Graphical abstract

Keywords: SARS-CoV-2, COVID-19, Coronavirus, Interferon, Immune evasion, Innate immunity
Abstract
The on-going pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to unprecedented medical and socioeconomic crises. Although the viral pathogenesis remains elusive, deficiency of effective antiviral interferon (IFN) responses upon SARS-CoV-2 infection has been recognized as a hallmark of COVID-19 contributing to the disease pathology and progress. Recently, multiple proteins encoded by SARS-CoV-2 have been shown to act as potential IFN antagonists with diverse possible mechanisms. Here, we summarize and discuss the strategies of SARS-CoV-2 for evasion of innate immunity (particularly the antiviral IFN responses), understanding of which will facilitate not only the elucidation of SARS-CoV-2 infection and pathogenesis but also the development of antiviral intervention therapies.
1. Introduction
The emergence of a novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) quickly caused the global pandemic of coronavirus disease 2019 (COVID-19). As of April 29, 2021, more than 149 million confirmed cases with over 3 million deaths have been recorded throughout the world. SARS-CoV-2 is an enveloped positive-sense single-stranded RNA virus, belonging to Coronaviridae family, Betacoronaviruses genus which also contains two other notorious life-threatening pathogens, SARS-CoV and MERS-CoV (Middle East respiratory syndrome coronavirus). The genome of SARS-CoV-2 is approximately 29.9-kb long with at least 14 open reading frames (ORFs) encoding viral proteins [1]. Two large overlapping ORFs in the 5′-proximal two-third of the genome, ORF1a and ORF1b, encode continuous polypeptides pp1a and pp1ab, which are cleaved by viral proteases into 16 nonstructural proteins (nsp1-16), making up the replicase. Other ORFs encode four structural proteins (S [spike], E [envelop], M [membrane], and N [nucleocapsid]) that are assembled to the virion, and a number of accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9a, ORF9b, ORF10, etc.). SARS-CoV-2 infection starts with the attachment of the S protein to the cellular receptor angiotensin-converting enzyme 2 (ACE2) and other entry cofactors, followed by virus-cell membrane fusion, genome release, RNA transcription/replication, protein production, and virion assembly and budding [2]. Upon viral infection, host cells generally respond by recognizing particular molecular structures (called pathogen-associated molecular patterns, PAMPs, e.g. foreign viral RNAs) introduced or produced in viral life cycle to trigger innate immune responses [3]. As the first line of host defense, innate immunity and especially the antiviral interferon (IFN) system restrict viral replication and spread, promote tissue repair, and aid in development of the subsequent adaptive immunity, eventually facilitating viral clearance [4], [5], [6]. However, extraordinarily, IFN responses seem to be weak during SARS-CoV-2 infection, indicating efficient counteraction of the antiviral system by SARS-CoV-2 [7], [8], [9]. This deficiency of IFN responses likely leads to productive viral replication and contributes to COVID-19 pathology and severity [8], [10], [11], [12]. Therefore, elucidation of the interactions of SARS-CoV-2 with IFN system will not only provide pivotal insights into SARS-CoV-2 infection and pathogenesis but also benefit design of prophylaxis and treatment against COVID-19. In this review, we summarize the recent progress on the evasion strategies of SARS-CoV-2 from host innate immunity and in particular the antiviral IFN system and discuss significant future directions for further study.
2. Antiviral IFN responses to SARS-CoV-2
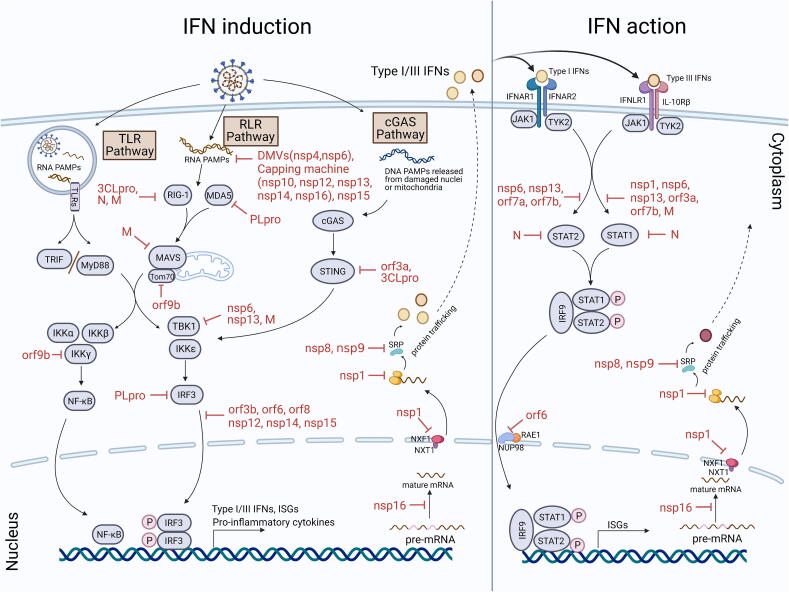
Antiviral IFN system based on type I IFNs (especially IFN-α and IFN-β) and type III IFNs (IFN-λ) plays essential roles in host defense against viral infections by acting as a primary component of innate immunity and promoting the induction of adaptive immunity [4], [5], [6]. Responses of the antiviral IFN system comprise two phases, IFN induction and IFN action (also called IFN signaling) (Fig. 1).
