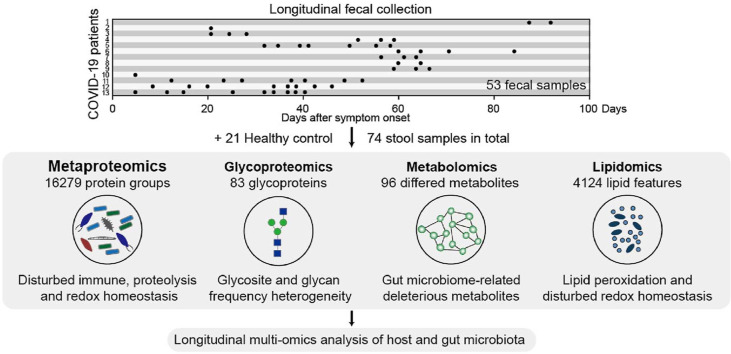
Abstract
Gut ecosystem has profound effects on host physiology and health. Gastrointestinal (GI) symptoms were frequently observed in patients with COVID-19. Compared with other organs, gut antiviral response can result in more complicated immune responses because of the interactions between the gut microbiota and host immunity. However, there are still large knowledge gaps in the impact of COVID-19 on gut molecular profiles and commensal microbiome, hindering our comprehensive understanding of the pathogenesis of SARS-CoV-2 and the treatment of COVID-19. We performed longitudinal stool multi-omics profiling to systemically investigate the molecular phenomics alterations of gut ecosystem in COVID-19. Gut proteomes of COVID-19 were characterized by disturbed immune, proteolysis and redox homeostasis. The expression and glycosylation of proteins involved in neutrophil degranulation and migration were suppressed, while those of proteases were upregulated. The variable domains of Ig heavy chains were downregulated and the overall glycosylation of IgA heavy chain constant regions, IgGFc-binding protein, and J chain were suppressed with glycan-specific variations. There was a reduction of beneficial gut bacteria and an enrichment of bacteria derived deleterious metabolites potentially associated with multiple types of diseases (such as ethyl glucuronide). The reduction of Ig heave chain variable domains may contribute to the increase of some Bacteroidetes species. Many bacteria ceramide lipids with a C17-sphingoid based were downregulated in COVID-19. In many cases, the gut phenome did not restore two months after symptom onset. Our study indicates widely disturbed gut molecular profiles which may play a role in the development of symptoms in COVID-19. Our findings also emphasis the need for ongoing investigation of the long-term gut molecular and microbial alterations during COVID-19 recovery process. Considering the gut ecosystem as a potential target could offer a valuable approach in managing the disease.
Keywords: Metaproteomics, Metabolomics, Glycoproteomics, Gut microbiome, COVID-19
Graphical abstract
1. Introduction
The ongoing coronavirus disease 2019 (COVID-19) outbreak caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to pose a threat to human beings. In addition to fever and cough, gastrointestinal (GI) symptoms were frequently observed in patients with COVID-19 [[1], [2], [3]]. We have provided the first clinical and molecular evidence for GI infection of SARS-CoV-2 with viral RNA detected in both GI tissues (esophagus, duodenum and rectum) and stool samples [1,4]. Furthermore, viable virus have been isolated in patient stool samples [5]. Our studies revealed that 11.6% of patients have GI symptoms on admission, and 49.5% of patients developed GI symptoms during hospitalization [1]. Importantly, 11.6% of patients did not exhibit any imaging features of COVID-19 pneumonia but only showed GI symptoms. However, little is known about why and how SARS-CoV-2 affects GI tract. Current evidence suggests that SARS-CoV-2 uses the host receptor angiotensin converting enzyme 2 (ACE2) for cell entry and the transmembrane serine protease 2 (TMPRSS2) for Spike protein (S) priming [6].
Compared with other organs, gut antiviral response can result in more complicated immune responses because of the interactions between the gut microbiota and host immunity. It is known that a normal gut ecosystem plays an important role in maintaining host health and immune homeostasis. Gut microbiota may be associated with the COVID-19 susceptibility, severity, treatment and outcomes in many different ways. Metagenomics has revealed that COVID-19 patients had significant altered fecal microbiome compared with controls [7,8]. A recent study revealed that severe COVID-19 infection is associated with systemic release of bacterial products [9]. Previous research has demonstrated the gut bacterial translocation to the lung in post-stroke pneumonia due to increased gut barrier permeability [10]. Gut microbiota can also regulate the intestinal infection of certain virus such as norovirus [11]. Furthermore, gut microbiome perturbation can alters immunity to vaccines [12], thus may affect the efficacy of SARS-CoV-2 vaccines.
Currently, there are still large knowledge gaps in the impact of COVID-19 on gut molecular profiles and commensal microbiome, hindering our understanding of the pathogenesis of SARS-CoV-2 and the treatment of COVID-19. Here, we investigate the comprehensive molecular phenomic signatures, including metaproteome, glycoproteome, metabolome, and lipidome, to deepen our comprehension of the gut molecular features related to SARS-CoV-2 infection.
2. Materials and methods
2.1. Subject details
This study was approved by the Ethics Committee of The Fifth Affiliated Hospital, Sun Yat-sen University (K161-1). This study involved 13 patients with COVID-19 and 21 healthy controls. The associated clinical data and metadata are provided in Table S1 and Fig. 1 A. SARS-CoV-2 infection was confirmed by two consecutive real-time reverse transcription PCR (RT-PCR) tests. These patients were classified into three groups according to the severity of their symptoms: mild (mild clinical symptoms without pneumonia manifestations in CT imaging (7 patients)); moderate (respiratory symptoms, fever, and imaging features of COVID-19 pneumonia (5 patients); severe (respiratory distress (respiratory rate ≥30 breaths/min), oxygen saturation ≤93% and arterial oxygen tension (PaO2)/fractional inspired oxygen (FiO2) ratio ≤300 mm Hg (1 patient)). We collected 53 stool samples from COVID-19 patients with a range of one to nine longitudinal time-points that occurred 1–94 days post symptom onset (Fig. 1A, Table S1). Stool samples from 21 healthy subjects severed as controls. A total of 74 stool samples were subjected to multi-omics analysis.
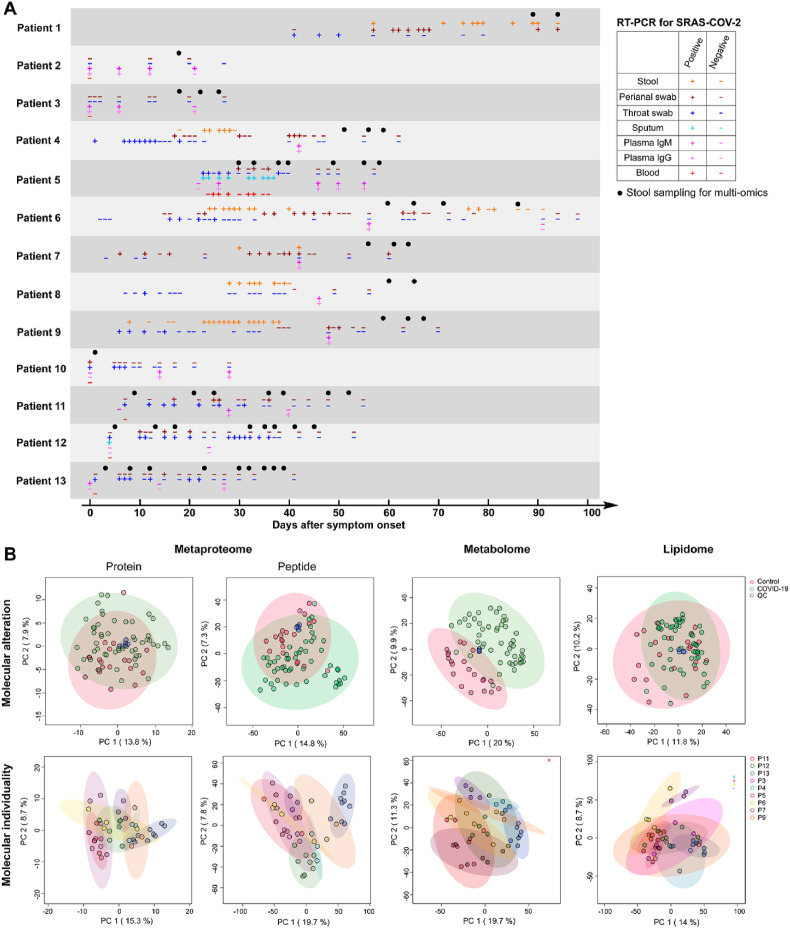
Fig. 1.
Overview of the multi-omics characterization of gut ecosystem in COVID-19 patients. A. Longitudinal SARS-COV-2 RT-PCR test results and stool sampling. B. Principle component analysis of metaproteomic (protein and peptide level), metabolomic, and lipidomic profiles.
2.2. Metaproteomics sample preparation
Fecal sample (∼150 mg) was lysed by boiling for 5 min at 95 °C in 800 μL of lysis buffer (6 M Guanidinium hydrochloride (GdmCl), 10 mM tris(2-carboxyethyl)phosphine (TCEP), 40 mM chloroacetamide, 100 mM Tris pH 8.5) in Eppendorf protein LoBind tubes. The lysate was then sonicated for 15 min using a waterbath sonicator. The crude protein extract was centrifuged at 16,000 g for 5 min with the clarified lysate subjected to ultrafiltration (cut off 30 kD, Millipore), diluted 1:10 with dilution buffer (10% (v/v) acetonitrile (ACN), 25 mM Tris pH 8.5) containing 1 μg sequencing grade trypsin (1/50, w/w), and digested overnight at 37 °C. The digest was acidified to an end-concentration of 1% trifluoroacetic acid (TFA) and debris were removed after centrifugation at 16,000 g for 5 min. Finally, the peptides were desalted on StageTips assembled by Empore C18 disk, dried using a SpeedVac centrifuge at 45 °C, and suspended in 2% ACN and 0.1% formic acid (FA).
