Abstract
Pseudorabies virus (PRV) is an economically significant swine infectious agent. A PRV outbreak took place in China in 2011 with novel virulent variants. Although the association of viral genomic variability with pathogenicity is not fully confirmed, the knowledge concerning PRV genomic diversity and evolution is still limited. Here, we sequenced 54 genomes of novel PRV variants isolated in China from 2012 to 2017. Phylogenetic analysis revealed that China strains and US/Europe strains were classified into two separate genotypes. PRV strains isolated from 2012 to 2017 in China are highly related to each other and genetically close to classic China strains such as Ea, Fa, and SC. RDP analysis revealed 23 recombination events within novel PRV variants, indicating that recombination contributes significantly to the viral evolution. The selection pressure analysis indicated that most ORFs were under evolutionary constraint, and 19 amino acid residue sites in 15 ORFs were identified under positive selection. Additionally, 37 unique mutations were identified in 19 ORFs, which distinguish the novel variants from classic strains. Overall, our study suggested that novel PRV variants might evolve from classical PRV strains through point mutation and recombination mechanisms.
Keywords: pseudorabies virus, genome, phylogeny, recombination, selection pressure
1. Introduction
Pseudorabies virus (PRV), belonging to the family Herpesviridae, the subfamily Alphaherpesvirinae, is an economically important and devastating viral pathogen for the pig industry [1,2]. Pigs are natural and reservoir hosts of PRV [3,4]. PRV infection in pigs causes a wide range of clinical signs, including central neural disorder and high mortality in newborn piglets, neural disorder and respiratory diseases in nursery pigs, and abortion/stillbirth in sows [5]. It was documented that PRV infects a wide variety of hosts, except for higher primates and humans [3]. However, several recent studies reported that multiple human encephalomyelitis cases were linked to PRV in China, implying that PRV might be a zoonotic pathogen [6,7,8,9,10].
Because of the wide application of live-attenuated vaccines, PRV has been well controlled in most countries, including China [11,12,13]. Nevertheless, novel PRV variants with enhanced virulence broke out in pig farms in China in 2011 [14,15,16,17,18,19]. Animal experiments demonstrated that classic live vaccines failed to provide complete protection against novel PRV variants [10,12,14,17,20,21].
Comparative genome analysis revealed that virulent variant PRV acquired considerable mutations [14,15,16,18,22,23,24,25,26]. One study demonstrated that the genomic changes in gB contributed to the enhanced viral virulence of novel PRV variants [27]. However, the detailed genetic mechanisms of the enhanced virulence of PRV variants are still unclear.
Intermolecular recombination is one of the major driving forces for herpesvirus evolution [28]. The genomic intermolecular recombination between two PRV strains has been reported in natural infection conditions by two studies [29,30]. Recently, several studies revealed the potential recombination between Bartha-K61 and virulent PRV strains, suggesting that recombination might have involved PRV evolution during the pandemic in China [31,32]. Additionally, recombination between virulent field viruses and the live vaccine strain was recorded in bovine herpesvirus 1 and infectious laryngotracheitis viruses [33,34]. These studies raised significant biosafety concerns about the application of live-attenuated vaccines. Most of these studies only identified recombination events in one strain. Whether recombination has been frequently involved in the evolution of other prevalent PRV strains is still unknown.
Phylogenetic analysis showed that novel PRV variants are genetically close to China classic strains such as Ea, Fa, and SC. The mutations were found in various ORFs [15,22,23,24,25,35]. Furthermore, the phylogenetic and selection analysis of gB, gC, and gE revealed the significant enhancement of genetic diversity since 2011 and disclosed several adaptive mutation sites in gC and gB [36]. However, these studies mainly involved limited PRV genomes or only focused on several glycoproteins such as gB, gC, gE, and gD. The genomic sequence diversity of prevalent PRV strains and the selection pressure of most ORFs are still largely unknown. In summary, there are still several questions waiting to be addressed: (1) In recent years, the genome diversity of prevalent PRV strains in China is still unknown. Is there a new genotype that evolved during the PRV pandemic in China? (2) What is the genetic mechanism of PRV virulence enhancement? (3) What is the overall frequency of recombination in prevalent PRV strains? (4) The comprehensive profile of genetic diversity and selection pressure of each ORF of PRV is still unknown.
To address these questions, we employed the high-throughput sequencing (HTS) technique to sequence the genomes of 54 PRV strains isolated from 2011 to 2017. Together with 19 PRV genomes in GenBank, a total of 73 PRV genomes were included in the present study to analyze the PRV genomic diversity and evolution. At the full-genome level, we analyzed the genomic diversity, phylogenetic relationship, and the recombination of PRV strains from China, Europe, and the US. Additionally, we conducted ORF diversity and selection pressure analysis through all 67 ORFs. The selection pressure analysis of each ORF showed strong purifying selection pressure and identified multiple amino acid residues under positive selection. Furthermore, 37 distinct mutations, capable of differentiating novel PRV variants from classic strains, were identified in 19 ORFs, which would be useful for further genotype–phenotype studies.
2. Material and Methods
2.1. Viruses and Sequencing
Between 2012 and 2017, a total of 54 PRV strains were isolated from clinical samples of PRV-infected pigs in China. These samples were mainly abortive fetus or neonatal piglets with fatal neural disorders. These PRV strains came from 14 provinces of China. The geographical background of each PRV strain is listed in the Supplementary Materials (Table S1). All the PRV strains were isolated by PK-15 cells, and the passage times were strictly constrained within 5 times. Viral genome DNA was extracted as described previously [37]. Briefly, monolayers of PK-15 cells were infected with the PRV at an MOI of 10 and cultivated at 37 °C until a complete cytopathic effect was observed. Subsequently, the culture medium was collected without disrupting the cells and clarified by centrifugation at 4000× g for 10 min. Next, the viruses in the supernatant fluids were sedimented on a 30% sucrose cushion by ultracentrifugation at 77,000 × g for 2 h. The sedimented virion pellets were resuspended in sodium Tris-EDTA buffer. After the addition of proteinase-K (100 μg/mL final concentration) and sodium dodecyl sulfate (SDS; 0.5% final concentration), the lysate was incubated at 37 °C for 1 h, which was followed by phenol–chloroform extraction and ethanol precipitation. The white precipitation was viral genome DNA. The viral genome DNA was dissolved in TE buffer (pH = 8.0) and stored at −80 °C. Subsequently, the genome DNA of each PRV strain was subjected to DNA library preparation for high-throughput genomic sequencing (Illumina Hiseq 2500).
2.2. Assembly and Annotation of Genome Sequences
DNA library preparation was performed following the manual of the NexteraTM DNA Flex Library Prep Kit (Illumina, CA). Libraries were sequenced on the HiSeq2500 (Illumina) platform. The paired-end sequencing gave a 150 bp reading length from each terminal. Full-genome consensus sequences were assembled using a pipeline reported previously, with some modifications [38,39]. Firstly, raw FastQ files were assembled de novo by using IDBA version 1.1.0 [40]. The contigs were further oriented and assembled by aligning the contigs against the genomic sequence of PRV reference strain NIA3 (KU900059.1), resulting in a draft genome sequence. The reads were mapped to this draft by using Maq [41], and the quality of the final assemblies was inspected by visualizing the alignment in Tablet [42] and manually corrected if necessary. At this stage, some genomes still showed a problematic assembly in the repetitive regions. To solve this problem, we used the iterative mapping approach [43]. Briefly, these regions were cut out from assemblies, and the separate contigs were extended and finally joined by iterative mapping of sequencing reads. The PRV genome contains two unique regions: unique long (UL) and unique short (US). The US region is flanked by large inverted and terminal repeat sequences (IR and TR). Considering that the length of each read (150 bp/reads) is much shorter than the inverted repeat sequences among the IR and TR regions, it is impossible to distinguish the reads from one copy within the IR region from the other identical copy located in the TR region. Hence, the sequencing data were assembled into a trimmed version of the genome, which only kept IR in the middle of the genome. These trimmed genome sequences were deposited in GenBank, and the accession numbers are listed in Table S1. The number of total clean reads, average sequencing depth, and coverage are summarized in Table S1. All the gapped regions are listed in the Supplementary Materials (Table S6).
2.3. Multiple Sequence Alignment and Genome Annotation
A multiple sequence alignment of 54 in-house-assembled viral genomes plus 19 genome sequences from GenBank (Table S2) was constructed by MAFFT version 7.221.3, option FFT-NS-I (maximum of 1000 cycles) [44]. Online PRV genomes used in this study are listed in Table S2. The TR regions of 19 reference genome sequences were also trimmed before all the analyses in the current study.
