Abstract
Differentiation of sex chromosomes is thought to have evolved with cessation of recombination and subsequent loss of genes from the degenerated partner (Y and W) of sex chromosomes, which in turn leads to imbalance of gene dosage between sexes. Based on work with traditional model species, theory suggests that unequal gene copy numbers lead to the evolution of mechanisms to counter this imbalance. Dosage compensation, or at least achieving dosage balance in expression of sex-linked genes between sexes, has largely been documented in lineages with male heterogamety (XX/XY sex determination), while ZZ/ZW systems are assumed to be usually associated with the lack of chromosome-wide gene dose regulatory mechanisms. Here, we document that although the pygopodid geckos evolved male heterogamety with a degenerated Y chromosome 32–72 Ma, one species in particular, Burton's legless lizard (Lialis burtonis), does not possess dosage balance in the expression of genes in its X-specific region. We summarize studies on gene dose regulatory mechanisms in animals and conclude that there is in them no significant dichotomy between male and female heterogamety. We speculate that gene dose regulatory mechanisms are likely to be related to the general mechanisms of sex determination instead of type of heterogamety.
This article is part of the theme issue ‘Challenging the paradigm in sex chromosome evolution: empirical and theoretical insights with a focus on vertebrates (Part II)’.
Keywords: dosage compensation, Gekkota, reptiles, RNA-seq, sex chromosomes, sex determination
1. Introduction
Differentiated sex chromosomes evolved independently in numerous animal and plant lineages [1]. The differentiation is connected with cessation of recombination and subsequent loss of functional genes from the Y or W sex chromosomes, which leads to gene dose differences between sexes. Selection will favour the evolution of mechanisms that regulate these differences at the cellular level, as alterations in gene copy number generally alter gene expression, ultimately impacting cell physiology and organismal fitness [2–5]. Different taxa have evolved distinct strategies to regulate the unequal gene copy numbers and the associated gene dosage imbalances between the sexes related to differentiated sex chromosomes [6]. The most well-known mechanism is dosage compensation, which restores the expression of X- or Z-specific genes in the heterogametic sex to the ancestral expression levels [7–9]. Dosage compensation usually leads to dosage balance, i.e. equal expression levels of the X- or Z-specific genes between the sexes; however, some animal lineages can reach dosage balance in the expression between sexes without keeping the ancestral expression level. Other animal lineages do not compensate and balance expression in the majority of the sex-linked genes at either the level of transcription or translation [10,11]. Dosage compensation or at least dosage balance between sexes was documented largely in lineages with male heterogamety (XX/XY sex determination) such as in several insect lineages, nematode worms, the green anole and eutherian mammals, with sticklebacks, basilisks and platypus being exceptions [6,12,13]. On the contrary, ZZ/ZW systems are usually associated with the lack of chromosome-wide gene dose regulatory mechanisms, often referred to as ‘partial’ or ‘incomplete’ dosage compensation. In such cases, it is assumed that the epigenetic mechanisms regulating gene expression in the heterogametic sex are restricted to a few dosage-sensitive genes on the Z chromosome where changes in gene dosage are tied to deleterious fitness effects or lethality, whereas the majority of the genes display different expression levels in males and females [6,14]. This implies that some genes are dosage-sensitive (low heterozygote fitness or lethality), whereas others are less so. The lack of chromosome-wide dosage compensation and dosage balance has been documented in parasitic blood flukes, tonguefish, caenophidian snakes, birds, a trionychid turtle and the Komodo dragon, with lepidopteran insects and Artemia franciscana representing the only known exceptions here [6,11,15–19].
It is assumed that a dichotomy in the gene dose regulatory mechanisms between male and female heterogamety occurs, and several, mostly adaptive explanations have been suggested to explain this pattern [20–25]. The hypothesis of differences in gene dose regulation mechanisms between male and female heterogamety is supported from studies of a limited number of lineages across animals (i.e. mainly nematodes, insects, vertebrates), with notably different embryonic (and mainly gonadal) development, highly dissimilar sex chromosome gene content and genome organization. We argue that this conclusion was premature. To study this phenomenon effectively, we need to explore patterns within a single, phylogenetically coherent lineage with variable sex determining modes. Amniotes (mammals and sauropsids) evolved sex chromosomes independently around 40 times, with geckos representing about half of the recorded transitions [26,27]. Currently, we know genes linked to sex chromosomes in only 27 amniote lineages with putative independently evolved sex chromosomes (reviewed in [13,28]) and gene dose regulatory mechanisms were studied in just eight of these lineages (table 1). In our quest for understanding the evolution of sex determination and gene dose regulatory mechanisms, we focus here on the pygypodid geckos (family Pygopodidae).
Table 1.
Summary of the current knowledge on presence/absence of dosage balance across animals. Animal species are split to groups reflecting putative independent origins of sex chromosomes (see [29–32] for evidence on homology of sex chromosomes in dipteran insects). Most evidence was taken from the review by Gu & Walters [6], supplemented by newer data (references by the individual species in the table).
