Abstract
Objectives
The emergence of new variants of concern (VOCs) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) around the world significantly complicated the exit from Coronavirus disease 2019 (COVID-19) pandemic. The aim of this study was to evaluate the serum neutralizing activity of three cohorts.
Methods
BNT162b2-elicited serum (N = 103), candidates as hyper-immune plasma donors (N = 90) and patients infected with the SARS-CoV-2 P1 variant (N = 22) were enrolled. Three strains of SARS-CoV-2 have been tested: 20A.EU1, B.1.1.7 (alpha) and P.1 (gamma). Neutralizing antibodies (NT-Abs) titers against SARS-CoV-2 were evaluated.
Results
B.1.1.7 and P.1 are less efficiently neutralized by convalescent wild-type infected serums if compared to 20A.EU1 strain (mean titer 1.6 and 6.7-fold lower respectively). BNT162b2 vaccine-elicited human sera show an equivalent neutralization potency on the B.1.1.7 but it is significantly lower for the P.1 variant (mean titer 3.3-fold lower). Convalescent P.1 patients are less protected from other SARS-CoV-2 strains with an important reduction of neutralizing antibodies against 20A.EU1 and B.1.1.7, about 12.2 and 10.9-fold, respectively.
Conclusions
BNT162b2 vaccine confers immunity against all the tested VOCs, while previous SARS-CoV-2 infection may be less protective.
Keywords: COVID-19, SARS-CoV-2, Vaccine, Variant, Antibodies, Neutralizing
Introduction
More than one year after the declaration of Coronavirus disease 2019 (COVID-19) as a pandemic,1 severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is still spreading around the world causing serious public health concerns. A number of effective vaccines are being administered at an unprecedented pace, decreasing the incidence and severe consequences of SARS-CoV-2 infection. However, massive and prolonged worldwide replication let SARS-CoV-2 rapidly explore its genetic space and new variants of concern (VOCs) eventually emerged by the end of 2020. Briefly, VOC indicates a variant for which there is evidence of increased transmissibility and possibly more severe disease as well as decreased neutralization by antibodies elicited by natural infection or vaccination with the original lineage, reduced effectiveness of treatments and diagnostic detection failures. The most relevant variants emerged in United Kingdom (known as 20I/501Y.V1, VOC 202,012/01, B.1.1.7, or alpha), South Africa (known as 20H/501Y.V2, B.1.351, or beta) and Brazil (known as P.1, or gamma) during 2020 but started to spread all around the world between December 2020 and January 2021.2 These variants possess different mutations on the receptor-binding domain (RBD) of the spike protein responsible for binding to the ACE2 receptor on the human cell surface. Since the RBD is a major target for neutralizing antibodies and all the available vaccines have been produced based on the “original” spike protein, the efficacy of vaccine-induced immune response is being questioned. Furthermore, convalescent people previously infected and recovered from SARS-CoV-2 infection may no longer be protected against SARS-CoV-2 reinfection.3
In Italy, the SARS-CoV-2 lineage B.1, clade 20A.EU1 circulating across Europe was first identified in March 2020 and remained dominant up to November 2020.4 By the end of February 2021, surveillance data of Umbria region, Italy, revealed the SARS-CoV-2 variants B.1.1.7 and P.1 as accounting for 36.2% and 51.1% of the total cases analyzed, respectively.5
The vaccination campaign started by the end of December 2020 with the administration of BNT162b2 (Comirnaty® - BioNTech / Pfizer) COVID-19 mRNA vaccine, firstly to healthcare workers. The original aim of this study was to evaluate the dynamics and duration of SARS-CoV-2 neutralizing activity of BNT162b2-elicited antibody. The subsequent emergence of the different VOCs allowed us to expand the study design and analyze the cross-neutralization pattern with the different variants by using sera from subjects vaccinated with BNT162b2 or infected with different virus lineages. In particular, samples from patients infected with the SARS-CoV-2 P.1 variant have been included in the study together with the ones having a history of SARS-CoV-2 infection from June to October 2020 and candidates as hyper-immune plasma donors.
Materials and methods
Design, setting and participants
This research was approved by the Ethics Committee of the Umbria Region (protocol number 20,686/21/OV) and it was conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent. The study used a longitudinal cohort design, with three separate cohorts: healthcare workers of Perugia Hospital vaccinated with BNT162b2 (Comirnaty® - BioNTech/Pfizer) COVID-19 mRNA vaccine; patients with a history of SARS-CoV-2 infection from June to October 2020 and candidates as hyper-immune plasma donors; patients with documented SARS-CoV-2 P1 variant infection.