2.3. Metaproteomics data acquisition
Peptides were trapped onto an Acclaim PepMap 100C18 column (75 μm × 20 mm, 3 μm, 100 Å, Thermo Scientific) at a flow rate of 8 μL/min and separated using an Acclaim PepMap 100C18 column (75 μm × 250 mm, 2 μm, 100 Å, Thermo Scientific) at 300 nL/min on a Thermo Scientific Dionex UltiMate 3000 RSLCnano LC system. Mobile phase solvents were 0.1% formic acid in water (A) and 0.1% formic acid in 80% acetonitrile (B). The separation gradient was as follows: 15% B rising to 30% B at 130 min, rising to 98% B at 137 min, and keeping at 98% B for 27 min. Finally, the separation column was equilibrated using 15% B for 6 min. The trap column was switched online with the separation column at 3 min and switched back to load position at 168 min. Data were acquired on an Orbitrap Fusion Lumos Tribrid mass spectrometer with a Nanospray Flex ion source in positive ionization mode with a spray voltage of +2600 V using Xcalibur software (Thermo Scientific, San Jose, CA, USA). The ion transfer tube temperature was 300 °C, the vaporized temperature was 325 °C, the sheath gas flow was 40 units, the auxiliary gas flow was 15 arbitrary units, and the sweep gas was 1 unit. Full scan MS spectra was acquired in the 400−1,600 m/z range with an AGC target of 5 × 104, a maximum injection time of 50 ms, and a resolution of 60 K at m/z 200. MS/MS spectra were acquired using higher-energy collisional dissociation (HCD) with a normalized collision energy (NCE) of 30% and a resolution of 15 K with an AGC target of 5 × 104 and a maximum ion injection time of 22 ms.
2.4. Glycopeptide enrichment
Glycopeptides were enriched using hydrophilic interaction liquid chromatography (HILIC) cartridges packed with the C18 plug followed by microcrystalline cellulose resins [13]. The resin was washed with 300 μL of 0.1% TFA and initialized using 300 μL of 0.1% TFA in 80% acetonitrile. After loading ∼200 μg of peptides in 300 μL 80% acetonitrile/0.1% TFA), the resin was washed with 80% acetonitrile/0.1% TFA three times to remove non-specific peptides. Then glycopeptides were eluted by 300 μL of H2O, followed by 200 μL of 80% acetonitrile. Peptides were dried using a SpeedVac centrifuge at 45 °C, and suspended in 2% ACN and 0.1% formic acid (FA).
2.5. Glycoproteomics data acquisition
Peptides were trapped onto an Acclaim PepMap 100C18 column (75 μm × 20 mm, 3 μm, 100 Å, Thermo Scientific) at a flow rate of 8 μL/min and separated using an Acclaim PepMap 100C18 column (75 μm × 250 mm, 2 μm, 100 Å, Thermo Scientific) at 300 nL/min. Mobile phase solvents were 0.1% formic acid in water (A) and 0.1% formic acid in 50% acetonitrile and 40% isopropanol (B). The separation gradient was as follows: 5% B at 0–15 min, 20%–30% B at 90–100 min, and 98% B at 107 min and kept for 20 min. Data were acquired on an Orbitrap Fusion Lumos Tribrid mass spectrometer with a Nanospray Flex ion source in positive ionization mode with a spray voltage of +2600 V using Xcalibur software (Thermo Scientific, San Jose, CA, USA). The ion transfer tube temperature was 300 °C, the vaporized temperature was 325 °C, the sheath gas flow was 40 units, the auxiliary gas flow was 15 arbitrary units, and the sweep gas was 1 unit. Full scan MS spectra was acquired in the 400−1,600 m/z range with a maximum injection time of 50 ms and a resolution of 60 K at m/z 200. MS/MS spectra were acquired using HCD with stepped NCE at 20%, 30%, and 40% to generate fragment ions of both glycan and peptide of a glycopeptide in a single spectrum and MS/MS spectra. The resolution of HCD was 15 K with a maximum ion injection time of 22 ms.
2.6. Metabolomics sample preparation
Metabolite extraction was performed by adding 1 mL of ice-old 80% methanol to ∼150 mg stool samples, vortexing for 30 s, and centrifuging (16,000g) at 4 °C for 10 min. The supernatants were evaporated to dryness under nitrogen, reconstituted in 150 μL of 0.1% formic acid in 5% acetonitrile, and kept at −80 °C until analysis.
2.7. Metabolomics data acquisition
Metabolic extracts were separated on a Thermo Scientific Dionex UltiMate 3000 Rapid Separation LC (RSLC) using an ACQUITY UPLC HSS T3 analytical column (2.1 × 150 mm, 1.8 μm, 100 Å, Waters) protected by an ACQUITY UPLC HSS T3 VanGuard pre-column (2.1 × 5 mm, 1.8 μm, 100 Å, Waters). Mobile phase solvents for positive ionization mode were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B); mobile phase solvents for negative ionization mode were 0.01% formic acid in water (A) and acetonitrile (B). The following gradient elution was used: 0–3 min, 95% A; 5–13 min, 80%–30% A; 15–18 min, 2% A; 18.1–22 min, 5% A. The flow rate was 0.3 mL/min, the injection volume was 2 μL and the column oven was set at 35 °C. Data were acquired on an Orbitrap Fusion Lumos Tribrid mass spectrometer fitted with a HESI source in both positive and negative ionization modes with an independent run for each polarity and a spray voltage of +3500 V and −3500 V, respectively (Thermo Scientific, San Jose, CA, USA). The ion transfer tube temperature was 300 °C, the vaporized temperature was 350 °C, the sheath gas flow was 40 units, the auxiliary gas flow was 15 arbitrary units, and the sweep gas was 1 unit. Metabolite profiling was profiled in full scan mode using a mass range of m/z 100–1000 with a resolution of 120 K at m/z 200, an AGC target of 5 × 104, and a maximum injection time of 50 ms. For metabolite identification, data dependent MS/MS data were acquired on quality control samples (QC) containing equally volumes of all samples used in this study. In-depth MS/MS was performed using nine staggered gas-phase fractionations (sGPFs) to allow more homogeneous selection of precursor ions in low, medium, and high m/z ranges [14]. This was achieved in nine separated LC-MS runs: (run 1) 100–110, 200–210, 300–310, 400–410, 500–510, 600–610, 700–710, 800–810; (run 2) 110–120, 210–220, 310–320, 410–420, 510–520, 610–620, 710–720, 810–820; (run 3) 120–130, 220–230, 320–330, 420–430, 520–530, 620–630, 720–730, 820–830; (run 4) 130–140, 230–240, 330–340, 430–440, 530–540, 630–640, 730–740, 830–840; (run 5) 140–150, 240–250, 340–350, 440–450, 540–550, 640–650, 740–750, 840–850; (run 6) 150–160, 250–260, 350–360, 450–460, 550–560, 650–660, 750–760, 850–860; (run 6) 160–170, 260–270, 360–370, 460–470, 560–570, 660–670, 760–770, 860–870; (run 7) 170–180, 270–280, 370–380, 470–480, 570–580, 670–680, 770–780, 870–880; (run 8) 180–190, 280–290, 380–390, 480–490, 580–590, 680–690, 780–790, 880–890; (run 9) 190–200, 290–300, 390–400, 490–500, 590–600, 690–700, 790–800, 890–900. Each sGPF LC-MS run was performed twice. Quadrupole isolation window was 1.4 m/z and dynamic exclusion was enabled for 10 s. The stepped NCE at 10%, 25%, and 40% was employed to obtain information-rich MS/MS spectra. The run order was the blank first (0.1% formic acid in 5% acetonitrile), pooled QC samples for DDA-MS/MS, and a pooled QC every 12 randomized clinical samples.
2.8. Lipidomics sample preparation
Extraction of lipids started with the addition of 1 mL of methanol to 150 mg of fecal samples and the tube was vigorously shaken with a vortex for 30 s [15]. Subsequently, 5 mL of methyl tertbutyl ether was added, vortexed for another 30 s, and shaken for 20 min at 200 rpm at room temperature. Next, phase separation was induced by adding 3 mL of ultrapure water with 2.5% trichloroacetic acid (w/v) and centrifugation for 5 min at 3000 rpm. Thereafter, 1 mL of the upper layer (consisting of methyl tert-butyl ether) was transferred and evaporated to dryness at 37 °C under a gentle stream of nitrogen. The residue was sequentially resuspended in 250 μL of chloroform and 650 μL of methanol.
2.9. Lipidomics data acquisition
Lipid extracts (2 μL) were separated on a Thermo Scientific Dionex UltiMate 3000 Rapid Separation LC (RSLC) using an ACQUITY UPLC HSS T3 analytical column (2.1 × 150 mm, 1.8 μm, 100 Å, Waters) protected by an ACQUITY UPLC HSS T3 VanGuard pre-column (2.1 × 5 mm, 1.8 μm, 100 Å, Waters). Mobile phase solvents A and B were ACN: H2O (6:4 v/v) and isopropanol: ACN (9:1 v/v), respectively, both contained 10 mM ammonium acetate and 0.1% acetic acid. The separation was performed at 55 °C with a flow rate of 0.35 mL/min using the following gradient: 0–3.0 min, 30%–35% A; 5.0–14.0 min, 65%–98% A; 18.0–18.1 min, 98%–30% A; 18.1–22.0 min, 30% A. Data were acquired on an Orbitrap Fusion Lumos Tribrid mass spectrometer fitted with a HESI source in both positive and negative ionization modes with an independent run for each polarity and a spray voltage of +3500 V and −3500 V, respectively (Thermo Scientific, San Jose, CA, USA). The ion transfer tube temperature was 300 °C, the vaporized temperature was 350 °C, the sheath gas flow was 40 units, the auxiliary gas flow was 15 arbitrary units, and the sweep gas was 1 unit. Lipid profiling was profiled in DDA mode using a full MS scan range of m/z 150–2000 (resolution was 60 K at m/z 200) with top ranked precursor ions subjected to DDA-MS/MS using a maximum injection time of 22 ms. The stepped normalized collision energy (NCE) at 25, 30, and 35 was employed to obtain information-rich MS/MS spectra with a resolution of 15 K at m/z 200. Quadrupole isolation window was 1.6 m/z and dynamic exclusion was enabled for 10s. To promote lipid identification, in-depth DDA MS/MS of QC sample was performed using the following four sGPFs which was performed in four separated runs [14]: (run 1) 150–250, 550–650, 950–1050, 1350–1450, 1750–1850; (run 2) 250–350, 650–750, 1050–1150, 1450–1550, 1850–1950; (run 3) 350–450, 750–850, 1150–1250, 1550–1650; (run 4) 450–550, 850–950, 1250–1350, 1650–1750.
