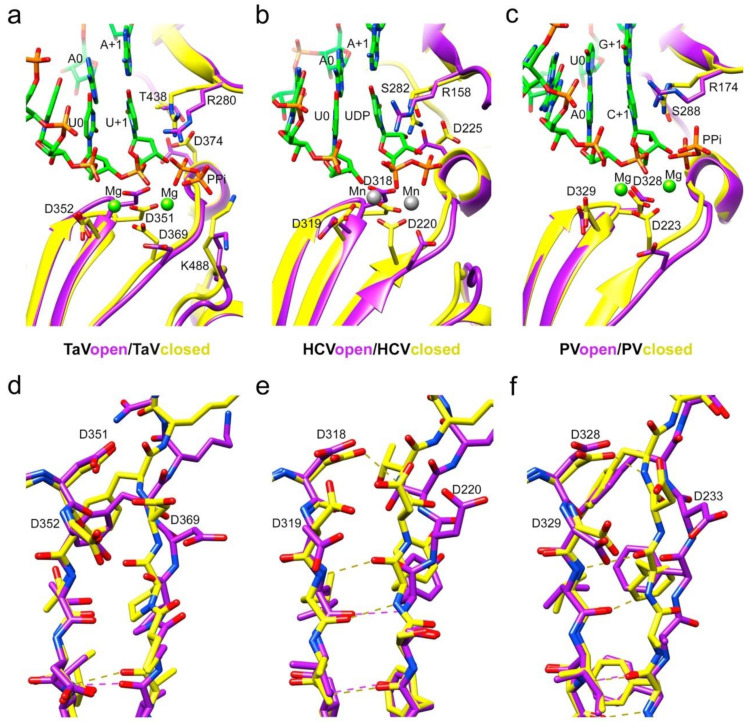
Figure 3.
Local rearrangements of the TaVpol active site on substrate binding. Comparison with the open and closed active site conformations in Flavivirus and Picornavirus RdRPs. (a) Ribbon representation of the TaVpol, apo form, showing an open conformation of the active site (magenta) compared with the TaVpolΔ607-624/5′CAAAAUUU3′ complex after UMP incorporation (closed conformation; yellow). Only the base pair A+1:U+1, A0:U0 and A-1:U-1 are shown in the stick representation in the atom type color: C green, P orange, O red and N blue. The pyrophosphate byproduct (PPi), present in the catalytic site, is also shown as sticks. The two Mg+2 ions present in the active site of the TaVpolΔ607-624/5′CAAAAUUU3′ complex are shown as green spheres. Changes in the main chain conformation and reorientation of side chains of residues: D351 (active site; motif C), D369 (active site; motif A) and D374 (NTP 2′OH recognition; motif A) and K448 (general acid; motif D) are apparent. (b) Ribbon representation of the HCV RdRP (closed, PDB id 4WTA, yellow) in complex with a template/primer RNA. Only the base A+1 and A0 in the template and U0 in the primer are shown as sticks as in A. The incoming UDP substrate (atom type sticks), base paired with A+1 and the two Mn2+ ions (grey spheres) are also shown and explicitly labeled. The HCV open conformation (purple) corresponds to the apo enzyme (PDB id: 2XI2). The side chains of the RNA-interacting residues in the catalytic site, motifs A to D, are shown as sticks and explicitly labeled. (c) Ribbon representation of PV the RdRP (PV closed, PDB id: 3OPL7, yellow) in complex with a template/primer/product RNA (with the base pairs G+1:C+1 and A0:U0 shown and explicitly labeled), and two Mg2+ ions (green spheres). The PPi byproduct is also shown as sticks. PV open (yellow) corresponds to a PV RdRP–RNA complex after reaction and translocation (PDB id: 3OL6). Panels d to f show stick representations of the superimpositions of the β-strands containing motifs C and A in TaVpol (d), HCV (e) and PV (f), with the main chain hydrogen bonds indicated in dashed lines and the side chains of the catalytic aspartic acid residues explicitly labeled.