Samples of vaccinated patients were withdrawn after 14–21 days from the second dose.
Furthermore, the subgroup of candidate plasma donors with high neutralizing antibodies (NT-Abs) titers (≥1:160) was also analyzed separately to establish if the high titer would be confirmed for the B.1.1.7 and P.1 strains.
SARS-CoV-2 strains and Vero E6 cell cultures
All experiments were performed using three SARS-CoV-2 strains isolated in our Biosafety Level 3 (BSL3) virology laboratory at Santa Maria della Misericordia Hospital, Perugia, Italy, as previously described.6 Briefly, the transport medium (UTM) of a nasopharyngeal swab was incubated with a 1:1 nystatin (10,000 U/mL) and penicillin-streptomycin (10,000 U/mL) mixture for 1 h at 4 °C to remove bacterial/fungal contamination. The suspension was centrifuged at 400 × g for 10 min, and the supernatant was inoculated on an African green monkey kidney clone E6 (Vero E6) cells monolayer maintained in Eagle's minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37 °C with 5% CO2. The viral titer in the supernatant was determined by Half-maximal Tissue Culture Infectious Dose (TCID50) endpoint dilution assay7 and stock aliquots were stored at −80 °C. Whole-genome sequencing of multiple isolates was used to identify a SARS-CoV-2 genome belonging to clade 20A.EU1 (lineage B.1) and clustered with viruses circulating in Italy in spring 2020, a SARS-CoV-2 genome belonging to clade B.1.1.7 (better known as alpha variant), and a SARS-CoV-2 genome belonging to clade P.1 (also known as gamma variant).8 The SARS-CoV-2 clade 20A.EU1 (lineage B.1) strain was isolated in May 2020 from a symptomatic patient during the first wave of infections. SARS-CoV-2 clade B.1.1.7 and P.1 were isolated on January 2021 during the third wave. Single virus stock aliquots were thawed immediately before each experiment and discarded after use.
SARS-CoV-2 neutralization test
Neutralizing antibodies (NT-Abs) titers against SARS-CoV-2 were evaluated using flat-bottom tissue culture 96-well microtiter plates as previously published.9 One day prior to the experiment, Vero E6 cells (2.5 × 104/well) were cultivated in MEM + 10% FBS in a 96-well plate incubated at 37 °C + 5% CO2. Serum samples of each study cohort were heat-inactivated for 30 min at 56 °C before to be two-fold serially diluted from 1:10 to 1:640 with MEM + 2% FBS and seeded into 96-well microtiter plates. Each sample was tested in duplicate and mixed with an equal volume of medium containing 50 TCID50 of SARS-CoV-2 selected strains and the plates were incubated for 30 min at 37 °C. Sera with known neutralization titer and medium were used as positive and negative control, respectively. Subsequently, the mixtures were transferred to Vero E6 cell containing plates and incubated for 72 h at 37 °C + 5% CO2. Then, the medium was removed and the cells were fixed and stained with 0.25% crystal violet and 10% formalin for 30 min at room temperature. The plates were washed to remove excess staining and the absorbance at 595 nm was measured using a microtiter plate reader (Multiskan Fc Photometer, Thermo Fisher Scientific). The neutralizing titer was determined as the maximum dilution showing reduction ≥ 90% of CPE respect to the virus control.