2.10. Metaproteomics data analysis
Peptide identifications were performed using the search engine PEAKS DB combined with PEAKS de novo sequencing [16] (De Novo ALC(%) threshold was 15). False discovery rate (FDR) was set to 1% using the decoy fusion approach. Raw files were refined by precursor ion mass correction and resolving chimeric MS/MS spectra. The precursor mass tolerance was set to 15 ppm and the fragment mass tolerance to 0.03 Da. Enzyme specificity was set to trypsin and up to three missed cleavage sites were allowed. The maximum number of variable posttranslational modifications per peptide was three, including acetylation of protein N-terminus, carbamidomethylation of Cys, oxidation of Met, deamidation of Asn and Gln as well as Pyro-glu from Gln. PEAKS PTM search tool [17] was used to search for peptides with unspecified modifications (313 built-in post-translational modifications), and the SPIDER [18] search tool was used for exploring novel peptides that are homologous to peptides in the protein database.
Database search was performed using a comprehensive meta-database containing human, microbial, and dietary organism sequences [19]. The gut microbial protein database was generated by combining the following parts: (1) the integrated gene catalog of 1,267 human fecal metagenomes [20]; (2) the 1,520 reference genomes of >6,000 cultivated human fecal bacteria isolates [21]; (3) the genomes of 215 human fecal bacteria isolates [22]; (4) all Archaea, Bacteria, and Fungi sequences in NCBI RefSeq (Release 90) and UniProtKB (Release 2017_06). The microbial database was appended by the SARS-COV-2 protein sequences [23], an UniProt human reference proteome (downloaded on 2017_06), and a food database of common dietary organisms. A total number of 130,975,891 non-redundant sequences were obtained after dereplicating at 100% amino acid identity using USEARCH v11.0.667 (–fastx_uniques) [24]. Proteins identified by at least one unique peptides (1% false discovery rate (FDR) using the decoy fusion approach) was considered for further analysis. Label-free quantification of protein groups was performed based on the number of peptide spectrum matches (PSM).
2.11. Taxonomy and functional analysis of gut microbiota
Taxonomy and functional analysis of peptides was performed with UniPept (version 4.3.7) [25] based on the lowest common ancestor (LCA) algorithm using the following parameters: Equate I and L, Advanced missing cleavage handling. Peptide functional annotations were performed using Gene Ontology (GO) terms and Enzyme Commission (EC) numbers. The relative abundance of microbial taxonomic and functional groups were determined using the normalized number of corresponding peptides. Functions of the unannotated microbial proteins were predicted using protein-protein BLAST (BlastP, https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the non-redundant protein sequences (nr) with an E-value threshold of 1e-10.
2.12. Glycoproteomics data analysis
High-confidence identification of intact N-glycopeptides was performed by pGlyco 2.0 [26]. Sequences of proteins identified in the above metaproteomics analysis as well as the SARS-COV-2 protein sequences were used in glycopeptide identification. The precursor mass tolerance was set to 10 ppm and the fragment mass tolerance to 20 ppm. For N-glycopeptide analysis, a FDR of 5% at the glycan level and a FDR of 1% at the peptide level was used. Match between run and intensity based label-free quantification was not supported by pGlyco. To calculate the frequency of N-glycosylation at a specific site, any sample containing a glycopeptide bearing an N-glycan at that site was considered positive regard less of the variations of glycopeptide sequences and glycans. To calculate the frequency of a specific N-glycan at a specific site, any sample containing a glycopeptide bearing that glycan at that site was considered positive regard less of glycopeptide sequence variations. The frequency was defined as the number of positive samples relative to the total number of samples in each group (18 for the control and 49 for the COVID-19 groups, respectively).
Global identification of both O-glycopeptides and N-glycopeptides were performed using PEAKS. Acetylation (protein N-term), carbamidomethylation, deamidation (NQ), pyro-glu from Q, oxidation (M), HexNAcylation (ST), Hex1HexNAc1, Hex1HexNAc2, Hex2HexNAc2, Hex3HexNAc2, Hex1HexNAc(3), Hex(2)HexNAc(3), Hex(3)HexNAc(3), Hex(4)HexNAc(2), Hex(6)HexNAc(2), Hex(7)HexNAc(2), Hex(8)HexNAc(2), Hex(9)HexNAc(2), and dHexHex(3)HexNAc(2) were set as variable posttranslational modifications. The precursor mass tolerance was set to 10 ppm and the fragment mass tolerance to 0.02 Da. The PEAKS output file gave the monosaccharide composition of the attached glycan. Label-free quantification was performed using match between runs with a mass error tolerance of 20 ppm and a retention time shift tolerance of 1 min.
2.13. Metabolomics data analysis
Metabolomics features were extracted, aligned, identified and quantified using Compound Discoverer (v3.1, Thermo Fisher Scientific). The analysis employed the following major steps and parameters: retention time alignment (alignment model = adaptive curve, mass tolerance = 5 ppm, maximum shift = 2 min), unknown compound detection (mass tolerance = 5 ppm, intensity threshold = 30%, S/N threshold = 3, minimum peak intensity = 1 × 106, adducts ions = [M+H]+1, [M + H–H2O]+1, [M + H–NH3]+1, [M+K]+1, [M+Na]+1, [M + NH4]+1, [2 M + H]+1, [2 M + K]+1, [2 M + Na]+1, [2 M + NH4]+1, [M+2H]+2, [M − H]−1, [M − 2H]−2, [M-H + HAc]−1, [M-H-H2O]−1, [M − H + FA]−1, [M+Cl]−1, [2M − H]−1, [2M-H + HAc]−1), compound grouping (mass tolerance = 5 ppm, RT tolerance = 0.2 min), prediction of elemental compositions (mass tolerance = 5 ppm, maximum element counts = 90 × C, 190 × H, 10 × N, 15 × O, 5 × S and 3 × P), filling gaps across all samples (mass tolerance = 5 ppm, S/N threshold = 1.5), chemical background subtraction (using blank samples), identifying compounds by searching ChemSpider (by formula or mass, https://www.chemspider.com/), mzVault and mzCloud (by MS and MS/MS data, precursor mass tolerance = 10 ppm, fragment mass tolerance = 10 ppm, match factor threshold = 60, https://www.mzcloud.org), and QC-based batch normalization (regression model = Cubic Spline). The mzCloud and mzVault match were performed base on Similarity Forward method and HighChem-HighRes search algorithm, respectively. Extracted ion chromatogram (EIC) and MS/MS spectra of all metabolites of interests were manually inspected.
2.14. Lipidomics data analysis
Raw data files were processed using the LipidSearch software (version 4.1) (Thermo Fisher Scientific) to identify and quantify lipid molecular species. Peak detection was performed as follows: Recalc Isotope, on; RT interval (min), 0.01. Lipid identification was as follows: Search type, Product; Exp type, LC-MS; Precursor tolerance, 10 ppm; Product tolerance, 10 ppm; Intensity threshold, 1.0%; Target class, ALL lipid classes; Ion adducts (positive ion mode) of +H, +NH4, +Na, +H–H2O, and +2 H; Ion adducts (negative Ion mode) of –H, +HCOO, +CH3COO, +Cl, and −2H; Top rank filter, On; Main node filter, Main isomer peak; m-Score threshold, 5.0; FA priority, On. ID Quality Filter, Check A, B, C, D (A: lipid class and FA are identified, B: lipid class and some FA were identified, C: Lipid class or FA were identified, D: Lipid identified by other fragment ions (H2O loss, and other non-specific neutral losses). Quantitation was performed using a m/z tolerance of −/+ 5.0 ppm and a RT range of −0.5/+ 0.5 min. Peak alignment was performed using the following parameters: Alignment Method, Max; RT Tolerance, 0.25 min; Calculate unassigned peak area, On; Filter type, New filter; Top rank filter, On; Main node filter, Main isomer peak; m-Score threshold, 5.0; ID quality filter, A, B and C.
2.15. Statistical analysis
The raw quantification data matrix of different omics was imported to MetaboAnalyst [27] for further processing and analysis. Data filtering was performed using interquantile range (IQR) to remove baseline noises. Missing values were imputed using KNN. Quantile normalization and pareto scaling were employed. Unsupervised multivariate data analysis was performed using principal component analysis (PCA) and hierarchical cluster analysis (HCA). Significantly differentiated omics features between COVID-19 and control groups (should present in at least 50% of samples) were detected using Wilcoxon's rank sum test (FDR adjusted p value (q) < 0.05). Microbial taxonomic and functional groups were normalized by total abundance. Statistical significance of microbial taxonomic groups was calculated using Mann-Whitney U test (p value < 0.05). Protein-microbiome and metabolite-microbiome correlations were determined using Pearson correlation (q < 0.05) using R. The association between differentiating omics features with categorical confounding variables (gender and medicine) were determined using Wilcoxon's rank sum test. The association between differentiating omics features with continuous confounding variables (age and Body Mass Index (BMI)) were determined using Pearson correlation.
3. Results
We collected a total of 53 stool samples from 13 COVID-19 patients with a range of one to nine longitudinal time-points that occurred 1–94 days post symptom onset (Fig. 1A, Table S1). Stool samples from 21 healthy subjects severed as controls. Positive RT-PCR results for SARS-COV-2 were observed in stool and/or perianal swab samples even 3 months post symptom onset in a patient with diabetes (patient 1). Furthermore, SARS-CoV-2 viral RNA can persist in stool and/or perianal swab samples long after respiratory samples have tested negative in patients 1, 4, 6, 7, 8, and 9, highlighting the susceptibility of GI tract to SARS-CoV-2 infection.
Multi-omics profiling was performed on each sample to study the alterations of molecular phenomics of the gut ecosystem in COVID-19. Principal component analysis (PCA) showed partially or largely separated multi-omics profiles between COVID-19 patients and controls (Fig. 1B). Quality control (QC) samples of different omics data closely clustered together indicating reproducible MS measurements. Samples from the same patients tended to cluster together indicating greater individual similarity even in the presence of the SARS-COV-2 perturbation.