Fig. 1.
The antiviral IFN system and its antagonism by SARS-CoV-2. Two phases of IFN system, IFN induction and IFN action, and the viral counteraction against these antiviral responses at various levels are depicted. See text for details.
Innate immune response (including the IFN induction) is initiated by host recognition of viral PAMPs (mainly specific viral nucleic acids or some other particular products of viral infection) via cellular pattern recognition receptors (PRRs). PRRs sensing RNA virus infections mainly include cytosolic RIG-I-like receptors (RLRs) and transmembrane toll-like receptors (TLRs, residing on cell surface or in endosomes). For immune recognition of coronavirus infections, RLRs, melanoma differentiation-associated protein 5 (MDA5) and retinoic acid-inducible gene I (RIG-I), and TLRs, TLR3 and TLR7, are usually considered to play notable roles in sensing various viral RNAs and leading to type I and III IFN production [13], [14], [15], [16]. TLR3 in endosomes can detect viral double-stranded RNA (dsRNA) and then activate the downstream adaptor protein Toll/IL-1 receptor (TIR) domain-containing adaptor (TRIF), while viral single-stranded RNA (ssRNA) can be recognized by TLR7, thus triggering the downstream adaptor protein myeloid differentiation primary response gene 88 (MyD88) [15], [17]. In the cytoplasm, viral RNA PAMPs activate RIG-I and MDA5 which subsequently induce the activation of their common adaptor, mitochondrial antiviral signaling protein (MAVS) [18], [19]. These PRR-adaptor signaling cascades continue with the downstream kinases, TANK binding kinase 1 (TBK1) and inhibitor of κB kinase ε (IKKε), and IKKα, IKKβ and IKKγ. TBK1 and IKKε phosphorylate and hence activate transcription factors IFN regulatory factor 3 (IRF3) and IRF7, while IKKα, IKKβ and IKKγ direct the activation of transcription factor nuclear factor-κB (NF-κB). Then activated transcription factors translocate to the nucleus, inducing expression of type I and III IFNs, some IFN-stimulated gene (ISGs), and proinflammatory cytokines [18]. In addition, the cGAS (cyclic GMP-AMP synthase)-STING (stimulator of interferon genes) pathway that is commonly associated with sensing of cytosolic DNA PAMPs may be also activated by SARS-CoV-2 infections [20], [21], perhaps due to the DNA release from damaged mitochondria or nuclei, although to what extent it would contribute to the anti-SARS-CoV-2 immune responses is uncertain. In this pathway, cGAS senses and is activated by DNA PAMPs [22], [23]. Subsequently, activated cGAS catalyzes the synthesis of 2′3′-cyclic GMP–AMP (2′3′-cGAMP) that binds to STING as the secondary messenger to activate TBK1 and other IKKs, then directing similar downstream signaling cascades to induce expression of the antiviral IFNs and other immune regulatory genes [22], [23], [24].
In the following IFN action phase, type I and III IFNs establish the cellular state of viral resistance in infected cells and adjacent cells through autocrine and paracrine pathways, respectively. The expressed and secreted IFNs bind to IFN receptors on the cell surface, activating Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2). The activated kinases immediately phosphorylate and activate signal transducer and activator of transcription proteins (STAT1 and STAT2) which then combine with IRF9 to form a heterotrimeric transcription factor complex called IFN-stimulated gene factor 3 (ISGF3) [14], [25], [26]. ISGF3 translocates into the nucleus and binds to interferon-stimulated response elements (ISREs), thus rapidly inducing the systematic expression of hundreds of ISGs that can restrict infection at almost every steps of the viral life cycle [14], [26]. Many ISGs themselves are signaling molecules or regulatory proteins of innate and adaptive immunity, induction of which can, in turn, lead to further amplification and development of immune responses (including IFN responses) [27], [28].