Genome annotation for 54 PRV genomic sequences was transferred from a genetically intact reference strain NIA3 (KU900059.1) by using RATT with a word size of 30, a cluster size of 400, a maximum extend cluster of 500, and an identity cutoff of 40 [45]. Annotation was manually inspected and corrected if the transfer was failed due to assembly gaps, sequence variability, or disruptive mutations. The annotated genomes of the newly sequenced PRV isolates were deposited in GenBank as partial genome sequences.
2.4. Genomic Diversity
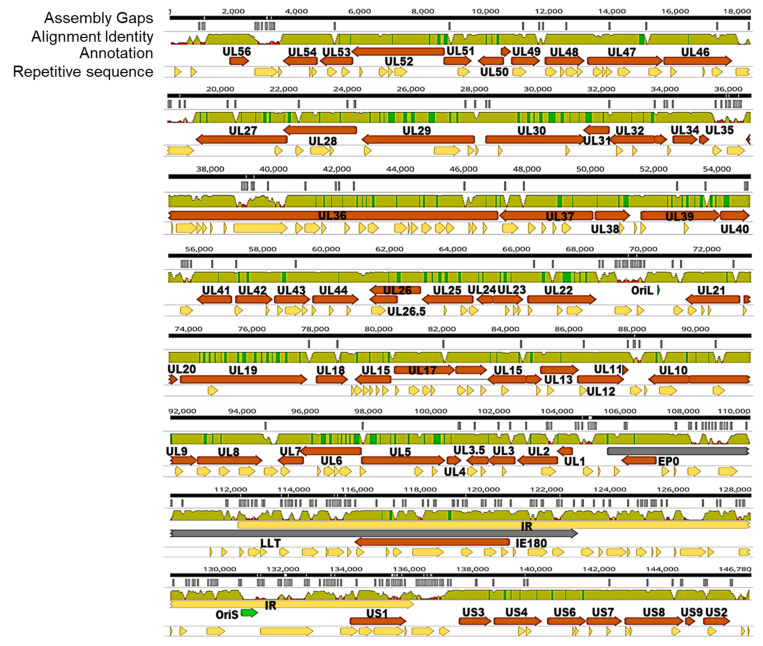
Genome alignment was visualized by Geneious Prime 2020.02.1 (www.geneious.com, accessed on 5 March 2020), including genome annotation, alignment identity curve, gapped regions, and the regions of repetitive sequence (Figure 1). All the gapped regions were removed from alignment by Geneious Prime. The overall mean distance, indicating the arithmetic mean of all individual pairwise distances between each item in alignment, was measured by MEGA X (Jukes–Cantor model) with the pairwise deletion of gapped sites [46].
Figure 1.
Overview of the sequence diversity in the alignment of 73 PRV genomic sequences. The alignment of 73 PRV genomic sequences was visualized by Geneious Prime. The black bar at the top represents a consensus sequence drawn from the alignment of 73 PRV genomic sequences. The white lines in the second row indicate the distribution of assembly gaps. In the third row, the curve of identity level is plotted from this alignment, which is colored as follows: green, 100% identity; green-brown, 30 to <100% identity; red, <30% identity. The fourth row is the annotation of the PRV genome. The bottom row shows the region of the repetitive sequence of the PRV genome.
2.5. Phylogenetic and Recombination Analysis
A maximum likelihood (ML) phylogenetic tree was constructed by using MEGA X with the general time-reversible (GTR) model and 100 bootstrap replications [46]. Phylogenetic network analysis was performed using SplitsTree version 4.14.6 with the Jukes–Cantor model [47]. Recombination analysis was assessed by the Phi test in SplitsTree and further evaluated by the RDP 4 package [48]. All the PRV strains isolated in China plus Bartha-K61 were selected for RDP analysis. Considering that PRV was eradicated in domestic pig populations in most European and North American countries decades ago, classic Europe/US strains, except the live vaccine strain Bartha-K61, are highly impossible to show up in China pig farms. Therefore, we removed the Europe/US strains, except Bartha-K61, from the RDP recombination analysis. In the RDP analysis, 7 algorisms were considered, which are RDP, GENECONV, BootScan, Maxchi, Chimaera, Siscan, and 3seq. The recombination events were considered significant when at least 4 out of 7 algorisms showed p < 0.001.
2.6. ORF Alignment and Divergence
Corresponding nucleotide or amino acid sequences of each ORF of every strain were extracted from the annotated genomes and pooled into one fasta file. Among the 67 ORFs, several ORFs contained gapped regions (Table S6). Those ORFs were excluded from the analysis. The sequences of each ORF were aligned at the codon level and the amino acid level by T-Coffee [49]. Nucleotide diversity was calculated by using MEGA X with all positions in alignment (Tamura–Nei model), and standard errors were calculated with a bootstrap procedure (100 replicates). Amino acid diversity was calculated by using MEGA X with the Poisson correction model [50].
2.7. Selection Pressure of Each ORF
In this analysis, nonsynonymous substitutions per nonsynonymous site (dN), synonymous substitutions per synonymous site (dS), and the dN/dS ratio were calculated by using the FUBAR algorism in the Datamonkey web server [51,52,53]. The individual site of each ORF under positive selection was evaluated by FUBAR [51], MEME [54], and CodeML of PAML [55]. Positive selection residues were at least confirmed by 2 out of 3 algorisms. The significance level of FUBAR is a posterior probability of >0.90. The significance level of MEME is p < 0.1. The significance level of CodeML is p < 0.05.
2.8. Data Availability
The 54 PRV genomic sequences in our study were submitted to GenBank, and the accession numbers are listed in Table S1. The accession numbers of the reference PRV sequences are listed in Table S2.
3. Result and Discussion
3.1. Genomic Sequencing, Assembly, and Alignment
To explore the genomic diversity of PRV-prevalent strains, 54 PRV strains were isolated from clinical samples from pig farms in China from 2012 to 2017. These samples were collected from 14 provinces in China. The viral genomic DNA samples of 54 PRV strains were subjected to high-throughput sequencing. The G+C% of PRV reference strain NIA3 was 74.0%, while the G+C% of 54 PRV strains ranged from 73.9% to 74.1%. The full genomic sequences of 54 PRV genomes and 19 online reference PRV genomes were aligned by MAFFT. Figure 1 shows a schematic diagram containing the PRV genome annotations, alignment identity curve, assembly gaps, and repetitive region. As shown in Figure 1, the distribution of nucleotide variation is uneven, and the highly variable regions usually are the regions containing repetitive sequences. Previous studies also showed a similar distribution in the genome diversity of HSV-1 and HCMV, indicating that the repetitive regions are highly variable [38,39,56].
3.2. Phylogenetic Analysis Indicates Strong Geographic Clustering
In our study, phylogenetic analysis divided the PRV strains into two genotypes: Genotype I and Genotype II (Figure 2A). The Europe/US strains were clustered together forming Genotype I, while Chinese strains were all clustered as Genotype II, which is consistent with previous studies [14,24,25,35,57]. The mean genetic distance, representing the evolutionary distance of different genomes or genes from various strains, was measured by the number of nucleotide or amino acid substitutions between them. The overall mean distance of the whole alignment is 0.65%, and the genetic distance between Genotype I and Genotype II is 2.2%. The genetic distances between each genetic clade range from 0.36% to 2.6% (Figure 2B). Novel PRV strains prevalent in China are genetically close to local classic strains such as Ea and Fa, whereas newly sequenced Europe strains such as Herculus/Kolchis and ADV32751 are also close to Europe/US classic strains such as Becker and NIA3 (Figure 2A). The mean distance between two genotypes is substantially higher than the inner genotype mean distance (0.5–1%). This result is similar to previous findings, showing that the overall mean distances of PRV, BHV-1, EHV-1, EHV-4, and VZV are 1.65%, 0.81%, 0.79%, 0.16%, and 0.136%, respectively [57].
Figure 2.
Phylogenetic relationship of PRV genome sequences. (A) A maximum likelihood (ML) tree was constructed by using MEGA X, based on the alignment of 73 PRV genome sequences (Tables S1 and S2). Gapped regions, which are labeled in Figure 1 within the multisequence alignment, were removed from all sequences before phylogenetic analysis. The ML tree is a rooted tree, and all branch lengths were drawn to a scale of nucleotide substitutions per site. Bootstrap resampling (100 replication) was performed. The branches of Genetic Clade 2.1 are collapsed. (B)The mean distance between each genetic clade was calculated by MEGA X with the Jukes–Cantor model. (C) Expansion of the collapsed clade (Clade 2.1) in the panel A of this figure.