| male heterogamety | female heterogamety | |||
|---|---|---|---|---|
| dosage balance | viviparous mammals | Bos taurus | butterflies/moths | Bombyx mori |
| Gorilla gorilla | Cydia pomonella | |||
| Homo sapiens | Danaus plexippus [17] | |||
| Macaca mulatta | Heliconius melpomene | |||
| Mus musculus | Manduca sexta | |||
| Ovis aries [33] | Plodia interpunctella | |||
| Pan paniscus | brine shrimps | Artemia franciscana [18] | ||
| Pan troglodytes | ||||
| Monodelphis domestica | ||||
| green anole | Anolis carolinensis | |||
| swamp guppy | Poecilia picta [34] | |||
| fruitflies | Drosophila melanogaster | |||
| Drosophila miranda | ||||
| Drosophila pseudoobscura | ||||
| stalk-eyed flies | Teleopsis dalmanni | |||
| Australian sheep blowfly | Lucilia cuprina | |||
| mosquitos | Anopheles gambiae | |||
| Anopheles stephensi | ||||
| hemipteran insects | Acyrthosiphon pisum | |||
| Halyomorpha halys | ||||
| Homalodisca vitripennis | ||||
| Oncopeltus fasciatus | ||||
| beetle + strepsipteran insect | Tribolium castaneum | |||
| Xenos vesparum | ||||
| roundworms | Caenorhabditis elegans | |||
| Pristionchus pacificus | ||||
| lack of dosage balance | platypus | Ornithorhynchus anatinus | birds | Charadrius alexandrinus |
| brown basilisk | Basiliscus vittatus [12,13] | Corvus corone | ||
| Burton's legless lizard | Lialis burtonis (this study) | Ficedula albicollis | ||
| three-spined stickleback | Gasterosteus aculeatus | Gallus gallus | ||
| Taeniopygia guttata | ||||
| Florida softshell turtle | Apalone ferox [19] | |||
| Komodo dragon | Varanus komodoensis [16] | |||
| caenophidian snakes | Sistrurus miliarius | |||
| Thamnophis elegans | ||||
| tongue sole | Cynoglossus semilaevis | |||
| blood flukes | Schistosoma haematobium | |||
| Schistosoma japonicum | ||||
| Schistosoma mansoni | ||||
Pygopodids (legless or flap-footed lizards) are a small family of 45 species of gecko lizards [35] native to Australia and New Guinea. Pygopodids are the only lineage within the gekkotan radiation that possess an attenuate, snake-like body plan lacking limbs and digits, retaining only small flaps where rear legs would normally be [36]. Up until now, information on their sex determination has been limited to largely cytogenetic evidence in four species: XX/XY sex chromosomes were reported in Aprasia parapulchella [37] and Delma butleri [38], and the X1X1X2X2/X1X2Y sex chromosomes in Lialis burtonis and L. jicari likely evolved via a fusion of an ancestral X with an autosome [39,40]. Male heterogamety in L. burtonis was confirmed by finding several male-specific anonymous molecular markers in RAD sequencing [26]. However, the homology of sex chromosomes among pygopodids and with sex chromosomes in other amniote lineages remains unknown.
In order to expand our knowledge on the evolution of sex chromosomes and gene dose regulatory mechanisms in amniotes, we tried to identify the sex chromosome gene content of the pygopodid Burton's legless gecko (Lialis burtonis), where XX/XY sex determination was previously identified by cytogenetic methods. Here, we used an mRNA-seq-based pipeline to identify genes located on the X chromosome and a real-time quantitative PCR (qPCR) method to validate the candidate X-specific genes. Subsequently, the qPCR approach was further used to explore the homology of sex chromosomes among pygopodid geckos, while mRNA-seq data were used to explore the gene dose regulatory mechanism regulating the gene dose imbalance between sexes of X-specific genes in L. burtonis.
2. Material and methods
(a) . Animal sampling and DNA/RNA isolation
Tissue or blood samples were collected from both sexes of five species of pygopodids: Aprasia parapulchella, Delma inornata, Lialis burtonis, Lialis jicari and Pygopus nigriceps (electronic supplementary material, table S1). The processing of the biological material was carried out by accredited researchers and under the supervision and with the approval of the Ethics Committee of the Faculty of Science, Charles University in Prague followed by the Ministry of Education, Youth and Sports of the Czech Republic (permission 8604/2019-7).
Genomic DNA from all specimens was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA). Total RNA from the blood of two females and four males of L. burtonis and one male of L. jicari was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer protocols. The quantity and purity of the extracted DNA and RNA samples were estimated using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific Inc, Waltham, MA, USA).
(b) . RNA sequencing and identification of X-specific genes in L. burtonis
Barcoded stranded mRNA-sequencing libraries were constructed from the total RNA samples from six individuals of L. burtonis and one individual of L. jicari by GeneCore (EMBL, Heidelberg, Germany) using the Illumina TruSeq mRNA v2 sample preparation kit (Illumina, San Diego, CA, USA) with poly-A mRNA enrichment. The libraries were pooled in equimolar amounts and loaded on the Illumina NextSeq 500 sequencer and 85 base pairs (bp) were sequenced bidirectionally. The raw Illumina reads were deposited in GenBank database under the BioProject PRJNA623146.
The raw Illumina reads were trimmed for adapters and low quality bases in Trimmomatic [41], according to the default parameters. Reads with a size of less than 50 bp were removed from the dataset, resulting in a final dataset of 40–80 million reads per specimen. Trimmed reads were checked for quality in FastQC [42] and MultiQC [43].
Trimmed reads from a single male of L. burtonis were assembled de novo with Trinity v. 2.8.5 [44]. The assembled transcripts were compared with BLASTn [45] to the reference transcriptomes of Anolis carolinensis, Chrysemys picta, Gallus gallus, Gekko japonicus, Pelodiscus sinensis, Pogona vitticeps and Python molurus. Transcript sequences of L. burtonis with higher than 70% similarity spanning over 150 bp of homologous sequences to a reference transcriptome were selected for further analyses, resulting in a final dataset of 64 432 annotated transcripts. The Illumina reads from all five male pygopodid specimens were independently mapped to our L. burtonis reference transcriptome using Geneious Prime. Consensus sequences from the assembly were exported, treating polymorphic sites (for example SNPs) in all sequences as ambiguous bases. Transcript regions with coverage below 10× and size less than 500 bp were removed from the dataset.