3.1. Disturbed host proteome
Using untargeted metaproteomics, a total of 16279 protein groups (including 268 human proteins) with at least one unique peptide and a total of 435632 peptides were identified. No SARS-COV-2 protein was detected due to the sensitivity limitation of metaproteomics. Metaproteomics revealed suppressed expression of host proteins involved in immune regulation in COVID-19 (q < 0.05, Fig. 2 A, Table S2), including IGHV3-64D (immunoglobulin (Ig) heavy variable 3-64D), IGHV3-74 (Ig heavy variable 3–74), and IGLL1 (Ig lambda-like polypeptide 1). Meanwhile, two members of the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family that belong to the immunoglobulin superfamily (CEACAM5 and CEACAM6), were also down-regulated in COVID-19. Significant reduction of CEACAM 5 has been reported in intestinal epithelial cells (IECs) from inflammatory bowel disease (IBD) patients and this defect correlated with the inability of IBD IECs to activate CD8+ T cells [[28], [29], [30]], the main T cell population that can kill virus-infected cells. Hence, the reduced CEACAM 5 in COVID-19 may impair cytotoxic CD8+ T cell response in GI tract. Serpin B6, an inhibitor of chymotrypsin-like proteases decreased in COVID-19, while proteases CELA3A (chymotrypsin-like elastase family member 3A), CTRC (Chymotrypsin-C), and MEP1A (Meprin A subunit alpha) increased in COVID-19 (Fig. 2A, Tables S2 and S3). Serpin B6 can prevent cathepsin G-dependent neutrophil death [31] and protect dendritic cells from cytotoxic T lymphocyte induced apoptosis [32]. Serpin deficiency may lead to high inflammation via cathepsin G and gasdermin D. In contrast, intestinal alkaline phosphatase (ALPI), which inhibits host inflammatory responses by detoxifying gut bacterial lipopolysaccharide [33], and PLA2G2A (phospholipase A2, membrane associated), which participates in host antimicrobial defense and inflammatory response, were upregulated in COVID-19. These differentiating host proteins did not exhibit significant associations to other typical confounding variables (e.g. age, gender, and medicine, Table S2), suggesting they represented COVID-19-associated gut pathologies.
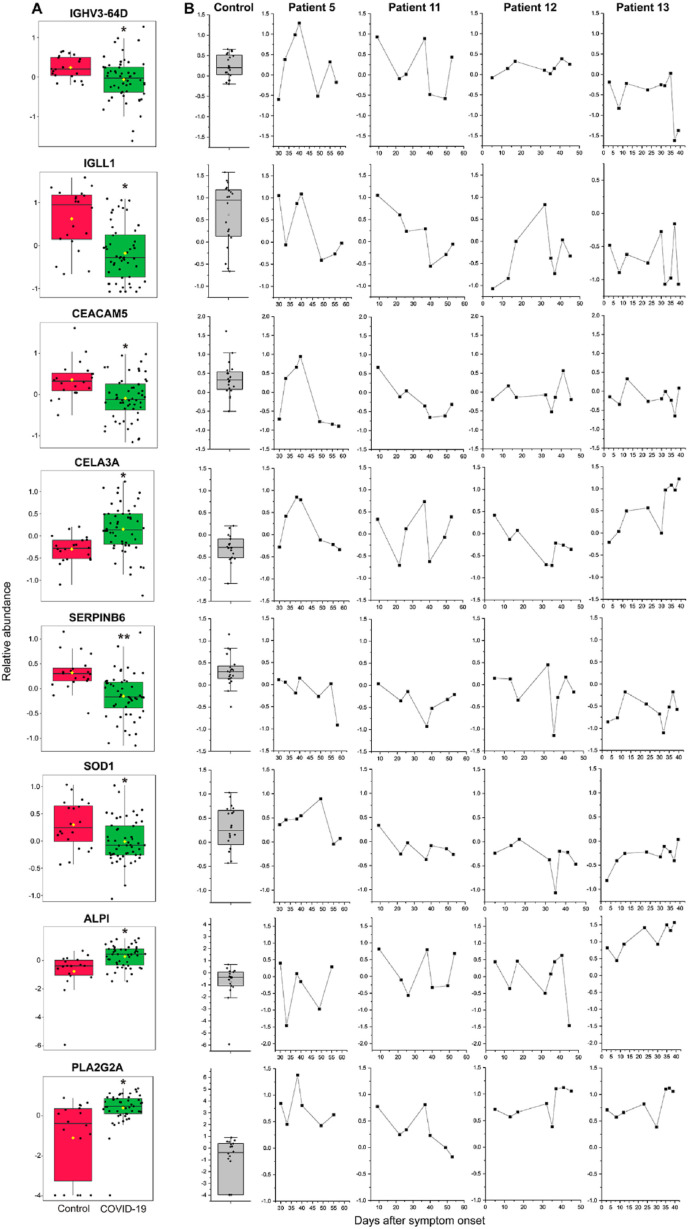
Fig. 2.
Disturbed host proteome homeostasis in COVID-19 patients. A. The abundance changes of global protein expression of IGHV3-64D, IGLL1, CEACAM5, CELA3A, SERPINB6, SOD1, ALPI, and PLA2G2A in the control and COVID-19 samples. Significance is indicated by: (Wilcoxon's rank sum test, ∗q < 0.05, ∗∗q < 0.01). B. The longitudinal changes of IGHV3-64D, IGLL1, CEACAM5, CELA3A, SERPINB6, SOD1, ALPI, and PLA2G2A in patients 5, 11, 12 and 13.
We then investigated the longitudinal changes of these altered proteins in patients 5, 11, 12 and 13, who had serial stools displaying positive to negative stool SARS-CoV-2 infection. Overall, all patients showed considerable protein abundance variations, indicating an unstable gut proteome during the disease course of COVID-19 (Fig. 2B). In many cases, the protein abundance did not restore to the normal levels even several weeks after symptom onset. For patient 5, IGHV3-64D, IGLL1, CEACAM5, and CELA3A showed similar trajectory changes, whose abundance reached a high level 38–40 days after symptom onset but reduced dramatically on day 49. For patient 11, IGHV3-64D and CELA3A elevated on day 37 but decreased sharply on day 40. The longitudinal proteome changes of patient 13 were characterized by a steep fall of IGHV3-64D on days 37 and 39 and a gradually increase of CELA3A, ALPI, and PLA2G2A.
3.2. Glycosylation insight into mucosal immunological pathogenesis
To further investigate the phenomics alterations of COVID-19, we studied the protein glycosylation which plays a key role immunological regulation [34] by HILIC based enrichment (Tables S3 and S4). We first analyzed the intact N-glycopeptides using pGlyco 2.0 because this search engine improves the identification accuracy by comprehensive quality control at all three levels of glycans, peptides, and glycopeptides [26]. In total, 4960 glycopeptide-spectrum matches (GPSMs) derived from 54 human proteins were identified with a 1% GPSM FDR (combing peptide FDR and glycan FDR) (Table S4.1). Only 3 microbial N-glycopeptides were identified probably because of the lower abundance of microbial glycoproteins compared with the dominant human glycoproteins (such as IgA) and the incomplete microbial N-glycan database of pGlyco 2.0 (currently only for human and mouse glycans). With the less stringent criteria (1% peptide FDR and 5% glycan FDR), we retrieved significantly more glycosylation features with 8423 GPSMs corresponding to 486 distinct site-specific N-glycans on 177 glycosylation sites from 83 human glycoproteins (Table S4.1). Frequency is important to understand the pathology of the different post-translational modifications (PTMs). The glycosylation frequency (merged from different glycan types (Fig. 3 A, Table S4.2) or calculated separately (Fig. 3B, Table S4.2)) of major N-glycosylated sites of proteins involved in neutrophil degranulation (including ANPEP, AZU1, MGAM, CEACAM6, CEACAM8, LCN2, OLFM4, and SERPINA1) and neutrophil migration (GP2) were decreased by up to 81.6% in COVID-19. The N-glycosylation of mucins was dominated by Hex1Fuc1, the frequency of which reduced by approximately 25% (Fig. 3B). The N-glycosylation of proteases was dominated by Hex1Fuc1 and Hex3HexNAc2Fuc1 and the frequency of both glycans was reduced in COVID-19.
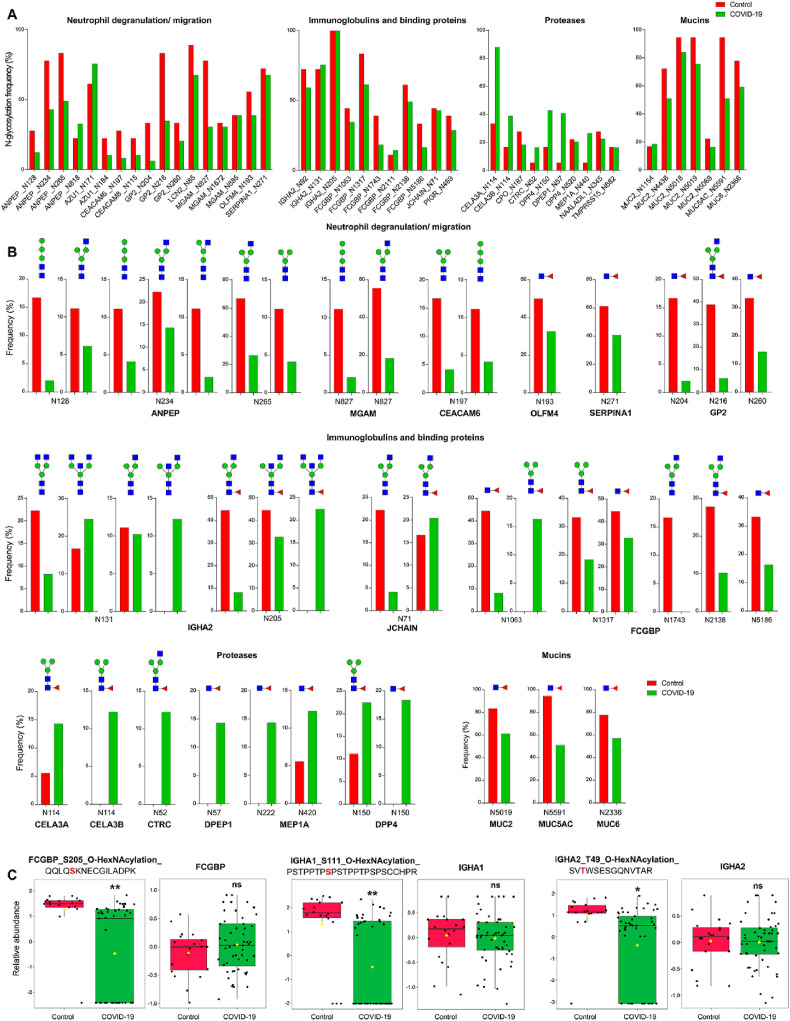
Fig. 3.
