Abstract
Sex chromosomes are a great example of a convergent evolution at the genomic level, having evolved dozens of times just within amniotes. An intriguing question is whether this repeated evolution was random, or whether some ancestral syntenic blocks have significantly higher chance to be co-opted for the role of sex chromosomes owing to their gene content related to gonad development. Here, we summarize current knowledge on the evolutionary history of sex determination and sex chromosomes in amniotes and evaluate the hypothesis of non-random emergence of sex chromosomes. The current data on the origin of sex chromosomes in amniotes suggest that their evolution is indeed non-random. However, this non-random pattern is not very strong, and many syntenic blocks representing putatively independently evolved sex chromosomes are unique. Still, repeatedly co-opted chromosomes are an excellent model system, as independent co-option of the same genomic region for the role of sex chromosome offers a great opportunity for testing evolutionary scenarios on the sex chromosome evolution under the explicit control for the genomic background and gene identity. Future studies should use these systems more to explore the convergent/divergent evolution of sex chromosomes.
This article is part of the theme issue ‘Challenging the paradigm in sex chromosome evolution: empirical and theoretical insights with a focus on vertebrates (Part II)’.
Keywords: sex chromosomes, amniotes, co-option, vertebrates
1. Why should the emergence of sex chromosomes be non-random?
Sex determination, the process that controls whether an individual develops as a male or a female, is of vital importance at both individual and population levels. Gonochoristic vertebrates show extensive variability in sex determination systems ranging from environmental sex determination (ESD), where sexes do not differ in genotype, to genotypic sex determination (GSD) with either XX/XY (heterogametic males) or ZZ/ZW (heterogametic females) sex chromosomes [1–3]. The classical model postulates that sex chromosomes evolve from a pair of autosomes, after one chromosome acquires a sex-determining locus [4,5]. According to this widely accepted model, restricting recombination between the sex chromosomes facilitates the linkage of the sex-determining locus with nearby sexually antagonistic alleles [6,7], but it can be caused by other, more neutral processes [8]. Over time, the cessation of recombination leads to the accumulation of repetitive elements, heterochromatin, deleterious mutations and degradation of gene content on the Y and W chromosomes. However, some lineages such as medaka fishes, ranid frogs and cichlids have rapid turnovers of poorly differentiated sex chromosomes [9–11] and yet other lineages, for example sturgeons and skinks, have old, poorly differentiated sex chromosomes with little degeneration for a long evolutionary time [12,13].
An interesting extension of the classical model of sex chromosome evolution came with ongoing discovery of sex chromosome gene content in vertebrates. It appeared that the same chromosomes or syntenic blocks were sex chromosomes in distantly related lineages [14]. This has been explained using two discrete hypotheses. The first hypothesis proposes that different parts of the ancient sex chromosome have retained a sex-determining role in several modern vertebrate lineages [14–18]. The second hypothesis proposes that the same autosomes repeatedly and independently evolved into sex chromosomes in different lineages with certain genomic regions possibly having a non-randomly higher chance to be co-opted for the role [14,17,19,20]. It was suggested that gene content can ‘predispose some chromosomes to a specialized role in sex determination’ [17, p. 7]. In other words, it was assumed that a limited pool of genes involved in gonadal development such as amh, ar, dmrt1 or sox3 have a greater chance of becoming a master sex-determining gene in vertebrates turning their syntenic blocks into sex chromosomes. This hypothesis received some support [14,17] based on limited data available at that time. Recently, our knowledge of vertebrate sex chromosome homology has expanded owing to the increase of studies applying next-generation sequencing methodologies and other molecular approaches (electronic supplementary material, table 1), although it is still very far from being complete. Here, we present an up to date overview of the data on sex determination and sex chromosome genomic content in amniotes, and we briefly summarize knowledge on evolutionary history of sex determination in this group and evaluate more rigorously the hypothesis of non-random emergence of sex chromosomes.
2. Variability in sex determination in amniotes: mistakes of the past in data, independent origins and sex chromosome stability
Amniotes encompass over 27 000 species, and as a whole possess varied sex-determining systems; however, this variability is unequally distributed among amniote clades. Viviparous mammals and birds are well known for having a long-term evolutionary stability of sex chromosomes [21,22], but ‘reptiles’, the paraphyletic group of ectothermic amniotes, were suggested to have labile, rapidly evolving sex determination [23,24]. Nevertheless, high stability of sex chromosomes for dozens of million years has been documented in several reptile lineages (figure 1). Furthermore, it has been demonstrated that the estimations of the variability in reptile sex determination and numbers of reconstructed transitions among particular sex determination systems were biased owing to the inclusion of imprecise data, mostly claiming ESD in species with GSD, which concerned, e.g. skinks, chameleons, varanids, anguids, lacertids and iguanas [12,29–35]. On the other hand, sex chromosomes, and therefore GSD, were misidentified in some cases. For example, ZZ/ZW sex chromosomes were repeatedly reported in the past 40 years for the brown roofed turtle (Pangshura smithii) until re-examination by more sensitive molecular cytogenetic techniques did not reveal sex chromosomes and demonstrated that the original report was based on incorrect pairing of chromosomes in karyogram [36]. The reported turnovers of sex chromosomes within lacertids [19] based on data by Srikulnath et al. [37] were also not supported by later re-examinations and as far as known all examined species of this family share homologous ZZ/ZW sex chromosomes [30,38] In the same context, snakes were another prominent case of erroneous identification of sex determination. Poorly differentiated sex chromosomes homologous with differentiated ZZ/ZW sex chromosomes of the caenophidian snakes were reported in the past in boas and pythons [4,39,40]. Recent investigations showed a more complex history of snake sex chromosomes with XX/XY sex chromosomes evolving independently in Neotropical boas and pythons and ZZ/ZW sex chromosomes in Malagasy boas [41,42].
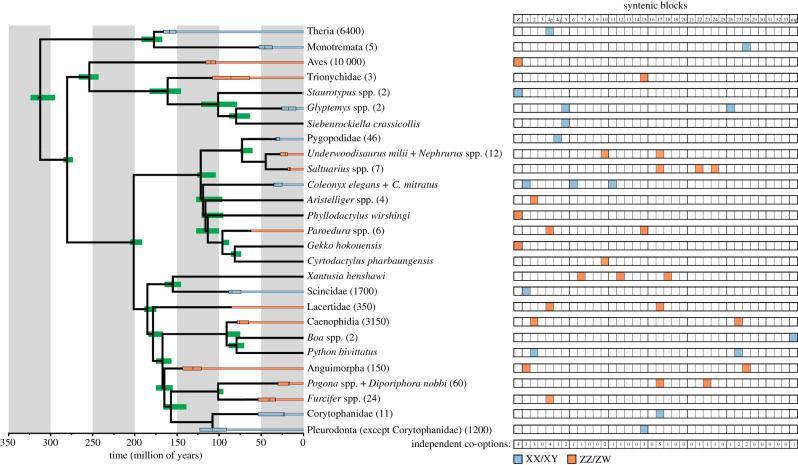
Figure 1.
Synteny and age of sex chromosomes across amniote lineages. The data on synteny are presented in the electronic supplementary material, table 1. The age of divergence between clades and the minimal age of sex chromosomes (estimated as the split between the phylogenetically most distant members of a clade with evidence for homology of sex chromosomes) are taken from Kumar et al. [25], available at http://timetree.org. The numbers next to the clade names represent the estimated number of species putatively sharing the given sex chromosomes. The phylogenetic tree follows Shen et al. [26] for the topology of major lineages and Pyron et al. [27] for squamates. Corytophanids might form an inner group of Pleurodonta, but this question is not resolved yet [28]. mg, ‘missing genes’ in chicken. (Online version in colour.)