The Y chromosomes in both species of the genus Lialis contain extensive heterochromatic blocks and accumulations of repetitive motifs, indicating a high degree of degeneration of the Y chromosome [40]. Comparative genome hybridization showed that the Y and X chromosomes differ significantly in sequence content [40]. Degenerated Y and W sex chromosomes have usually lost, in their non-recombining region, most of the genes present on their X or Z counterparts, respectively. Single-copy X- and Z-specific loci should contain just a single allele in the genome of the individuals from the heterogametic sex. Therefore, we can uncover candidates for such hemizygous loci based on the constant lack of SNPs in homologous transcripts from all specimens of the heterogametic sex. However, homozygous autosomal and pseudoautosomal loci might also not possess SNPs in their transcripts. To differentiate between these categories, we took advantage of the high level of conservation in chromosome synteny across sauropsids [46,47]. We assume that genuine X-specific (hemizygous) genes from male individuals should form a syntenic chromosome block enriched in loci without SNPs, but false positive (homozygous) genes should be scattered randomly across chromosomes [28]. We assigned as many transcripts of L. burtonis as possible to putative syntenic blocks according to chromosomal position of their orthologous genes in the chicken (Gallus gallus, GGA) genome (www.ncbi.nlm.nih.gov/genome). We used the chicken genome because it is well assembled and annotated compared to other avian and reptile genomes. We determined which syntenic blocks defined by chicken chromosomes are unusually enriched in loci without SNPs. Such blocks were identified as significant outliers from the linear regression between the number of genes without SNPs in a given putative syntenic block and the total number of expressed genes in a given block. For this analysis, we filtered out all transcripts with less than 500 bp in length and all gene duplicates, i.e. each gene was represented by a single transcript in the dataset. Genes that lacked SNPs in all five males were considered candidate X-specific genes (i.e. those on the X chromosome but absent in the degenerated part of the Y chromosome). The differences in gene copy numbers between sexes triggered by the degeneration of the Y chromosome can also be directly measured by qPCR applied to genomic DNA [16,28,48,49]. In L. burtonis, we used this approach for the validation of X-specificity in a subset of loci from the candidate putative syntenic blocks. Primer pairs were designed for the amplification of the 120–200 bp exon fragments of autosomal control genes and candidate X-specific genes in the Primer-BLAST software [50] using Primer3 approach [51]. The qPCR with DNA template was carried out in a LightCycler II 480 (Roche Diagnostics, Basel, Switzerland) with all samples run in triplicates (for the list of genes and primers see electronic supplementary material, table S2). The qPCR protocol and the formula for the calculation of the relative gene dose between sexes have been presented previously [52]. A relative male-to-female gene dose ratio (r) of 0.5 is expected for X-specific genes and of 1.0 for autosomal and pseudoautosomal genes, and genes with poorly differentiated gametologs. We recently used similar methodology to discover sex-linked genes in lacertid and anguimorphan lizards and in the gecko genus Paroedura [16,28,49].
(c) . qPCR test of homology of sex chromosomes in pygopodid geckos
Candidate hemizygous genes in L. burtonis were tested for X-specificity in four additional pygopodid species (A. parapulchella, D. inornata, L. jicari and P. nigriceps) using the qPCR technique (for the list of genes and primers see electronic supplementary material, table S2) to explore sex chromosome homology. The tests and calculations were performed as described above.
(d) . Test of dosage balance in L. burtonis
We used transcriptome data from two females and four males of L. burtonis to test for dosage balance of the X-specific genes. FPKM (Fragments Per Kilobase Million mapped reads) expression values were independently calculated for each transcript with average read coverage greater than 10 across all specimens, from data provided by the Geneious Prime ‘map to reference’ assembler. Subsequently, we computed the average sex-specific FPKMs for each transcript as the mean value from the two females and the four males, respectively. Gene expression may vary significantly between individuals of the same sex not owing to gene copy number, but to physiological parameters (e.g. age, fitness, sickness, reproductive stage). Therefore, we excluded from further analysis all transcripts that had high variation among specimens of the same sex (i.e. variation more than 30% of the mean standard deviation). The duplicities in gene identity were filtered out. For the analysis, we kept only the transcript with the smallest FPKM value in males in each gene. However, the results of the following analyses led to the same conclusion even without such strict filtering of transcripts.
We applied the analysis of covariance (ANCOVA) with log10-transformed average male FPKM as the dependent variable, chicken chromosome as the factor representing the grouping of genes to putative syntenic blocks and log10-transformed average female FPKM as the covariate. We also compared ratios of average male FPKM to average female FPKM between the putative syntenic block determined as X-specific and other syntenic blocks (putative autosomes) by analysis of variance (ANOVA).
3. Results
(a) . Candidate sex chromosome genes in L. burtonis
We confidently assigned 7718 individual genes of L. burtonis to chicken chromosomes (electronic supplementary material, table S3). The total number of genes linked to individual chicken chromosomes correlates tightly with the expressed genes of L. burtonis we assigned to them (Pearson's r = 0.98, n = 34, p < 0.0001; electronic supplementary material, figure S1), which demonstrates that individual chicken chromosomes are more or less uniformly represented in the pygopodid transcriptomes.
