Abstract
Introduction
COVID-19 is a worldwide public health threat. Diagnosis by RT-PCR has been employed as the standard method to confirm viral infection. Sample pooling testing can optimize the resources by reducing the workload and reagents shortage, and be useful in laboratories and countries with limited resources. This study aims to evaluate SARS-CoV-2 detection by sample pooling testing in comparison with individual sample testing.
Materials and methods
We created 210 pools out of 245 samples, varying from 4 to 10 samples per pool, each containing a positive sample. We conducted detection of SARS-CoV-2-specific RdRp/E target sites.
Results
Pooling of three samples for SARS-CoV-2 detection might be an efficient strategy to perform without losing RT-PCR sensitivity.
Conclusions
Considering the positivity rate in Dominican Republic and that larger sample pools have higher probabilities of obtaining false negative results, the optimal sample size to perform a pooling strategy shall be three samples.
Keywords: COVID-19, Real-time PCR, Pool testing, SARS-CoV-2, RdRp/E genes
Abstract
Introducción
La COVID-19 es una amenaza de salud pública mundial. La RT-PCR es el método estándar para confirmar la infección. La estrategia de pruebas de muestras agrupadas puede reducir la carga de trabajo y la escasez de reactivos, y ser útil en países con escasos recursos. Evaluamos la detección del SARS-CoV-2 mediante esta estrategia en comparación con pruebas individuales.
Materiales y métodos
Creamos 210 grupos de 245 muestras, de 4 a 10 muestras por grupo, cada uno con una muestra positiva. Realizamos extracción de ARN y qRT-PCR para detectar la presencia de la diana RdRp/E.
Resultados
La combinación de hasta 3 muestras para la detección del SARS-CoV-2 podría ser una estrategia eficaz sin perder la sensibilidad.
Conclusiones
Considerando la tasa de positividad en República Dominicana y que los grupos con más muestras tienen mayor probabilidad de obtener resultados falsos negativos, el tamaño óptimo para realizar esta estrategia es de 3 muestras.
Palabras clave: COVID-19, PCR en tiempo real, Pruebas agrupadas, SARS-CoV-2, Genes RdRp/E
Introduction
After the first reports of a new Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), low-middle income countries (LMIC) like the Dominican Republic started facing the urgency of extensive rapid testing to limit the impact of this public health threat. SARS-CoV-2 is the causing agent of Coronavirus Disease 2019 (COVID-19), which has been spread all over the world. After a year, more than 70 million cases have been confirmed, and it is estimated that a million of them were detected across the Americas.1 A crucial part of the public health response to COVID-19 response is prompt diagnosis, tracing, and isolation of infected individuals to prevent further spreading.2
The standard diagnostic test, the real-time reverse transcriptase-polymerase chain reaction (qRT-PCR), has been implemented worldwide to confirm SARS-CoV-2 infection, allowing results with satisfactory levels of sensitivity and specificity.3 However, due to the high demand, there has been a limited number of reagents, requiring researchers to consider pool testing strategies.3, 4, 5 Pool testing is a diagnostic strategy used to identify viral infections, such as viral hepatitis.6 Several samples must be combined, analyzed in a single reaction, and if test results are negative, all the samples are considered undetected.7 Whereas, if the results are positive, all the individual samples in the pool must be analyzed to elucidate the positive cases.3, 4, 6
Pooling strategies have proven to prevent wasting SARS-CoV-2 testing resources and time, improving the testing capability in countries with an incidence rate of SARS-CoV-2 infection of 10% or less. However, the role of sample titration has not been fully elucidated.8, 9 Scaling up of this methodology may be convenient in countries experiencing a shortage of resources and an increased demand throughout the pandemic.10, 11 Dominican Republic response has been substantially affected by implementing a rapid deployment of test and tracing strategy in transmission hot spots throughout the country, mainly due to the limitations in importing and cold chains in the Caribbean customs system.12
This study aims to evaluate the effectiveness of the SARS-CoV-2 detection by pooled sample testing compared to individual testing in the context of COVID-19 response in a Caribbean country.