Sanger sequencing and variant assignation
Viral RNA was extracted from 150 µl of viral stock using the ZR Viral RNA Kit (Zymo Research), according to the manufacturer's protocol. The cDNA was generated by random hexamer-driven reverse transcription using 10 µl of heat-denaturated RNA extract, 660 µM dNTPs, 6 µl 5X ImProm-II TM Reaction Buffer, 50 ng hexanucleotides, 1.5 mM MgCl2, 20 U RNasin® Plus RNase Inhibitor and 1 U of ImProm-II™ Reverse Transcriptase (Promega) in a final volume of 30 µl. Reactions were run for 30 min at 37 °C followed by enzyme inactivation for 5 min at 80 °C. The cDNA was used as the template to amplify a spike region of about 2000 nucleotides spanning the mutations involved in variant designation. To design conserved primers, the alignment of reference genomes available at Gisaid site was used (https://www.gisaid.org/). The PCR mixture included 3 µl cDNA, 5 µl 5X Q5 Reaction buffer (NEB), 4 pmol P1004-Fwd (5′-AATTAGAGAAAACAACAGAGT-3′) and P976-Rev (5′- AAATTTGTGGGTATGGCAATAGAGTTA-3′), 150 µM dNTPs and 0.5 U Q5 Hot Start High-Fidelity DNA Polymerase (NEB) in a final volume of 25 µl. Reactions were performed with an initial denaturation step at 98 °C for 3 min followed by 40 cycles each including 30 s at 58 °C, 1 min and 15 s at 72 °C and 10 s at 98 °C and a final step at 72 °C for 5 min. Bidirectional DNA sequencing reactions were performed using the BrilliantDye TM Terminator Kit v1.1 (Nimagen) with four different primers spanning the spike region of interest. Briefly, 3 µl of PCR products, diluted at final concentration of 1–3 ng/µl, were mixed with 3.2 pmol/µl of each sequencing primer, 0.5 µl of BrilliantDyeTM Terminator Ready Reaction Sequencing and 2 µl of 5x Sequencing Buffer in a final volume of 10 µl. The reactions were denatured at 96 °C for 1 min followed by 25 cycles at 50 °C for 5 s, 60 °C for 4 min and 96 °C for 10 s. Sequencing reactions were treated with X-Terminator® Purification kit (Applied Biosystems) and then resolved by capillary electrophoresis with the 3130 XL Genetic Analyzer (Applied Biosystems). Chromatograms were assembled and edited with the DNAStar 7.1.0 SeqMan module and imported in the clade assignment tool (https://clades.nextstrain.org/) to determine the variant.
Statistical analysis
Statistical analysis was performed using Graphpad 8.3. Data were tested for normality using the Kolmogorov-Smirnov test and were presented as mean with the respective standard deviation (SD) or median with interquartile range (IQR), as appropriate. Data with normal distribution were analysed with one-way repeated measures analysis of variance (ANOVA) and Bonferroni's multiple-comparison test. For nonparametric variables, the Friedman test and Dunn's multiple comparison test were performed. The relationship between NT-Abs titre and time passed since diagnosis was evaluated by Spearman r correlation test. A p-value <0.05 was considered significant.
Results
Design, setting and participants
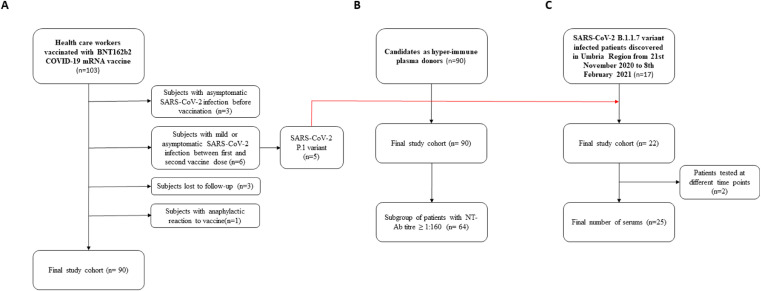
As shown in Fig. 1 A-C, three cohorts of subjects were enrolled in this study.
Fig. 1.
Patients selection flowchart. Coronavirus disease 2019, COVID-19; Severe Acute Respiratory Syndrome Coronavirus 2, SARS-CoV-2; Neutralizing antibodies, NT-Abs.
Vaccinated healthcare workers
This cohort was composed of 103 healthcare workers vaccinated with the BNT162b2 COVID-19 mRNA vaccine (Fig. 1A).
The mean age was 44.7 ± 10.1 years and 18.1% were male. All subjects were tested for SARS-CoV-2 antibody before vaccination. Ninety subjects had no SARS-CoV-2 infection, either before or within 21 days from completed vaccination.
Three subjects had asymptomatic SARS-CoV-2 infection before vaccination. Six subjects had mild or asymptomatic SARS-CoV-2 infection between the first and second vaccine dose. Three subjects were lost to follow-up. Another participant had an anaphylactic reaction to the first dose of the vaccine and did not undergo the second one. Consequently, these 13 patients were analyzed separately.
For subjects completing the two-dose vaccination schedule, serum samples were drawn in median 16 days (IQR 15–18 days) after the second dose.