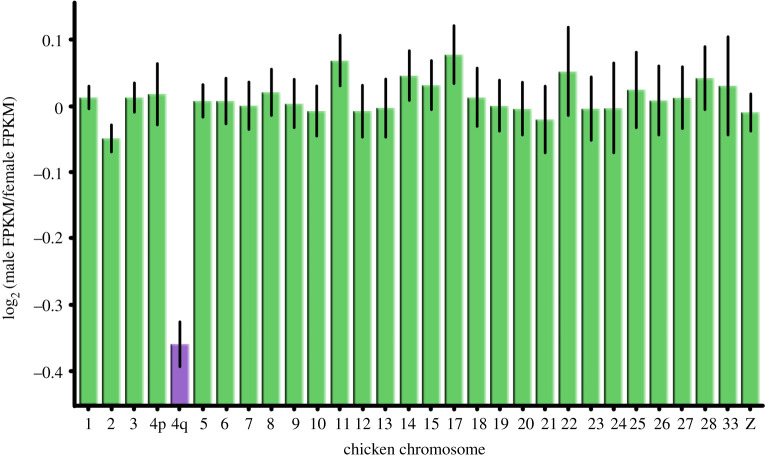
The number of L. burtonis genes per individual chicken chromosome correlates well with the number of L. burtonis genes without SNPs across the same chromosomes. There is only one significant outlier (the fourth largest chicken chromosome, GGA4) from this relationship. As GGA4 emerged relatively recently in the chicken ancestor via fusion of two ancestral chromosomes that now largely form the small (p) and large (q) arms of GGA4 [53], we further analysed genes from GGA4p and GGA4q separately to resolve the gene content of pygopodid sex chromosomes. The residual analysis showed that GGA4q is the only significant outlier from an otherwise tight relationship (r = 0.83, n = 35, p < 0.0001; electronic supplementary material, figure S2) between the number of L. burtonis genes without SNPs versus those assigned to individual chicken chromosomes. The standard residual of GGA4q from this linear regression is very large (4.57), suggesting that this putative syntenic block is exceptionally enriched for genes without SNPs. The residuals of all the other chicken chromosomes including GGA4p are in the range between −1.40 and 1.39.
(b) . Sex chromosome homology in pygopodid geckos
We tested four candidate X-linked genes in L. burtonis (elf2, maml3, noct, rab33b) with synteny to the q arm of chicken chromosome 4 using qPCR. The genes cabin1, derl3, fbxo33, rag1, ubr5 and usp12 were used as positive autosomal controls; the gene mecom was used for the normalization of the qPCR values (electronic supplementary material, table S2). Our qPCR experiments confirmed that the tested loci from syntenic block GGA4q are X-specific in all five tested pygopodid species (figure 1). Our results demonstrate that pygopodid geckos have homologous sex chromosomes, probably derived from their common ancestor.
Figure 1.
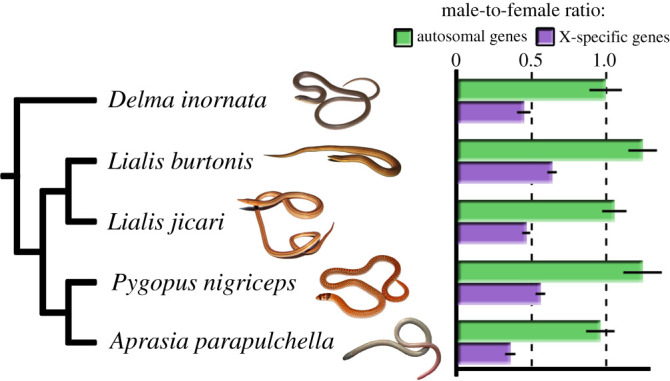
Average relative gene dose ratios between sexes for autosomal genes and X-specific genes of Lialis burtonis examined across five species of pygopodid lizards. The value 1.0 is expected for autosomal or pseudoautosomal genes, whereas 0.5 is consistent with X-specificity. Standard errors are indicted by black bars. (Online version in colour.)
(c) . Gene dose regulatory mechanism in L. burtonis
ANCOVA showed that log-transformed average male FPKM is highly predictable by log-transformed average female FPKM (covariate: F1,5057 = 203,786, p < 0.00001). However, at the same time, the syntenic blocks defined by chicken chromosomes strongly differ in male expression in comparison to female expression (F29,5057 = 15.80, p < 0.00001), with chromosome GGA4q being the only very significant outlier (figure 2) showing that the genes linked to this syntenic block homologous to the pygopodid X chromosome are transcribed less in males. Chicken chromosomes 16 and 29–32 were represented by less than 10 genes in our L. burtonis dataset and were excluded from the analyses.
Figure 2.
Comparison of sex-specific expression of genes from the Burton's legless lizard among putative syntenic blocks defined by linkage of orthologues to chicken chromosomes. Note that GGA4q has exceptional sex-specific expression, which suggests that there is no dosage balance in this species. (Online version in colour.)
ANOVA confirmed that the putative syntenic blocks defined by chicken chromosomes significantly differ in the log2-transformed ratios of average male FPKM to average female FPKM in L. burtonis (F29,5058 = 15.80, p < 0.00001) and that the ratios are significantly lower in genes with orthologues linked to the chromosome GGA4q than in genes linked to other chicken chromosomes (figure 3).
Figure 3.
