Abstract
The B.1.1.7 SARS-CoV-2 strain that has emerged in the UK in early December presents seven mutations and three deletions on S-protein structure that could lead to a more infective strain. The P681H mutation in the “PRRAR” furin cleavage site might affect the binding affinity to furin enzyme and hence its infectivity.
Therefore, in this study, various structural bioinformatics approaches were used to model the S-protein structure with the B.1.1.7 variant amino acid substitutions and deletions. In addition to modelling the binding of furin to the cleavage site of the wild-type and the B.1.1.7 variant.
Conclusively the B.1.1.7 variant resulted in dynamic stability, conformational changes and variations in binding energies in the S-protein structure, resulting in a more favourable binding of furin enzyme to the SARS-CoV-2 S-protein.
Keywords: SARS-CoV-2, B.1.1.7 variant, Furin enzyme, Infectivity, Molecular modelling
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is associated with COVID-19 disease that emerged in Wuhan, China, in late 2019 is closely related to the SARS-CoV (2002) and MERS (2012) (Wang et al., 2020a). Despite the proofreading capacity during viral replication, the fast spread of the SARS-CoV-2 virus has led to a high rate of mutations in the viral proteins (Moelling, 2021; Plante et al., 2021). Consequently, resulting in evolved viral variants/strains that are more efficient in host cell entry (Shang et al., 2020) and evasion of the immune system (Wang et al., 2020b). The current SARS-CoV-2 strain (B.1.1.7) that has emerged in the UK (United Kingdom) in early December and extended to other parts of the world has created an alarming situation (Kirby, 2021; O'Toole et al., 2021). The B.1.1.7 strain consists of seven mutations and three residue deletions on the spike protein (S-protein) (Davies et al., 2021; Ostrov, 2021) compared to the initial SARS-CoV-2 strain that was reported in Wuhan, China (Wu et al., 2020), and the D614G strain that originated in Europe (Haddad et al., 2021; Phan, 2020).
The S-protein facilitates the SARS-CoV-2 viral invasion of the host cell by binding to angiotensin-converting enzyme 2 (ACE2), mainly expressed in the lungs, testes, and kidneys (Mohammad et al., 2020b; Wrapp et al., 2020). For the binding process to occur, the S-protein has to be cleaved by proteases. The SARS-CoV-2 S-protein has a unique polybasic “PRRAR” cleavage site, recognized by the furin enzyme (Coutard et al., 2020). Mutations and deletions in the S-protein sequence can alter the structure, affecting its stability and function, further exacerbating SARS-CoV-2 infectivity (Berger and Schaffitzel, 2020; Korber et al., 2020). Increased SARS-CoV-2 infectivity was observed with the S-protein-D614G mutation initially found in the European population (Phan, 2020; Zhang et al., 2020). Patients infected with SARS-CoV-2 D614G mutation manifested higher viral RNA levels and demonstrated higher titers with pseudoviruses in vitro infection assays, indicating that the D614G mutation is more permissive than the wild type (WT) virus first discovered in Wuhan, China(Korber et al., 2020). Furthermore, the structural analysis showed that the amino acid substitution aspartic acid (D) to glycine (G) at position 614 altered the structure of the S-protein, making it easy for the furin to be cleaved (Eaaswarkhanth et al., 2020; Jackson et al., 2021; Mohammad et al., 2020a; Zhang et al., 2020).
The B.1.1.7 SARS-CoV-2 variant strain exhibits missense mutations (N501Y, A570D, P681H, D614G, T716I, S982A, D1118H) and three deletions in residues H69, V70, Y144 (Davies et al., 2021). Notably, the N501Y mutation is located in the receptor-binding domain (RBD), which interacts with ACE2 and the deletions present on the N-terminal domain (NTD) of the S-protein (Khan et al., 2021b). One of the notable mutations in the B.1.1.7 variant is the P681H, which is a substitution of proline (P) to histidine (H) on the S-protein “PRRAR” at furin cleavage site. Since furin proteolysis occurs at a specific multi-basic sequence (R-[X]-(R/K)-R) (Tian et al., 2011), the substitution of P681H on the furin cleavage site may affect the structure of S-protein, hence enhancing the infectivity of the new SARS-CoV-2 variant. Therefore, it would be of interest to ascertain if all the mutations and deletions present in the B.1.1.7 variant affect the binding to furin or just the P681H mutation.