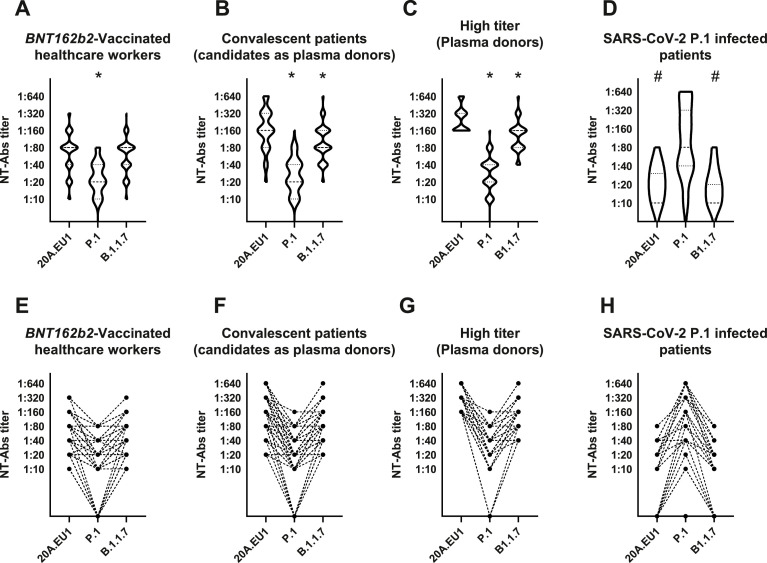
The distribution of the 90 vaccinated healthcare workers according to NT-Abs titers is shown in Fig. 2 A, E. In detail, the median NT-Abs titer was 1:80 (1:40–1:80) when serum was tested with 20A.EU1 and B.1.1.7 strains. However, when the same sera were tested with P.1 strain the median NT-Abs titer was 1:20 (1:10–1:40). The median NT-Abs titer was significantly higher for 20A.EU1/B.1.1.7 than for P.1 (p<0.0001). The mean titer was 3.3-fold higher for 20A.EU1 than P.1 strain.
Fig. 2.
Distribution of neutralizing antibodies (NT-Abs) titers of BNT162b2-vaccinated healthcare workers (N = 90) (A, E), convalescent candidates as plasma donors (N = 90) (B, F), high titers plasma donors (N = 64) (C, G) and SARS-CoV-2 P1 infected patients (N = 25) (D, H). NT-Abs titers against SARS-CoV-2 were evaluated using flat-bottom tissue culture 96-well microtiter plate serum dilution assay. Serums have been tested against 20A.EU1, B.1.1.7 and P.1 strains isolated from symptomatic patients with Coronavirus disease 2019 (COVID-19). Panels A, B, C and D show NT-Abs titers distribution for the three strains. Panels E, F, G and H show how NT-Abs serum titers of each patient change for the three variants. Data were presented as median with interquartile range (IQR). The Friedman test and Dunn's multiple comparison test were performed. A p-value <0.05 was considered significant. *Significant if compared to 20A.EU1 strain. #Significant if compared to P.1 strain.
All the three subjects with previous SARS-CoV-2 infection had an NT-Abs titer of 1:640 for 20A.EU1 and B.1.1.7, and 1:160 to 1:640 for the P.1 variant.
Among patients with SARS-CoV-2 infection occurring between the first and the second vaccine dose, 5/6 were from P.1 strain and 1/6 strain was not identified. Four of the five patients with P.1 infection were analyzed among the P.1 patients’ cohort, one was lost to follow-up.
Only one participant developed SARS-CoV-2 infection after two weeks from completed vaccination: she was a young woman with an NT-Abs titer of 1:20 for 20A.EU1 and B.1.1.7 and 1:10 for P.1. She contracted the B.1.1.7 variant and complained mild cough and rhinorrhea without fever.
Candidates as hyper-immune plasma donors
We postulated that patients with a history of SARS-CoV-2 infection acquired from June to October 2020 were infected with SARS-CoV-2 lineage 20A.EU1, the highly predominant variant in Italy at that time.4 We enrolled 90 convalescent patients as candidates as hyper-immune plasma donors (Fig. 1B).
Patients were enrolled in a median of 67 days (IQR 43–87, range 22–246) after diagnosis of SARS-CoV-2 infection.