Corrections of these inaccuracies together with the recent reports of sex determination in previously unstudied lineages (electronic supplementary material, table 1) such as Gila monsters and beaded lizards [43,44], xantusiids [45] and several lineages of geckos [46–49] and skinks [12] led to a hopefully more reliable dataset to reconstruct the evolutionary history of sex determination in amniotes. We should keep in mind that we have more or less reliable direct data on sex determination in likely less than 5% out of roughly 11 000 current species of non-avian reptiles [47]. Nevertheless, Kostmann et al. [12] recently estimated that about 60% of reptiles are members of five highly diversified clades with molecular evidence for stability of XX/XY (skinks [12]; iguanas with the exception of corytophanids [50,51]) and ZZ/ZW (lacertids [30]; varanids [44] and caenophidian snakes [52]) sex chromosomes. Each of these five groups has sex chromosomes non-homologous with each other (electronic supplementary material, table 1). The variability in sex determination is thus largely concentrated to the remaining 40% of reptile species, but even among them, we can find smaller lineages with ancient sex chromosomes such as softshell turtles (Trionychidae) [53], the gecko families Pygopodidae [54] and Carphodactylidae [55], the gecko genera Paroedura (although a likely turnover to another GSD system was uncovered within the genus [20]) and Aristelliger [48], the chameleons of the genus Furcifer (M. Rovatsos and L. Kratochvíl 2021, personal communication) and the agamids of the genus Pogona and its relatives [56]. Some estimations expected that about 25% of reptile species exhibit ESD [57]. Nevertheless, it seems that the number of reptile species with ESD was greatly overestimated, and recently, Kostmann et al. [12] suggested that only 5% of non-avian reptiles have ESD. A detailed list of reptile species with documented ESD can be extracted from [47,58] (lizards), [59] (turtles) and [60] (crocodiles).
3. Evolutionary history of sex determination in amniotes: critical overview of current models
As reviewed above, it seems that for many amniote lineages: (i) the sex determination is, in fact, stable over the long term, (ii) ESD is relatively rare in comparison to GSD, and (iii) transitions among particular sex determination systems are less frequent than previously expected. The situation is still too complex and poorly investigated to allow straightforward reconstruction of the deep evolutionary history of sex determination. It is obvious that sex chromosomes are non-homologous among many amniote lineages, but from the perspective of the test of non-random co-option it is crucial to decide whether in some cases sex chromosomes of distant lineages were inherited from a common ancestor, or evolved independently, and what was the ancestral sex determination for amniotes. In this respect, there are different opinions in the literature.
The observation that sex chromosomes in birds and monotremes [61,62] and the gecko Gekko hokouensis [63] contain the same, orthologous genomic region led to the hypothesis that the last common amniote ancestor possessed bird-like proto-sex chromosomes [15,17,18,64]. Nevertheless, this hypothesis is unlikely as these three lineages are phylogenetically separated by numerous lineages with different sex determination mechanisms [65]. Also, a detailed analysis of monotreme sex chromosomes showed that the ancestral sex chromosomes of this lineage are homologous with an avian autosome (chicken chromosome 28) and that the material homologous with the avian Z was added into monotreme sex chromosomes much later [21]. Finally, the ZZ/ZW sex chromosomes of G. hokouensis are poorly differentiated, with no apparent loss of genes on the W, indicating a relatively recent independent origin [63].
The hypothesis of an ancestral bird-like sex chromosome has been further expanded to suggest the common ancestor of amniotes possessed a ‘super-sex chromosome’, i.e. that certain regions forming the extant sex chromosomes in various lineages were linked in the amniote ancestor in a large chromosome that subsequently underwent multiple fissions [19,66–68]. These previously linked genomic regions are now sex chromosomes in different extant lineages. Part of this hypothetical ‘super-sex chromosome’ should include avian Z and parts of X of therian mammals [19]. It was argued that this hypothesis is supported by several lines of evidence. First, the mammalian X-conserved region, the X-added region incorporated to the sex chromosomes in the ancestor of eutherian mammals, and the chicken Z chromosomes are all linked on a large autosome in the Mexican axolotl (Ambystoma mexicanum) [69]. However, this huge chromosome probably evolved within amphibians and contains these unrelated segments just by chance [14,17,70]. Second, some putative W-specific loci in a caenophidian snakes (namely the Siamese cobra, Naja kaouthia) showed partial homology with genes on sex chromosomes of several amniote lineages [68]. However, although chromosomes in sauropsids generally share high synteny conservation (see evidence further in the text), translocations of genes are expected to occur during millions of years of independent evolution. Such translocations of small chromosome fragments are common and well documented in amniotes with sequenced genomes. In fact, the identified genes from the cobra W chromosome have homologues scattered across the chicken (Gallus gallus, GGA) genome, i.e. linked to chromosomes GGA 1q, GGA 2p, GGA 3, GGA 5, GGA 6, GGA 7, GGA 8, GGA 10, GGA 11, GGA 15, GGA 17, GGA 23, GGA 27 and GGA Z [68]. The interpretation that few genes homologous to several chicken chromosomes might indicate that the corresponding blocks were once part of a single ‘super-sex chromosome’ in the ancestral amniote is not very convincing to us. Translocations of these genes to snake sex chromosomes by minor rearrangements during hundreds of millions of years of amniote evolution, e.g. during 280 Myr since the separation of snake and avian lineages, seem more plausible to us. Such insertions of minor chromosomal regions can be also detected in autosomes by cross-species comparisons between the genomes of birds and snakes with chromosome-level assemblies [71]. Furthermore, sex chromosomes are enriched for genes with sex-specific function. For example, the mammalian X and avian Z are enriched for testis-expressed genes often from multi-copy families, most of these changes in gene content occurring after they became sex chromosomes [72,73]. The trafficking of genes onto the X and Z is probably a feature common to differentiated sex chromosomes, and the functional property of genes could lead in some cases to convergent acquisition of homologous genes and may explain the shared sequences found among species.
The observation that the non-homologous W chromosomes of birds and caenophidian snakes share similar repetitive content has been claimed as supporting the ancestral ‘super-sex chromosome’ hypothesis [17,66]. However, shared repetitive content is a rather poor evidence for homology of genomic regions. Repeat content of degenerated sex chromosomes represents one of the most dynamic genome parts as evidenced by comparative studies, for instance, in oplurid, varanid and lacertid lizards [74–76], and frequent homoplasies can be expected in this respect. Indeed, the same repetitions accumulated independently on clearly non-homologous sex chromosomes several times, as well as on autosomes [16,77–79]. Moreover, differentiated sex chromosomes with degenerated W is a clear apomorphy of caenophidian snakes [52], and the other snakes have generally poorly differentiated sex chromosomes without notable accumulations of repeats [80] and at least in some cases non-homologous to those of caenophidian sex chromosomes [41].
Previous studies argued that the Anolis chromosome 2 (occasionally referred as the ‘lizard chromosome 2’) is composed of a fragment orthologous with avian chromosome Z and a small part of human X, suggesting that the multiple sex chromosomes of platypus and avian sex chromosomes may have been a part of the hypothetical ancestral amniote chromosome, fission of which ‘gave rise to reptilian, monotreme, and therian sex chromosomes' (depicted in fig. 2 of their study, [19, p. 99]). Apart from the fact that owing to the diversity in karyotypes it is difficult to decide what are ‘reptilian sex chromosomes’ and ‘lizard chromosomes’, as pointed above, the part orthologous with the avian Z was added to monotreme sex chromosomes only later [21] and was not present in the ancestral monotreme sex chromosomes. The hypothesis on the ‘super-sex chromosome’ also does not address the underlying molecular mechanisms of sex determination and their evolution. As far as we know, sex determination is mostly driven by one (or exceptionally, a few) locus, e.g. by a sex-determining gene whose evolutionary origin gave rise to sex chromosomes [81]. The molecular mechanism of sex determination linked to the ancestral ‘super-sex chromosome’ is not clear. Birds, monotremes and therian mammals have different sex-determining genes working on different mechanisms. Sex determination is based on a difference in copy numbers of the sex-determining gene dmrt1 linked to Z and missing on W between sexes in birds, on the dominant Y-linked genes in therian mammals and probably on a homologue of amh in monotremes [21,22]. Evidence suggests that each of these systems is an apomorphy of the particular lineage. It is not clear how fissions of the ancestral ‘super-sex chromosome’ would give rise to an array of descendent ZZ/ZW and XX/XY sex chromosomes with these and probably other sex-determining loci. It thus seems that there is a little support for a ‘super-sex chromosome’ in the amniote ancestor.