Altered glycoproteome in COVID-19 patients. A. Altered glycosylation frequency of major N-glycosylated sites. Different types of glycans were merged. B. Altered glycosylation frequency of major N-glycans in major N-glycosylated sites. C. Reduced O-glycosylated peptide abundance of FCGBP, IGHA1, and IGHA2. The identifier of glycopeptide (left panel) is presented using the format: gene name_modification site_glycan_peptide sequence (the modification sites are highlighted in red). The protein abundance is shown in the corresponding right panel (Wilcoxon's rank sum test, ∗q < 0.05, ∗∗q < 0.01, ns, not significant). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In contrast to the above proteins, Ig related proteins including IGHA2, FCGBP, and JCHAIN exhibited greater glycosylation heterogeneity. On the glycosite N131 of IGHA2, the frequency of glycan Hex3HexNAc4 decreased by 63.3% but that of analogue Hex3HexNAc5 (with an additional HexNAc) increased by 46.9% in COVID-19 (Fig. 3B). On the same glycosite, the frequency of glycan Hex3HexNAc3 was comparable between two groups but the analogue Hex3HexNAc4 (with an additional HexNAc) was only detected in COVID-19. On the glycosite N205 of IGHA2, the relative frequency of glycans increased as the number of HexNAc increased. These results suggest the N-glycosylation alterations of gut IGHA2 are characterized by the conjugation of more complex glycans through the attachment of more HexNAc. The glycan specific alteration was also observed in JCHAIN (N71), where glycan Hex3HexNAc3Fuc, with an additional Fuc compare to its counterpart, exhibited higher frequency in COVID-19. On the other hand, the frequency of the same glycan on different sites can be quite different. For instance, while Hex3HexNAc2Fuc was only detected in COVID-19 on N1063 of FCGBP, the frequency of this glycan was decreased in COVID-19 on N1317. Taken together, the overall N-glycosylation of IGHA2, FCGBP, and JCHAIN was suppressed with glycan-specific and site-specific variations.
We also extended our analysis to O-glycosylation and performed intensity based label-free quantification (Tables S4.3). Similar to N-glycoproteome, O-glycoproteome also revealed increased glycosylation of proteases and reduced glycosylation of IGHA2, FCGBP, ANPEP, and GP2. As shown in Fig. 3C, the relative abundance of O-HexNAcylated peptides QQLQS205KNECGILADPK from FCGBP, PSTPPTPS111PSTPPTPSPSCCHPR from IGHA1, and SVTWSESGQNVT49AR from IGHA2 were significantly decreased in COVID-19 (q < 0.05), while the corresponding protein abundance did not change.
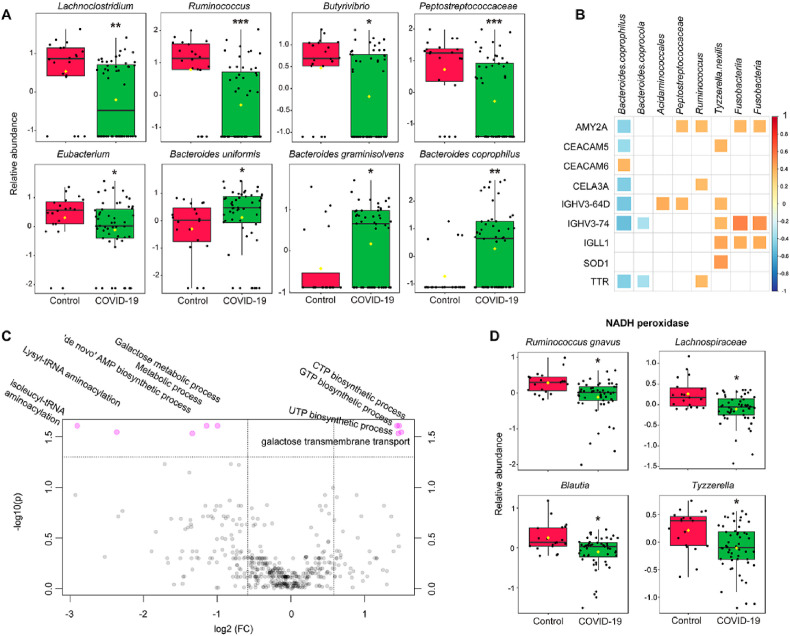
3.3. Reduction of beneficial gut bacteria and potential host-bacteria interactions
We used the metaproteomics approach, which is more accurate than sequencing methods for biomass estimates, to investigate microbial community structure and activity [35]. The relative abundance of 34 bacterial taxa were significantly changed between healthy subjects and patients with COVID-19, most of which were from the Firmicutes phylum (20 out of 34, 58.8%) followed by the Bacteroidetes phylum (10 out of 34, 29.4%) (Table S5). Strikingly, the relative abundance of all 20 altered members in the Firmicutes phylum significantly decreased in COVID-19 (p < 0.05), the majority of which were butyrate-producers [36] belonging to the Lachnospiraceae family, such as genera Lachnoclostridium, Ruminococcus, Butyrivibrio, and Dorea, and species Blautia hansenii, Ruminococcus lactaris, and Tyzzerella nexilis (Fig. 4 A). There was also a significant depletion of butyrate-producing genus Eubacterium in COVID-19, which also carry out bile acid and cholesterol transformations in the gut, contributing to gut and hepatic homeostasis through modulation of bile acid metabolism [37]. In addition, a recent study has found that several species of the phylum Firmicutes (such as genera Clostridium, Ruminococcus, and Eubacterium) were positively associated with memory scores, while species from the phylum Bacteroidetes mainly presented negative associations with memory scores [38]. Taken together, these data suggest a significant reduction of beneficial gut bacteria in COVID-19.
Fig. 4.
Taxonomical and functional alterations of gut microbiome. A. The abundance changes of representative taxonomical groups. Significance is indicated by: (Mann-Whitney U test, ∗q < 0.05, ∗∗q < 0.01, ∗∗∗q < 0.01). B. Associations between altered host proteins and gut microbiome taxonomical groups. Only correlations with q < 0.05 were indicated with colored squares. Positive correlations were shown in red and negative correlations were shown in blue, with sizes of squares representing the magnitude of the correlations. C. Volcano plots illustrate biological processes of microbial proteome that were significantly different (q < 0.05) indicated on top of the plot. D. Reduced NADH peroxidase in bacterial species and genera belonging to order Clostridiales (Wilcoxon's rank sum test, ∗q < 0.05, ∗∗q < 0.01, ∗∗∗q < 0.001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The relative abundance of all altered members in the Bacteroidetes phylum significantly increased in COVID-19 (p < 0.05), such as Bacteroides coprophilus, Bacteroides coprocola, Bacteroides graminisolvens, Bacteroides uniformis, and Bacteroides stercoris (Fig. 4A). Importantly, it has been shown Bacteroidetes and Firmicutes bacteria mainly down-regulate and up-regulate ACE2 expression in the murine gut, respectively [39]. Therefore, the enrichment of Bacteroidetes and the reduction of Firmicutes may potentially inhibit SARS-CoV-2 entry by down-regulating intestinal ACE2 expression.
Association analysis of altered host proteins and bacteria revealed potential host-microbiome interactions. Overall, bacteria groups increased in COVID-19 including B. coprophilus and B. coprocola exhibited negative correlations with host proteins, while those increased in COVID-19 such as Ruminococcus and Fusobacteria exhibited positive correlations (Fig. 4B). An exception was CEACAM6, which was positively associated with B. coprophilus. A recent study has shown CEACAM6 is critical for pathogen enterotoxigenic Escherichia coli adhesion [40]. The reduction of host proteins such as IGHV3-73 and IGHV3-64D may potentially contribute to the enrichment of Bacteroidetes phylum because of the reduced anti-bacteria Igs.
3.4. Functional alteration of gut microbiome
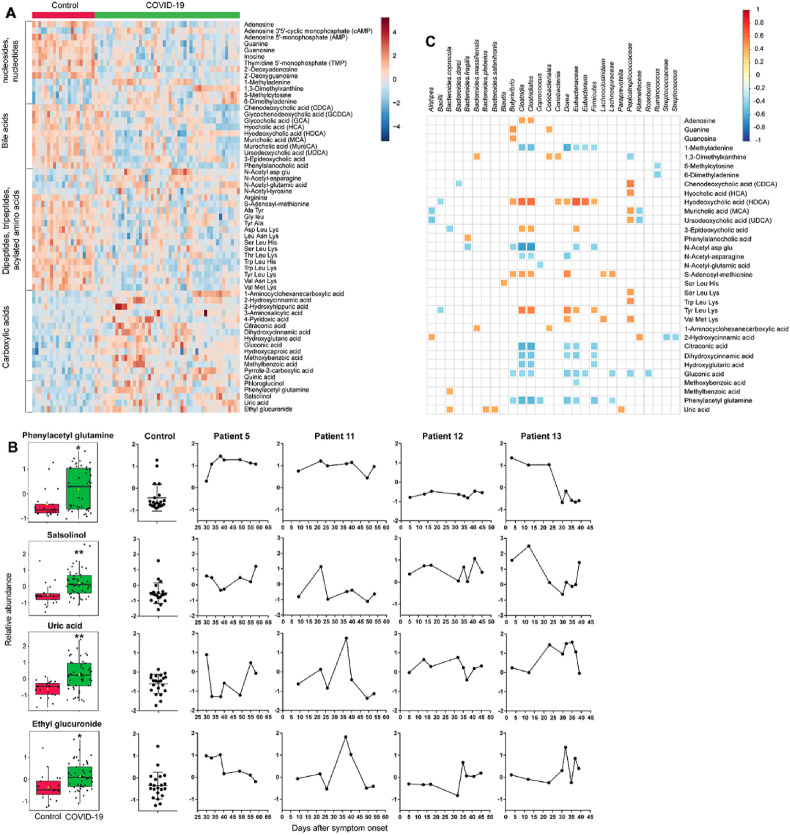
Gene ontology (GO) analysis of metaproteomics data revealed that 9 biological processes of microbial proteome exhibited significant difference between healthy subjects and patients with COVID-19 (q < 0.05) (Fig. 4C). Among them, CTP biosynthetic process, GTP biosynthetic process, and UTP biosynthetic process reduced in COVID-19, while de novo' AMP biosynthetic process increased in COVID-19. Untargeted metabolomics revealed that nucleobase (guanine), nucleosides (adenosine, guanosine, 2′-deoxyadenosine, and inosine) and nucleotides (adenosine 5′-monophosphate (AMP), thymidine 5′-monophosphate (TMP), 2′-deoxyguanosine 5′-monophosphate (dGMP)) decreased in COVID-19 (q < 0.05) (Fig. 5 A), while cyclic AMP (cAMP), methylated purines (1-methyladenine, 6-dimethyladenine) and methylated pyrimidine (5-methylcytosine, 1,3-dimethylxanthine) increased in COVID-19. Association analysis of microbial and metabolomics data revealed that adenosine was positively associated with class Clostridia and order Clostridiales, and guanine and guanosine were positively associated with genus Butyrivibrio (Fig. 4C). In contrast, 1-methyladenine was negatively correlated with genus Dorea, order Clostridiales and class Clostridia, and 5-methylcytosine and 6-dimethyladenine were negatively correlated with genus Ruminococcus. Consistent with the metabolomics findings, GO analysis indicated that there was a 1.8-fold increase in the protein abundance of DNA methylation process in the COVID-19 group, although this difference only reached a relaxed statistical significance threshold (raw p = 0.02). On the other hand, the process of tRNA aminoacylation (lysyl-tRNA aminoacylation, isoleucyl-tRNA aminoacylation) an essential step of protein synthesis, increased in COVID-19.
