports the clinical outcomes in two CoronaVac vaccinated patients who acquired a breakthrough SARS-COV-2 infection. Although both patients required hos-pitalization, the disease presentation was mild and did not progress in severity requiring invasive mechanical ventilation (IMV) or death. We believe that their quick recovery and absence sequelae were directly related to their recent immunization against COVID-19, as has been previously suggested [38]. Although the vaccine did not prevent SARS-CoV-2 infection by the P1 variant, there was no need for IMV, and both patients were recovered at discharge. In a retrospective study based on the first 250,000 COVID-19 hospitalizations in Brazil, the rates of IMV and death were 23% and 38%, respectively [39]. An important limitation of our study was a lack of serological, cytokine or cellular data to account for their re-infection. However, caution should be exercised in inferences derived from serological assays for the protective efficacy of SARS-CoV-2 vaccination against VOCs, as the correlates of protection are still not determined. The outcomes suggest that CoronaVac may be contributing to the reduction of severe clinical outcomes in SARS-COV-2 P.1 infections [35].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v13071237/s1, Table S1: Information of SARS-CoV-2 sequences included in the dataset.
Author Contributions
Conceptualization, C.F.E. and M.L.N.; methodology, C.A.B., G.R.F.C., B.d.C.M., L.S., C.B., L.S.U., F.S.P., J.P.A.J., H.L.F. and P.R.; validation, C.A.B.; formal analysis, C.F.E. and C.A.B.; investigation, C.F.E., L.F.M., J.D.S. and S.M.A.L.; resources, M.L.N.; data curation, C.F.E. and C.A.B.; writing—original draft preparation, C.F.E., C.A.B. and C.B.; writing—review and editing, C.F.E., C.A.B., N.V. and M.L.N.; supervision, M.L.N.; funding acquisition, N.V. and M.L.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the FAPESP-COVID Program (Grant # 2020/04836-0 to MLN) and partly by a Centers for Research in Emerging Infectious Diseases (CREID) 1U01AI151807 grant awarded to NV by the National Institutes of Health (NIH). MLN is supported by FAPESP (Grant # 2013/21719/3). JPAJ and MLN are CNPq Research Fellows. The funders had no role in the design of the study, collection, analyses, or interpretation of data, writing of the manuscript, or in the decision to publish the results.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of FAMERP (protocol code 34634620.1.2003.5415, approved on 14 July 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
All SARS-CoV-2 genomes generated and analyzed in this study are available in the EpiCov database in GISAID at: https://www.gisaid.org/, accessed on 25 June 2021. Accession IDs are listed in Table S1.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO . Weekly Epidemiological Update on COVID-19—27 April 2021. WHO; Geneva, Switzerland: 2021. [Google Scholar]
- 2.WHO . Draft Landscape and Tracker of COVID-19 Candidate Vaccines. World Health Organization; Geneva, Switzerland: 2021. [Google Scholar]
- 3.Palacios R., Batista A.P., Albuquerque C.S.N., Patiño E.G., Santos J.d.P., Conde M.T.R.P., Piorelli R.d.O., Pereira Júnior L.C., Raboni S.M., Ramos F. Efficacy and SAFETY of a COVID-19 Inactivated Vaccine in Healthcare Professionals in Brazil: The PROFISCOV Study. [(accessed on 25 June 2021)];2021 Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3822780.
- 4.CDC SARS-CoV-2 Variant Classifications and Definitions. [(accessed on 10 April 2021)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html.
- 5.O’Toole A., Scher E., Underwood A., Jackson B., Hill V., McCrone J., Ruis C., Abu-Dahab K., Taylo B., Yeats C., et al. Pangolin: Lineage Assignment in An Emerging Pandemic as An Epidemiological Tool. [(accessed on 10 April 2021)]; doi: 10.1093/ve/veab064. Available online: https://cov-lineages.org. [DOI] [PMC free article] [PubMed]
- 6.COVID C., Team V.B., Birhane M., Bressler S., Chang G., Clark T., Dorough L. COVID-19 Vaccine Breakthrough Infections Reported to CDC—United States, 1 January–30 April 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:792–793. doi: 10.15585/mmwr.mm7021e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symptom. Ser. 1999;41:4. [Google Scholar]
- 9.Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z.L., Hou Y.L., Li D.T., Li F.Z. Laboratory findings of COVID-19: A systematic review and meta-analysis. Scand. J. Clin. Lab. Investig. 2020;80:441–447. doi: 10.1080/00365513.2020.1768587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong C.K.H., Wong J.Y.H., Tang E.H.M., Au C.H., Wai A.K.C. Clinical presentations, laboratory and radiological findings, and treatments for 11,028 COVID-19 patients: A systematic review and meta-analysis. Sci. Rep. 2020;10:19765. doi: 10.1038/s41598-020-74988-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV) [(accessed on 26 March 2020)]; Available online: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov)
- 15.WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. [(accessed on 26 March 2020)]; Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020.