The distribution of NT-Abs titers is shown in Fig. 2B, F. In detail, the median NT-Abs titer was 1:160 (1:80–1:320), 1:80 (1:80–1:160) and 1:20 (1:10–1:40) when serum was tested with 20A.EU1, B.1.1.7, and P.1 strains, respectively. The median NT-Abs titer was significantly higher for 20A.EU1 than for B.1.1.7 (mean titer 1.6-fold higher) and P.1 (mean titer 6.7-fold higher) (p = 0.0002 and p<0.0001 respectively). Furthermore, the median NT-Abs titer was significantly higher for B.1.1.7 than for P.1 (mean titer 4.2-fold higher) (p<0.0001).
A subgroup of 64 patients had a high NT-Abs titer (≥1:160) and, consequently were considered eligible as hyper-immune plasma donors. This subgroup had a median NT-Abs titer of 1:240 (1:160–1:320), 1:160 (1:80–1:160) and 1:30 (1:20–1:40) when serum was tested with 20A.EU1, B.1.1.7, and P.1 strains, respectively (Fig. 2C, G). The median NT-Abs titer was significantly higher for 20A.EU1 than for B.1.1.7 (mean titer 1.8-fold higher) and P.1 (mean titer 8.5-fold higher) (p<0.0001) and it was higher for B.1.1.7 than for P.1 (mean titer 4.3-fold mean titer) (p<0.0001).
SARS-CoV-2 P.1 variant infection cohort
Twenty-two patients with ascertained SARS-CoV-2 P.1 infection acquired from 21st November 2020 to 8th February 2021 in the Umbria region were enrolled, including 5 healthcare workers who already had a dose of BNT162b2 vaccine (Fig. 1C). Two patients were tested at multiple time points, one 2 and the other 3 times. Each measure was considered separately, therefore, a total of 25 samples were considered for the analysis. The mean age was 61.1 ± 19.8 years and 13 (59.1%) were male. Patients were enrolled a median of 21.0 days (IQR 15.3–34.0, range 12–94) after diagnosis of SARS-CoV-2 infection.
The distribution of the 25 sera according to NT-Abs titers is shown in Fig. 2D, H. In detail, the median NT-Abs titer was 1:10 (IQR <1:10–1:30, range <1:10–1:80), 1:10 (IQR 1:5–1:20, range <1:10–1:80) and 1:80 (IQR 1:40–1:320, range <1:10–1:640) when serum was tested with 20A.EU1, B.1.1.7, and P.1 strains, respectively. The median NT-Abs titer was significantly higher for P.1 than for 20A.EU1 (mean titer 12.2-fold higher) and B.1.1.7 (mean titer 10.9-fold higher) (p<0.0001). The median NT-Abs titer was not significantly different for 20A.EU1 and B.1.1.7.
No correlation between NT-Abs titre and time passed since diagnosis was found for any of the three lineages.
Discussion
The emergence of VOCs could significantly complicate SARS-CoV-2 pandemic. Starting from January 2021, in the Umbria region, a small area in the center of Italy, SARS-CoV-2 B.1.1.7 and P.1 variants widely spread causing an important increase of cases during the third wave. In particular, the prevalence of P.1 was disproportionally higher, reaching 36.2% as compared to 4.1% of the whole Italy, while the prevalence of B.1.1.7 was in line with the rest of the country (51.1% versus 54.0%, respectively).5 It must be noted that P.1 outbreaks involved healthcare workers and thus may have been easier to detect within surveillance programs.
Few studies have so far investigated how different mutations/variants could impact the immunologic response both of vaccinated and convalescent people. Due to different complexity of the assays, neutralizing antibody titration has been used far more often than analysis of cell mediated response and most studies have used pseudoviruses instead of live virus. However, neutralizing antibodies are of primary importance as a key component of immune memory providing sterilizing immunity. Even sub-sterilizing NT-Abs titers would limit infection and COVID-19 severity.10 In this study, we only evaluated the role of neutralizing antibodies but, also CD4+ and CD8+ T cell responses play important roles in COVID-19 course modulating disease severity and contrasting viral replication. Tarke et al. demonstrated that T cell responses elicited by either SARS-CoV-2 natural infection or vaccination with mRNA vaccines were not affected by the B.1.1.7, B.1.351, P.1 and CAL.20C variants.11 However, SARS-CoV-2 specific CD4+ T cells and CD8+ T cells showed an earlier decline with a half-life of 3–5 months compared to IgG to the spike protein that was relatively stable over 6 months.10 Sub-sterilizing immunity and T cell responses probably contribute together to limit COVID-19 severity in patients with reinfection or infection after vaccination due to VOCs.