Fig. 5.
Fecal metabolome alterations in COVID-19 patients. A. Heatmap showing the relative abundance of representative fecal metabolites differentiating between COVID-19 and controls (Wilcoxon's rank sum test, q < 0.05). B. Abundance and longitudinal changes of phenylacetyl glutamine, salsolinol, uric acid, and ethyl glucuronide. Significance is indicated by: (Wilcoxon's rank sum test, ∗q < 0.05, ∗∗q < 0.01). C. Correlations of gut microbial and metabolite abundance. Nodes are sized according to the Pearson correlation coefficient (only correlations with q < 0.05 are shown).
3.5. Enrichment of bacterial related deleterious metabolites
Using untargeted metabolomics, we identified 96 fecal metabolites significantly differed between control subjects and COVID-19 patients, mainly including nucleosides, nucleotides, bile acids, carboxylic acids, dipeptides, tripeptides, and acylated amino acids (Table S6). Notably, we detected an enrichment of several gut microbiome-related deleterious metabolites in COVID-19 (Fig. 5B and S1), including phenylacetyl glutamine (q = 0.01), which promotes cardiovascular disease such as platelet thrombosis [41], and salsolinol (q = 0.003), which is a potential gut bacterial neurotoxin contributing to the development of neurodegenerative diseases [42,43]. The reduction of Firmicutes phylum (such as class Clostridia, order Clostridiales, and genus Dorea) may be at least partially responsible for the increment of phenylacetyl glutamine because they were correlated inversely with each other (Fig. 5C). Longitudinal analysis indicated that the phenylacetyl glutamine level was sustained at high levels in sever patient 5 and in patient 11 who exhibited significant GI symptoms two months after symptom onset. In contrast, this metabolite was kept at a steady and normal level in patient 12 throughout the course of disease and restored to a normal level in patient 13 after one month following symptom onset (Fig. 5B).
We also observed elevated levels of uric acid (q = 0.002) in COVID-19, a uremic toxin playing an important role in several kidney diseases such as lithiasis, gout nephropathy, and preeclampsia. One third of endogenous uric acid is extrarenally excreted via the gut lumen, where it undergoes uricolysis by gut microbiota [44,45]. Increased fecal uric acid was positively associated with several Bacteroides species (Fig. 5C). Interestingly, although all COVID-19 patients involved in this study were non-drinkers, a significantly higher abundance of ethyl glucuronide in COVID-19 (q = 0.01), a metabolite of ethanol formed by glucuronidation, was observed in the COVID-19 group, which indicates a higher susceptibility of ethanol toxicity. Recent studies have demonstrated that certain gut bacteria (such as Klebsiella pneumoniae) contribute to endogenous ethanol production and promote the development of non-alcoholic fatty liver disease [[46], [47], [48]]. Furthermore, gut microbial (such as E. coli and Clostridum sordellii) β-glucuronidases could hydrolyze ethyl glucuronide, which may increase the retention of ethanol in the body by enterohepatic circulation [49]. For patient 11, both ethyl glucuronide and uric acid climbed sharply on day 35 of disease onset, when the discriminative proteins IGHV3-64D, CELA3A, ALPI, and PLA2G2A also dramatically increased (Fig. 5B).
Bile acids are critical for lipid absorption, antibacterial defense and immune regulation [50]. Gut microbiome mediates the primary-to-secondary bile acid conversion. Primary bile acids (chenodeoxycholic acid and muricholic acid), two glycine conjugates (glycochenodeoxycholic acid and glycocholic acid), and secondary bile acids (ursodeoxycholic acid and hyodeoxycholic acid) were decreased (q < 0.05) in fecal samples from participants with COVID-19, compared with control samples (Fig. 5A). Furthermore, a newly discovered conjugated bile acid phenylalanocholic acid [51] was also decreased in COVID-19. Hyodeoxycholic acid exhibited significant positive associations with many taxonomic groups including class Clostridia, order Clostridiales, family Eubacteriaceae, and genera Butyrivibrio, Dorea, and Eubacterium (Fig. 5C).
3.6. Alerted microbial lipidome profiles
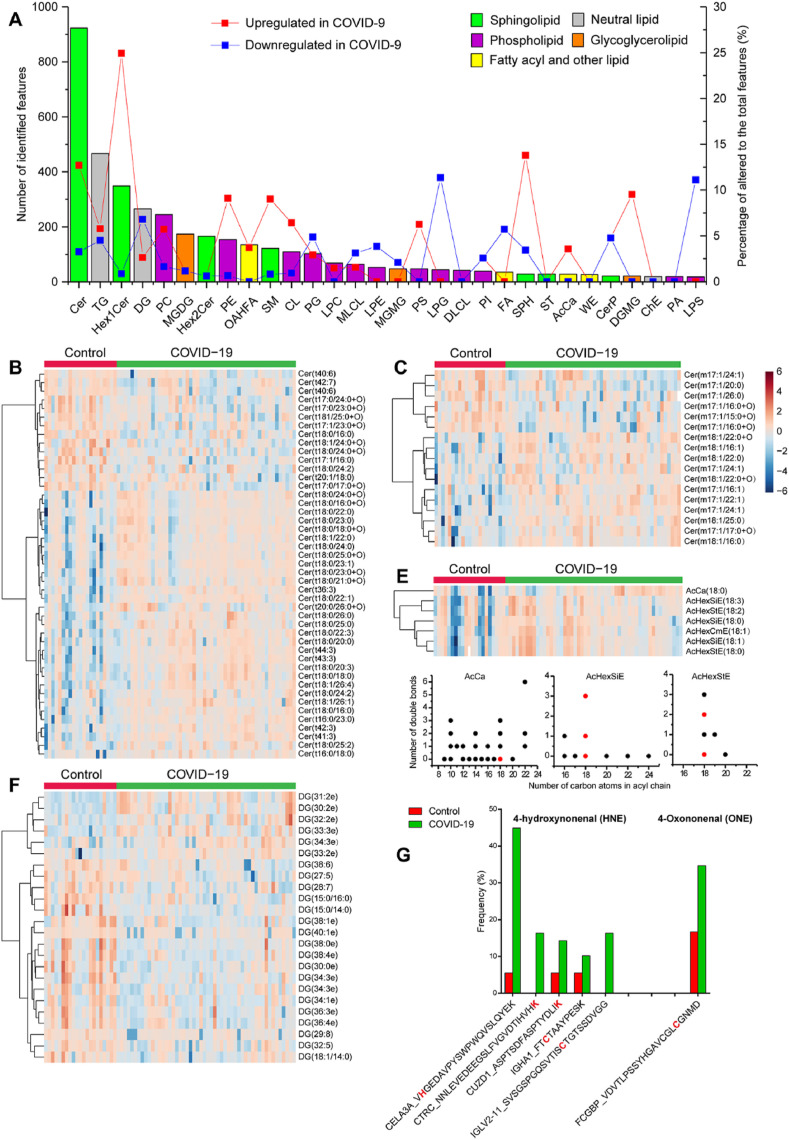
A total of 4,124 lipid features covering 5 lipid categories (sphingolipid, phospholipid, neutral lipid, glycoglycerolipid, fatty acyl and other lipid subclasses) and 67 lipid subclasses (Table S7) were identified based on diagnostic fragment ions along with associated acyl chain fragment information. The most commonly identified lipid species in the fecal lipidome belonged to the ceramide (Cer) subclass with 923 identifications, followed by the triacylglycerol (TG) and monohexosylceramides (Hex1Cer) subclasses with 467 and 349 identifications, respectively (Fig. 6 A). Other frequently identified lipid species included the diradylglycerol (DG, 265 identifications), phosphatidylcholine (PC, 245), monogalactosyldiacylglycerol (MGDG, 174), dihexosylceramide (Hex2Cer, 166), phosphatidylethanolamine (PE, 154), OAcyl-(gamma-hydroxy) fatty acid (OAHFA, 135), and sphingomyelin (SM, 122) subclasses. Among the top 30 identified lipid species, Hex1Cer, SPH, and Cer, all of which belong to the sphingolipid category, underwent the greatest amount of change, with 24.9%, 17.2%, 15.9% significantly increased (q < 0.05) in the COVID-19 group compared to the control group, whereas only 0.9%, 3.5%, and 3.3% significantly decreased (q < 0.05) in the same comparison, respectively (Fig. 6A, Table S7). Within the phospholipid category, the proportions of upregulated lipids were much greater than those of downregulated lipids for the PC (5.7 vs. 1.6%), PE (9.1 vs. 0.7%), and cardiolipin (CL, 6.4 vs. 0.9%) species, while the proportions of upregulated lipids were lower than those of downregulated lipids for lysophosphatidylglycerol (LPG, 0 vs. 11.4%), lysophosphatidylserine (LPS, 0 vs. 11.1%), lysophosphatidylethanolamine (LPE, 0 vs. 3.9%), phosphatidylglycerol (PG, 2.9 vs. 4.9%). Within the neutral lipid category, the proportions of upregulated lipids were comparable to or lower than those of downregulated lipids for the DG (2.6 vs. 6.8%) and TG (5.8 vs. 4.5%) species.