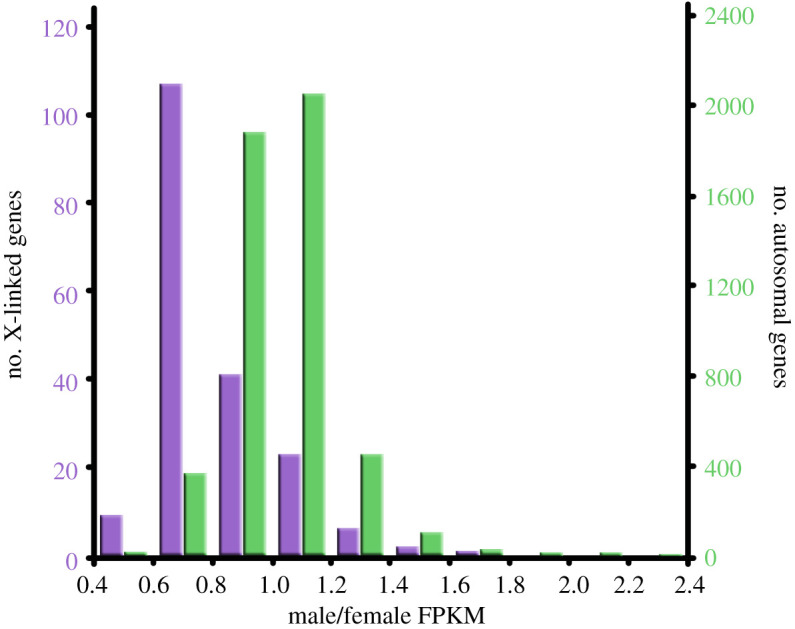
Histograms of the ratios of male to female measures of expression (FPKM) for genes linked to chicken chromosome 4q syntenic to pygopodid X chromosome and to other chromosomes in Lialis burtonis. (Online version in colour.)
4. Discussion
We identified the partial gene content of X-specific region of L. burtonis (i) based on the analysis of the distribution of SNPs across genes validated by the measurement of differences in gene copy numbers between sexes and (ii) by analysing expression differences of those genes between sexes. We show that the same X-specific region is shared by all sampled pygopodid species in the genera Aprasia, Delma, Lialis and Pygopus, despite the differences in morphology of their sex chromosomes and origin (e.g. the fusion of the ancestral sex chromosomes with an autosome leading to multiple neo-sex chromosomes in the common ancestor of L. burtonis and L. jicari) [40]. It seems that the differentiated XX/XY sex chromosomes in pygopodids are ancient and can be dated to the last common ancestor of living pygopodids, i.e. to at least 32–50 Myr [54]. As female heterogamety is known in the sister group to pygopodids, the family Carphodactylidae [55], the XX/XY sex chromosomes in pygopodids might be as old as 55 to 78 Myr, the estimated time when these two families split [54,56,57]. The pygopodid sex chromosomes are homologous to chromosome 4q of the chicken and the human chromosome 4. It seems that sex chromosomes in the pygopodid ancestor evolved independently from sex chromosomes of other amniotes, as no amniote group studied to date with known partial gene content of sex chromosomes shares sex-linked gene content with pygopodids [28], including three other gekkotan lineages: Phyllodactylus wirshingi (its ZZ/ZW chromosomes are syntenic with chicken Z; GGAZ), Gekko hokouensis (GGAZ as well, but likely independently derived), and the geckos of the genus Paroedura (GGA4p and GGA15) [28,58,59]. It should be noted that previously reported synteny of amniote sex chromosomes with chromosome GGA4 in lacertid lizards, geckos of the genus Paroedura, and therian mammals involved the small arm (GGA4p) not the larger arm (GGA4q) of the fourth chicken chromosome.
Genes linked to sex chromosomes in L. burtonis are expressed in blood cells significantly less in males in comparison to females (figures 2 and 3), suggesting lack of dosage balance between sexes in the expression of X-specific genes, and likely also of the global dosage compensation mechanism. Although dosage balance is lacking in all four amniote lineages with independently evolved ZZ/ZW sex chromosomes (i.e. birds, caenophidian snakes, a trionychid turtle, and the Komodo dragon), it is present in only two (i.e. eutherian mammals and the green anole) out of five studied lineages of amniotes with male heterogamety (reviewed in table 1). This study adds Burton's legless lizard to platypus and basilisks as another exception to the rule concerning differences in gene dose regulatory mechanisms between male and female heterogamety in amniotes.
To test whether male heterogamety is strictly linked to dosage balance, we summarized the current state of knowledge concerning dosage balance across animals (table 1). Dosage balance was studied in 22 lineages representing equal numbers of putative independent origins of sex chromosomes. In contrast to the classical models for the evolution of gene dose regulatory mechanisms, lineages with male heterogamety are not significantly more likely to possess dosage balance between sexes in the expression of genes linked to sex chromosomes than lineages with female heterogamety (Fisher's exact test: p = 0.074). Moreover, the ratio can even be biased in favour of the tested hypothesis; e.g. nematodes in fact do not represent the difference in expression between males and females, but between males and hermaphrodites [60,61]. Also, we grouped species according to putative independent evolution of their sex chromosomes based on sex-linked gene content, but owing to gaps in knowledge we were not able to separate independent origins of gene dose regulatory mechanisms. This is especially important in insects, which are overrepresented in the studies on gene dose regulatory mechanisms (table 1). Most insect lineages have male heterogamety and the origin of an epigenetic mechanism ensuring dosage balance of X-linked genes could be ancient and independently co-opted for regulation of expression of sex-linked genes even after turnover of sex chromosomes. On the other hand, sex chromosomes of marsupial and placental mammals are likely homologous, but their dosage compensating mechanisms are probably not [62]. Going forward, the sampling of lineages should be increased and we should focus on testing the homology of gene dose regulatory mechanisms and sex chromosomes. However, it seems that the earlier recognized pattern of a dichotomy in gene dose regulatory mechanisms between male and female heterogamety could be the result of limited sampling instead of a systematic difference.