Neutralizing activity of BNT162b2-elicited sera against B.1.1.7 variant was widely tested. All studies concluded that neutralizing activity to this VOC is largely preserved. Some authors demonstrated roughly equivalent NT-Abs activity to B.1.1.7 and the wild-type strain,12, 13, 14 while others detected less efficient neutralization against the B.1.1.7 variant.15, 16, 17, 18, 19 In particular, different studies found a reduction of B.1.1.7 neutralizing titer from 1.9 to 3.3-fold respect to wild-type strain.20, 21, 22, 23 Edara et al. found significant differences only from A.1 to B.1 and B.1.1.7, but not from B.1 and B.1.1.7.16 In our study, no differences were found in post-vaccination sera for NT-Abs titers to the 20A.EU1 and B.1.1.7 strains.
Fewer studies are available for P.1 variant. Wanwisa et al. demonstrated that neutralization titers against P.1 and B.1.1.7 were similarly reduced in BNT162b2-elicited sera, while P.1 is significantly less resistant to naturally acquired or vaccine induced antibody responses compared with B.1.351. This difference may be considered unexpected because P.1 and B.1.351 share the key E484K and N501Y spike mutations.20 Chen et al. studied the in vitro impact of different SARS-CoV-2 spike protein mutations on serum neutralizing activity by introducing individual point mutations in the spike gene into an infectious complementary DNA clone of the 2019n-CoV/USA_WA1/2020. They found similarly reduced inhibitory activity against viruses containing the E484K spike mutation, such as P.1 and B.1.351.24
Other studies reported the relative resistance of P.1 to neutralization by multiple therapeutic monoclonal antibodies, convalescent plasma, and sera from vaccine recipients with respect to the wild-type strain.12 , 25 , 26 Conversely, Liu et al., studying the neutralizing activity of BNT162b2-elicited serum, did not find a significant difference in neutralization activity across wild-type, B.1.1.7 and P.1 spike proteins, while neutralization of B.1.351-spike protein was still robust but lower,14 a finding corroborated by other studies.12 , 14 , 16 Overall, data on P.1 neutralization by antibody elicited by natural or artificial exposure to the wild type spike protein remain equivocal, likely due to the use of different methods and the lack of reference isolates at this time.
In our study, the lower neutralizing activity of BNT162b2-elicited sera on the P.1 variant may explain the five cases of P.1 variant infection in vaccine recipients.
Our vaccine recipients with previous SARS-CoV-2 infection had a neutralizing activity significantly higher for all the three strains compared to the uninfected vaccinees. These results, despite the low number of samples, are in line with the published data.27 , 28
Studies on SARS-CoV-2 convalescent patients who previously contracted wild-type SARS-CoV-2 infection demonstrated neutralizing activity to B.1.1.7 and P.1 VOCs but with significantly lower titers compared with the homologous virus (2–4.5-fold and 3.1-fold, respectively).15 , 20, 21, 22, 23 , 25 , 26 However, few studies did not detect a reduction of activity on B.1.1.7, highlighting limited consistency of literature data also with convalescent sera.13 , 16
Similar to our findings, Colier et al., demonstrated that vaccinated patient serum has higher neutralizing activity against B.1.1.7 compared to convalescent ones (about 3.6-fold of difference).23 Indeed, in our study, vaccine sera neutralized both 20A.EU1 and B.1.1.7 strains with the same efficacy. We separately analyzed a subgroup of high NT-Abs titer convalescent patients (NT-Abs≥1:160) and we found that these sera could not be considered as hyperimmune to B.1.1.7 and P.1 VOCs because they were significantly less effective in neutralizing these variants. Thus, the emergence of VOCs can abrogate this therapeutic option when the SARS-CoV-2 lineage of donors and recipient do not match.
To the best of our knowledge, this is the first study that evaluated neutralizing activity of sera from P.1 variant infected patients to other SARS-CoV-2 lineages. As expected, we found an important reduction of NT-Abs against 20A.EU1 and B.1.1.7 strains (12.2 and 10.9-fold, respectively). Considering that patients with a previous wild-type infection had a titer reduction of 6.7 and 1.58-fold on P.1 and B.1.1.7, it appears that patients with a previous P.1 infection are less protected from further SARS-CoV-2 reinfections from other variants.