Within Genotype II, three strains (SC, Ea, and Fa) were isolated from the 1980s to 1990s in China [13]. As shown in Figure 2A, the strain SC formed a unique genetic clade (Clade 2.3), while classic strains Ea and Fa formed the major genetic clade (Clade 2.1) with other PRV strains isolated after 2011 in China, except strain HuB1/CHN2017. For a close observation, we collapsed Genetic Clade 2.1 in Figure 2A and displayed the phylogenetic tree of Clade 2.1 in Figure 2C. Interestingly, strain HuB1/CHN2017 formed a unique genetic clade (Clade 2.2), showing a substantial genetic distance against all other genetic clades in Genotype II (Figure 2B).
Overall, these results indicate that the prevalent PRV strains in China are highly homologous, suggesting these strains might evolve from the same ancestor. However, the unique genetic clade was still detected in this study, which implied that PRV-prevalent strains continuously evolved during the PRV pandemic in China. The evolutionary mechanisms that drive novel genetic clades to emerge remain unknown.
3.3. Recombination between PRV Strains Is Robust
It has been well established that recombination contributes significantly to alphaherpesvirus evolution [57,58]. However, due to limited genome sequence availability, the profile of recombination within PRV-prevalent strains has not yet been clarified. In this study, we further investigated recombination between PRV strains by the SplitsTree and RDP package (Figure 3). The Phi test, which gives the statistical significance of the occurrence of recombination, strongly supported the existence of recombination (P = 0.00) (Figure 3A). Phylogenetic network analysis showed that recombination events between each genetic clade within Genotype II were more robust than Genotype I (Figure 3A). Recombination between two genotypes was also detected (Figure 3A).
Figure 3.
Recombination occurring pervasively in PRV newly sequenced strains. Recombination between PRV strains was determined by the Neighbor-Net split network and BootScan. (A) The Neighbor-Net split network was drawn by SplitsTree version 4.16 of 73 genomic sequences. The reticulate connections between each clade indicate events of recombination. Each clade was covered by a color scheme. The name of each strain is not displayed. (B) RDP analysis was applied to detect the recombination events in newly sequenced PRV genomes. All the recombination events are plotted in the schematic diagram. Each strip represents one recombination event and is labeled with one color. There are 4 colors in total, each representing different combinations of parental strains, as follows: red = Bartha-K61 + novel PRV variants; blue = Bartha-K61 + China classic strains, green = China classic strains + novel variants; yellow = novel variants + novel variants.
To further determine the distribution of recombination events within the newly sequenced strains, all the PRV strains isolated in China plus Bartha-K61 were selected for RDP analysis. All the recombination events are exhibited in Figure 3B, and detailed information concerning the location of recombination events, recombination parental strains, and the significance of each recombination event is listed in Tables S3–S5. In total, 23 recombination events were identified, of which only four events were recombination between two newly sequenced PRV strains (Events 24, 36, 40, and 52), whereas the others were recombination between two classic strains or recombination between a classic strain and a newly sequenced strain (Table S3). Additionally, the distribution of recombination events was not even. Clearly, the UL and US regions showed fewer recombination events, while the EP0 to IR regions contained significantly more recombination events (Figure 3B). One previous study also showed similarly that the IR and TR regions are recombination hot spots in HSV-1 [59].
It is worth noticing that Recombination Event 37, located in the IR region, was identified in 59 strains, while Recombination Event 17, spanning from the IR to US regions, was detected in 13 strains (Table S4). Other recombination events were only recognized in one strain (Table S3). Most recombination events occurred within the fragments shorter than 6 kb (Figure 3B); however, Event 47 detected in HuB1/CHN2017 showed a notably long recombination region starting from UL6 to the entire IR region (33kb), implying a different recombination mechanism in the evolutionary process of the novel genetic clade (Figure 3B).
Among all 23 recombination events, Bartha-K61 was involved in 16 events, including Event 17 and Event 37 (Table S3). The analysis results that showed the recombination between wild PRV strains and Bartha-K61 is possible, and more direct field evidence is ideally needed to support these results. Previous studies have demonstrated the high possibility of recombination between vaccine and clinically prevalent strains in different herpesviruses such as pseudorabies virus, varicella-zoster virus, bovine herpesvirus 1, infectious laryngotracheitis virus, and Marek’s disease virus [31,32,60,61]. Overall, these results indicate that recombination contributes to the evolution of novel PRV variants. It should be noted that recombination analysis based on software such as SplitsTree and RDP can only predict the potential recombination events among different viral strains and estimate the potential contribution of recombination during viral evolution in certain periods. To demonstrate the existence and the contribution of certain recombination events, more direct evidence such as detecting both recombination parental strains from the same animal is necessary.
3.4. Diversity of Protein-Coding Sequences
A smaller number of genomic sequences available limits the analysis of amino acid diversity of most ORFs. Previous studies only focused on several ORFs, such as UL44 (gC) and US6 (gD) [22,36,62]. In this study, we assessed the global diversity of PRV protein-coding sequences, including all 67 ORFs.
As shown in Table 1, most ORFs are conservative, while only 11 ORFs show over 3% diversity. The relatively more conservative ORFs are UL30, UL19, UL22, UL5, and UL7, with diversity below 0.5% (Table 1). Within these proteins, UL30 and UL5 are key proteins of viral genome DNA synthesis [4]. UL19, also named VP5, is the major capsid component [4,62]. It is not surprising that those proteins are highly conservative, as they play critical roles during viral lytic replication. UL7 shows the lowest diversity. Although the exact function of UL7 remains unclear, it has been demonstrated that UL7 is involved in viral egress and virion release from the cytoplasmic membrane [63]. Viral glycoproteins are the major targets of neutralizing antibodies. Consequently, viruses with mutations in glycoproteins could evade host-neutralizing antibodies. In our results, only gE and gI exhibit a relatively high level of divergence, while other glycoproteins show relatively conservative. Nevertheless, care should be taken when comparing the diversity level of different ORFs, because the diversity at certain amino acid residues may truly reflect the functional diversity or epitope diversity, while the diversity at certain regions may not affect the neutralization epitope or protein function at all.
Table 1.
Nucleotide and amino acid diversity of PRV strains in open reading frames.
