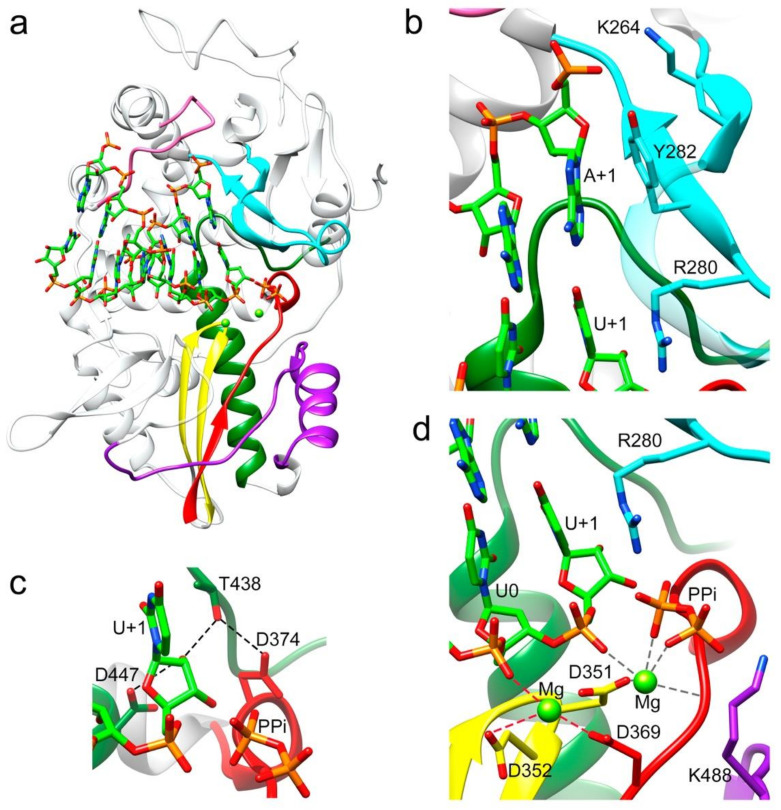
Figure 2.
Interactions between the TaV RdRP and the template/primer/product RNA in the RdRP catalytic cavity. (a) Side view of the overall structure of the Δ10-TaVpolΔ607-624/5′CAAAAUUU3′/UTP complex with the UMP molecule incorporated in a pretranslocation state. The polymerase molecule is shown as grey ribbons, with the conserved structural motifs highlighted in different colors: G (pink) F (cyan), A (red), B (green), C (yellow) and D (purple). The dsRNA molecule bound to the central cavity is represented as sticks in the atom type color: C green, P orange, O red and N blue. Panels b to d show different close up views of the RNA–protein interactions. Molecules are colored as in A and selected protein side chains are represented as sticks and explicitly labeled. (b) The template acceptor base A+1 appears to always be stacked to the Y282 side chain, whereas its triphosphate moiety appears salt bridged to K264 in motif F. A second stacking interaction was established between the guanidinium group of R280 (motif F) and the newly incorporated U in position +1. (c) The hydrogen bonding network involved in the 2′-hydroxyl recognition of the recently incorporated U+1 nucleotide. Hydrogen bonds are shown as black dashed lines. (d) Organization of the metal cluster after nucleotide incorporation into RNA, the product before translocation. Coordination distances < 2.5 Å for Mg2+ (B) are shown as dark grey dashed lines. In this structure, Mg2+ (A) is loosely coordinated as would be expected of a complex trapped in post-catalysis (distances < 3 Å are shown as red dashed lines). The liberated pyrophosphate (PPi) is present in the catalytic site, coordinated with Mg2+ (B) and salt bridged with residue R280.