Recently it was shown that the D614G (European origin) substitution influenced the S-protein stability and enhanced binding of furin to S-protein “PRRAR” cleavage site as compared to the WT discovered in Wuhan, China (Eaaswarkhanth et al., 2020; Mohammad et al., 2020). Hence, in this study, we compared the S-protein-B.1.1.7 variant structure in complex with furin with the S-protein P681H-furin complex to determine if the B.1.1.7 variant or the P681H mutation affected furin cleavage. Therefore, we utilized structural bioinformatics tools to decipher the binding differences and structural-dynamic variations of S-protein-P681H and the B.1.1.7 variant. Furthermore, to understand the stability of the S-protein P681H mutation, compared the S-protein B.1.1.7 variant upon interaction with furin, both states were subjected to 100ns molecular dynamic (MD) simulations. In addition, we calculated the binding affinity of furin to S-protein-P681H and S-protein-B.1.1.7 variant using Generalized-Born surface area molecular mechanics (MM/GBSA). We show that the B.1.1.7 variant allows a tighter binding of the S-protein with furin, which justifies the higher infectivity than the WT strain of SARS-CoV-2.
2. Methods
2.1. Structural stability analysis
The structures of spike protein (PDB ID: 6VSB) (Wrapp et al., 2020) and furin (PDB ID: 4Z2A) (Pearce et al., 2019) were used as models for structural analysis in this report. SWISS-Model (Waterhouse et al., 2018) was used to model the missing amino acids that were not visible in the Cryo-EM structure of the S-protein for both WT-P681H and the B.1.1.7 variant. The DynaMut (Rodrigues et al., 2018) webserver was used to introduce mutations on the S-protein structure.
2.2. Protein-protein docking
Protein-protein complexes of WT-P681H and B.1.1.7 variant S-protein (PDB ID: 6VSB) with furin (PDB ID: 4Z2A) were modelled by employing a protein-protein docking approach using the HDOCK server (Yan et al., 2020; Yan et al., 2017), which is based on a hybrid algorithm of template-based modelling and ab initio free docking. Restraint docking was performed by defining the critical residues 682, 683, 684, and 685 of the RRAR site as binding site residues of the S-protein structure (PDB ID: 6VSB). Model 1, with the lowest docking energy score and the highest ligand RMSD (Xue et al., 2016).
2.3. MD simulations
Solvated complexes of WT-P681H and B.1.1.7 neutralized by counter ions were subjected to all atoms molecular dynamics simulation using force field (ff18) in AMBER20 simulation program (Salomon-Ferrer et al., 2013; Salomon‐Ferrer et al., 2013). Two-steps each 12000 and 6000 conjugate gradient energy minimization cycles were completed to relax the complexes and remove the bad clashes. The heating each complex was heated with default parameters set as 300 K for 200 ps. For density equilibration, using weak restraint for 2ns at constant pressure was executed. GPU accelerated, 100ns MD for each complex using constant pressure. To control the temperature Langevin thermostat with 1 atm pressure and 300K employed (Zwanzig, 1973), while Particle Mesh Ewald (PME) algorithm to evaluate long-range interactions, respectively, keeping the cutoff distances 10 Å. To treat the covalent interactions involving hydrogens SHAKE algorithm was used (Ryckaert et al., 1977).
Structural-dynamics features were estimated to understand the impact of S-protein-P681H and B.1.1.7 variant on the stability, flexibility, compactness of the protein, motion using CPPTRAJ and PTRAJ(Roe and Cheatham III, 2013). Thermodynamic stability of each complex was calculated as the root mean square deviation (RMSD), while the residual flexibility was evaluated as root mean square fluctuation (RMSF).
2.4. Binding energy differences estimation
To compute the binding differences elicited on the protein structures of S-protein-P681H and B.1.1.7, structural frames from the conformational dynamics were used to estimate binding free energy. MM/GBSA methods were employed to calculate the contributed Van der Waal, electrostatic and total binding energies of each complex (Hou et al., 2011). These methods are widely used and reported to have a strong correlation with experimental results (Khan et al., 2020; Khan et al., 2021a; Ylilauri and Pentikäinen, 2013). Each energy term such as vdW, electrostatic, GB and SA was calculated as a part of the total binding energy.