| Nucleotide | Amino Acid | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ORF | Common Name | Core Gene | Mean Distance (%) |
Standard Error | Variable Sites | Length * (Nucleotide) |
Ratio of Variable Sites | Mean Distance (%) |
Standard Error | Ratio of Variable Sites |
| UL1 | gL | Yes | 2.66% | 0.006 | 22 | 468 | 4.70% | 3.10% | 0.011 | 5.13% |
| UL2 | UNG | Yes | 0.72% | 0.002 | 47 | 969 | 4.85% | 0.89% | 0.002 | 6.81% |
| UL3 | No | 2.00% | 0.002 | 79 | 720 | 10.97% | 3.79% | 0.007 | 17.50% | |
| UL3.5 | No | 1.64% | 0.003 | 45 | 666 | 6.76% | 2.80% | 0.006 | 13.06% | |
| UL4 | No | 0.84% | 0.002 | 18 | 435 | 4.14% | 1.27% | 0.004 | 6.21% | |
| UL5 | Yes | 0.23% | 0.000 | 43 | 2505 | 1.72% | 0.24% | 0.001 | 2.16% | |
| UL6 | Yes | 0.42% | 0.001 | 44 | 1935 | 2.27% | 0.72% | 0.002 | 3.26% | |
| UL7 | Yes | 1.13% | 0.002 | 24 | 798 | 3.01% | 0.19% | 0.005 | 5.26% | |
| UL8 | Yes | 1.60% | 0.001 | 114 | 2064 | 5.52% | 1.63% | 0.003 | 7.70% | |
| UL9 | OBP | No | 0.53% | 0.001 | 104 | 2532 | 4.11% | 0.83% | 0.001 | 6.40% |
| UL10 | gM | Yes | 0.84% | 0.002 | 34 | 1179 | 2.88% | 1.80% | 0.004 | 6.11% |
| UL11 | Yes | 1.67% | 0.006 | 10 | 189 | 5.29% | 2.94% | 0.014 | 9.52% | |
| UL12 | AN | Yes | 1.32% | 0.002 | 79 | 1449 | 5.45% | 2.15% | 0.004 | 8.90% |
| UL13 | PK | Yes | 1.28% | 0.002 | 51 | 1173 | 4.35% | 2.29% | 0.004 | 7.42% |
| UL14 | Yes | 1.30% | 0.003 | 16 | 477 | 3.35% | 2.20% | 0.008 | 5.66% | |
| UL15 | Yes | 1.38% | 0.002 | 119 | 2226 | 5.35% | 1.90% | 0.002 | 7.68% | |
| UL16 | Yes | 2.00% | 0.003 | 48 | 984 | 4.88% | 2.80% | 0.006 | 7.01% | |
| UL17 | Yes | 1.25% | 0.002 | 83 | 1800 | 4.61% | 1.82% | 0.003 | 6.67% | |
| UL18 | VP23 | Yes | 0.76% | 0.002 | 24 | 888 | 2.70% | 0.58% | 0.003 | 2.36% |
| UL19 | VP5 | Yes | 0.45% | 0.001 | 84 | 3990 | 2.11% | 0.47% | 0.001 | 2.56% |
| UL20 | No | 1.90% | 0.005 | 20 | 495 | 4.04% | 3.10% | 0.010 | 6.06% | |
| UL21 | Yes | 1.48% | 0.002 | 101 | 1599 | 6.32% | 2.20% | 0.004 | 9.57% | |
| UL22 | gH | Yes | 0.22% | 0.000 | 44 | 2064 | 2.13% | 0.44% | 0.001 | 4.22% |
| UL23 | TK | 0.49% | 0.001 | 21 | 960 | 2.19% | 0.83% | 0.003 | 3.44% | |
| UL24 | Yes | 0.83% | 0.003 | 9 | 513 | 1.75% | 0.84% | 0.005 | 1.75% | |
| UL25 | Yes | 0.72% | 0.001 | 54 | 1608 | 3.36% | 1.00% | 0.002 | 4.85% | |
| UL26 | VP24 | Yes | 0.77% | 0.001 | 62 | 1599 | 3.88% | 0.92% | 0.002 | 5.63% |
| UL26.5 | VP22 | Yes | 1.19% | 0.002 | 40 | 861 | 4.65% | 1.20% | 0.004 | 5.92% |
| UL27 | gB | Yes | 0.96% | 0.001 | 105 | 2742 | 3.83% | 1.67% | 0.003 | 6.67% |
| UL28 | ICP18.5 | Yes | 0.71% | 0.001 | 66 | 2166 | 3.05% | 0.98% | 0.002 | 4.99% |
| UL29 | ICP8 | Yes | 0.51% | 0.001 | 87 | 3537 | 2.46% | 0.56% | 0.001 | 3.14% |
| UL30 | Yes | 0.47% | 0.001 | 52 | 3144 | 1.65% | 0.48% | 0.001 | 2.10% | |
| UL31 | Yes | 0.96% | 0.002 | 16 | 813 | 1.97% | 1.44% | 0.005 | 2.95% | |
| UL32 | Yes | 0.80% | 0.002 | 33 | 1413 | 2.34% | 0.88% | 0.003 | 2.76% | |
| UL33 | Yes | 1.30% | 0.003 | 11 | 351 | 3.13% | 2.23% | 0.010 | 5.98% | |
| UL34 | Yes | 2.06% | 0.004 | 45 | 783 | 5.75% | 3.20% | 0.008 | 8.81% | |
| UL35 | VP26 | Yes | 1.45% | 0.004 | 10 | 309 | 3.24% | 3.60% | 0.013 | 7.77% |
| UL36 | VP1/2 | Yes | 1.87% | 0.001 | 993 | 9489 | 10.46% | 2.70% | 0.001 | 14.64% |
| UL37 | Yes | 0.70% | 0.001 | 100 | 2757 | 3.63% | 1.32% | 0.002 | 6.96% | |
| UL38 | VP19c | Yes | 0.93% | 0.002 | 32 | 1104 | 2.90% | 0.77% | 0.002 | 3.26% |
| UL39 | RR1 | Yes | 0.82% | 0.001 | 81 | 2364 | 3.43% | 1.18% | 0.002 | 5.58% |
| UL40 | RR2 | No | 0.82% | 0.001 | 25 | 909 | 2.75% | 0.90% | 0.003 | 3.30% |
| UL41 | VHS | No | 1.02% | 0.002 | 34 | 1095 | 3.11% | 1.08% | 0.003 | 4.11% |
| UL42 | Yes | 0.81% | 0.001 | 52 | 1155 | 4.50% | 1.69% | 0.004 | 9.09% | |
| UL43 | No | 0.93% | 0.001 | 45 | 1119 | 4.02% | 1.46% | 0.004 | 5.63% | |
| UL44 | gC | No | 2.38% | 0.003 | 108 | 1461 | 7.39% | 4.20% | 0.005 | 13.35% |
| UL46 | VP11/12 | No | 1.63% | 0.002 | 125 | 2085 | 6.00% | 2.63% | 0.003 | 9.64% |
| UL47 | VP13/14 | No | 1.89% | 0.002 | 150 | 2217 | 6.77% | 2.60% | 0.003 | 9.47% |
| UL48 | VP16 | No | 0.97% | 0.002 | 41 | 1239 | 3.31% | 1.52% | 0.004 | 5.33% |
| UL49 | VP22 | No | 1.37% | 0.003 | 40 | 729 | 5.49% | 2.06% | 0.005 | 8.23% |
| UL49.5 | gN | Yes | 4.10% | 0.008 | 25 | 297 | 8.42% | 7.56% | 0.022 | 15.15% |
| UL50 | dUTPase | Yes | 1.40% | 0.003 | 37 | 807 | 4.58% | 2.10% | 0.006 | 5.95% |
| UL51 | Yes | 2.06% | 0.004 | 40 | 729 | 5.49% | 4.38% | 0.009 | 11.11% | |
| UL52 | Yes | 1.81% | 0.001 | 329 | 2910 | 11.31% | 2.56% | 0.002 | 14.74% | |
| UL53 | gK | No | 1.60% | 0.003 | 39 | 936 | 4.17% | 2.38% | 0.006 | 6.09% |
| UL54 | ICP27 | Yes | 2.00% | 0.003 | 73 | 1083 | 6.74% | 2.80% | 0.005 | 11.08% |
| UL56 | No | 2.87% | 0.004 | 47 | 621 | 7.57% | 2.93% | 0.008 | 8.70% | |
| US1 | ICP22 | No | 2.49% | 0.003 | 235 | 1332 | 17.64% | 3.34% | 0.005 | 24.55% |
| US2 | 28K | No | 2.30% | 0.004 | 40 | 768 | 5.21% | 3.50% | 0.009 | 8.59% |
| US3 | No | 0.83% | 0.002 | 28 | 1002 | 2.79% | 1.33% | 0.004 | 4.19% | |
| US4 | gG | No | 1.29% | 0.001 | 142 | 1497 | 9.49% | 1.70% | 0.003 | 12.63% |
| US6 | gD | No | 0.83% | 0.001 | 44 | 1206 | 3.65% | 1.50% | 0.003 | 6.72% |
| US7 | gI | No | 1.98% | 0.002 | 99 | 1098 | 9.02% | 3.00% | 0.005 | 13.11% |
| US8 | gE | No | 1.78% | 0.001 | 233 | 1734 | 13.44% | 2.68% | 0.004 | 16.26% |
| US9 | 11K | No | 1.97% | 0.005 | 12 | 294 | 4.08% | 2.07% | 0.011 | 4.08% |
| EP0 | ICP0 | No | 0.79% | 0.001 | 44 | 1104 | 3.99% | 1.19% | 0.003 | 5.98% |
| IE180 | ICP4 | No | 0.64% | 0.001 | 214 | 4425 | 4.84% | 0.98% | 0.001 | 6.98% |
*: The ORF nucleotide sequence lengths of each ORF were collected from the reference genome of PRV Ea strain.