Fig. 6.
Fecal lipidome alterations in COVID-19 patients. A. Distribution of top 30 lipid subclasses identified in untargeted fecal lipidomics. The bar plots indicate number of all identified lipids (left y-axis) and the solid line correspond to the proportion of altered lipids (right y-axis) within each subclass. B-E. Heatmap depicting the relative abundance of differentially expressed lipid subclasses (Wilcoxon's rank sum test, q < 0.05) in COVID-19. Several lipid subclasses exhibited chain-length dependent alterations including ceramide (Cer) lipids with trihydroxy (B) or monohydroxy (C) sphingoid bases, as well as fatty acyl lipids and neutral lipids (D and F). F. Distribution of chain length and number of double bonds of upregulated (red dots) and unchanged (black dots) fatty acyl lipids and neutral lipids (no downregulated feature was observed). Degree-of-unsaturation dependent alterations were observed in Cer with trihydroxy bases (B) and DG lipids (E). G. Increased frequency of protein modification by lipid peroxidation products 4-hydroxynonenal (HNE) and 4-oxononenal (ONE). The identifier of features is presented using the format: gene name_peptide sequence (the modification sites are highlighted in red). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Gut bacterial sphingolipids like Cer, although less well characterized than their mammalian counterparts, are increasingly understood to play important roles in microbial-host interactions [[52], [53], [54]]. The sphingoid backbones and attached fatty acyl chains of bacterial sphingolipids are often odd-chain length, hydroxylated or methylated, while the sphingoid bases for mammals are predominantly even chained and linear backbones [55]. We found many Cer lipids downregulated in the COVID-19 group have a C17-sphingoid base, which probably derived from gut bacteria (based on the odd number of carbon atoms). Specifically, five C17-Cer lipids with trihydroxy sphingoid bases, including Cer(t17:0/17:0+O), Cer(t17:0/23:0+O), Cer(t17:0/24:0+O), Cer (t17:1/16:0), and Cer (t17:1/23:0+O) were significantly reduced in COVID-19 (q < 0.05), but no C17-Cer lipids with trihydroxy bases were increased in COVID-19 (Fig. 6B). In addition, a total of 6 Cer lipids with monohydroxy sphingoid bases, including Cer(m17:1/24:1), Cer(m17:1/20:0), Cer(m17:1/26:0), Cer(m17:1/15:0+O), and 2 Cer(m17:1/16:0+O) isomers, were significantly reduced in COVID-19 (q < 0.05), all of which have a C17-sphingoid base (Fig. 6C). Unlike C17-Cer species with monohydroxy or trihydroxy sphingoid bases, C17-Cer species with dihydroxy sphingoid bases did not shown significant difference in COVID-19. Association analysis of microbial and lipidomics data revealed that Cer(m17:1/22:1), Cer(d18:1/23:0+O), Cer(t18:0/22:0), and Cer(t44:3) were associated with species Bacteroides coprocola, genus Collinsella, class Fusobacteria, and family Peptostreptococcaceae, respectively (Fig. S2).
The chain-length-dependent alteration was also observed for fatty acyl lipid (acyl carnitine (AcCa)) and neutral lipid (acylGlcSitosterol, acylGlcStigmasterol, acylGlcCampesterol, (AcHexSiE, AcHexStE, AcHexZyE, AcHexCmE) subclasses, for which a total of 7 lipids were significantly upregulated in COVID-19 (Fig. 6D). All of them have a C18 acyl chain regardless of the number of double bonds (Fig. 6F).
In addition to chain length, the degree of unsaturation also influenced the behavior of certain lipid species. Highly unsaturated Cer lipids with trihydroxy bases carrying 6 or 7 double bonds (Cer(t40:7) and Cer(t40:6)) were downregulated in COVID-19, while those upregulated Cer lipids have no more than 3 double bonds (Fig. 6B). Similarly, many highly unsaturated DG lipids carrying 5–8 double bonds, such as DG(29:8), DG(28:7), DG(38:6), DG(27:5), and DG(32:5) were downregulated in COVID-19, while those upregulated DG lipids in COVID-19 only have 2 or 3 double bonds (Fig. 6E).
3.7. Increased lipid peroxidation and disturbed redox homeostasis in host and microbiome
We also observed proteomics level evidence of altered lipid features by open database search which allows mining modified peptides. The increased frequency of protein modification by reactive lipid peroxidation products 4-hydroxynonenal (HNE) and 4-oxononenal (ONE) suggests oxidative stress in COVID-19 (Fig. 6E). Indeed, human superoxide dismutase (SOD1), the major antioxidant enzyme for superoxide removal and the first line of defense against oxidative stress, was significantly downregulated in COVID-19 (Fig. 2A). Meanwhile, NADH peroxidase, which reduces peroxides, of several bacterial species and genera belonging to order Clostridiales were also down regulated (Fig. 4D). These results indicate a redox homeostasis disruption for both host and gut bacteria.
4. Discussion
GI tract is susceptible to SARS-COV-2 infection due to the high expression of ACE2 receptor. GI symptoms are frequently observed in patients with COVID-19. Gut's immune responses to SARS-CoV-2 necessitate greater attention because they can alter the commensal microbiome and the crosstalk between microbiota and extra-intestinal organ immunity. However, little is known about the importance of the enteric SARS-CoV-2 for the development of COVID-19-associated pathologies. Increasing evidence has shown that COVID-19 can promote cardiovascular disorders such as myocardial injury, acute coronary syndrome, and thromboembolism [56], neurologic symptoms such as myalgias, encephalopathy, and dizziness [57], and kidney manifestations such as proteinuria and dipstick hematuria [58]. A recent study revealed that harmful metabolites, such as oxalate, were enriched in COVID-19 patients fecal. Moreover, some metabolites (e.g., sucrose) have the potential to predict COVID-19 severity [59]. Our study revealed an enrichment of gut bacteria related deleterious metabolites including phenylacetylglutamine (capable of causing cardiovascular diseases), neurotoxin salsolinol, and uremic toxin uric acid. In addition to metabolites, we observed a larger number of altered host and bacterial lipids (predominated by sphingolipids such as ceramide and hexosylceramide). Sphingolipids produced by gut bacteria can enter host metabolic pathways and impact host ceramide level [40]. Our study may provide an alternative microbiome-based molecular mechanism to explain how the gut ecosystem may play a role in the development of symptoms in COVID-19 and impact the host metabolome and lipidome.
The anti-viral response may impose an immunological off-target effects on gut microbiome in COVID-19 patients. Indeed, we observed disturbed mucosal immunological defense. The reduction of host Igs such as IGHV3-73 and IGHV3-64D may potentially contribute to the enrichment of Bacteroidetes phylum because of the reduced anti-bacteria Igs. The suppressed expression of proteins involved in neutrophil degranulation and migration can also impair the gut anti-bacteria defense system. Furthermore, there is an increased risk of colonic mucosal damage and therefore greater risk of viral and bacterial infection in COVID-19 because of the increased intestinal protease and glycosylation (indicating potential higher activity) and suppressed mucin glycosylation (important for mucin protection function). A major limitation of our study of is the limited sample size and further larger scale studies are needed. Nevertheless, our study has demonstrated widely disturbed gut molecular profiles which may play a role in the development of symptoms in COVID-19. Considering the gut ecosystem as a potential target could offer a valuable approach in managing the disease.
5. Conclusions
Using metaproteomics, metabolomics, glycoproteomics, and lipidomics, our study has demonstrated widely disturbed gut molecular profiles and microbial structure in COVID-19 characterized by disturbed immune, proteolysis and redox homeostasis. Our findings suggest that considering the gut ecosystem as a potential target could offer a valuable approach in managing the disease.
Availability of data and materials
The multi-omics data generated for this manuscript have been deposited in ProteomeXchange Consortium (https://www.iprox.org/) under the following identifier: Metaproteomics (IPX0002453001), Metabolomics (IPX0002453002), Lipidomics (IPX0002453003), and Glycoproteomics (IPX0002453004).
Funding
This work was supported by the Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai, SML2020SP003), Guangdong Basic and Applied Basic Research Foundation (2019A1515011771), National Natural Science Foundation of China (31900070), the National Key Research and Development Program of China (2020YFC082400), Science and Technology Development Fund of Macau SAR (FDCT0017/2020/A), the Task-Force Project on the Prevention and Control of Novel Coronavirus of Guangdong Province (20201113), the Three Major Constructions of Sun Yat-sen University (the Task-Force Project on the Prevention and Control of Novel Coronavirus of Sun Yat-sen University), the Emergency Task-Force of SARS-CoV-2 research of Guangzhou Regenerative Medicine and Health Guangdong Laboratory, the Emergency Task-Force Project on the Prevention and Control of Novel Coronavirus of Zhuhai 2020.
Ethics statement
This study was approved by the Ethics Committee of The Fifth Affiliated Hospital, Sun Yat-sen University (K161-1).
CRediT authors contribution statement
Conceptualization, Z.Y.; Data analysis, Z.Y.; Methodology, Z.Y., F.H.; Sample coordination and preparation, F.H., T.Z., K.X., and F.X.; Sample collection, Z.F., S.H., Z.G., H.S., Z.Z., and H.Z.; Clinical data collection, Z.Y., H.Z., L.L., and J.L.; Clinical laboratory tests, G.J., and K.L.; Writing - Original Draft, Z.Y.; Writing - Review & Editing, Z.Y., X.L., H.S., and R.Y.; Work supervised by Z.Y., X.L., and R.Y.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2021.338881.