The important remaining question is what (if anything) besides male and female heterogamety determines whether a lineage would evolve global dosage balance in the expression of X- and Z-specific genes or not. We suggest that it is related to the general mechanisms of sex determination, which generally work in two ways: sex determination might be controlled either by the copy number of X- or Z-linked loci per cell (i.e. gene dosage), or by a dominant W or Y locus [63]. We hypothesize that the dosage-dependent sex determination can work only in the absence of global dosage balance between sexes, at least at the time of the expression of the sex-determining locus. By contrast, the sex determination based on a dominant factor on Y and W chromosomes is compatible with both presence and lack of a dosage balance influencing chromosome-wide expression of X- and Z-linked genes. Unfortunately, our knowledge on the identity and function of sex-determining loci together with information on gene dose regulatory mechanisms is sporadic and restricts the testing of our hypothesis, yet the limited existing information is in agreement with this hypothesis. Only lineages with sex determination controlled by the gene dose of X- or Z-linked loci per cell are informative for the testing. In support of this hypothesis, both studied lineages with female heterogamety likely relying on the dosage-dependent mechanism, i.e. birds and caenophidian snakes [64,65], have a lack of global dosage balance [10,15]. At first sight, two model organisms, the fruit fly Drosophila melanogaster and the nematode worm Caenorhabditis elegans, represent a contradictory case, since it is textbook knowledge that their sex determination primarily relies on the number of copies of the X chromosome (in correlation to autosomes ratio), but at the same time they have global dosage compensation [66,67]. However, when inspected more closely, these cases in fact do not contradict our hypothesis: dosage compensation in fruit flies and worms is triggered only later in development, and thus does not interfere with the earlier sex-determination mechanisms based on copy numbers [67,68], which illustrates that detailed knowledge on molecular machinery and timing of particular steps will often be needed for testing mechanistic hypothesis on the evolution of gene dose regulatory mechanisms.
Acknowledgement
The authors express their gratitude to Jana Thomayerová for technical support.
Ethics
The processing of the biological material was carried out by accredited researchers and under the supervision and with the approval of the Ethics Committee of the Faculty of Science, Charles University in Prague followed by the Ministry of Education, Youth and Sports of the Czech Republic (permission 8604/2019-7).
Data accessibility
The raw Illumina reads were deposited in GenBank database under the BioProject PRJNA623146.
Authors' contributions
M.R.: experimental part and bioinformatics; L.K.: statistics; T.G., S.V.N., A.G., T.E.: provided part of the material and useful consultations; M.R., L.K.: conceived the project and wrote the first draft. All authors contributed to the final form of the manuscript and take responsibility for its content.
Competing interests
We declare we have no competing interests.
Funding
M.R. and L.K. were supported by Czech Science Foundation (GAČR) project no. 19-19672S, M.R. also by Charles University PRIMUS/SCI/46 and Research Centre program (204069).
References
- 1.Beukeboom LW, Perrin N. 2014. The evolution of sex determination. Oxford, UK: Oxford University Press & British Academy. [Google Scholar]
- 2.Birchler JA, Riddle NC, Auger DL, Veitia RA. 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21, 219-226. ( 10.1016/j.tig.2005.02.010) [DOI] [PubMed] [Google Scholar]
- 3.Birchler JA. 2014. Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma 123, 459-469. ( 10.1007/s00412-014-0478-5) [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Oliver B. 2007. Dosage compensation goes global. Curr. Opin Genet. Dev. 17, 113-120. ( 10.1016/j.gde.2007.02.002) [DOI] [PubMed] [Google Scholar]
- 5.Dürrbaum M, Storchová Z. 2016. Effects of aneuploidy on gene expression: implications for cancer. FEBS J. 283, 791-802. ( 10.1111/febs.13591) [DOI] [PubMed] [Google Scholar]
- 6.Gu L, Walters JR. 2017. Evolution of sex chromosome dosage compensation in animals: a beautiful theory, undermined by facts and bedeviled by details. Genome Biol. Evol. 9, 2461-2476. ( 10.1093/gbe/evx154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller HJ. 1918. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3, 422-499. ( 10.1093/genetics/3.5.422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin, Germany: Springer. [Google Scholar]