Another hypothesis on the evolution of sex determination is that ESD is the ancestral sex determination system in amniotes, i.e. that the amniote ancestor did not have any sex chromosomes at all [3,4,47,58,65]. This scenario is based on several observations: (i) the stability of GSD in many lineages in amniotes (figure 1), but also in other lineages such as amphibians or insects, where no ESD species have been ever found [82]; (ii) a lack of well-supported evolutionary transitions from GSD to ESD within amniotes: the earlier reported transitions appeared to be based on misclassification of GSD species as ESD such as in skinks, varanids and lacertids [12,30,34,44], or are reconstructed deeply in the evolutionary history, e.g. in turtles [59], and it is thus difficult to verify them [65]; (iii) often non-homology of sex chromosomes among GSD lineages phylogenetically separated by a ESD lineage [3,65]; (iv) variability in sex chromosomes largely in lineages with the presence of ESD [47] (turtles, geckos and agamids; e.g. [56,59]), which would be in agreement with multiple independent origins of GSD from the ancestral ESD; (v) phylogenetical distribution of ESD reported to be possibly ancestral for several major amniote groups (crocodiles, tuatara, turtles and squamates; [3,65]); and newly also (vi) on the at least partial sharing of the molecular mechanism of environmentally sensitive molecular machinery among phylogenetically distant ESD lineages [83–85].
While the scenario of ancestral ESD could be considered as a working hypothesis, the molecular machinery of ESD is still only poorly known (although the recent progress is huge; [83–85]) and more direct test of the (non)homology of ESD based on the comparison of molecular mechanisms across amniote lineages is not possible yet. Notably, GSD-to-ESD transition was induced easily in the laboratory in the bearded dragon (Pogona vitticeps) [86], which suggests that such transitions could occur also in nature. Nevertheless, comparative evidence suggests that sex chromosomes in the lineage including the genus Pogona have been stable for around 25 Myr [56,87–89]. Thus, transitions from GSD to ESD might not occur regularly in nature, even in such a lineage with seemingly all the necessary conditions. To us a viable alternative to the ancestral ESD in amniotes is ancestral GSD with unknown sex chromosomes followed by many turnovers of sex chromosomes accompanied with a few, probably quite ancient origins of ESD. Future research should try to differentiate between these alternatives. For the purposes of the current paper, we will assume that sex chromosomes evolved independently within amniotes and we will take putative turnovers of sex chromosomes as independent origins.
4. How strong is non-randomness in the origin of sex chromosomes?
Despite differences in genome size and occasional whole-genome duplications in some lineages, a major feature of the vertebrate evolution is the structural conservation of the gene order on chromosomes, known as synteny [70,90], with a pattern of conserved syntenic associations dating back 360 Myr [91], and even further in other metazoans [92]. ‘Conserved synteny’ describes a block of orthologous genes that are physically linked on a chromosome, though not necessarily in the same gene order, among genomes of related species. Various vertebrate lineages differ in the rate of chromosomal evolution, i.e. in the frequency of chromosome rearrangements altering the chromosome structure and morphology; however, sauropsids (non-mammalian amniotes) generally exhibit remarkable conserved synteny as shown by chromosome painting and other molecular cytogenetic approaches [93] as well as comparative genomics [94–97]. Viviparous mammals exhibit a much higher rate of interchromosomal rearrangements, but still maintain syntenic sex chromosomes (so-called Ohno's Law [4,98]). In addition, whole-genome comparisons among birds and mammals point to regions in genomes where the order of orthologous sequences has been maintained for tens of millions of years [99,100]. The high conservation of chromosomes across sauropsids and sex chromosomes in mammals facilitates testing the non-random co-option of genomic regions for the role of sex chromosomes. There is also a question of how to define syntenic blocks used for the test. Following former attempts and tradition of reporting homology of sex chromosomes across amniotes [19,20,45,53,54,101], we decided to focus on syntenic blocks represented by chicken (G. gallus) chromosomes. Chicken genome is adequately sequenced at chromosome level and well annotated, and seems to maintain many syntenic genomic blocks intact as whole chromosomes (e.g. Ensembl database) [102,103]. It is thus probably a good approximation of the ancestral avian/amniote karyotype, although we understand that the lineage leading to chicken went through several chromosome rearrangements [104]. Two chromosomes now forming the fourth chicken largest chromosome (GGA 4) fused relatively recently in the chicken ancestor [105]. Therefore, we keep the small (GGA 4p) and the long (GGA 4q) arms of GGA 4 as two different syntenic blocks in our analyses. In addition, the chicken genome is missing in 232 protein-coding genes, many of them are largely conserved in most other vertebrate lineages, including non-avian reptiles. In comparison to the genome of Anolis carolinensis, chicken is lacking gene content located in several chromosome regions, particularly from the linkage group F (ACA-Lg F) [106]. Since ACA-Lg F is syntenic with the part of sex chromosomes in Boa spp. [41], we added ‘missing genes in chicken’ as an additional syntenic block to our analysis for accuracy. The exclusion of this artificial block from the analyses does not change the significance of the results. Also, chromosome GGA 29 is not well assembled and contains no annotated genes in the current chicken genome assembly. Therefore, it has not been checked if its orthologues evolved into sex chromosomes in other amniote lineages and we do not include it in our analyses here. In total, we assumed that there were 35 syntenic blocks (GGA 4p + GGA 4q + 31 other chicken autosomes excluding GGA 29 + chicken Z chromosome + ‘missing genes in chicken’) that had chance to become sex chromosomes in the history of amniotes. For each block, we take data on the number of linked protein-coding genes and physical length in Mb from NCBI Gallus gallus assembly GRCg6a. In the case of the ‘missing genes in chicken’ block, we assumed 232 genes and the physical length 10.19 Mb, which is roughly proportional to the number of genes in other blocks.
Based on previous surveys [3,47,58,65], we estimate that sex chromosomes might have evolved independently within amniotes over 40 times. However, we have knowledge on the partial gene content allowing an inference about orthology of their sex chromosomes to a syntenic block defined by chicken chromosomes only in 27 of them representing putative independent origins of sex chromosomes (figure 1; electronic supplementary material, table 1). In non-mammalian and non-avian amniotes, we have direct knowledge on homology of sex chromosomes in 245 species (electronic supplementary material, table 1). As we were interested in the origin of sex chromosomes, wherever known, we included into the analysis only the syntenic blocks forming the ancestral sex chromosomes, e.g. in monotremes we count only the GGA 28 syntenic block which has the amh sex-determining locus and not the GGA Z, GGA 2, GGA 3, GGA 11, GGA 12, GGA 13, GGA 16, GGA 17, GGA 25 and GGA 30 blocks which probably represent later fusions to the ancestral sex chromosomes [21]. However, in some cases, e.g. in lacertids and caenophidian snakes, current sex chromosomes are composed of more blocks (chicken chromosomes), and we do not know which of them is the ancestral one. Therefore, we performed two alternative analyses. In the first, we included into the dataset all syntenic blocks forming such a sex chromosome (e.g. GGA 4p and GGA 17 for lacertids), which gave us in total 41 blocks representing actual sex chromosomes. In the alternative test, we included only 17 lineages with known ancestral syntenic block, although their sex chromosomes include more neo-parts (e.g. GGA 28 for monotremes) and lineages where only a single syntenic block was reported to be largely orthologous with their sex chromosomes (e.g. birds and skinks). From these restricted analyses, we hence excluded all lineages such as lacertids. Each putative independent origin of sex chromosomes was taken as a single sample (observation) for the statistical tests; therefore, the tests do not include any pseudoreplications caused by phylogenetic dependence.