Materials and methods
We analyzed saliva and nasopharyngeal samples from several hospitals in the Dominican Republic. Experiments were conducted at the Institute for Tropical Medicine and Global Health from October 5 to November 23, 2020. Samples used had a previously known diagnosis for COVID-2019.
We evaluated 210 pools out of 245 samples, among which SARS-CoV-2 was detected in 31% (n = 77) and not detected in 69% (n = 168). We distributed the positive samples according to the threshold cycle value (Ct), being the specific ranges <30Ct, ≥30Ct–≤33Ct, and >33Ct–≤36Ct. Positive samples were diluted with negative sample sets in 2 ml viral transport media (VTM) to perform the pooling dilution. Each group included a single positive sample and the remaining volume of negative samples, depending on group size. Hence, the total volume of the smaller group (n = 4) was 800 ml, whereas the largest group (n = 10) was 2 ml.
We used the AdvanSure™ Nucleic Acid kit for RNA extraction following manufacturers’ protocol. Afterwards, we conducted a 1step RT-qPCR assay to detect the SARS-CoV-2-specific RdRp/E genes (RNA-dependent RNA polymerase), employing the PowerChek™ 2019-nCoV PCR kit and using FAST ABI 7500 Real-Time PCR (Applied Biosystems, Foster City, CA). A pool with a Ct value ≤37 was considered positive for SARS-CoV-2. Once we obtained the results, we evaluated the concordance, the rate of false negatives, the estimated increase in Ct obtained by the sample dilution, and the use of positive samples with low viral load. Identificators were eliminated from samples to protect patients’ identities, the Institutional Review Board (IRB) at Universidad Iberoamericana (CEI# 2020-16) approved this study.
Results
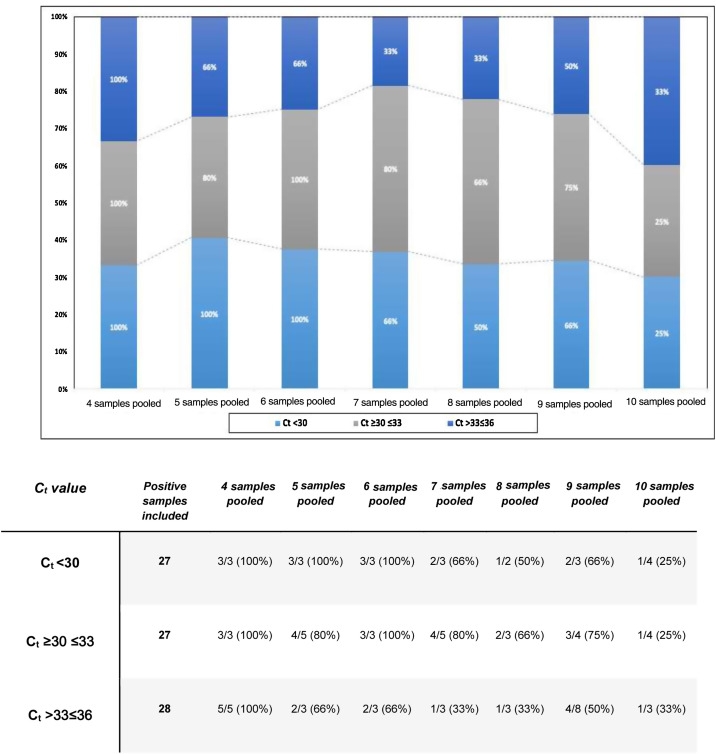
To estimate the effectiveness of the pooling technique according to the group size, we calculated the concordance rate between the pooled tested and individual samples, as shown in Fig. 1 .
Fig. 1.
Concordance of testing between pooling of samples and individual samples.
Afterwards, the comparison between the Ct values of the positive samples used with the Ct obtained from the pools, enable us to identify how the dilution of the samples and the positive samples used with low viral load directly affect the analytical sensitivity of RT-PCR. As a result, for the groups of 4, 5, 6, and 7 samples, an estimated increase of 2Ct was obtained. Concerning the groups of 8 and 9, we attained an estimated increase of 3Ct. Finally, for the groups of 10 samples, an estimated increase of 4Ct was obtained. In addition, the rate of false negative results was obtained by dividing the number of negative tests and the total samples analyzed, as shown in Table 1 .