- 9.Brockdorff T, Turner BM. 2015. Dosage compensation in mammals. Cold Spring Harb. Perspect. Biol. 7, a019406. ( 10.1101/cshperspect.a019406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uebbing S, Konzer A, Xu L, Backström N, Brunström B, Bergquist J, Ellegren H. 2015. Quantitative mass spectrometry reveals partial translational regulation for dosage compensation in chicken. Mol. Biol. Evol. 32, 2716-2725. ( 10.1093/molbev/msv147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picard MAL, Cosseau C, Ferré S, Quack T, Grevelding CG, Couté Y, Vicoso B. 2018. Evolution of gene dosage on the Z-chromosome of schistosome parasites. eLife 7, e35684. ( 10.7554/eLife.35684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acosta A, Suárez-Varón G, Rodríguez-Miranda LA, Lira-Noriega A, Aguilar-Gómez D, Gutiérrez-Mariscal M, Hernández-Gallegos O, Méndez-de-la-Cruz F, Cortez D. 2019. Corytophanids replaced the pleurodont XY system with a new pair of XY chromosomes. Genome Biol. Evol. 11, 2666-2677. ( 10.1093/gbe/evz196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen SV, Guzmán-Méndez IA, Gamble T, Blumer M, Pinto BJ, Kratochvíl L, Rovatsos M. 2019. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol. Lett. 15, 20190498. ( 10.1098/rsbl.2019.0498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mank JE. 2009. The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet. 25, 226-233. ( 10.1016/j.tig.2009.03.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11, e1001643. ( 10.1371/journal.pbio.1001643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rovatsos M, Rehák I, Velenský P, Kratochvíl L. 2019. Shared ancient sex chromosomes in varanids, beaded lizards and alligator lizards. Mol. Biol. Evol. 36, 1113-1120. ( 10.1093/molbev/msz024) [DOI] [PubMed] [Google Scholar]
- 17.Gu L, Reilly PF, Lewis JJ, Reed RD, Andolfatto P, Walters JR. 2019. Dichotomy of dosage compensation along the neo Z chromosome of the monarch butterfly. Curr. Biol. 29, 4071-4077. ( 10.1016/j.cub.2019.09.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huylmans AK, Toups MA, Macon A, Gammerdinger WJ, Vicoso B. 2019. Sex-biased gene expression and dosage compensation on the Artemia franciscana Z-chromosome. Genome Biol. Evol. 11, 1033-1044. ( 10.1093/gbe/evz053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rovatsos M, Kratochvíl L. 2021. Evolution of dosage compensation does not depend on genomic background. Mol. Ecol. 30, 1836-1845. ( 10.1111/mec.15853) [DOI] [PubMed] [Google Scholar]
- 20.Vicoso B, Bachtrog D. 2009. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res. 17, 585-602. ( 10.1007/s10577-009-9053-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson SMA, Makova KD. 2011. Genome analyses substantiate male mutation bias in many species. BioEssays 33, 938-945. ( 10.1002/bies.201100091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mank JE, Vicoso B, Berlin S, Charlesworth B. 2010. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution 64, 663-674. ( 10.1111/j.1558-5646.2009.00853.x) [DOI] [PubMed] [Google Scholar]
- 23.Naurin S, Hansson B, Bensch S, Hasselquist D. 2012. Why does dosage compensation differ between XY and ZW taxa? Trends Genet. 26, 15-20. ( 10.1016/j.tig.2009.11.006) [DOI] [PubMed] [Google Scholar]
- 24.Mank JE. 2013. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29, 677-683. ( 10.1016/j.tig.2013.07.005) [DOI] [PubMed] [Google Scholar]
- 25.Mullon C, Wright AE, Reuter M, Pomiankowski A, Mank JE. 2015. Evolution of dosage compensation under sexual selection differs between X and Z chromosomes. Nat. Commun. 6, 7720. ( 10.1038/ncomms8720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. 2015. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 32, 1296-1309. ( 10.1093/molbev/msv023) [DOI] [PubMed] [Google Scholar]
- 27.Johnson PM, Kratochvíl L. 2016. What was the ancestral sex-determining mechanism in amniote vertebrates? Biol Rev. 91, 1-12. ( 10.1111/brv.12156) [DOI] [PubMed] [Google Scholar]
- 28.Rovatsos M, Farkačová K, Altmanová M, Johnson Pokorná M, Kratochvíl L. 2019. The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 28, 3042-3052. ( 10.1111/mec.15126) [DOI] [PubMed] [Google Scholar]
- 29.Jiang X, Biedler JK, Qi Y, Hall AB, Tu Z. 2015. Complete dosage compensation in Anopheles stephensi and the evolution of sex-biased genes in mosquitoes. Genome Biol. Evol. 7, 1914-1924. ( 10.1093/gbe/evv115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linger RJ, Belikoff EJ, Scott MJ. 2015. Dosage compensation of X-linked Muller element F genes but not X-linked transgenes in the Australian sheep blowfly. PLoS ONE 10, e0141544. ( 10.1371/journal.pone.0141544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicoso B, Bachtrog D. 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13, e1002078. ( 10.1371/journal.pbio.1002078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pal A, Vicoso B. 2015. The X chromosome of hemipteran insects: conservation, dosage compensation and sex-biased expression. Genome Biol. Evol. 7, 3259-3268. ( 10.1093/gbe/evv215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan J, et al. 2019. Dosage compensation and gene expression of the X chromosome in sheep. G3 9, 305-314. ( 10.1534/g3.118.200815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darolti I, et al. 2019. Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc. Natl Acad. Sci. USA 116, 19 031-19 036. ( 10.1073/pnas.1905298116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uetz P, Freed P, Hošek J. 2020. The Reptile Database. See http://www.reptile-database.org (accessed 8 March 2020).