We performed the multinomial test in R [107] using the package XNomial [108]. Because the tested hypotheses required too many combinations to be checked, we used a test based on ‘Monte Carlo’ simulations. We compared the goodness of fit of the observed distribution with the expected frequencies by the log-likelihood ratio test. First, we tested whether the observed blocks representing actual sex chromosomes are distributed among these 35 categories randomly, i.e. we compared the observed data with the frequencies, assuming that each syntenic block has equal chance to become a sex chromosome. Subsequently, we tested whether the observed distribution can be explained by differences in the number of genes among the syntenic blocks, i.e. assuming that the probability of each syntenic block to evolve to sex chromosome is proportional to the number of protein-coding genes it contains. Lastly, we tested whether the observed distribution reflects differences in the physical length among the syntenic blocks, i.e. assuming that the probability of each syntenic block to evolve to sex chromosome is proportional to its physical size, because gene density differs significantly among chromosomes.
The multinomial test based on the dataset, including all 41 syntenic blocks potentially involved in the origin of sex chromosomes in amniote lineages, supported that the syntenic blocks differ in the probability to be co-opted for the function of sex chromosomes (p = 0.021). Some syntenic blocks never evolved into the sex chromosomes although they are very large (GGA 3) or medium-sized chromosomes (GGA 8, 9, 13 and 14). On the other hand, the syntenic block GGA 17 was probably involved in the evolution of sex chromosomes five times, GGA Z and GGA 4p four times, GGA 1, 2 and 15 three times, and GGA 5, 10, 27 and 28 twice each. The syntenic blocks GGA 4q, 6, 7, 11, 12, 18, 22, 23, 24, 26 and the block ‘missing genes in chicken’ turned into sex chromosomes once each. The non-random probability of turning into sex chromosomes cannot be explained by the differences in the gene numbers (p = 0.01) and the physical length (p < 0.0001) among the syntenic blocks. The analyses including only 17 lineages where we can tentatively assign a single ancestral syntenic block orthologous to their sex chromosomes largely confirmed these results. The origins of sex chromosomes among these lineages from the restricted datasets were still significantly non-randomly distributed among 35 syntenic blocks (p = 0.02), and their distribution did not reflect the physical size of the syntenic blocks (p = 0.006). However, the observed distribution did not significantly depart from the expectation based on the number of genes in each syntenic block any more (p = 0.057). We tend to attribute this marginally insignificant result to the lower test power owing to much more restricted sample size. Although we believe that our analysis is as comprehensive as possible at the current stage of knowledge, the above comparisons have several important limitations. As already mentioned, we lack data on the genetic content of sex chromosomes in many amniote lineages with putatively independently evolved sex chromosomes. Moreover, knowledge on the genetic content of sex chromosomes in many lineages included into the analysis is limited too, particularly in cases when the homology is inferred from the sex linkage of loci found from reduce representation approaches (e.g. in Xantusia and Aristelliger) [45,48]. It is possible that the syntenic blocks forming the oldest evolutionary strata of sex chromosomes were missed and we included into the analysis a block that was added into sex chromosomes only later. We should also keep in mind that the same syntenic blocks playing the role of sex chromosomes do not necessarily mean homologous sex determination systems. The same block could be repeatedly and independently co-opted for the function of sex chromosomes within a lineage where we assume a single common origin of sex chromosomes. The number of repeated independent origins would then be underestimated. For example, we assume a single origin of sex chromosomes orthologous to GGA 28 in anguimorphan lizards, but it seems that at least some anguids do not share the same sex chromosomes [41]. The chromosomes orthologous to GGA 28 might have evolved independently in the families Varanidae and Helodermatidae. On the other hand, some categories of sex chromosomes counted by us as independent origins can represent a single origin instead. It could apply to the Burmese pythons and caenophidian snakes (both these lineages have sex chromosomes orthologous to GGA 2 and GGA 27, but differ in heterogamety [41]), and the carphodactylid lineages Nephrurus–Underwoodisaurus and Saltuarius, where their respective ZZ/ZW sex chromosomes slightly overlap in a syntenic block with orthologues linked to GGA 17 [55]. Future studies should test whether our assumptions on homology are correct based on deeper knowledge of genetic content, particularly based on the identification of sex-determining loci.
5. Why should we care about non-randomness?
In summary, it seems that there is indeed a non-randomness in the origin of sex chromosomes in amniotes with several blocks turning independently repeatedly into the sex chromosomes. Nevertheless, many blocks representing sex chromosomes are unique. We can conclude that it is quite difficult to predict the origin of sex chromosomes from a particular genomic region, although there is a significantly higher chance that a ‘popular’ block will turn into sex chromosomes. Why should some blocks have a higher chance to become sex chromosomes? It seems that the physical size and the number of protein-coding genes do not explain this non-randomness. In some of the blocks that repeatedly evolved into sex chromosomes, there are well-known genes included in gonad differentiation pathways. It was demonstrated that a mutation in some of them, such as sox3 and ar included in GGA 4p, dmrt1 included in GGA Z, nr5a1 included in GGA 17 and amh included in GGA 28, can cause a sex reversal or disrupt sexual development [109–113], which is a basic predisposition to become a sex-determining locus. However, mutations in some other genes such as follicle-stimulating hormone receptor (fshr) and luteinizing hormone receptor gene (lhcgr) led to female-to-male sex reversal as well [114,115], but the syntenic block orthologous to GGA 3 where both genes are located have never become a sex chromosome among amniotes investigated thus far (electronic supplementary material, table 1). Probably not all genes with a potential to cause sex reversal can act as sex-determining genes, for instance because mutations in some of them can have severe negative pleiotropic effects. We should also keep in mind that in some cases, a sex-determining gene can be a paralogue linked to different linkage groups than the original copy and it can even jump through transposition throughout a genome [81,116]. In such cases, information about other genes linked to sex chromosomes can be misleading in the identification of the sex-determining locus. For the more robust test of the hypothesis that syntenic blocks with genes with higher potential to become sex-determining genes have a higher chance to become sex chromosomes, we will need a better knowledge on the identity of sex-determining genes in amniotes. Up to date knowledge on sex-determining genes is limited to viviparous mammals [117] and to strongly supported candidates in birds and monotremes [21,118,119].
Marshall Graves & Peichel [14] also pointed that some syntenic blocks have higher chance to become a part of sex chromosomes as they can ‘make better sex chromosomes', i.e. that their involvement in sex chromosomes can contribute to the resolution of sexual conflict between sexes for the expression of traits under sexually antagonistic selection. This hypothesis suggests that selection would favour a location of a sex-determining gene in a region containing loci with sexually antagonistic effects [120] or a fusion of ancestral sex chromosome with autosomes enriched for genes with sex-related function [101]. In our review, we focused on the origin of sex chromosomes from autosomes. We still have only limited information about identity and location of sex-determining genes and content of parts newly added to sex chromosomes. Emerging evidence in birds, monotremes and lizards suggests more frequent involvement of certain chromosomes in neo-sex chromosome formations [21,101,121,122]. Particularly multiple neo-sex chromosomes formed by a fusion of an autosome with a Y chromosome evolved many times in amniotes, e.g. around 20 times in mammals and 10 times in iguanas [123,124], but such autosome-sex chromosome fusions are rare in birds [124,125]. Nevertheless, previously autosomal material was added to both Z and W chromosomes in birds in the Sylvioidea superfamily [101] and in the Australian songbird Eopsaltria australis [126]. Future studies should focus on the determination of syntenic blocks involved in these rearrangements and should test whether the pattern is indeed non-random and uncover its drivers.