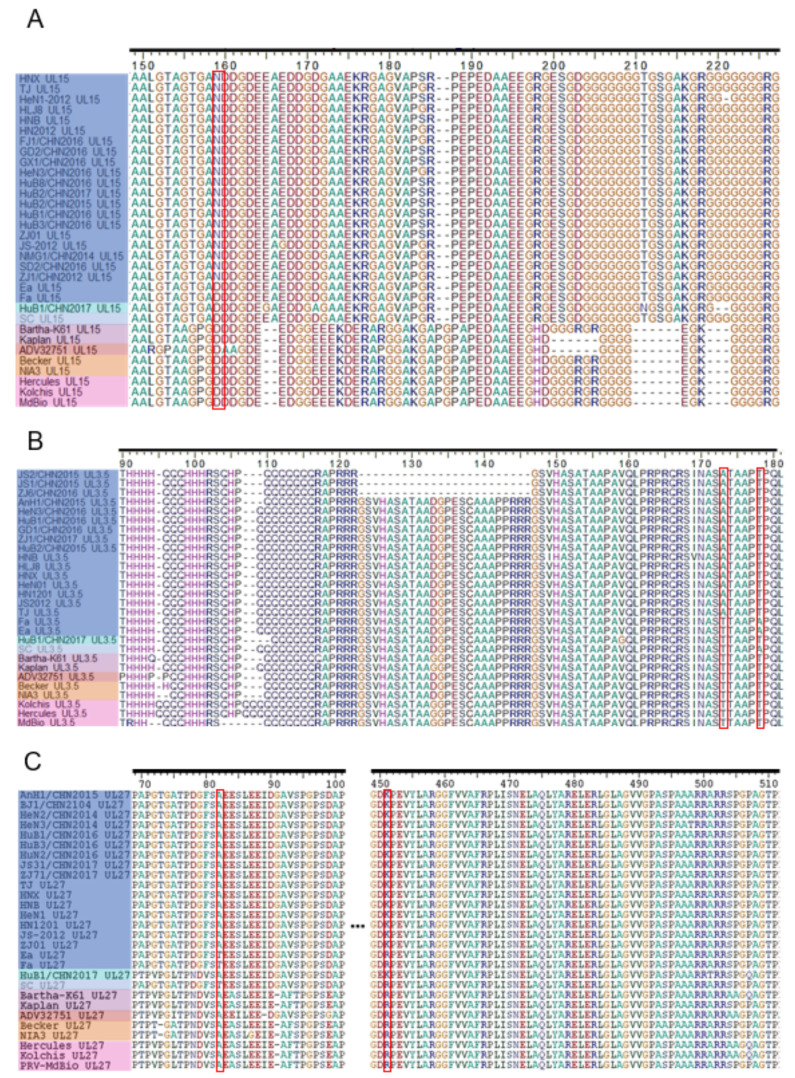
The alignments in UL27, UL15, and UL3.5 chosen as their corresponding ORFs exhibit the variations seen in most PRV proteins in Table 1, including single amino acid variations, short insertion/deletion, and simple sequence repeat (SSR)-based variations. To easily view variations in the alignment, we removed the identical sequences among newly sequenced ORFs in this study (Figure 4). In the alignment of UL15, there are multiple mutations that distinguish all PRV isolates into Genotype I and Genotype II, such as GTGA155-158AGPG, AEDD165-169ΔΔDG, R185A, and RG198HD (Figure 4A). There are also multiple mutations representing the unique genetic background of HuB1/CHN2017 such as E165G, D171A, and T211N. Although most residues show more conservative in each genotype, aa184 shows variation within Genetic Clade 2.1 of Genotype II (Figure 4A). Only one unique mutation at aa159 was identified in UL15, changing from aspartate in classic strains to asparagine in novel PRV variants (Figure 4A).
Figure 4.
Presentative amino acid (AA) variation in UL15, UL3.5, and UL27. Partial regions of the amino acid (AA) alignment of UL15 (A), UL3.5 (B), and UL27 (C) are displayed in the three panels. To make the alignment easy to display, the redundancy of newly sequenced ORFs is reduced, while all the reference sequences are maintained. The panel of the strain name is covered by colored boxes, which represent their genetic clades with the same color scheme used in Figure 3A. The distinguishing mutations that distinguish PRV strains from classic strains are highlighted by red boxes.
In the UL3.5 alignment, the pattern of mutation was different. The major mutations occurred within the SSR region (aa90-146) (Figure 4B). Particularly, two distinct mutations were identified at aa 173 and aa 178 (Figure 4B). Interestingly, ZJ1/CHN2016, JS1/CHN2015, and JS2/CHN2015 showed short deletion at aa123-146 (Figure 4B).
In the alignment of UL27, Genotype I showed different amino acid sequences in Region aa70-91 from Genotype II, while a distinguishing mutation was located at aa82 (Figure 4C). Interestingly, UL27 of HuB1/CHN2017 showed a different amino acid sequence in this region compared with other strains in Genotype II. One study showed that replacing the UL27 of the PRV JS-2012 strain with Bartha-K61 caused the attenuation of viral virulence [27]. Therefore, we speculated that the variations in UL27 might contribute to both antigenic drift and virulence variation. This hypothesis is worth further investigation in the future.
Several studies have demonstrated that, compared with classic strains isolated decades ago, novel PRV variants possess enhanced virulence, including a larger plaque size, severer clinical signs, and higher mortality in pigs [14,16,24,35]. Although the correlation of viral genome variability with pathogenicity enhancement is highly possible, the exact causative mutation sites are hard to locate in herpesvirus due to limited genome sequences and many single mutations in a large genome. We speculated that the causative mutations leading to virulence enhancement in variant PRV strains might be included in the unique mutations, which are collectively different from all other classic strains. Therefore, identifying unique mutations is crucial to understanding viral evolution.
In the present study, we determined the unique mutations that distinguish novel PRV variants in China except HuB1/CHN2017 from classic strains in China (Ea, Fa, SC) or classic strains isolated in both China and Europe/US. Surprisingly, these unique mutations are sparse and sporadic (Table 2). Among the listed proteins, UL5, UL15, and UL42 are the key components for viral genomic DNA replication and packages. UL36, UL46, and UL47 are important tegument proteins, essential for virion assembly and egress. UL27, UL44, UL49.5, and US4 are envelope glycoproteins [4]. UL13 is a serine/threonine protein kinase that promotes viral egress and neural virulence [4]. UL27 (gB) is the receptor-binding protein, containing the most important neutralizing epitopes and strongly associated with PRV virulence [4,27]. Previous studies have identified several neutralizing epitopes of gB, such as aa59-126 and aa204-223 [64,65]. In our study, four distinguishing mutations (T82A, R451K, H560Q, T737A, V895A) were identified in gB (Table 2), of which was T82A located within epitope aa59-126, which may cause antigenic drift.
Table 2.
Distinguishing mutations in newly sequenced PRV strains.
| ORF Name | Mutation Position $ | Function |
|---|---|---|
| UL2 | V107A * | UNG, DNA repair; Uracil-DNA glycosylase |
| UL3.5 | T173A &, A178T, | Viral egress (secondary envelopment); membrane-associated protein |
| UL5 | H250R | DNA replication; UL5 is helicase subunit of UL5/UL8/UL52 helicase/primase complex; |
| UL9 | A283T, W500R, P696T, | OBP, sequence-specific ori-binding protein, ATP-dependent helicase motif |
| UL13 | T169A | VP18.8, protein-serine/threonine kinase |
| UL14 | G145E | Virion tegument protein, |
| UL15 | D163N | Interacts with UL33, UL28 & UL6; DNA viral concatemeric DNA cleavage/encapsidation; terminase subunit of the UL15/UL28 complex; |
| UL27 | T82A, R451K, H560Q, T737A, V895A | Viral entry (fusion); cell–cell spread; glycoprotein B; type I membrane protein |
| UL36 | P/S292A, P/S/Q293A, R2286Q, A/G2068R, A3059T/G | Viral egress (capsid tegumentation); major tegument scaffold; interacts with UL37 and capsid |
| UL37 | M782V | Viral egress (capsid tegumentation); interacts with UL36 |
| UL42 | T270A | DNA replication; polymerase accessory subunit of UL30/UL42 holoenzyme |
| UL44 | G194E | Glycoprotein C (gC), viral entry (virion attachment); type I membrane protein; binds to heparan sulfate |
| UL47 | G67E, A219T | VP13/14, viral egress (secondary envelopment); tegument protein |
| UL49.5 | A49T | Glycoprotein N (gN), immune evasion (TAP inhibitor); type I membrane protein; complexed with gM |
| US1 | E211G, E260G | ICP22, acts as a regulator of gene expression |
| US4 | S82P | Glycoprotein G (secreted) |
| US6 | V338A, | Glycoprotein D (gD), viral entry (cellular receptor-binding protein); type I membrane protein |
| US8 | G54D, P403A S517P |
Glycoprotein E (gE), cell–cell spread; glycoprotein E; type I membrane protein; complexed with gI; C-terminus interacts with UL49; protein sorting in axons |
| IE180 | ΔΔ349-350RG, A841P, A842S, | ICP4, gene regulation (transcription activator); immediate-early protein |
*: The mutation sites listed in the table indicate that certain amino acid residues of every PRV strain isolated after 2011 except HuB1/CHN2017 are collectively different from the classic Chinese PRV strains Ea/Fa and SC, which distinguish the novel PRV variants from the classic Chinese PRV strains (Ea, Fa, and SC). &: The mutation sites with bold letters indicate that certain amino acid residues of every PRV strain isolated after 2011 except HuB1/CHN2017 are collectively different from the classic Chinese PRV strains Ea, Fa, and SC and all EU/US strains. $: Each amino acid residue number is the codon number in the corresponding ORF of reference genome Ea.