- 36.Vitt LJ, Caldwell JP. 2013. Herpetology: An introductory biology of amphibians and reptiles, 4th edn. New York, NY: Academic Press. [Google Scholar]
- 37.Matsubara K, Knopp T, Sarre SD, Georges A, Ezaz T. 2013. Karyotypic analysis and FISH mapping of microsatellite motifs reveal highly differentiated XX/XY sex chromosomes in the pink-tailed worm-lizard (Aprasia parapulchella, Pygopodidae, Squamata). Mol Cytogenet. 6, 60. ( 10.1186/1755-8166-6-60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King M. 1987. Chromosomal evolution in the Diplodactylinae (Gekkonidae: Reptilia). I. Evolutionary relationships and patterns of change. Aust. J. Zool. 35, 507-531. ( 10.1071/ZO9870507) [DOI] [Google Scholar]
- 39.Gorman GC, Gress F. 1970. Sex chromosomes of a pygopodid lizard, Lialis burtonis. Experientia 26, 206-207. ( 10.1007/bf01895586) [DOI] [PubMed] [Google Scholar]
- 40.Rovatsos M, Johnson Pokorná M, Altmanová M, Kratochvíl L. 2016. Mixed-up sex chromosomes: identification of sex chromosomes in the X1X1X2X2/X1X2Y system of the legless lizards of the genus Lialis (Squamata: Gekkota: Pygopodidae). Cytogenet Genome Res. 149, 282-289. ( 10.1159/000450734) [DOI] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. See https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 43.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047-3048. ( 10.1093/bioinformatics/btw354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644-652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 46.Alföldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477, 587-591. ( 10.1038/nature10390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokorná M, Giovannotti M, Kratochvíl L, Caputo V, Olmo E, Ferguson-Smith MA, Rens W. 2012. Conservation of chromosomes syntenic with avian autosomes in squamate reptiles revealed by comparative chromosome painting. Chromosoma 121, 409-418. ( 10.1007/s00412-012-0371-z) [DOI] [PubMed] [Google Scholar]
- 48.Nguyen P, Sýkorová M, Šíchová J, Kůta V, Dalíková M, Čapková Frydrychová R, Neven LG, Sahara K, Marec F. 2013. Neo-sex chromosomes and adaptive potential in tortricid pests. Proc. Natl Acad. Sci. USA 110, 6931-6936. ( 10.1073/pnas.1220372110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rovatsos M, Vukić J, Altmanová M, Johnson Pokorná M, Moravec J, Kratochvíl L. 2016. Conservation of sex chromosomes in lacertid lizards. Mol. Ecol. 25, 3120-3126. ( 10.1111/mec.13635) [DOI] [PubMed] [Google Scholar]
- 50.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 134. ( 10.1186/1471-2105-13-134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3--new capabilities and interfaces. Nucleic Acid Res. 40, e115. ( 10.1093/nar/gks596) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rovatsos M, Altmanová M, Pokorná M, Kratochvíl L. 2014. Conserved sex chromosomes across adaptively radiated Anolis lizards. Evolution 68, 2079-2085. ( 10.1111/evo.12357) [DOI] [PubMed] [Google Scholar]
- 53.Griffin DK, Robertson LB, Tempest HG, Skinner BM. 2007. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 117, 64-77. ( 10.1159/000103166) [DOI] [PubMed] [Google Scholar]
- 54.Skipwith PL, Bi K, Oliver PM. 2019. Relicts and radiations: Phylogenomics of an Australasian lizard clade with east Gondwanan origins (Gekkota: Diplodactyloidea). Mol. Phylogenet. Evol. 140, 106589. ( 10.1016/j.ympev.2019.106589) [DOI] [PubMed] [Google Scholar]
- 55.Augstenová B, Pensabene E, Veselý M, Kratochvíl L, Rovatsos M. 2021. Are geckos special in sex determination? Independently evolved differentiated ZZ/ZW sex chromosomes in carphodactylid geckos. Genome Biol. Evol. evab119. ( 10.1093/gbe/evab119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gamble T, Greenbaum E, Jackman TR, Bauer AM. 2015. Into the light: diurnality has evolved multiple times in geckos. Biol. J. Linn. Soc. Lond. 115, 896-910. ( 10.1111/bij.12536) [DOI] [Google Scholar]
- 57.Zheng Y, Wiens JJ. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 94, 537-547. ( 10.1016/j.ympev.2015.10.009) [DOI] [PubMed] [Google Scholar]
- 58.Nielsen SV, Daza JD, Pinto BJ, Gamble T. 2019. ZZ/ZW sex chromosomes in the endemic Puerto Rican leaf-toed gecko (Phyllodactylus wirshingi). Cytogenet Genome Res. 157, 89-97. ( 10.1159/000496379) [DOI] [PubMed] [Google Scholar]
- 59.Kawai A, Ishijima J, Nishida C, Kosaka A, Ota H, Kohno S, Matsuda Y. 2009. The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118, 43-51. ( 10.1007/s00412-008-0176-2) [DOI] [PubMed] [Google Scholar]
- 60.Wheeler BS, Anderson E, Frøkjær-Jensen C, Bian Q, Jorgensen E, Meyer BJ. 2016. Chromosome-wide mechanisms to decouple gene expression from gene dose during sex-chromosome evolution. eLife 5, e17365. ( 10.7554/eLife.17365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albritton SE, Kranz A-L, Rao P, Kramer M, Dieterich C, Ercan S. 2014. Sex-biased gene expression and evolution of the X chromosome in nematodes. Genetics 197, 865-883. ( 10.1534/genetics.114.163311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schütz F, Daish T, Grützner F, Kaessmann H. 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10, e1001328. ( 10.1371/journal.pbio.1001328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clinton M. 1998. Sex determination and gonadal development: a bird's eye view. J. Exp. Zool. 281, 457-465. () [DOI] [PubMed] [Google Scholar]
- 64.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267-271. ( 10.1038/nature08298) [DOI] [PubMed] [Google Scholar]
- 65.Rovatsos M, Augstenová B, Altmanová M, Sloboda M, Kodym P, Kratochvíl L. 2018. Triploid colubrid snake provides insight into the mechanism of sex determination in advanced snakes. Sex Dev. 12, 251-255. ( 10.1159/000490124) [DOI] [PubMed] [Google Scholar]
- 66.Deng X, et al. 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43, 1179-1185. ( 10.1038/ng.948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baker BS, Belote JM, 1983. Sex determination and dosage compensation in Drosophila melanogaster. Annu. Rev. Genet. 17, 345-393. ( 10.1146/annurev.ge.17.120183.002021) [DOI] [PubMed] [Google Scholar]
- 68.Zanetti S, Puoti A, 2013. Sex determination in the Caenorhabditis elegans germline. In Germ cell development in C. elegans. Advances in experimental medicine and biology (ed. Schedl T), vol. 757, pp. 41-69. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw Illumina reads were deposited in GenBank database under the BioProject PRJNA623146.