Although the current data on the origin of sex chromosomes in amniotes suggest that their evolution is non-random only to a certain degree, we would like to stress that repeatedly co-opted chromosomes are an excellent model system. Independent co-option of the same genomic region for the role of sex chromosome hence offers a great opportunity for testing evolutionary scenarios on the sex chromosome evolution under the explicit control for the genomic background and gene identity. For example, Rovatsos & Kratochvíl [127] compared the expression of genes linked to sex chromosomes between the green anole (XX/XY) and the Florida softshell turtle (ZZ/ZW). Sex chromosomes in both these systems are orthologous to GGA 15 and thus contain the same genes. However, the X-specific genes (genes linked to X but missing on the Y chromosome) are fully dosage compensated at the level of transcription in the anole, while Z-specific genes have approximately only half expression in females in comparison to males in the turtle. This comparison shows that exactly the same genes can or cannot be dosage compensated in independently evolved sex chromosomes, and thus the evolution of dosage compensation does not depend upon the genomic background. Among amniotes, more lineages than the iguanas and the softshell turtles co-opted the same syntenic block for sex chromosomes (electronic supplementary material, table 1), as shown, for instance, by our ongoing research on lacertid lizards, chameleons from the genus Furcifer and geckos from the genus Paroedura (ZZ/ZW) and therian mammals (XX/XY) [20,30,128] (M. Rovatsos and L. Kratochvíl 2021, personal communication). Future studies should further use these systems to explore the convergent/divergent evolution of sex chromosomes.
Authors' contributions
All three authors developed the major idea and contributed to writing.
Competing interests
We declare we have no competing interests.
Funding
The project was supported by the Czech Science Foundation project 17-22604S, Charles University project PRIMUS/SCI/46 and Charles University Research Centre programme 204069.
References
- 1.Bull JJ. 1983. Evolution of sex determining mechanisms. Menlo Park, CA: Benjamin/Cummings. [Google Scholar]
- 2.Capel B (ed.). 2019. Sex determination in vertebrates. Current topics in developmental biology, vol. 134. Amsterdam, The Netherlands: and New York, NY: Elsevier. [Google Scholar]
- 3.Straková B, Rovatsos M, Kubička L, Kratochvíl L. 2020. Evolution of sex determination in amniotes: did stress and sequential hermaphroditism produce environmental determination? BioEssays 42, e2000050. ( 10.1002/bies.202000050) [DOI] [PubMed] [Google Scholar]
- 4.Ohno S. 1967. Sex chromosomes and sex-linked Genes. Berlin, Germany: Springer-Verlag. [Google Scholar]
- 5.Charlesworth D, Charlesworth B, Marais G. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118-128. ( 10.1038/sj.hdy.6800697) [DOI] [PubMed] [Google Scholar]
- 6.Fisher RA. 1931. The evolution of dominance. Biol. Rev. 6, 345-368. ( 10.1111/j.1469-185X.1931.tb01030.x) [DOI] [Google Scholar]
- 7.Rice WR. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38, 735-742. ( 10.1111/j.1558-5646.1984.tb00346.x) [DOI] [PubMed] [Google Scholar]
- 8.Furman BLS, Metzger DCH, Darolti I, Wright AE, Sandkam BA, Almeida P, Shu JJ, Mank JE. 2020. Sex chromosome evolution: so many exceptions to the rules. Genome Biol. Evol. 12, 750-763. ( 10.1093/gbe/evaa081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M. 2015. Turnover of sex chromosomes in celebensis group Medaka fishes. G3: Genes Genom. Genet. 5, 2685-2691. ( 10.1534/g3.115.021543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffries DL, et al. 2018. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat. Commun. 9, 4088. ( 10.1038/s41467-018-06517-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lichilín N, El Taher A, Böhne A. 2021. Sex-biased gene expression and recent sex chromosome turnover. Phil. Trans. R. Soc. B 376, 20200107. ( 10.1098/rstb.2020.0107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostmann A, Kratochvíl L, Rovatsos M. 2021. Poorly differentiated XX/XY sex chromosomes are widely shared across skink radiation. Proc. R. Soc. B 288, 20202139. ( 10.1098/rspb.2020.2139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhl H, et al. 2021. A 180 Myr-old female-specific genome region in sturgeon reveals the oldest known vertebrate sex determining system with undifferentiated sex chromosomes. Phil. Trans. R. Soc. B 376, 20200089. ( 10.1098/rstb.2020.0089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall Graves JA, Peichel CL. 2010. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 11, 205. ( 10.1186/gb-2010-11-4-205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall Graves JA. 2009. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 42, 565-586. ( 10.1146/annurev.genet.42.110807.091714) [DOI] [PubMed] [Google Scholar]
- 16.O'Meally D, Patel HR, Stiglec R, Sarre SD, Georges A, Marshall Graves JA, Ezaz T. 2010. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 18, 787-800. ( 10.1007/s10577-010-9152-9) [DOI] [PubMed] [Google Scholar]
- 17.O'Meally D, Ezaz T, Georges A, Sarre SD, Graves JA. 2012. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 20, 7-19. ( 10.1007/s10577-011-9266-8) [DOI] [PubMed] [Google Scholar]
- 18.Livernois AM, Marshall Graves JA, Waters PD. 2012. The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity 108, 50-58. ( 10.1038/hdy.2011.106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ezaz T, Srikulnath K, Marshall Graves JA. 2017. Origin of amniote sex chromosomes: an ancestral super-sex chromosome, or common requirements? J. Heredity 108, 94-105. ( 10.1093/jhered/esw053) [DOI] [PubMed] [Google Scholar]
- 20.Rovatsos M, Farkačová K, Altmanová M, Johnson Pokorná M, Kratochvíl L. 2019. The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 28, 3042-3052. ( 10.1111/mec.15126) [DOI] [PubMed] [Google Scholar]
- 21.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488-493. ( 10.1038/nature13151) [DOI] [PubMed] [Google Scholar]
- 22.Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MTP, Zhang G. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346, 1246338. ( 10.1126/science.1246338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarre SD, Georges A, Quinn A. 2004. The ends of a continuum: genetic and temperature-dependent sex determination in reptiles. BioEssays 26, 639-645. ( 10.1002/bies.20050) [DOI] [PubMed] [Google Scholar]
- 24.Grossen C, Neuenschwander S, Perrin N. 2011. Temperature-dependent turnovers in sex-determination mechanisms: a quantitative model. Evolution 65, 64-78. ( 10.1111/j.1558-5646.2010.01098.x) [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812-1819. ( 10.1093/molbev/msx116) [DOI] [PubMed] [Google Scholar]
- 26.Shen XX, Liang D, Wen JZ, Zhang P. 2011. Multiple genome alignments facilitate development of NPCL markers: a case study of tetrapod phylogeny focusing on the position of turtles. Mol. Biol. Evol. 28, 3237-3252. ( 10.1093/molbev/msr148) [DOI] [PubMed] [Google Scholar]
- 27.Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 13, 93. ( 10.1186/1471-2148-13-93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen SV, Guzmán-Méndez IA, Gamble T, Blumer M, Pinto BJ, Kratochvíl L, Rovatsos M. 2019. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol. Lett. 15, 20190498. ( 10.1098/rsbl.2019.0498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rovatsos M, Johnson Pokorná M, Altmanová M, Kratochvíl L. 2015. Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): differentiation of sex and neo-sex chromosomes. Sci. Rep. 5, 13196. ( 10.1038/srep13196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rovatsos M, Vukić J, Mrugała A, Suwala G, Lymberakis P, Kratochvíl L. 2019. Little evidence for switches to environmental sex determination and turnover of sex chromosomes in lacertid lizards. Sci. Rep. 9, 7832. ( 10.1038/s41598-019-44192-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Telemeco RS. 2015. Sex determination in southern alligator lizards (Elgaria multicarinata; Anguidae). Herpetologica 71, 8-11. ( 10.1655/Herpetologica-D-14-00033) [DOI] [Google Scholar]