For Free Energy calculation, the following equation was used:
Each component of the total free energy was estimated using the following equation:
Gbond, Gele, and GvdW denote bonded, electrostatic, and van der Waals interactions, respectively. G-pol and Gnpol are polar and nonpolar solvated free energies. The Gpol and Gnpol are calculated by the generalized Born (GB) implicit solvent method with the solvent-accessible surface area SASA term.
3. Results and discussion
3.1. S-protein P681H and B.1.1.7 mutations
Genome sequencing of the new B.1.1.7 SARS-CoV-2 strain presented missense mutations (N501Y, A570D, P681H, D614G, T716I, S982A, D1118H) and three deletions in residues H69, V70, Y144. One of the notable mutations in the B.1.1.7 variant is the P681H, which is a substitution of proline (P) to histidine (H) on the S-protein “PRRAR” furin cleavage site. Since the substitution of P681H on the furin cleavage site may affect S-protein structure, enhancing the infectivity of the new SARS-CoV-2 variant. Here, we compared the S-protein-P681H structure with the S-protein-B.1.1.7 variant structure (Fig. 1 ).
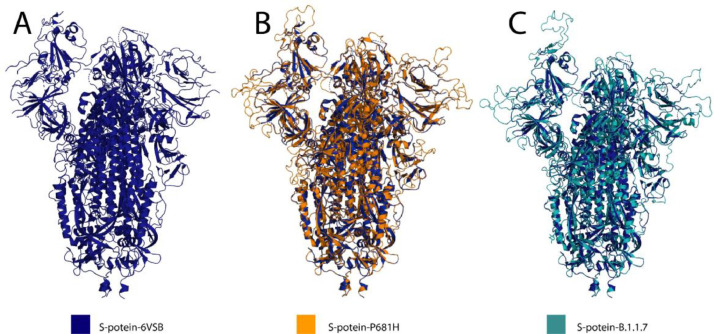
Fig. 1.
(A) SARS-CoV-2 S-protein trimeric structure PDB:6VSB (Blue). (B) Overlay of the modeled P681H S-protein (orange) and S-protein 6VSB (blue). (C) Overlay of the S-protein 6VSB (Blue) and modeled B.1.1.7 S-protein variant (teal).
The S-protein-P681H and S-protein-B.1.1.7 variant were modeled with the Swiss Model server (Waterhouse et al., 2018) using the 3.5-Å-resolution cryo-EM structure of the SARS-CoV-2 S-protein (PDB ID: 6VSB) (Wrapp et al., 2020) as a scaffold (Fig. 1A). The S-protein-P681H (Fig. 1B) structure showed a root-mean-square deviation (RMSD) of 0.25 Å between modeled and 6VSB structure. Whereby, the modeled B.1.1.7 (Fig. 1C) variant structure presented a 1.093Å RMSD difference with S-protein 6VSB, indicating that the mutations and deletions have induced substantial conformational changes to the S-protein structure, affecting the function of the S-protein.
3.2. Furin docking to P681H and B.1.1.7
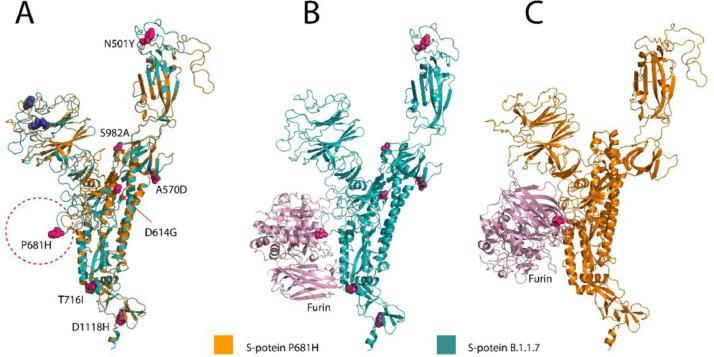
Furin-S-protein docking was conducted with HDOCK (Yan et al., 2020; Yan et al., 2017) to elucidate how the conformational changes of P681H and B.1.1.7 S-protein structures affect the binding to furin. Model 1 was chosen (Dudenhoeffer et al., 2019) as the best fit for structural analysis. The S-protein cleavage site is positioned in a surface-exposed loop region, making it accessible for furin binding. Structural analysis of the B.1.1.7 loop region demonstrated a conformational change compared to WT S-protein (Fig. 2 A). The introduction of conformational changes near the binding site can influence the furin cleavage process (Yang et al., 2014). Moreover, surface accessible surface area (SASA) analysis showed that the B.1.1.7 (P681H) HRRAR, in the furin cleavage site residues, demonstrated a SASA at the H (163.075), A (176.744), A (216.679), A (100.593), A (84.162), in contrast, the WT PRRAR furin site showed a SASA of P (137.106), A (119.285), A (115.560), A (7.003), A (71.406). Consequently, the higher exposure of the B.1.1.7 S-protein furin cleavage site, making it more accessible to furin cleavage.