UL36 is one of the major scaffold proteins in the tegument, which is also essential for viral replication and neural invasiveness [4]. It was shown that the most functional essential regions of UL36 are located at the N terminal, while aa 2087-2795 deletion only has a marginal effect on viral replication in vitro [66]. The only deletion region at aa6-225 caused a lower viral titer in PK15, while the PRV-virus-bearing deletion at aa226-299, aa2026-2970, and aa3055-3078 showed no viral replication in vitro [67]. As shown in Table 2, four distinguishing mutations (P/S292A, P/S/Q293A, A/G2068R, A3059/G), identified in UL36, are located in the indispensable region, while R2286Q is located in the dispensable region. Whether these mutations contribute to viral virulence enhancement is still unclear. Until now, the functional essential motifs or aa residues of most PRV ORFs are still unknown. Although it is unable to link the evolutionary diversity sites with biological functions in most ORFs, the ORF diversity profile identified in our study has shed the light on genotype–phenotype studies in the future.
3.5. Selection Pressure Act on Each ORF
Intermolecular recombination is an important mechanism to increase genomic diversity, while spontaneous mutation caused by polymerase infidelity is another important mechanism leading to genome diversity [58]. A spontaneous mutation is accumulated by positive selection pressure but erased by negative selection pressure, which is also called purifying selection [58]. The ability to maintain the lifespan of persistent infection and its constant interaction with the host immune system highlights the substantial selective pressure on genome diversity. To characterize the overall selection pressure on each ORF, we determined the nonsynonymous substitutions per nonsynonymous site (dN), synonymous substitutions per synonymous site (dS), and the dN/dS ratio as evidence of purifying, neutral, or positive selection (Figure 5).
Figure 5.
Selection pressure analysis. Selection pressure of each ORF of PRV was analyzed. ORF sequences were extracted from each annotated genome. By using FUBAR in the Datamonkey web server, nonsynonymous substitutions per nonsynonymous site (dN), synonymous substitutions per synonymous site (dS), and the dN/dS ratio of each ORF were calculated. The result is plotted as a bar graph. The dN and dN/dS ratio read the major y-axis at the left side, and dS takes the minor y-axis at the right side.
As shown in Figure 5, most PRV genes show evolutionary constraints. The average dN/dS ratio is 0.26. Only 7.5% of the 67 ORFs have a dN/dS ratio higher than 0.6, and 88% of the ORFs have a ratio less than 0.3. A recent study of herpes simplex virus 1 (HSV-1) also showed a predominance of the evolutionary constraint with an average dN/dS ratio at 0.27 [38], and similar results were found in beta- and gamma-herpesviruses [56,68]. In general, a low dN/dS ratio, representing evolutionary constraint or negative selection pressure, suggests the excellent adaptation of the parasite within their hosts, with mutations mostly leading to negative fitness effects and being removed effectively [56,58]. The evolutionary constraint possibly correlates with the low diversity of newly sequenced PRV genomes in this study.
Five proteins exhibit a remarkably higher dN/dS ratio than that of most proteins: UL3, UL3.5, UL6, UL36, and US8 (Figure 5). The functions of UL3 and UL3.5 are largely unknown. In PRV, US8 (gE) shows close to neural selection or drift (dN/dS = 0.64), while the dN/dS ratio of US8 of HSV-1 is substantially lower (dN/dS = 0.19) [38]. gE is an essential neural virulence factor of PRV and one of the major targets of neutralizing antibodies. A relatively higher dN/dS ratio and diversity on gE imply that gE mutations may lead to antigenic drift or virulence enhancement. Considering all the PRV vaccine strains applied in pig farms have gE deletion, a higher dN/dS ratio in gE may result from the host immune response against wild PRV strains but not vaccination. In addition, nine ORFs (UL18, UL24, UL26.5, UL38, UL19, UL26, UL30, UL32, UL56) exhibit a relatively lower dN/dS ratio (lower than 0.2) (Figure 5). As expected, most of them are key components of DNA replication complex or essential virion structure proteins.
In Table 3, we list the amino acid residues under positive selection determined by FUBAR, MEME, and CodeML of PAML. Interestingly, most ORFs bearing positive selection residues are essential for viral replication or virion assembly, while only UL23, US8, and EP0 are dispensable for viral replication in vitro [4]. In UL27 (gB), the Residues 75 and 505 show significant positive selection. Our alignment of UL27 shows that Residue 505 is proline in most strains, while four strains (Bartha-K61, Kolchis, Hercules, ADV32751) show alanine at this position (Figure 4C). A previous study demonstrated that region aa495–505 (PAAARRARRSP) is the furin cleavage site of gB [69]. The furin cleavage site of gB is not required for PRV viral replication in vitro but plays an essential role for the virus to spread between cells [69]. In human cytomegalovirus (HCMV), the positive selection of the furin cleavage site of gB leads to different furin cleavage efficacy [70]. However, the functional significance of positive selection at the furin cleavage site of gB in PRV is still unclear.
Table 3.
Position of each codon under positive selection.
| ORF Name | Codon Position $ | Detection Method |
|---|---|---|
| UL5 | 574 | F*, M& |
| UL6 | 462, 464 | F, C# |
| UL9 | 283 | F, C |
| UL12 | 474 | F, M |
| UL15 | 674 | F, M |
| UL19 | 502 | F, M |
| UL23 | 293 | M, C |
| UL25 | 472 | F, C |
| UL27 | 75 | F, M, C |
| 505 | F, C | |
| UL36 | 561 | F, M |
| UL39 | 4 | F, M, C |
| UL52 | 662 | F, C |
| 663 | M, C | |
| US8 | 575 | F, C |
| 578 | M, C | |
| EP0 | 212 | F, M |
| IE180 | 1462 | F, M |
F*: FUBAR, posterior probability >90%; M&: MEME, p < 0.1; C#: CodeML, p < 0.05. $: All amino acid residue positions are the codon number in the corresponding ORF of reference genome Ea.
The residues under positive selection of UL27 (gB), UL44 (gC), and US8 (gE) were previously determined [36]. In their study, four positive selection residues of gB were detected by FUBAR and MEME (aa43, aa75, aa848, aa922), of which aa75 was also detected in our study [36]. Additionally, two positive selection residues were detected in gE (aa348, aa578). In our study, aa578 was also determined as positive selection sites in US8. No positive selection residues were detected in UL44 in our study, while two residues (aa59, aa194) were determined under positive selection in He’s study [36].
It should be noted that Residue 283 of UL9 is not only under positive selection (Table 3) but also a distinguishing mutation (A283T) (Table 2). This concordance strongly suggests that Residue 283 of UL9 might play a critical role in viral virulence enhancement.
4. Conclusions
In the study, we analyzed PRV genomic diversity and evolution with 73 PRV genomes, of which 54 genomes were newly sequenced in this study. Phylogenetic analysis divided PRV strains into two genotypes, which strongly supports geographical clustering and suggests PRV strains evolve independently in Asia and Europe. However, recombination analysis indicates that recombination between different genetic clades is robust, and the classic vaccine strain Bartha-K61 might contribute to the evolution process of novel PRV variants. Several distinguishing mutations were identified in 19 ORFs, which provided potential targets for further phenotype–genotype studies. Selection pressure analysis revealed that most ORFs of PRV are under strong purifying selection. Additionally, 19 amino acid residues were determined under positive selection within 15 different ORFs.
Acknowledgments
We would like to acknowledge Zhuofei Xu at Shanghai MasScience Biotechnology Co., Ltd. for his technical support during the study.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v13071322/s1, Table S1. PRV strains sequenced in this study. Table S2. Previously published PRV strains used in this study. Table S3. RDP results summary. Table S4. Recombination events detected in multiple PRV strains. Table S5. P value of each recombination event detected by RDP. Table S6. Location of gapped regions in each newly sequenced PRV genome.