- 32.Nielsen SV, Banks JL, Diaz RE Jr, Trainor PA, Gamble T. 2018. Dynamic sex chromosomes in Old World chameleons (Squamata: Chamaeleonidae). J. Evol. Biol. 31, 484-490. ( 10.1111/jeb.13242) [DOI] [PubMed] [Google Scholar]
- 33.Iannucci A, et al. 2019. Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae). Heredity 123, 215-227. ( 10.1038/s41437-018-0179-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornejo-Páramo P, et al. 2020. Viviparous reptile regarded to have temperature-dependent sex determination has old XY chromosomes. Genome Biol. Evol. 12, 924-930. ( 10.1093/gbe/evaa104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiggins JM, Santoyo-Brito E, Scales JB, Fox SF. 2020. Gene dose indicates presence of sex chromosomes in collared lizards (Crotaphytus collaris), a species with temperature-influenced sex determination. Herpetologica 76, 27-30. ( 10.1655/Herpetologica-D-19-00036) [DOI] [Google Scholar]
- 36.Mazzoleni S, et al. 2019. Turtles of the genera Geoemyda and Pangshura (Testudines: Geoemydidae) lack differentiated sex chromosomes: the end of a 40-year error cascade for Pangshura. PeerJ 7, e6241. ( 10.7717/peerj.6241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srikulnath K, Matsubara K, Uno Y, Nishida C, Olsson M, Matsuda Y. 2014. Identification of the linkage group of the Z sex chromosomes of the sand lizard (Lacerta agilis, Lacertidae) and elucidation of karyotype evolution in lacertid lizards. Chromosoma 123, 563-575. ( 10.1007/s00412-014-0467-8) [DOI] [PubMed] [Google Scholar]
- 38.Lisachov AP, Giovannotti M, Pereira JC, Andreyushkova DA, Romanenko SA, Ferguson-Smith MA, Borodin PM, Trifonov VA. 2020. Chromosome painting does not support a sex chromosome turnover in Lacerta agilis Linnaeus, 1758. Cytogenet. Genome Res. 160, 134-140. ( 10.1159/000506321) [DOI] [PubMed] [Google Scholar]
- 39.Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl Acad. Sci. USA 103, 18 190-18 195. ( 10.1073/pnas.0605274103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11, e1001643. ( 10.1371/journal.pbio.1001643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gamble T, Castoe TA, Nielsen SV, Banks JL, Card DC, Schield DR, Schuett GW, Booth W. 2017. The discovery of XY sex chromosomes in a boa and python. Curr. Biol. 27, 2148-2153. ( 10.1016/j.cub.2017.06.010) [DOI] [PubMed] [Google Scholar]
- 42.Augstenová B, Johnson Pokorná M, Altmanová M, Frynta D, Rovatsos M, Kratochvíl L. 2018. ZW, XY, and yet ZW: sex chromosome evolution in snakes even more complicated. Evolution 72, 1701-1707. ( 10.1111/evo.13543) [DOI] [PubMed] [Google Scholar]
- 43.Pokorná MJ, Rovatsos M, Kratochvíl L. 2014. Sex chromosomes and karyotype of the (nearly) mythical creature, the Gila monster, Heloderma suspectum (Squamata: Helodermatidae). PLoS ONE 9, e104716. ( 10.1371/journal.pone.0104716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rovatsos M, Rehák I, Velenský P, Kratochvíl L. 2019. Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol. Biol. Evol. 36, 1113-1120. ( 10.1093/molbev/msz024) [DOI] [PubMed] [Google Scholar]
- 45.Nielsen SV, Pinto BJ, Guzmán-Méndez IA, Gamble T. 2020. First report of sex chromosomes in night lizards (Scincoidea: Xantusiidae). J. Hered. 111, 307-311. ( 10.1093/jhered/esaa007) [DOI] [PubMed] [Google Scholar]
- 46.Pokorná M, Rens W, Rovatsos M, Kratochvíl L. 2014. A ZZ/ZW sex chromosome system in the thick-tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet. Genome Res. 142, 190-196. ( 10.1159/000358847) [DOI] [PubMed] [Google Scholar]
- 47.Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. 2015. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 32, 1296-1309. ( 10.1093/molbev/msv023) [DOI] [PubMed] [Google Scholar]
- 48.Keating SE, Griffing AH, Nielsen SV, Scantlebury DP, Gamble T. 2020. Conserved ZZ/ZW sex chromosomes in Caribbean croaking geckos (Aristelliger: Sphaerodactylidae). J. Evol. Biol. 33, 1316-1326. ( 10.1111/jeb.13682) [DOI] [PubMed] [Google Scholar]
- 49.Pensabene E, Kratochvíl L, Rovatsos M. 2020. Independent evolution of sex chromosomes in eublepharid geckos, a lineage with environmental and genotypic sex determination. Life 10, 342. ( 10.3390/life10120342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rovatsos M, Pokorná M, Altmanová M, Kratochvíl L. 2014. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol. Lett. 10, 20131093. ( 10.1098/rsbl.2013.1093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altmanová M, Rovatsos M, Johnson Pokorná M, Veselý M, Wagner F, Kratochvíl L. 2018. All iguana families with the exception of basilisks share sex chromosomes. Zoology 126, 98-102. ( 10.1016/j.zool.2017.11.007) [DOI] [PubMed] [Google Scholar]
- 52.Rovatsos M, Vukić J, Lymberakis P, Kratochvíl L. 2015. Evolutionary stability of sex chromosomes in snakes. Proc. R. Soc. B 282, 20151992. ( 10.1098/rspb.2015.1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rovatsos M, Praschag P, Fritz U, Kratochvíl L. 2017. Stable cretaceous sex chromosomes enable molecular sexing in softshell turtles (Testudines: Trionychidae). Sci. Rep. 7, 42150. ( 10.1038/srep42150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rovatsos M, Gamble T, Nielsen SV, Georges A, Ezaz T, Kratochvíl L. 2021. Do male and female heterogamety really differ in expression regulation? Lack of global dosage balance in pygopodid geckos. Phil. Trans. R. Soc. B 376, 20200102. ( 10.1098/rstb.2020.0102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Augstenová B, Pensabene E, Veselý M, Kratochvíl L, Rovatsos M. In press. Are geckos special in sex determination? Independently evolved differentiated ZZ/ZW sex chromosomes in carphodactylid geckos. Genome Biol. Evol. ( 10.1093/gbe/evab119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ezaz T, Quinn AE, Sarre SD, O'Meally D, Georges A, Marshall Graves JA. 2009. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 17, 91-98. ( 10.1007/s10577-008-9019-5) [DOI] [PubMed] [Google Scholar]
- 57.Bókony V, Milne G, Pipoly I, Székely T, Liker A. 2019. Sex ratios and bimaturism differ between temperature-dependent and genetic sex-determination systems in reptiles. BMC Evol. Biol. 19, 57. ( 10.1186/s12862-019-1386-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pokorná M, Kratochvíl L. 2009. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 156, 168-183. ( 10.1111/j.1096-3642.2008.00481.x) [DOI] [Google Scholar]
- 59.Bista B, Valenzuela N. 2020. Turtle insights into the evolution of the reptilian karyotype and the genomic architecture of sex determination. Genes 11, 416. ( 10.3390/genes11040416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.González EJ, Martínez-López M, Morales-Garduza MA, García-Morales R, Charruau P, Gallardo-Cruz JA. 2019. The sex-determination pattern in crocodilians: a systematic review of three decades of research. J. Anim. Ecol. 88, 1417-1427. ( 10.1111/1365-2656.13037) [DOI] [PubMed] [Google Scholar]
- 61.Grützner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O'Brien PC, Jones RC, Ferguson-Smith MA, Marshall Graves JA. 2004. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432, 913-917. ( 10.1038/nature03021) [DOI] [PubMed] [Google Scholar]
- 62.Rens W, et al. 2007. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 8, R243. ( 10.1186/gb-2007-8-11-r243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawai A, Ishijima J, Nishida C, Kosaka A, Ota H, Kohno S, Matsuda Y. 2009. The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118, 43-51. ( 10.1007/s00412-008-0176-2) [DOI] [PubMed] [Google Scholar]
- 64.Zechner U, Hameister H. 2011. Sex chromosomes in vertebrates: XX/XY against ZZ/ZW. Sex Dev. 5, 266-271. ( 10.1159/000331233) [DOI] [PubMed] [Google Scholar]
- 65.Pokorná MJ, Kratochvíl L. 2016. What was the ancestral sex-determining mechanism in amniote vertebrates? Biol. Rev. 91, 1-12. ( 10.1111/brv.12156) [DOI] [PubMed] [Google Scholar]
- 66.Singchat W, et al. 2018. Chromosome map of the Siamese cobra: did partial synteny of sex chromosomes in the amniote represent ‘a hypothetical ancestral super-sex chromosome’ or random distribution? BMC Genomics 19, 939. ( 10.1186/s12864-018-5293-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singchat W, Ahmad SF, Laopichienpong N, Suntronpong A, Panthum T, Griffin DK, Srikulnath K. 2020. Snake W sex chromosome: the shadow of ancestral amniote super-sex chromosome. Cells 9, E2386. ( 10.3390/cells9112386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laopichienpong N, et al. 2020. Genome-wide SNP analysis of Siamese cobra (Naja kaouthia) reveals the molecular basis of transitions between Z and W sex chromosomes and supports the presence of an ancestral super-sex chromosome in amniotes. Genomics 13, 624-636. ( 10.1016/j.ygeno.2020.09.058) [DOI] [PubMed] [Google Scholar]