Table 1.
Rates of false negative results for every pool of samples evaluated.
| Ct value range | Total pools analyzed | 4 samples pooled | 5 samples pooled | 6 samples pooled | 7 samples pooled | 8 samples pooled | 9 samples pooled | 10 samples pooled |
|---|---|---|---|---|---|---|---|---|
| Ct < 30 | 70 | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 4/10 (40%) | 6/10 (60%) | 4/10 (40%) | 5/10 (50%) |
| Ct ≥ 30 ≤ 33 | 70 | 0/10 (0%) | 1/10 (10%) | 1/10 (10%) | 3/10 (30%) | 4/10 (40%) | 3/10 (30%) | 8/10 (80%) |
| Ct > 33 ≤ 36 | 70 | 0/10 (0%) | 3/10 (30%) | 6/10 (60%) | 5/10 (50%) | 9/10 (90%) | 6/10 (60%) | 8/10 (80%) |
Discussion
Sample pooling strategy has been implemented in different countries to increase the SARS-CoV-2 detection capacity and reduce reagent wasting.5 Our findings have important implications in countries with a high positivity rate. To date, Latin-America is an epicenter for new infections in America. The higher the positivity rate, the lower the number of samples chosen for pooling analysis to ensure the RT-PCR sensitivity and guarantee non-reagent exhaustions in limited settings. As shown in Fig. 1, for pools of 4 samples, we obtained a concordance of 100% for all Ct values. Additionally, the false negative rate was 0% even for pools with positive samples with Ct ≥ 36. These results suggest that the viral RNA concentration in the positive samples used was not affected after the dilution of the samples tested. Therefore, pooling of four samples for SARS CoV-2 detection by real-time RT-PCR may be an adequate strategy without the loss of sensitivity for low viral loads.
Praharaj et al.5 created 110 pools out of 5 samples and obtained a concordance of 100% for a Ct less than 30. Whereas concordance for values with Ct ≥ 30–≤33 and >33–≤36 was 95.5% and 69.7%, respectively. Their results affirm that concordance rates for samples with a Ct value greater than 33 cycles were considerably lower.5 When we compared our findings, we noticed the same concordance for Ct values less than 30. However, the concordance for Ct values with a Ct ≥ 30–≤33 and >33–≤36 was 80% and 66%, respectively. Our results suggest that pools of five samples are a viable size, in case the evaluated tests have a low positivity rate (less than 10%), being ideal to employ this strategy in asymptomatic individuals. For pools of 6 samples, concordance and false-negative results demonstrate how the efficiency of the strategy flattens out due to the dilution and viral load of the samples used.
This information is crucial for selecting the ideal pool size for the Dominican Republic, since using a pool of 5 samples or greater could fail to detect SARS-CoV-2 positive cases. In contrast with our findings, Bateman et al.13 obtained fewer false negatives for pools of 5 and 10 samples. Unlike us, they used Ct values between 14 and 35 instead of setting the limit on 36, which probably affected the sensitivity of RT-PCR for those pools that had such number of samples.13
One limitation of our study is the use of two types of samples, saliva and nasopharyngeal swabs. Although the use of a single sample type would have been more meticulous, we selected different types of samples due to the current restraints in the accessibility of collection tools in LMIC countries.
Based on official data provided by the Dominican Ministry of Health, the positivity rate in our country as of February 2021 was 17.34%.14 Aragón-Caqueo et al. suggested that the optimal number of samples per pool is three, since the positivity rate of cases is inversely proportional to the optimal pool size. Furthermore, the minimum net savings in detection kits for a positivity rate in the range of 8–20% is 17.9%. Choosing to perform pools of three samples entails evaluating all the subjects in the event of a positive pool sample to detect the cases that are positive. It implies an increase in the logistical complexity of this strategy.3
In conclusion, according to the positivity rate in the Dominican Republic, the optimal size to perform a pooling strategy shall be three samples. However, our results indicate that a pooling of four samples is an optimal strategy for SARS-CoV-2 detection that helps to reduce the amounts of reagents used. We do not recommend using pooling testing with more than five samples since the detection sensitivity of the RT-PCR is affected after the dilution of samples. This strategy might be a useful tool in pandemic responses, especially in countries with limited technical personnel and where investment on avant-garde technologies is not available.