Conclusions
The impact of VOCs on serum neutralization activity and protection from SARS-CoV-2 (re)infection needs to be further clarified. Various studies have shown different results and come to different conclusions. Our data corroborate the concept that B.1.1.7 and P.1 are less efficiently neutralized by convalescent sera from subjects infected by the original virus. However, BNT162b2 vaccine-elicited human sera have an equivalent neutralization potency on the B.1.1.7 but lower on the P.1 variant. Convalescent P.1 patients are less protected from other SARS-CoV-2 strains.
Ethical statements
The Vero E6 cell line was kindly provided by Istituto Zooprofilattico Sperimentale di Brescia, Brescia, Italy.
This research was approved by the Ethics Committee of the Umbria Region, protocol numbers 18,344/20/OV and 20,686/21/OV.
Disclaimers
Nothing to declare.
Acknowledgments
Thanks to the healthcare workers of Santa Maria della Misericordia Hospital that spontaneously participated to this study, once again their contribute was crucial.
Footnotes
ORCiD
Anna Gidari 0000-0002-6556-6553
Samuele Sabbatini 0000-0001-8334-8325
Claudia Monari 0000-0002-5027-9367
Filippo Dragoni 0000-0002-8148-9914
Elisabetta Schiaroli 0000-0002-3216-4144
Maurizio Zazzi 0000-0002-0344-6281
Daniela Francisci 0000-0001-8752-8278
References
- 1.World Health Organization. General's opening remarks at the media briefing on COVID-19. Available at https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020 2020.
- 2.Centers for Disease Control and Prevention. SARS-CoV-2 variant classifications and definitions. Available at https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fcases-updates%2Fvariant-surveillance%2Fvariant-info.html 2021.
- 3.Anna Gidari, Marco Nofri, Luca Saccarelli, Sabrina Bastianelli, Samuele Sabbatini, Silvia Bozza. Is recurrence possible in coronavirus disease 2019 (COVID-19)? Case series and systematic review of literature. Eur J Clin Microbiol Infect Dis. 2021;40(1):1–12. doi: 10.1007/s10096-020-04057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francesca Di Giallonardo, Ilaria Puglia, Valentina Curini, Cesare Cammà, Iolanda Mangone, Paolo Calistri. Emergence and spread of sars-cov-2 lineages b.1.1.7 and p.1 in Italy. Viruses. 2021;13(5):794. doi: 10.3390/v13050794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Istituto Superiore di Sanità. Prevalenza delle varianti VOC 202012/01 (lineage B.1.1.7), P.1, e 501.V2 (lineage B.1.351) in Italia Indagine del 18 febbraio 2021. Available at https://www.iss.it/documents/20126/0/Relazione+tecnica+terza+indagine+flash+per+le+varianti+del+virus+SARS-CoV-2+%282%29.pdf/a03f33e6-d775-9ab0-b0ce-9cdd289c711d?t=1614707205598 2021.
- 6.Anna Gidari, Samuele Sabbatini, Sabrina Bastianelli, Sara Pierucci, Chiara Busti, Desirèe Bartolini. SARS-CoV-2 survival on surfaces and the effect of UV-C light. Viruses. 2021;13(3):408. doi: 10.3390/v13030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27(3):493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 8.Alessia Lai, Annalisa Bergna, Sara Caucci, Nicola Clementi, Ilaria Vicenti, Filippo Dragoni. Molecular tracing of SARS-CoV-2 in Italy in the first three months of the epidemic. Viruses. 2020;12(8):798. doi: 10.3390/v12080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elena Percivalle, Giuseppe Cambiè, Irene Cassaniti, Vecchio Nepita Edoardo, Roberta Maserati, Alessandro Ferrari. Prevalence of SARS-CoV-2 specific neutralising antibodies in blood donors from the Lodi Red Zone in Lombardy, Italy, as at 06 April 2020. Eurosurveillance. 2020;25(24) doi: 10.2807/1560-7917.ES.2020.25.24.2001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dan Jennifer M., Mateus Jose, Kato Yu, Hastie Kathryn M., Yu Esther Dawen, Faliti Caterina E. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science (80-) 2021;371(6529):eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alison Tarke, John Sidney, Nils Methot, Yun Zhang, M Dan Jennifer, Benjamin Goodwin. Negligible impact of SARS-CoV-2 variants on CD4 + and CD8 + T cell reactivity in COVID-19 exposed donors and vaccinees. BioRxiv Prepr Serv Biol. 2021 doi: 10.1101/2021.02.27.433180. [DOI] [Google Scholar]