Fig. 2.
(A) Overlay of the P681H S-protein (orange) and B.1.1.7 variant (teal), residue mutations are depicted in magenta and deleted residues in blue. (B and B) Modeled furin enzyme (pink) bound to B.1.1.7 (teal) and P6816 (orange) S-protein furin cleavage site.
3.3. Molecular dynamic simulations of P681H and B.1.1.7 S-protein bound to furin
To characterize the structural stability and dynamic features at the atomic and spatial resolution, S-protein P681H-Furin and B.1.1.7–furin complexes, we ran 100 ns MD simulations using AMBER 20 package. The impact of S-protein-B.1.1.7 and P681H on the interaction with furin was examined by calculating the Cα-atoms root mean square deviation (RMSD) and root-mean-square fluctuation (RMSF) trajectories.
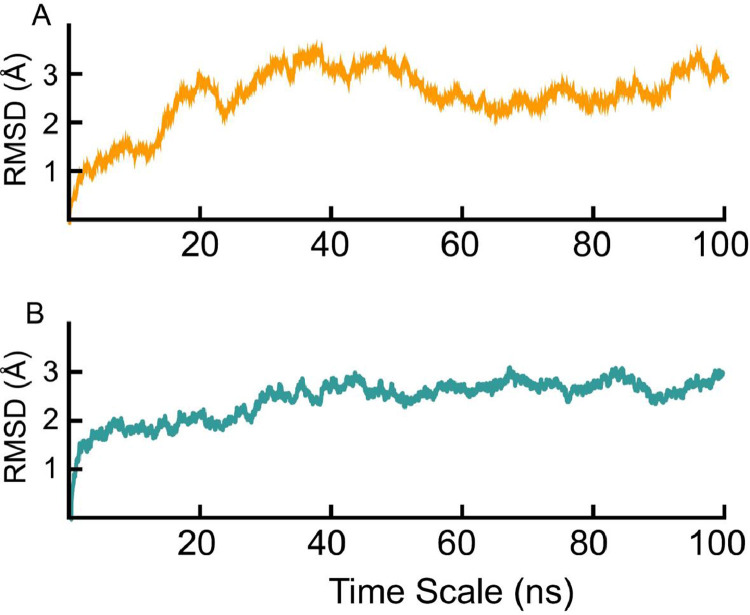
The stability and dynamics of P681H–furin and B.1.1.7–furin S-protein complexes are presented by RMSD values (Fig. 3 A and B). The P681H–furin complex demonstrated a deviation from 0 to 3 Å in the initial 20 ns. The RMSD, then decreased back to 2 Å at 24 ns, deviating to 4 Å during the simulation from 24-40 ns. Subsequently, from 40 to 100 ns, the S-protein-P681H-furin complex displayed high dynamic fluctuations presenting high structural perturbation, with the RMSD decreasing to 2 Å between 40-60 ns and then increased back to 3 Å at the end of the simulation. On the other hand, the S-protein-B.1.1.7-furin complex (Fig. 3B) exhibited a more stable system, where the complex converged to 2 Å at 5 ns, compared to P681H.Subsequentlythe complex stabilised at 2 Å from 5 to 30 ns. After which, the complex converged to 2.5 Å from 30 to 35 ns, where the B.1.1.7-furin complex fluctuated between 2.5 to 3 Å, for the remainder of the simulation (100 ns) indicating a more stable complex than P681H-furin complex.
Fig. 3.
Molecular dynamic simulations RMSD plots at 100 ns simulations of S-protein (A) P681H-furin (orange) and (B) S-protein B.1.1.7-furin (teal).
RMSD results for the B.1.1.7–furin complex revealed a stable system that affected the S-protein stability and, therefore, the binding to furin. For instance the global dynamics of the new variants revealed that mutations which increase the stability are strongly corelated with higher infectivity (Starr et al., 2020). This trend was also reported by a recent study on the B.1.617 variant, where the enhanced infectivity was correlated with the increased infection(Khan et al.).The dynamic structure, as a result of the seven mutations and three deletions present on the S-protein structure, has affected the conformational dynamics of the S-protein and consequently conformational optimization space interaction with furin. Consequently, the B.1.1.7-Furin complex remained stable, thus following the global stability trend and enhances the infectivity.