Conflict of interests
The authors declare that they have no conflicts of interest.
Acknowledgments
The authors would like to thank the laboratory team at Instituto de Medicina Tropical & Salud Global, specially to Nicole Guzman for all support received to accelerate our response to the pandemic at our center and provide insight on the data analysis.
References
- 1.WHO Coronavirus Disease (COVID-19) Dashboard. Available at: https://covid19.who.int [accessed 21.06.21]
- 2.Girum T., Lentiro K., Geremew M., Migora B., Shewamare S. Global strategies and effectiveness for COVID-19 prevention through contact tracing, screening, quarantine, and isolation: a systematic review. Trop Med Health. 2020;48:91. doi: 10.1186/s41182-020-00285-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aragón-Caqueo D., Fernández-Salinas J., Laroze D. Optimization of group size in pool testing strategy for SARS-CoV-2: a simple mathematical model. J. Med. Virol. 2020;92:1988–1994. doi: 10.1002/jmv.25929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres I., Albert E., Navarro D. Pooling of nasopharyngeal swab specimens for SARS-CoV-2 detection by RT-PCR. J. Med. Virol. 2020;92:2306–2307. doi: 10.1002/jmv.25971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Praharaj I., Jain A., Singh M., Balakrishnan A., Dhodapkar R., Borkakoty B., et al. Pooled testing for COVID-19 diagnosis by real-time RT-PCR: a multi-site comparative evaluation of 5- & 10-sample pooling. Indian J. Med. Res. 2020;152:88–94. doi: 10.4103/ijmr.IJMR_2304_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasomsub E., Watcharananan S.P., Watthanachockchai T., Rakmanee K., Tassaneetrithep B., Kiertiburanakul S., et al. Saliva sample pooling for the detection of SARS-CoV-2. J. Med. Virol. 2021;93:1506–1511. doi: 10.1002/jmv.26460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Žilinskas J., Lančinskas A., Guarracino M.R. Pooled testing with replication as a mass testing strategy for the COVID-19 pandemics. Sci Rep. 2021;11:3459. doi: 10.1038/s41598-021-83104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdalhamid B., Bilder C.R., McCutchen E.L., Hinrichs S.H., Koepsell S.A., Iwen P.C. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am. J. Clin. Pathol. 2020;153:715–718. doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanel R., Thurner S. Boosting test-efficiency by pooled testing for SARS-CoV-2-Formula for optimal pool size. PLoS One. 2020;15:e0240652. doi: 10.1371/journal.pone.0240652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabrera Alvargonzalez J.J., Rey Cao S., Pérez Castro S., Martinez Lamas L., Cores Calvo O., Torres Piñon J., et al. Pooling for SARS-CoV-2 control in care institutions. BMC Infect. Dis. 2020;20:745. doi: 10.1186/s12879-020-05446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan C.A., Sahoo M.K., Pinsky B.A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323:1967–1969. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulino-Ramirez R., Báez A.A., Vallejo Degaudenzi A., Tapia L. Seroprevalence of specific antibodies against SARS-CoV-2 from hotspot communities in the Dominican Republic. Am. J. Trop. Med. Hyg. 2020;103:2343–2346. doi: 10.4269/ajtmh.20-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman A.C., Mueller S., Guenther K., Shult P. Assessing the dilution effect of specimen pooling on the sensitivity of SARS-CoV-2 PCR tests. J. Med. Virol. 2021;93:1568–1572. doi: 10.1002/jmv.26519. [DOI] [PubMed] [Google Scholar]
- 14.Ministerio de Salud Pública de la República Dominicana Boletín COVID-19 no.343 24 de febrero 2021. 2021 https://www.msp.gob.do/web/?page_id=6948 [accessed 21.06.21] [Google Scholar]