Author Contributions
Conceptualization, R.H., Z.P. and B.W.; methodology, R.H., Q.L., L.H., X.H., Y.Z., J.F., H.C., W.S., W.L., N.D., Z.L., Z.D., X.T., Z.P.; writing—original draft preparation, L.W., Q.L., Z.P.; writing—review and editing, R.H., L.W., N.D., Z.L., Z.D., Z.P., B.W.; supervision, Z.P. and B.W.; project administration, Z.P. and B.W.; funding acquisition, B.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the National Key Research and Development Program of China (Grant Number: 2018YFD0500800), the Hubei Provincial Science and Technology Major Project (Grant Number: 2020ABA016), the Science and Technology Program of the Education Department of Jiangxi Province (Grant Number: GJJ190192), the Natural Science Foundation of Jiangxi Province (Grant Number: 20202BAB205004), and the Program for Jiangxi Agriculture Research System (Grant Number: JXARS-01).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in article and supplementary material.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mettenleiter T.C. Pseudorabies (Aujeszky’s disease) virus: State of the art. August 1993. Acta Vet. Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 2.Kluge J.P., Beran G.W., Hill H.T., Platt K.B. Pseudorabies (Aujeszky’s disease) In: Straw B.E., D’Allaire S., Mengeling W.L., Taylor D.J., editors. Diseases of Swine. Iowa State University Press; Ames, IA, USA: 1999. pp. 233–246. [Google Scholar]
- 3.Mettenleiter T.C. Aujeszky’s disease (pseudorabies) virus: The virus and molecular pathogenesis—State of the art, June 1990. Vet. Res. 2000;31:99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 4.Pomeranz L.E., Reynolds A.E., Hengartner C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mettenleiter T.C., Ehlers B., Müller T., Yoon K., Teifke J.P. Herpesviruses. In: Zimmerman J., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., editors. Diseases of Swine. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2012. pp. 421–446. [Google Scholar]
- 6.Ai J.W., Weng S.S., Cheng Q., Cui P., Li Y.J., Wu H.L., Zhu Y.M., Xu B., Zhang W.H. Human Endophthalmitis Caused By Pseudorabies Virus Infection, China, 2017. Emerg. Infect. Dis. 2018;24:1087–1090. doi: 10.3201/eid2406.171612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Nian H., Li Z., Wang W., Wang X., Cui Y. Human encephalitis complicated with bilateral acute retinal necrosis associated with pseudorabies virus infection: A case report. Int. J. Infect. Dis. 2019;89:51–54. doi: 10.1016/j.ijid.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Yang H., Han H., Wang H., Cui Y., Liu H., Ding S. A Case of Human Viral Encephalitis Caused by Pseudorabies Virus Infection in China. Front. Neurol. 2019;10:534. doi: 10.3389/fneur.2019.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X., Guan H., Li C., Li Y., Wang S., Zhao X., Zhao Y., Liu Y. Characteristics of human encephalitis caused by pseudorabies virus: A case series study. Int. J. Infect. Dis. 2019;87:92–99. doi: 10.1016/j.ijid.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Wong G., Lu J., Zhang W., Gao G.F. Pseudorabies virus: A neglected zoonotic pathogen in humans? Emerg. Microbes Infect. 2019;8:150–154. doi: 10.1080/22221751.2018.1563459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freuling C.M., Muller T.F., Mettenleiter T.C. Vaccines against pseudorabies virus (PrV) Vet. Microbiol. 2017;206:3–9. doi: 10.1016/j.vetmic.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y., Luo Y., Wang C.H., Yuan J., Li N., Song K., Qiu H.J. Control of swine pseudorabies in China: Opportunities and limitations. Vet. Microbiol. 2016;183:119–124. doi: 10.1016/j.vetmic.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Tong G.Z., Chen H.C. Pseudorabies epidemic status and control measures in China. Chin. J. Vet. Sci. 1999;19:1–2. (In Chinese) [Google Scholar]
- 14.An T.Q., Peng J.M., Tian Z.J., Zhao H.Y., Li N., Liu Y.M., Chen J.Z., Leng C.L., Sun Y., Chang D., et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 2013;19:1749–1755. doi: 10.3201/eid1911.130177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y., Li N., Cong X., Wang C.H., Du M., Li L., Zhao B., Yuan J., Liu D.D., Li S., et al. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet. Microbiol. 2014;174:107–115. doi: 10.1016/j.vetmic.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Tong W., Liu F., Zheng H., Liang C., Zhou Y.J., Jiang Y.F., Shan T.L., Gao F., Li G.X., Tong G.Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015;181:236–240. doi: 10.1016/j.vetmic.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Wu R., Bai C., Sun J., Chang S., Zhang X. Emergence of virulent pseudorabies virus infection in northern China. J. Vet. Sci. 2013;14:363–365. doi: 10.4142/jvs.2013.14.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang S., Zhou Z., Hu X., Li Y., Zhang C., Wang J., Li X., Tan F., Tian K. Complete Genome Sequence of a Variant Pseudorabies Virus Strain Isolated in Central China. Genome Announc. 2016;4:e00149-16. doi: 10.1128/genomeA.00149-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye C., Wu J., Tong W., Shan T., Cheng X., Xu J., Liang C., Zheng H., Li G., Tong G. Comparative genomic analyses of a virulent pseudorabies virus and a series of its in vitro passaged strains. Virol. J. 2018;15:195. doi: 10.1186/s12985-018-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu R.M., Zhou Q., Song W.B., Sun E.C., Zhang M.M., He Q.G., Chen H.C., Wu B., Liu Z.F. Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine. 2015;33:5733–5740. doi: 10.1016/j.vaccine.2015.09.066. [DOI] [PubMed] [Google Scholar]
- 21.Wang C.H., Yuan J., Qin H.Y., Luo Y., Cong X., Li Y., Chen J., Li S., Sun Y., Qiu H.J. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine. 2014;32:3379–3385. doi: 10.1016/j.vaccine.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 22.Fan J., Zeng X., Zhang G., Wu Q., Niu J., Sun B., Xie Q., Ma J. Molecular characterization and phylogenetic analysis of pseudorabies virus variants isolated from Guangdong province of southern China during 2013–2014. J. Vet. Sci. 2016;17:369–375. doi: 10.4142/jvs.2016.17.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y., Liang W., Liu Q., Zhao T., Zhu H., Hua L., Peng Z., Tang X., Stratton C.W., Zhou D., et al. Epidemiological and genetic characteristics of swine pseudorabies virus in mainland China between 2012 and 2017. PeerJ. 2018;6:e5785. doi: 10.7717/peerj.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X., Wu C.-X., Song X.-R., Chen H.-C., Liu Z.-F. Comparison of pseudorabies virus China reference strain with emerging variants reveals independent virus evolution within specific geographic regions. Virology. 2017;506:92–98. doi: 10.1016/j.virol.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Ye C., Zhang Q.Z., Tian Z.J., Zheng H., Zhao K., Liu F., Guo J.C., Tong W., Jiang C.G., Wang S.J., et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology. 2015;483:32–43. doi: 10.1016/j.virol.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Yu T., Chen F., Ku X., Zhu Y., Ma H., Li S., He Q. Complete Genome Sequence of Novel Pseudorabies Virus Strain HNB Isolated in China. Genome Announc. 2016;4:e01641-15. doi: 10.1128/genomeA.01641-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Z.-Q., Tong W., Zheng H., Li L.-W., Li G.-X., Gao F., Wang T., Liang C., Ye C., Wu J.-Q., et al. Variations in glycoprotein B contribute to immunogenic difference between PRV variant JS-2012 and Bartha-K61. Vet. Microbiol. 2017;208:97–105. doi: 10.1016/j.vetmic.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 28.Thiry E., Meurens F., Muylkens B., McVoy M., Gogev S., Thiry J., Vanderplasschen A., Epstein A., Keil G., Schynts F. Recombination in alphaherpesviruses. Rev. Med. Virol. 2005;15:89–103. doi: 10.1002/rmv.451. [DOI] [PubMed] [Google Scholar]
- 29.Christensen L.S., Lomniczi B. High frequency intergenomic recombination of suid herpesvirus 1 (SHV-1, Aujeszky’s disease virus) Arch. Virol. 1993;132:37–50. doi: 10.1007/BF01309842. [DOI] [PubMed] [Google Scholar]
- 30.Glazenburg K.L., Moormann R.J.M., Kimman T.G., Gielkens A.L.J., Peeters B.P.H. Genetic recombination of pseudorabies virus: Evidence that homologous recombination between insert sequences is less frequent than between autologous sequences. Arch. Virol. 1995;140:671–685. doi: 10.1007/BF01309957. [DOI] [PubMed] [Google Scholar]