- 69.Smith JJ, Voss SR. 2007. Bird and mammal sex-chromosome orthologs map to the same autosomal region in a salamander (Ambystoma). Genetics 177, 607-613. ( 10.1534/genetics.107.072033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voss SR, Kump DK, Putta S, Pauly N, Reynolds A, Henry RJ, Basa S, Walker JA, Smith JJ. 2011. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. 21, 1306-1312. ( 10.1101/gr.116491.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suryamohan K, et al. 2020. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 52, 106-117. ( 10.1038/s41588-019-0559-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bellott DW, et al. 2010. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612-616. ( 10.1038/nature09172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ellegren H. 2011. Emergence of male-biased genes on the chicken Z-chromosome: sex-chromosome contrasts between male and female heterogametic systems. Genome Res. 21, 2082-2086. ( 10.1101/gr.119065.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Altmanová M, Rovatsos M, Kratochvíl L, Johnson Pokorná M. 2016. Minute Y chromosomes and karyotype evolution in Madagascan iguanas (Squamata: Iguania: Opluridae). Biol. J. Linn. Soc. 118, 618-633. ( 10.1111/bij.12751) [DOI] [Google Scholar]
- 75.Pokorná MJ, Altmanová M, Rovatsos M, Velenský P, Vodička R, Rehák I, Kratochvíl L. 2016. First description of the karyotype and sex chromosomes in the Komodo dragon (Varanus komodoensis). Cytogenet. Genome Res. 148, 284-291. ( 10.1159/000447340) [DOI] [PubMed] [Google Scholar]
- 76.Suwala G, Altmanová M, Mazzoleni S, Karameta E, Pafilis P, Kratochvíl L, Rovatsos M. 2020. Evolutionary variability of W-linked repetitive content in lacertid lizards. Genes 11, 531. ( 10.3390/genes11050531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pokorná M, Kratochvíl L, Kejnovský E. 2011. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet. 12, 90. ( 10.1186/1471-2156-12-90) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rovatsos M, Kratochvíl L, Altmanová M, Johnson Pokorná M. 2015. Interstitial telomeric motifs in squamate reptiles: when the exceptions outnumber the rule. PLoS ONE 10, e0134985. ( 10.1371/journal.pone.0134985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsubara K, O'Meally D, Azad B, Georges A, Sarre SD, Graves JA, Matsuda Y, Ezaz T. 2016. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma 125, 111-123. ( 10.1007/s00412-015-0531-z) [DOI] [PubMed] [Google Scholar]
- 80.Augstenová B, Mazzoleni S, Kostmann A, Altmanová M, Frynta D, Kratochvíl L, Rovatsos M. 2019. Cytogenetic analysis did not reveal differentiated sex chromosomes in ten species of boas and pythons (Reptilia: Serpentes). Genes 10, 934. ( 10.3390/genes10110934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan Q, Kay T, Depincé A, Adolfi M, Schartl M, Guiguen Y, Herpin A. 2021. Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Phil. Trans. R. Soc. B 376, 20200091. ( 10.1098/rstb.2020.0091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beukeboom LW, Perrin N. 2014. The evolution of sex determination. Oxford, UK: Oxford University Press and British Academy. [Google Scholar]
- 83.Deveson IW, Holleley CE, Blackburn J, Marshall Graves JA, Mattick JS, Waters PD, Georges A. 2017. Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci. Adv. 14, e1700731. ( 10.1126/sciadv.1700731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge C, Ye J, Weber C, Sun W, Zhang H, Zhou Y, Cai C, Qian G, Capel B. 2018. The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 360, 645-648. ( 10.1126/science.aap8328) [DOI] [PubMed] [Google Scholar]
- 85.Weber C, Capel B. 2021. Sex determination without sex chromosomes. Phil. Trans. R. Soc. B 376, 20200109. ( 10.1098/rstb.2020.0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Holleley CE, O'Meally D, Sarre SD, Marshall Graves JA, Ezaz T, Matsubara K, Azad B, Zhang X, Georges A. 2015. Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature 523, 79-82. ( 10.1038/nature14574) [DOI] [PubMed] [Google Scholar]
- 87.Quinn AE, Ezaz T, Sarre SD, Marshall Graves JA, Georges A. 2010. Extension, single-locus conversion and physical mapping of sex chromosome sequences identify the Z microchromosome and pseudo-autosomal region in a dragon lizard, Pogona vitticeps. Heredity 104, 410-417. ( 10.1038/hdy.2009.133) [DOI] [PubMed] [Google Scholar]
- 88.Ehl J, Altmanová M, Kratochvíl L. In press. With or without W? Molecular and cytogenetic markers are not sufficient for identification of environmentally-induced sex reversals in the bearded dragon. Sex Dev. ( 10.1159/000514195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tonini JFR, Beard KH, Ferreira RB, Jetz W, Pyron RA. 2016. Fully-sampled phylogenies of squamates reveal evolutionary patterns in threat status. Biol. Conserv. 204, 23-31. ( 10.1016/j.biocon.2016.03.039) [DOI] [Google Scholar]
- 90.Catchen JM, Conery JS, Postlethwait JH. 2009. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 19, 1497-1505. ( 10.1101/gr.090480.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruiz-Herrera A, Farre M, Robinson TJ. 2012. Molecular cytogenetic and genomic insights into chromosomal evolution. Heredity 108, 28-36. ( 10.1038/hdy.2011.102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zimmermann B, Robert NSM, Technau U, Simakov O. 2019. Ancient animal genome architecture reflects cell type identities. Nat. Evol. Evol. 3, 1289-1293. ( 10.1038/s41559-019-0946-7) [DOI] [PubMed] [Google Scholar]
- 93.Pokorná M, Giovannotti M, Kratochvíl L, Caputo V, Olmo E, Ferguson-Smith MA, Rens W. 2012. Conservation of chromosomes syntenic with avian autosomes in squamate reptiles revealed by comparative chromosome painting. Chromosoma 121, 409-418. ( 10.1007/s00412-012-0371-z) [DOI] [PubMed] [Google Scholar]
- 94.Lind AL, et al. 2019. Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. Nat. Ecol. Evol. 3, 1241-1252. ( 10.1038/s41559-019-0945-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schield DR, et al. 2019. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 29, 590-601. ( 10.1101/gr.240952.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simison BW, Parham JF, Papenfuss TJ, Lam AW, Henderson JB. 2020. An annotated chromosome-level reference genome of the red-eared slider turtle (Trachemys scripta elegans). Genome Biol. Evol. 12, 456-462. ( 10.1093/gbe/evaa063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yurchenko AA, Recknagel H, Elmer KR. 2020. Chromosome-level assembly of the common lizard (Zootoca vivipara) genome. Genome Biol. Evol. 12, 1953-1960. ( 10.1093/gbe/evaa161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilcox SA, Watson JM, Spencer JA, Graves JA. 1996. Comparative mapping identifies the fusion point of an ancient mammalian X-autosomal rearrangement. Genomics 35, 66-70. ( 10.1006/geno.1996.0323) [DOI] [PubMed] [Google Scholar]
- 99.Nanda I, et al. 1999. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat. Genet. 21, 258-259. ( 10.1038/6769) [DOI] [PubMed] [Google Scholar]
- 100.Farré M, et al. 2019. Evolution of gene regulation in ruminants differs between evolutionary breakpoint regions and homologous synteny blocks. Genome Res. 29, 576-589. ( 10.1101/gr.239863.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sigeman H, Ponnikas S, Chauhan P, Dierickx E, Brooke ML, Hansson B. 2019. Repeated sex chromosome evolution in vertebrates supported by expanded avian sex chromosomes. Proc. R. Soc. B 286, 20192051. ( 10.1098/rspb.2019.2051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O'Connor RE, et al. 2018. Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat. Commun.. 9, 1883. ( 10.1038/s41467-018-04267-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Genomicus v100.01. See https://www.genomicus.biologie.ens.fr/ge/nomicu/s-100.01/cgi-bin/search.pl (accessed 8 January 2021).