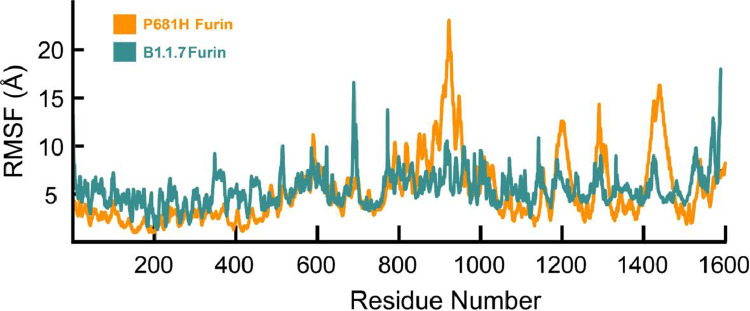
The RMSF values from the 100 ns simulations of S-protein-P681H and B.1.1.7 bound to furin are depicted in Fig. 4 . The P681H and B.1.1.7-furin complexes demonstrated a similar RMSF profile with the P681H-complex demonstrating higher residual fluctuations neat the loop region between residues 800 and 1000 where furin binds to S-protein. The mutations and deletions on the S-protein B.1.1.7 complex have resulted in a conformational change allowing a more stable complex with furin, as shown from the stable internal dynamics of the B.1.1.7-furin complex. In general, conformational changes that result in higher mobility of the structure might result in a more stable complex when interacting with an enzyme (Jernigan et al., 2020; Ruvinsky et al., 2012). The S-protein–furin binding interaction occurs through conformational selection or an induced-fit mechanism (Tsai et al., 2001, Yang et al., 2014), resulting in a more stable structure between B.1.1.7 and furin than the P681H-furin complex.
Fig. 4.
RMSF plot of S-protein P681H-furin (orange) and S-protein B.1.1.7-Furin complex (teal).
3.4. Binding energies of furin to P681H and B.1.1.7
The total binding free energy of the S-protein-furin complexes was calculated using the MM/GBSA method (Kollman et al., 2000) (Table 1 ). The binding affinity of furin to B.1.1.7 S-protein presented binding free energy of −57.08±0.73 (kcal/mol) compared with −12.4±0.45 kcal/mol for furin bound to P681H S-protein, indicating that the mutations and deletions present in the B.1.1.7 variant S-protein structure resulted in a tighter binding to furin than the S-protein-P681H mutation.
Table 1.
The MMGBSA binding free energy variables (Kcal/mol) of S-protein WT, P681H and B.1.1.7 variant in complex with furin.
| Complexes | vdW | Electrostatic | SASA | Total Binding Energy (ΔG) |
| WT-Furin* | 105.35±0.15 | 559.47±1.28 | 11.97±0.08 | -56.78±0.38 |
| G614-Furin* | -103.67±0.11 | -456.58±2.28 | -11.65±0.07 | -61.90±0.32 |
| P681H-Furin | -62.81±1.13 | 249.30±4.16 | -8.92±0.11 | -12.39±0.45 |
| B.1.1.7-Furin | -110.29±0.63 | -339.57±5.75 | -15.56±0.05 | -57.08±0.73 |
Data presented is taken from previously published results (Mohammad et al., 2020a).
In an earlier study, on the WT-S-protein-furin complex demonstrated a binding free energy of -56.78±0.38 (kcal/mol), whereas the S-protein-G614 showed binding energy of -61.90±0.32 (kcal/mol) to furin (Mohammad et al., 2020a) (Table 1). In the current study, the B.1.1.7-furin complex demonstrated binding energy of -57.08±0.78 (kcal/mol), which is tighter than the WT-furin complex by ~1 (Kcal/mol). However, the B.1.17-furin complex is weaker than the G614-furin complex by ~ 3 (Kcal/mol). As such, this would suggest that furin interaction to B.1.1.7 variant may result in more efficient cleavage of the S-protein and subsequent enhanced viral entry, resulting in a more virulent strain than WT but not the G614 strain. Nevertheless, the B.1.1.7 variant possess multiple mutations, especially on the RBD domain that interacts with ACE2. Whereby, a recent structural study involving B.1.17 variant suggested the N501Y variant in the RBD domain enhanced the binding to ACE2, resulting in a more infective virus strain (Khan et al., 2021b, Khan et al., 2021).