- 31.Ye C., Guo J.C., Gao J.C., Wang T.Y., Zhao K., Chang X.B., Wang Q., Peng J.M., Tian Z.J., Cai X.H., et al. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology. 2016;491:56–63. doi: 10.1016/j.virol.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Liu H., Shi Z., Liu C., Wang P., Wang M., Wang S., Liu Z., Wei L., Sun Z., He X., et al. Implication of the Identification of an Earlier Pseudorabies Virus (PRV) Strain HLJ-2013 to the Evolution of Chinese PRVs. Front. Microbiol. 2020;11:3068. doi: 10.3389/fmicb.2020.612474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Offay J.M., Fulton R.W., Fishbein M., Eberle R., Dubovi E.J. Isolation of a naturally occurring vaccine/wild-type recombinant bovine herpesvirus type 1 (BoHV-1) from an aborted bovine fetus. Vaccine. 2019;37:4518–4524. doi: 10.1016/j.vaccine.2019.06.059. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.W., Markham P.F., Coppo M.J., Legione A.R., Markham J.F., Noormohammadi A.H., Browning G.F., Ficorilli N., Hartley C.A., Devlin J.M. Attenuated vaccines can recombine to form virulent field viruses. Science. 2012;337:188. doi: 10.1126/science.1217134. [DOI] [PubMed] [Google Scholar]
- 35.Yu T., Chen F., Ku X., Fan J., Zhu Y., Ma H., Li S., Wu B., He Q. Growth characteristics and complete genomic sequence analysis of a novel pseudorabies virus in China. Virus Genes. 2016;52:474–483. doi: 10.1007/s11262-016-1324-z. [DOI] [PubMed] [Google Scholar]
- 36.He W., Auclert L.Z., Zhai X., Wong G., Zhang C., Zhu H., Xing G., Wang S., He W., Li K., et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019;219:1705–1715. doi: 10.1093/infdis/jiy731. [DOI] [PubMed] [Google Scholar]
- 37.Szpara M.L., Tafuri Y.R., Enquist L.W. Preparation of viral DNA from nucleocapsids. J. Vis. Exp. 2011:3151. doi: 10.3791/3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szpara M.L., Gatherer D., Ochoa A., Greenbaum B., Dolan A., Bowden R.J., Enquist L.W., Legendre M., Davison A.J. Evolution and Diversity in Human Herpes Simplex Virus Genomes. J. Virol. 2014;88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szpara M.L., Tafuri Y.R., Parsons L., Shamim S.R., Verstrepen K.J., Legendre M., Enquist L.W. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 2011;7:e1002282. doi: 10.1371/journal.ppat.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Y., Leung H.C.M., Yiu S.M., Chin F.Y.L. IDBA—A Practical Iterative de Bruijn Graph De Novo Assembler. In: Berger B., editor. Research in Computational Molecular Biology. Springer; Berlin/Heidelberg, Germany: 2010. pp. 426–440. [Google Scholar]
- 41.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne I., Bayer M., Stephen G., Cardle L., Marshall D. Tablet: Visualizing Next-Generation Sequence Assemblies and Mappings. Methods Mol. Biol. 2016;1374:253–268. doi: 10.1007/978-1-4939-3167-5_14. [DOI] [PubMed] [Google Scholar]
- 43.Tsai I.J., Otto T.D., Berriman M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 2010;11:R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otto T.D., Dillon G.P., Degrave W.S., Berriman M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 2011;39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huson D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 48.Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Notredame C., Higgins D.G., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 50.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murrell B., Moola S., Mabona A., Weighill T., Sheward D., Kosakovsky Pond S.L., Scheffler K. FUBAR: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pond S.L., Frost S.D. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 53.Weaver S., Shank S.D., Spielman S.J., Li M., Muse S.V., Kosakovsky Pond S.L. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35:773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murrell B., Wertheim J.O., Moola S., Weighill T., Scheffler K., Kosakovsky Pond S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 56.Sijmons S., Thys K., Mbong Ngwese M., Van Damme E., Dvorak J., Van Loock M., Li G., Tachezy R., Busson L., Aerssens J., et al. High-Throughput Analysis of Human Cytomegalovirus Genome Diversity Highlights the Widespread Occurrence of Gene-Disrupting Mutations and Pervasive Recombination. J. Virol. 2015;89:7673–7695. doi: 10.1128/JVI.00578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolb A.W., Lewin A.C., Moeller Trane R., McLellan G.J., Brandt C.R. Phylogenetic and recombination analysis of the herpesvirus genus varicellovirus. BMC Genomics. 2017;18:1–17. doi: 10.1186/s12864-017-4283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Renner D.W., Szpara M.L. Impacts of Genome-Wide Analyses on Our Understanding of Human Herpesvirus Diversity and Evolution. J. Virol. 2017;92:1–13. doi: 10.1128/JVI.00908-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee K., Kolb A.W., Sverchkov Y., Cuellar J.A., Craven M., Brandt C.R. Recombination Analysis of Herpes Simplex Virus 1 Reveals a Bias toward GC Content and the Inverted Repeat Regions. J. Virol. 2015;89:7214. doi: 10.1128/JVI.00880-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loncoman C.A., Vaz P.K., Coppo M.J., Hartley C.A., Morera F.J., Browning G.F., Devlin J.M. Natural recombination in alphaherpesviruses: Insights into viral evolution through full genome sequencing and sequence analysis. Infect. Genet. Evol. 2017;49:174–185. doi: 10.1016/j.meegid.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 61.Muir W.B., Nichols R., Breuer J. Phylogenetic analysis of varicella-zoster virus: Evidence of intercontinental spread of genotypes and recombination. J. Virol. 2002;76:1971–1979. doi: 10.1128/JVI.76.4.1971-1979.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verpoest S., Cay A.B., De Regge N. Molecular characterization of Belgian pseudorabies virus isolates from domestic swine and wild boar. Vet. Microbiol. 2014;172:72–77. doi: 10.1016/j.vetmic.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Fuchs W., Granzow H., Klopfleisch R., Klupp B.G., Rosenkranz D., Mettenleiter T.C. The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress. J. Virol. 2005;79:11291–11299. doi: 10.1128/JVI.79.17.11291-11299.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaripov M.M., Morenkov O.S., Siklodi B., Barna-Vetro I., Gyongyosi-Horvath A., Fodor I. Glycoprotein B of Aujeszky’s disease virus: Topographical epitope mapping and epitope-specific antibody response. Res. Virol. 1998;149:29–41. doi: 10.1016/S0923-2516(97)86898-7. [DOI] [PubMed] [Google Scholar]
- 65.Li X., Yang F., Hu X., Tan F., Qi J., Peng R., Wang M., Chai Y., Hao L., Deng J., et al. Two classes of protective antibodies against Pseudorabies virus variant glycoprotein B: Implications for vaccine design. PLoS Pathog. 2017;13:e1006777. doi: 10.1371/journal.ppat.1006777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bottcher S., Klupp B.G., Granzow H., Fuchs W., Michael K., Mettenleiter T.C. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 2006;80:9910–9915. doi: 10.1128/JVI.01247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J.I., Luxton G.W., Smith G.A. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: The carboxy terminus directs incorporation into capsid assemblons. J. Virol. 2006;80:12086–12094. doi: 10.1128/JVI.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palser A.L., Grayson N.E., White R.E., Corton C., Correia S., Ba Abdullah M.M., Watson S.J., Cotten M., Arrand J.R., Murray P.G., et al. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J. Virol. 2015;89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okazaki K. Proteolytic cleavage of glycoprotein B is dispensable for in vitro replication, but required for syncytium formation of pseudorabies virus. J. Gen. Virol. 2007;88:1859–1865. doi: 10.1099/vir.0.82610-0. [DOI] [PubMed] [Google Scholar]
- 70.Stangherlin L.M., de Paula F.N., Icimoto M.Y., Ruiz L.G.P., Nogueira M.L., Braz A.S.K., Juliano L., da Silva M.C.C. Positively Selected Sites at HCMV gB Furin Processing Region and Their Effects in Cleavage Efficiency. Front. Microbiol. 2017;8:934. doi: 10.3389/fmicb.2017.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in article and supplementary material.