- 104.Damas J, Kim J, Farré M, Griffin DK, Larkin DM. 2018. Reconstruction of avian ancestral karyotypes reveals differences in the evolutionary history of macro- and microchromosomes. Genome Biol. 19, 155. ( 10.1186/s13059-018-1544-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Griffin DK, Robertson LB, Tempest HG, Skinner BM. 2007. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 117, 64-77. ( 10.1159/000103166) [DOI] [PubMed] [Google Scholar]
- 106.Warren WC, et al. 2017. A new chicken genome assembly provides insight into avian genome structure. G3: Genes Genom. Genet. 7, 109-117. ( 10.1534/g3.116.035923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org/. [Google Scholar]
- 108.Engels B. 2015. XNomial: exact goodness-of-fit test for multinomial data with fixed probabilities, version 1.0.4. See https://CRAN.R-project.org/package=XNomial.
- 109.Sutton E, et al. 2011. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J. Clin. Invest. 121, 328-341. ( 10.1172/JCI42580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Masuyama H, Yamada M, Kamei Y, Fujiwara-Ishikawa T, Todo T, Nagahama Y, Matsuda M. 2012. Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res. 20, 163-176. ( 10.1007/s10577-011-9264-x) [DOI] [PubMed] [Google Scholar]
- 111.Oike A, Kodama M, Yasumasu S, Yamamoto T, Nakamura Y, Ito E, Nakamura M. 2017. Participation of androgen and its receptor in sex determination of an amphibian species. PLoS ONE 12, e0178067. ( 10.1371/journal.pone.0178067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Werner R, et al. 2017. New NR5A1 mutations and phenotypic variations of gonadal dysgenesis. PLoS ONE 12, e0176720. ( 10.1371/journal.pone.0176720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin Q, Mei J, Li Z, Zhang X, Zhou L, Gui JF. 2017. Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics 207, 1007-1022. ( 10.1534/genetics.117.300274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kumar TR. 2014. ‘Been hit twice’: a novel bi-allelic heterozygous mutation in LHCGR. J. Assist. Reprod. Genet. 31, 783-786. ( 10.1007/s10815-014-0284-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang ZW, Lau SW, Zhang LL, Ge W. 2015. Disruption of zebrafish follicle-stimulating hormone receptor (fshr) but not luteinizing hormone receptor (lhcgr) gene by TALEN leads to failed follicle activation in females followed by sexual reversal to males. Endocrinology 156, 3747-3762. ( 10.1210/en.2015-1039) [DOI] [PubMed] [Google Scholar]
- 116.Pan Q, et al. 2019. Identification of the master sex determining gene in northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet. 15, e1008013. ( 10.1371/journal.pgen.1008013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sinclair AH, et al. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240-244. ( 10.1038/346240a0) [DOI] [PubMed] [Google Scholar]
- 118.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267-271. ( 10.1038/nature08298) [DOI] [PubMed] [Google Scholar]
- 119.Ioannidis J, et al. 2020. Primary sex determination in birds depends on DMRT1 dosage, but gonadal sex does not determine secondary sexual characteristics. Proc. Natl Acad. Sci. USA 118, e2020909118. ( 10.1073/pnas.2020909118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Doorn G, Kirkpatrick M. 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449, 909-912. ( 10.1038/nature06178) [DOI] [PubMed] [Google Scholar]
- 121.Lisachov AP, Makunin AI, Giovannotti M, Pereira JC, Druzhkova AS, Barucchi VC, Ferguson-Smith MA, Trifonov VA. 2019. Genetic content of the neo-sex chromosomes in Ctenonotus and Norops (Squamata, Dactyloidae) and degeneration of the Y chromosome as revealed by high-throughput sequencing of individual chromosomes. Cytogenet. Genome Res. 157, 115-122. ( 10.1159/000497091) [DOI] [PubMed] [Google Scholar]
- 122.Lisachov AP, Tishakova KV, Romanenko SA, Molodtseva AS, Prokopov DY, Pereira JC, Ferguson-Smith MA, Borodin PM, Trifonov VA. 2021. Whole-chromosome fusions in the karyotype evolution of Sceloporus (Iguania, Reptilia) are more frequent in sex chromosomes than autosomes. Phil. Trans. R. Soc. B 376, 20200099. ( 10.1098/rstb.2020.0099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O'Brien SJ, Graphodatsky AS, Perelman PL (eds) 2020. Atlas of mammalian chromosomes, 2nd edn. Hoboken, NJ: Wiley-Blackwell. [Google Scholar]
- 124.Pokorná M, Altmanová M, Kratochvíl L. 2014. Multiple sex chromosomes in the light of female meiotic drive in amniote vertebrates. Chromosome Res. 22, 35-44. ( 10.1007/s10577-014-9403-2) [DOI] [PubMed] [Google Scholar]
- 125.Gunski RJ, Cañedo AD, Garnero ADV, Ledesma MA, Coria N, Montalti D, Degrandi TM. 2017. Multiple sex chromosome system in penguins (Pygoscelis, Spheniscidae). Comp. Cytogenet. 11, 541-552. ( 10.3897/CompCytogen.v11i3.13795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gan HM, Falk S, Morales HE, Austin CM, Sunnucks P, Pavlova A. 2019. Genomic evidence of neo-sex chromosomes in the eastern yellow robin. GigaScience 8, giz111. ( 10.1093/gigascience/giz111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rovatsos M, Kratochvíl L. 2021. Evolution of dosage compensation does not depend on genomic background. Mol. Ecol. 30, 1836-1845. ( 10.1111/mec.15853) [DOI] [PubMed] [Google Scholar]
- 128.Rovatsos M, Vukić J, Kratochvíl L. 2016. Mammalian X homolog acts as sex chromosome in lacertid lizards. Heredity 117, 8-13. ( 10.1038/hdy.2016.18) [DOI] [PMC free article] [PubMed] [Google Scholar]