- 12.Garcia-Beltran Wilfredo F., Lam Evan C., St. Denis Kerri, Nitido Adam D., Garcia Zeidy H., Hauser Blake M. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–2383. doi: 10.1016/j.cell.2021.03.013. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delphine Planas, Timothée Bruel, Ludivine Grzelak, Florence Guivel-Benhassine, Isabelle Staropoli, Françoise Porrot. Sensitivity of infectious SARS-CoV-2 B1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 14.Yang Liu, Jianying Liu, Hongjie Xia, Xianwen Zhang, Fontes-Garfias Camila R., Swanson Kena A. Neutralizing Activity of BNT162b2-Elicited Serum. N Engl J Med. 2021;384(15):1466–1468. doi: 10.1056/nejmc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates Timothy A, Leier Hans C., Lyski Zoe L., McBride Savannah K. Neutralization of SARS-CoV-2 variants by convalescent and vaccinated serum. Medrxiv. 2021 doi: 10.1101/2021.04.04.21254881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edara Venkata Viswanadh, H. Hudson William, Xuping Xie, Ahmed Rafi, Suthar Mehul S. Neutralizing antibodies against SARS-CoV-2 variants after infection and vaccination. JAMA Intern Med. 2021;325(18):1896–1898. doi: 10.1001/jama.2021.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zijun Wang, Fabian Schmidt, Yiska Weisblum, Frauke Muecksch, O. Barnes Christopher, Shlomo Finkin. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander Muik, Kathrin Wallisch Ann, Bianca Sänger, A. Swanson Kena, Julia Mühl, Wei Chen. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science (80-) 2021;371(6534):1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xuping Xie, Yang Liu, Jianying Liu, Xianwen Zhang, Jing Zou, Fontes-Garfias Camila R. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 20.Wanwisa Dejnirattisai, Daming Zhou, Piyada Supasa, Chang Liu, Mentzer Alexander J., Ginn Helen M. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184(11):2939–2954. doi: 10.1016/j.cell.2021.03.055. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiaoying Shen, Haili Tang, Charlene McDanal, Kshitij Wagh, William Fischer, James Theiler. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4):529–539. doi: 10.1016/j.chom.2021.03.002. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piyada Supasa, Daming Zhou, Wanwisa Dejnirattisai, Chang Liu, Mentzer Alexander J., Ginn Helen M. Reduced neutralization of SARS-CoV-2 B1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184(8):2201–2211. doi: 10.1016/j.cell.2021.02.033. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collier Dami A., De Marco, Anna Ferreira, Isabella A.T.M., Bo Meng, Rawlings Datir, Walls Alexandra C. Sensitivity of SARS-CoV-2 B1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593(7857):136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.E. Chen Rita, Xianwen Zhang, Brett Case James, S. Winkler Emma, Yang Liu, VanBlargan Laura A. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pengfei Wang, G. Casner Ryan, S. Nair Manoj, Maple Wang, Jian Yu, Gabriele Cerutti. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–751. doi: 10.1016/j.chom.2021.04.007. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markus Hoffmann, Prerna Arora, Rüdiger Groß, Seidel Alina Hörnich, Bojan F., Hahn Alexander S. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184(9):2384–2393. doi: 10.1016/j.cell.2021.03.036. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leier Hans C., Bates Timothy A., Lyski Zoe L., McBride Savannah K., Lee David X., Coulter Felicity J. Previously infected vaccinees broadly neutralize SARS-CoV-2 variants. MedRxiv Prepr Serv Heal Sci. 2021 doi: 10.1101/2021.04.25.21256049. [DOI] [Google Scholar]
- 28.Ilaria Vicenti, Francesca Gatti, Renzo Scaggiante, Adele Boccuto, Daniela Zago, Monica Basso. Single-dose BNT162b2 mRNA COVID-19 vaccine significantly boosts neutralizing antibody response in health care workers recovering from asymptomatic or mild natural SARS-CoV-2 infection. Int J Infect Dis. 2021;S1201-9712(21) doi: 10.1016/j.ijid.2021.05.033. 00435–5. [DOI] [PMC free article] [PubMed] [Google Scholar]