3. Evasion strategies of SARS-CoV-2
Most, if not all, pathogenic viruses evolve their evasion strategies against innate immune responses. Because of its rapidity and effectiveness in eliminating viral infection, the antiviral IFN pathway is often a prime target for evasion of innate immune. Upon SARS-CoV-2 infection, minimal expression of antiviral IFNs in cultured cells, experimental animals, and severe COVID-19 patients clearly suggest the extraordinarily efficient antagonism of the innate immune responses developed by the virus [8], [12]. Considering the potent and multiple antiviral and immunoregulatory activities of type I and III IFNs, SARS-CoV-2 inhibition of the IFN responses would not only directly facilitate the evasion from multifaceted antiviral actions of numerous ISGs but also impede the responses of various innate and adaptive immune cells (such as natural killer cells, dendritic cells, macrophages, and lymphocytes), including immune cell-mediated clearance of viruses and infected cells and antigen presentation [29], [30], [31], [32]. Based on the related findings of SARS-CoV-2 and combined with the knowledge from other coronaviruses (especially SARS-CoV), the innate immune evasion strategies of SARS-CoV-2 could be categorized as follows: (i) inhibiting IFN induction, including concealing or reducing PAMPs to evade host PRR sensing or disrupting the following signaling cascades of IFN induction; (ii) suppressing IFN action; (iii) globally interfering with production of host proteins including IFNs (Fig. 1).
3.1. Inhibition of IFN induction
3.1.1. Concealing or reduction of PAMPs
Recognition of viral PAMPs by host PRRs initiates innate immune responses including IFN induction. SARS-CoV-2 may have evolved strategies to counteract the host defense at this very early stage by concealing or decreasing potential PAMPs (Fig. 1). Like most positive-strand RNA viruses, SARS-CoV-2 induces cellular endomembrane remodeling to form double membrane vesicles (DMVs), which compartmentalize and facilitate viral RNA replication and thus likely prevents viral RNA PAMPs from exposure to cellular PRRs [33], [34], [35]. The N protein plays essential roles in SARS-CoV-2 genome packaging by encapsidation and intriguingly, seems to undergo liquid–liquid phase separation with RNA which might also contribute to the concealing of viral RNA PAMPs and merits further investigation [36], [37], [38]. Furthermore, SARS-CoV-2 can methylate the 5′-end of viral mRNA by nsp16/nsp10 heterodimer to mimic cellular mRNA, hijacking the host translation machinery and evading PRR recognition [39]. Like SARS-CoV, SARS-CoV-2 has a self-coded capping machinery composed of nsp10, nsp12, nsp13, nsp14, and nsp16 [39], [40], [41], [42], [43]. nsp13, the viral helicase, also has an RNA triphosphatase activity [44], [45] which initiates the formation of an RNA cap and moreover, likely reduces 5′-triphosphorylated viral RNA, an otherwise potential PAMP sensed by host PRRs (especially RIG-I). nsp12, RdRp-associated nucleotidyltransferase (NiRAN), possesses guanylyltransferase activity, catalyzing the formation of cap core structure (GpppA) [43]. nsp14, an mRNA cap guanine-N7-methyltransferase, produces a Cap-0 (me7GopppA1) [40], [42]. nsp16, a cap ribose 2′-O-methyltransferase, forms an obligatory complex with nsp10 to efficiently convert mRNA species from the Cap-0 to the Cap-1 (me7GopppA1m) [39]. Modified viral RNA can evade recognition by host PRRs, including RLRs and TLRs, and may also avoid the antiviral effects of some other ISGs such as interferon-induced proteins with tetratricopeptide repeats (IFIT) [46]. In addition, coronavirus nsp15 is an uridylate-specific endoribonuclease (designated EndoU) that seems to cleave 5′-polyuridines from negative-sense viral RNA (the product of polyA-templated RNA synthesis; potential PAMP triggering MDA5), significantly circumventing activation of MDA5 [47], [48], [49]. Considering the high conservativeness of the EndoU activity among coronaviruses, this evasion strategy previously demonstrated in the studies of other coronaviruses is very likely employed by SARS-CoV-2 as well, although direct testing of the SARS-CoV-2 nsp15 activity is required for validation. These possible strategies are summarized in Table 1.
Table 1.
