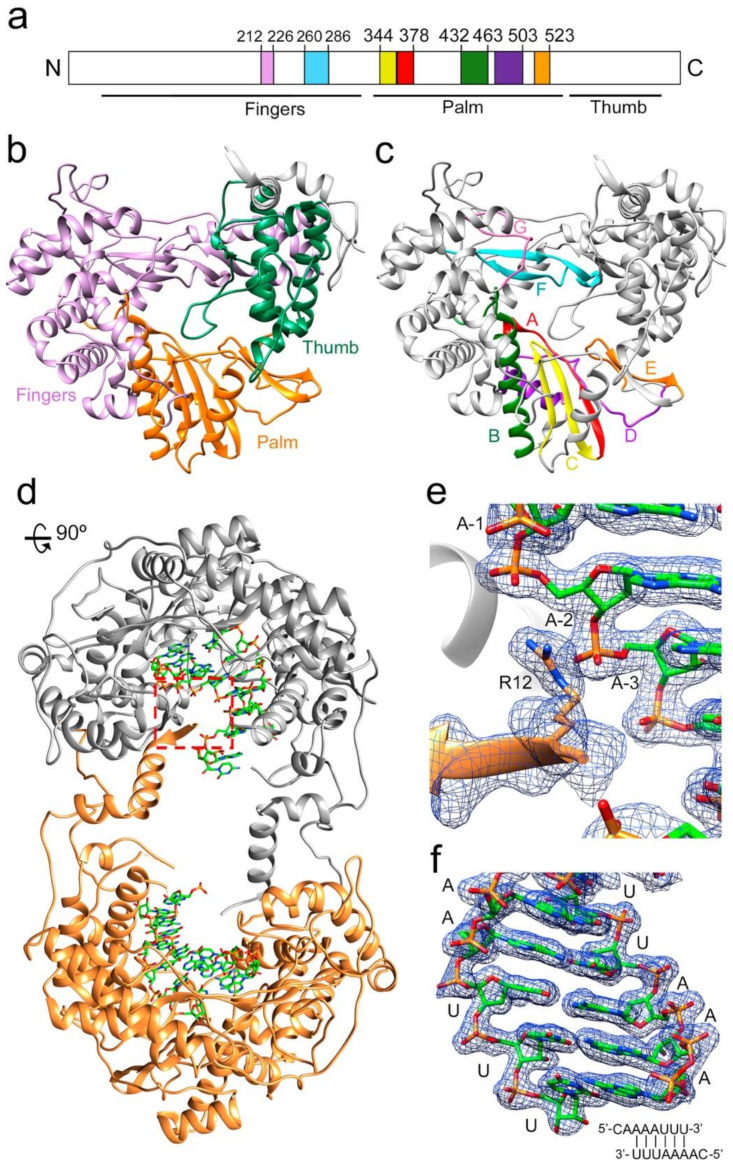
Figure 1.
Structure of the TaV RdRP and RdRP-RNA dimers. (a) linear representation of TaVpol subdomains (fingers, palm and thumb) and conserved structural motifs. Each motif is highlighted in different colors: G (pink) F (cyan), A(red), B (green), C (yellow), D (purple) and E (orange). (b) Ribbon diagram showing the overall architecture of the TaV RdRP with the finger, palm and thumb subdomains shown in different colors: fingers (violet), palm (orange) and thumb (green). (c) Ribbon diagram of the TaV enzyme showing the seven conserved motifs colored as in a. (d) The TaV RdRP-RNA dimers. The two associated molecules are shown as ribbons in grey and orange, respectively. Amino acids at the N- and C-terminal ends are responsible for stabilizing the dimeric structure of the enzyme that is maintained in all Δ10-TaVpol Δ607-624-RNA complexes analyzed. The RNA molecules bound to each subunit are shown as sticks in the atom type color: C, green; P, orange; O, red; N, blue. (e) Close-up of the squared region highlighting the interactions established between the N-terminal residue R12 of one molecule (orange) with the phosphates at positions -2 and -3 of the RNA template bound to the second RdRP molecule. The 2Fo-Fc electron density map around the region is displayed at a contour of 1.5σ (blue mesh). (f) The 2Fo-Fc electron density map shown around the dsRNA molecule. The sequence of the template-primer RNA is schematically represented at the panel bottom.