Abbreviations
- ACE2
angiotensin converting enzyme 2
- AcCa
acyl carnitine
- ACN
acetonitrile
- ALPI
alkaline phosphatase
- ANPEP
aminopeptidase N
- AZU1
azurocidin
- AMP
adenosine 5′-monophosphate
- BMI
body mass index
- COVID-19
coronavirus disease 2019
- CEACAM
carcinoembryonic antigen-related cell adhesion molecule
- CELA3A
chymotrypsin-like elastase family member 3A
- Cer
ceramide
- CTRC
Chymotrypsin-C
- dGMP
2′-deoxyguanosine 5′-monophosphate
- DG
diradylglycerol
- EC
Enzyme Commission
- EIC
extracted ion chromatogram
- FA
formic acid
- FDR
false discovery rate
- FiO2
fractional inspired oxygen
- FCGBP
IgGFc-binding protein
- GdmCI
Guanidinium hydrochloride
- GI
gastrointestinal
- GO
Gene ontology
- GPSMs
glycopeptide-spectrum matches
- HCA
hierarchical cluster analysis
- HCD
higher-energy collisional dissociation
- Hex2Cer
dihexosylceramide
- HILIC
hydrophilic interaction liquid chromatography
- HNE
hydroxynonenal
- IGHA2
Immunoglobulin heavy constant alpha 2
- IGHV3-64D
immunoglobulin heavy variable 3-64D
- IGHV3-74
Ig heavy variable 3-74
- IGLL1
Ig lambda-like polypeptide 1
- IECs
intestinal epithelial cells
- IBD
inflammatory bowel disease
- IQR
interquantile range
- JCHAIN
Immunoglobulin J chain
- LCA
lowest common ancestor
- LCN2
Neutrophil gelatinase-associated lipocalin
- LPE
lysophosphatidylethanolamine
- LPG
lysophosphatidylglycerol
- LPS
lysophosphatidylserine
- MEP1A
Meprin A subunit alpha
- MGAM
Maltase-glucoamylase
- MGDG
monogalactosyldiacylglycerol
- NCE
normalized collision energy
- OAHFA
OAcyl-(gamma-hydroxy) fatty acid
- OLFM4
Olfactomedin-4
- ONE
oxononenal
- PaO2
partial pressure of oxygen
- PE
phosphatidylethanolamine
- PCA
principal component analysis
- PLA2G2A
phospholipase A2
- PTMs
post-translational modifications
- PC
phosphatidylcholine
- PG
phosphatidylglycerol
- QC
quality control
- RT-PCR
reverse transcription polymerase chain reaction
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SERPINA1
Alpha-1-antitrypsin
- SM
sphingomyelin
- SOD1
superoxide dismutase
- sGPFs
staggered gas-phase fractionations
- TCEP
tris(2-carboxyethyl)phosphine
- TFA
trifluoroacetic acid
- TMPRSS2
transmembrane serine protease 2
- TMP
thymidine 5′-monophosphate
- TG
triacylglycerol
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lin L., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69(6):997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 2.Jin X., et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69(6):1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Y., et al. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69(6):1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- 4.Xiao F., et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833 e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y C.C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Feng Z., Wu G., et al. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Weekly. 2020;2(8):123–124. [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo T., et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159(4):1302–1310 e5. doi: 10.1053/j.gastro.2020.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo T., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159(3):944–955 e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arunachalam P.S., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanley D., et al. Translocation and dissemination of commensal bacteria in post-stroke infection. Nat. Med. 2016;22(11):1277–1284. doi: 10.1038/nm.4194. [DOI] [PubMed] [Google Scholar]
- 11.Grau K.R., et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat Microbiol. 2020;5(1):84–92. doi: 10.1038/s41564-019-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagan T., et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178(6):1313–1328 e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta Y., et al. Rapid glycopeptide enrichment using cellulose hydrophilic interaction/reversed-phase StageTips. Mass Spectrom. 2017;6(1):A0061. doi: 10.5702/massspectrometry.A0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan Z., Yan R. Improved data-dependent acquisition for untargeted metabolomics using gas-phase fractionation with staggered mass range. Anal. Chem. 2015;87(5):2861–2868. doi: 10.1021/ac504325x. [DOI] [PubMed] [Google Scholar]
- 15.Van Meulebroek L., et al. Holistic lipidomics of the human gut phenotype using validated ultra-high-performance liquid chromatography coupled to hybrid Orbitrap mass spectrometry. Anal. Chem. 2017;89(22):12502–12510. doi: 10.1021/acs.analchem.7b03606. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., et al. Peaks DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics. 2012;11(4) doi: 10.1074/mcp.M111.010587. M111 010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X., et al. PeaksPTM: mass spectrometry-based identification of peptides with unspecified modifications. J. Proteome Res. 2011;10(7):2930–2936. doi: 10.1021/pr200153k. [DOI] [PubMed] [Google Scholar]
- 18.Han Y., Ma B., Zhang K. SPIDER: software for protein identification from sequence tags with de novo sequencing error. J. Bioinf. Comput. Biol. 2005;3(3):697–716. doi: 10.1142/s0219720005001247. [DOI] [PubMed] [Google Scholar]
- 19.Yan Z., et al. A semi-tryptic peptide centric metaproteomic mining approach and its potential utility in capturing signatures of gut microbial proteolysis. Microbiome. 2021;9(1):12. doi: 10.1186/s40168-020-00967-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J., et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014;32(8):834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 21.Zou Y., et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 2019;37(2):179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browne H.P., et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533(7604):543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 25.Gurdeep Singh R., et al. Unipept 4.0: functional analysis of metaproteome data. J. Proteome Res. 2019;18(2):606–615. doi: 10.1021/acs.jproteome.8b00716. [DOI] [PubMed] [Google Scholar]
- 26.Liu M.Q., et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 2017;8(1):438. doi: 10.1038/s41467-017-00535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong J., Wishart D.S., Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics. 2019;68(1):e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- 28.Toy L.S., et al. Defective expression of gp180, a novel CD8 ligand on intestinal epithelial cells, in inflammatory bowel disease. J. Clin. Invest. 1997;100(8):2062–2071. doi: 10.1172/JCI119739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roda G., et al. Defect in CEACAM family member expression in Crohn's disease IECs is regulated by the transcription factor SOX9. Inflamm. Bowel Dis. 2009;15(12):1775–1783. doi: 10.1002/ibd.21023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roda G., et al. Characterizing CEACAM5 interaction with CD8alpha and CD1d in intestinal homeostasis. Mucosal Immunol. 2014;7(3):615–624. doi: 10.1038/mi.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgener S.S., et al. Cathepsin G inhibition by Serpinb1 and Serpinb6 prevents programmed necrosis in neutrophils and monocytes and reduces GSDMD-driven inflammation. Cell Rep. 2019;27(12):3646–3656 e5. doi: 10.1016/j.celrep.2019.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medema J.P., et al. Expression of the serpin serine protease inhibitor 6 protects dendritic cells from cytotoxic T lymphocyte-induced apoptosis: differential modulation by T helper type 1 and type 2 cells. J. Exp. Med. 2001;194(5):657–667. doi: 10.1084/jem.194.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bates J.M., et al. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2(6):371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zauner G., et al. Glycoproteomic analysis of antibodies. Mol. Cell. Proteomics. 2013;12(4):856–865. doi: 10.1074/mcp.R112.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleiner M., et al. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun. 2017;8(1):1558. doi: 10.1038/s41467-017-01544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vital M., Karch A., Pieper D.H. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. 2017;2(6) doi: 10.1128/mSystems.00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Udayappan S., et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. NPJ Biofilms Microbiomes. 2016;2:16009. doi: 10.1038/npjbiofilms.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnoriaga-Rodriguez M., et al. Obesity impairs short-term and working memory through gut microbial metabolism of aromatic amino acids. Cell Metabol. 2020;32(4):548–560 e7. doi: 10.1016/j.cmet.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Geva-Zatorsky N., et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168(5):928–943 e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheikh A., et al. CEACAMs serve as toxin-stimulated receptors for enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2020;117(46):29055–29062. doi: 10.1073/pnas.2012480117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemet I., et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell. 2020;180(5):862–877 e22. doi: 10.1016/j.cell.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villageliu D.N., Borts D.J., Lyte M. Production of the neurotoxin salsolinol by a gut-associated bacterium and its modulation by alcohol. Front. Microbiol. 2018;9:3092. doi: 10.3389/fmicb.2018.03092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurnik-Lucka M., et al. Salsolinol: an unintelligible and double-faced molecule-lessons learned from in vivo and in vitro experiments. Neurotox. Res. 2018;33(2):485–514. doi: 10.1007/s12640-017-9818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorensen L.B. Role of the intestinal tract in the elimination of uric acid. Arthritis Rheum. 1965;8(5):694–706. doi: 10.1002/art.1780080429. [DOI] [PubMed] [Google Scholar]
- 45.Hosomi A., et al. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PloS One. 2012;7(2) doi: 10.1371/journal.pone.0030456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L., et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57(2):601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 47.Michail S., et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol. Ecol. 2015;91(2):1–9. doi: 10.1093/femsec/fiu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan J., et al. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metabol. 2019;30(4):675–688 e7. doi: 10.1016/j.cmet.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 49.Baranowski S., et al. In vitro study of bacterial degradation of ethyl glucuronide and ethyl sulphate. Int. J. Leg. Med. 2008;122(5):389–393. doi: 10.1007/s00414-008-0229-3. [DOI] [PubMed] [Google Scholar]
- 50.Shapiro H., et al. Bile acids in glucose metabolism in health and disease. J. Exp. Med. 2018;215(2):383–396. doi: 10.1084/jem.20171965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinn R.A., et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579(7797):123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An D., et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156(1–2):123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown E.M., et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. 2019;25(5):668–680 e7. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson E.L., et al. Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun. 2020;11(1):2471. doi: 10.1038/s41467-020-16274-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoffel W., Dittmar K., Wilmes R. Sphingolipid metabolism in bacteroideaceae. Hoppe Seylers Z Physiol Chem. 1975;356(6):715–725. doi: 10.1515/bchm2.1975.356.s1.715. [DOI] [PubMed] [Google Scholar]
- 56.Madjid M., et al. Potential effects of coronaviruses on the cardiovascular system: a Review. JAMA Cardiol. 2020;5(7):831–840. doi: 10.1001/jamacardio.2020.1286. [DOI] [PubMed] [Google Scholar]
- 57.Liotta E.M., et al. Frequent neurologic manifestations and encephalopathy-associated morbidity in Covid-19 patients. Ann Clin Transl Neurol. 2020;7(11):2221–2230. doi: 10.1002/acn3.51210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong D., et al. Kidney manifestations of mild, moderate and severe coronavirus disease 2019: a retrospective cohort study. Clin Kidney J. 2020;13(3):340–346. doi: 10.1093/ckj/sfaa083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lv L., et al. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal. Chim. Acta. 2021;1152:338267. doi: 10.1016/j.aca.2021.338267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The multi-omics data generated for this manuscript have been deposited in ProteomeXchange Consortium (https://www.iprox.org/) under the following identifier: Metaproteomics (IPX0002453001), Metabolomics (IPX0002453002), Lipidomics (IPX0002453003), and Glycoproteomics (IPX0002453004).