4. Conclusion
Here in this study, we used biomolecular docking and simulation approaches to compare the binding affinities of furin to S-protein P681H and B.1.1.7 variant structures. The B.1.1.7 variant demonstrated seven mutations and three deletions, with mutation P681H on the furin cleavage site. Henceforth, we analyzed the S-protein-B.1.1.7 structure in complex with furin and compared it with the S-protein P681H complex to ascertain if the variant affected the furin binding P681H mutation affected furin cleavage using computational modelling and structural-dynamics approaches. S-protein- B.1.1.7 variant resulted in a conformational change in the furin cleavage site, subsequently affecting the binding affinity of furin to the S-protein. As a result, the S-protein B.1.1.7 variant may enhance viral entry to the host cell causing higher infectivity. A larger-scale investigation will be critical to build a deep understanding of the furin-S-protein interactions and their functional consequences on the spread of the new B.1.1.7 SARS-CoV-2 variant.
CRediT authorship contribution statement
Anwar Mohammad: Conceptualization, Formal analysis, Methodology, Writing – original draft. Jehad Abubaker: Formal analysis, Writing – original draft, Writing – review & editing. Fahd Al-Mulla: Writing – review & editing, Conceptualization.
Declaration of Competing Interest
All authors have no conflict of interest to declare.
Acknowledgement
This study was supported by the Kuwait Foundation for the Advancement of Sciences.
References
- Berger I., Schaffitzel C. The SARS-CoV-2 spike protein: balancing stability and infectivity. Cell Res. 2020;30(12):1059–1060. doi: 10.1038/s41422-020-00430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., van Zandvoort K., Silverman J.D., Group C.C.-W., Consortium C.-G.U., Diaz-Ordaz K., Keogh R., Eggo R.M., Funk S., Jit M., Atkins K.E., Edmunds W.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudenhoeffer B.R., Schneider H., Schweimer K., Knauer S.H. SuhB is an integral part of the ribosomal antitermination complex and interacts with NusA. Nucleic. Acids. Res. 2019;47(12):6504–6518. doi: 10.1093/nar/gkz442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaaswarkhanth M., Madhoun A.A., Al-Mulla F. Could the D614 G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad D., John S.E., Mohammad A., Hammad M.M., Hebbar P., Channanath A., Nizam R., Al-Qabandi S., Al Madhoun A., Alshukry A., Ali H., Thanaraj T.A., Al-Mulla F. SARS-CoV-2: Possible recombination and emergence of potentially more virulent strains. PLoS One. 2021;16(5) doi: 10.1371/journal.pone.0251368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.B., Zhang L., Farzan M., Choe H. Functional importance of the D614G mutation in the SARS-CoV-2 spike protein. Biochem. Biophys. Res. Commun. 2021;538:108–115. doi: 10.1016/j.bbrc.2020.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan R.L., Sankar K., Jia K., Faraggi E., Kloczkowski A. Computational ways to enhance protein inhibitor design. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.607323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Khan M.T., Saleem S., Junaid M., Ali A., Ali S.S., Khan M., Wei D.-Q. Structural Insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020 doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Wei D.-Q., Kousar K., Abubaker J., Ahmad S., Ali J., Al-Mulla F., Ali S.S., Nizam-Uddin N., Sayaf A.M. Preliminary Structural Data Revealed that the SARS-CoV-2 B. 1.617 Variant's RBD binds to ACE2 receptor stronger than the Wild Type to Enhance the Infectivity. ChemBioChem. 2021 doi: 10.1002/cbic.202100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cellular Physiol. 2021 doi: 10.1002/jcp.30367. n/a(n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9(2):e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D.A., Cheatham T.E. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelling K. Within-host and between-host evolution in SARS-CoV-2-new variant's source. Viruses. 2021;13(5) doi: 10.3390/v13050751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Alshawaf E., Marafie S.K., Abu-Farha M., Abubaker J., Al-Mulla F. Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Marafie S.K., Alshawaf E., Abu-Farha M., Abubaker J., Al-Mulla F. Structural analysis of ACE2 variant N720D demonstrates a higher binding affinity to TMPRSS2. Life Sci. 2020;259 doi: 10.1016/j.lfs.2020.118219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole A., Hill V., Pybus O.G., Watts A., II Bogoch, Khan K., Messina J.P., consortium C.-G.U., South Network for Genomic Surveillance in, A. Brazil, U.K.C.G.N. Tegally, H. Lessells, R.R. Giandhari, J. Pillay, S. Tumedi, K.A. Nyepetsi, G. Kebabonye, M. Matsheka, M. Mine, M. Tokajian, S. Hassan, H. Salloum, T. Merhi, G. Koweyes, J. Geoghegan, J.L. de Ligt, J. Ren, X. Storey, M. Freed, N.E. Pattabiraman, C. Prasad, P. Desai, A.S. Vasanthapuram, R. Schulz, T.F. Steinbruck, L. Stadler, T. Swiss Viollier Sequencing, C. Parisi, A. Bianco, A. Garcia de Viedma, D. Buenestado-Serrano, S. Borges, V. Isidro, J. Duarte, S. Gomes, J.P. Zuckerman, N.S. Mandelboim, M. Mor, O. Seemann, T. Arnott, A. Draper, J. Gall, M. Rawlinson, W. Deveson, I. Schlebusch, S. McMahon, J. Leong, L. Lim, C.K. Chironna, M. Loconsole, D. Bal, A. Josset, L. Holmes, E. St George, K. Lasek-Nesselquist, E. Sikkema, R.S. Oude Munnink, B. Koopmans, M. Brytting, M. Sudha Rani, V. Pavani, S. Smura, T. Heim, A. Kurkela, S. Umair, M. Salman, M. Bartolini, B. Rueca, M. Drosten, C. Wolff, T. Silander, O. Eggink, D. Reusken, C. Vennema, H. Park, A. Carrington, C. Sahadeo, N. Carr, M. Gonzalez, G. Diego. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Wellcome Open Res. 2021;6:121. doi: 10.12688/wellcomeopenres.16661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrov D.A. Structural consequences of variation in SARS-CoV-2 B.1.1.7. J. Cell Immunol. 2021;3(2):103–108. doi: 10.33696/immunology.3.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce K.H., Overton L.K., Gampe R.T., Barrett G.B., Taylor J.D., McKee D.D., Campobasso N., Nolte R.T., Reid R.A. BacMam production and crystal structure of nonglycosylated apo human furin at 1.89 A resolution. Acta Crystallogr. F Struct. Biol. Commun. 2019;75(Pt 4):239–245. doi: 10.1107/S2053230X19001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19′s next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic. Acids. Res. 2018;46(W1):W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe D.R., Cheatham III T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Ruvinsky A.M., Kirys T., Tuzikov A.V., Vakser I.A. Structure fluctuations and conformational changes in protein binding. J. Bioinform. Comput. Biol. 2012;10(2) doi: 10.1142/S0219720012410028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23(3):327–341. [Google Scholar]
- Salomon-Ferrer R., Götz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscipl. Rev. 2013;3(2):198–210. [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Huang Q., Fang Y., Wu J. FurinDB: a database of 20-residue furin cleavage site motifs, substrates and their associated drugs. Int. J. Mol. Sci. 2011;12(2):1060–1065. doi: 10.3390/ijms12021060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Hozumi Y., Yin C., Wei G.W. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. J. Chem. Inf. Model. 2020 doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic. Acids. Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32(23):3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- Yan Y., Tao H., He J., Huang S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020;15(5):1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- Yan Y., Zhang D., Zhou P., Li B., Huang S.Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic. Acids. Res. 2017;45(W1):W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.Q., Sang P., Tao Y., Fu Y.X., Zhang K.Q., Xie Y.H., Liu S.Q. Protein dynamics and motions in relation to their functions: several case studies and the underlying mechanisms. J. Biomol. Struct. Dyn. 2014;32(3):372–393. doi: 10.1080/07391102.2013.770372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylilauri M., Pentikäinen O.T. MMGBSA as a tool to understand the binding affinities of filamin–peptide interactions. J. Chem. Informat. Model. 2013;53(10):2626–2633. doi: 10.1021/ci4002475. [DOI] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., Li W., Izard T., Rader C., Farzan M., Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11(1):6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwanzig R. Nonlinear generalized Langevin equations. J. Statistical Phys. 1973;9(3):215–220. [Google Scholar]