- 16.da Cunha C., Cimerman S., Weissmann L., Chebabo A., Bellei N. Informativo da Sociedade Brasileira de Infectologia: Primeiro Caso Confirmado de Doença Pelo Novo Coronavírus (COVID-19) No Brasil–26/02/2020. [(accessed on 10 April 2021)];Soc. Bras. Infectol. Sao Paulo Brasil. 2020 Available online: https://cbc.org.br/wp-content/uploads/2020/02/Informativo-CoV-26-02-2020.pdf.pdf-1.pdf. [Google Scholar]
- 17.Orellana J.D.Y., de Cunha G.M., Marrero L., Horta B.L., da Costa Leite I. Explosion in mortality in the Amazonian epicenter of the COVID-19 epidemic 19. Cad. Saude Publ. 2020;36:e00120020. doi: 10.1590/0102-311x00120020. [DOI] [PubMed] [Google Scholar]
- 18.Buss L.F., Prete C.A., Abrahim C.M.M., Mendrone A., Salomon T., de Almeida-Neto C., França R.F.O., Belotti M.C., Carvalho M.P.S.S., Costa A.G., et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371:288–292. doi: 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okell L.C., Verity R., Watson O.J., Mishra S., Walker P., Whittaker C., Katzourakis A., Donnelly C.A., Riley S., Ghani A.C., et al. Have deaths from COVID-19 in Europe plateaued due to herd immunity? Lancet. 2020;395:e110–e111. doi: 10.1016/S0140-6736(20)31357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faria N.R., Claro I.M., Candido D., Moyses Franco L., Andrade P.S., Coletti T.M., Silva C.A., Sales F.C., Manuli E.R., Aguiar R.S. Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings. [(accessed on 3 June 2021)]; Available online: https://virological.org/
- 21.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brasil . Semana Epidemiológica 15. 1st ed. Ministério da Saúde; Brazilia, Brazil: 2021. Boletim Epidemiológico Especial. Doença pelo coronavírus COVID-19; p. 107. [Google Scholar]
- 23.Gomez C.E., Perdiguero B., Esteban M. Emerging SARS-CoV-2 Variants and Impact in Global Vaccination Programs against SARS-CoV-2/COVID-19. Vaccines. 2021;9:243. doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., Garcia-Sastre A., Schotsaert M. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv. 2021 2021.01.26.21250543. [Google Scholar]
- 26.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diamond M., Chen R., Xie X., Case J., Zhang X., VanBlargan L., Liu Y., Liu J., Errico J., Winkler E., et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res. Sq. 2021;10:rs.3.rs-228079. doi: 10.21203/rs.3.rs-228079/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brasil . Plano Nacional da Operacionalização da Vacinação Contra a COVID-19. 5th ed. Ministério da Saúde; Brazilia, Brazil: 2021. p. 188. [Google Scholar]
- 34.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Zeng G., Pan H., Li C., Hu Y., Chu K., Han W., Chen Z., Tang R., Yin W., et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hitchings M.D.T., Ranzani O.T., Scaramuzzini Torres M.S., de Oliveira S.B., Almiron M., Said R., Borg R., Schulz W.L., de Oliveira R.D., da Silva P.V., et al. Effectiveness of CoronaVac in the setting of high SARS-CoV-2 P.1 variant transmission in Brazil: A test-negative case-control study. medRxiv. 2021 doi: 10.1016/j.lana.2021.100025. 2021.04.07.21255081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasell J., Mathieu E., Beltekian D., Macdonald B., Giattino C., Ortiz-Ospina E., Roser M., Ritchie H. A cross-country database of COVID-19 testing. Sci. Data. 2020;7:345. doi: 10.1038/s41597-020-00688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogliani P., Chetta A., Cazzola M., Calzetta L. SARS-CoV-2 Neutralizing Antibodies: A Network Meta-Analysis across Vaccines. Vaccines. 2021;9:227. doi: 10.3390/vaccines9030227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranzani O.T., Bastos L.S.L., Gelli J.G.M., Marchesi J.F., Baião F., Hamacher S., Bozza F.A. Characterisation of the first 250,000 hospital admissions for COVID-19 in Brazil: A retrospective analysis of nationwide data. Lancet Respir. Med. 2021;9:407–418. doi: 10.1016/S2213-2600(20)30560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SARS-CoV-2 genomes generated and analyzed in this study are available in the EpiCov database in GISAID at: https://www.gisaid.org/, accessed on 25 June 2021. Accession IDs are listed in Table S1.