Antagonism of IFN induction and action by SARS-CoV-2.
| Antagonist | Function or mechanism | Cellular interaction target | Phase targeted | Refs |
|---|---|---|---|---|
| Double membrane vesicles (DMVs) | Compartmentalize viral RNAs to prevent their exposure to PRRs. nsp4 and nsp6 may be involved in DMV formation. | – | IFN induction | [33], [34] |
| nsp10, nsp12, nsp13, nsp14, nsp16 | Act as the viral capping machinery to modify viral mRNA, diminishing recognition by PRRs. | – | IFN induction | [39], [40], [43], [44] |
| nsp15 | Cleaves viral RNA polyuridine sequences to avoid the recognition by MDA5. | – | IFN induction | [47] |
| PLpro | Acts as deISGylase that directly remove ubiquitin-like ISG15 modifications from IRF3 and MDA5; Directly cleaves IRF3. | IRF3, MDA5 | IFN induction | [61], [62], [63], [64], [65] |
| 3CLpro | Interacts with RIG-I and thus obstruct K63-linked ubiquitination and activation of RIG-I by TRIM25; Might inhibit K63-ubiquitin modification of STING to disrupt the recruitment of TBK1 and IKKβ. | RIG-I, STING | IFN induction | [20], [57] |
| nsp1 | Suppresses STAT1 phosphorylation and nuclear translocation. | unclear | IFN action | [71] |
| nsp6 | Interacts with TBK1 to inhibit IRF3 activation. | TBK1 | IFN induction | [71] |
| Suppresses STAT1 and STAT2 phosphorylation. | unclear | IFN action | [71] | |
| nsp12 | Inhibits IRF3 nuclear import. | unclear | IFN induction | [74] |
| nsp13 | Interacts with TBK1 to disrupt TBK1-mediated IRF3 phosphorylation. | TBK1 | IFN induction | [69], [70], [71] |
| Suppresses STAT1 and STAT2 phosphorylation. | unclear | IFN action | [71] | |
| nsp14 | Inhibit IRF3 nuclear localization. | unclear | IFN induction | [70] |
| nsp15 | Inhibit IRF3 nuclear localization. | unclear | IFN induction | [70] |
| M | Interacts with RIG-I, MAVS, and TBK1, thus preventing the formation of the multiprotein complex, impeding IRF3 phosphorylation and nuclear translocation. | RIG-I, MAVS, TBK1 | IFN induction | [52], [53] |
| Suppresses STAT1 phosphorylation and nuclear translocation. | unclear | IFN action | [71] | |
| N | Binds to the DExD/H domain of RIG-I, thus impeding RIG-I signaling. | RIG-I | IFN induction | [50], [51] |
| Might bind to STAT1 and STAT2, suppressing STAT1 and STAT2 phosphorylation. | STAT1, STAT2 | IFN action | [80] | |
| ORF3a | Interacts with STING and blocks the nuclear accumulation of NF-κB, thus likely impeding IFN promoter activation. | STING | IFN induction | [20] |
| Suppresses STAT1 phosphorylation and nuclear translocation. | unclear | IFN action | [71] | |
| ORF3b | Inhibits IRF3 nuclear localization. | unclear | IFN induction | [78], [79] |
| ORF6 | Might interact with KPNA2 to block IRF3 nuclear accumulation but not activation. | KPNA2 | IFN induction | [4], [67], [70], [71], [72] |
| Interacts with NUP98-RAE1 complex to block STAT1 nuclear translocation. | NUP98-RAE1 complex | IFN action | [4], [67], [71], [73] | |
| ORF7a | Suppresses STAT2 phosphorylation. | unclear | IFN action | [71] |
| ORF7b | Suppresses STAT1 and STAT2 phosphorylation. | unclear | IFN action | [71] |
| ORF8 | Inhibits IRF3 nuclear localization. | unclear | IFN induction | [75], [77] |
| ORF9b | Interacts with Tom70, perhaps thus inhibiting type I IFN induction; Targets IKKγ and specifically interrupts IKKγ K63-linked polyubiquitination, thereby inhibiting NF-κB signaling and IFN promoter activation. | TOM7, IKKγ | IFN induction | [66], [67], [68] |
3.1.2. Disruption of the signaling cascades of IFN induction.
Aside from the passive stashing of PAMPs, SARS-CoV-2 also has been equipped with multiple active immune antagonists that can directly target to the PRR-triggered signaling cascades to dampen IFN induction (Table 1 and Fig. 1). The N protein of SARS-CoV-2 may bind to the DExD/H domain of RIG-I, which has ATPase activity and is important for the binding of PAMP RNAs, thus impeding RIG-I signalling [50], [51] The stress granule protein G3BP1 that positively regulates innate immune responses including RIG-I signaling was observed to be recruited in the phase separated condensates of SARS-CoV-2N [37]; it will be interesting to investigate whether RIG-I could be also recruited into SARS-CoV-2 N condensates and whether the recruitment of the host proteins in the condensates could contribute to viral inhibition of IFN induction. Another SARS-CoV-2 structural protein, M, also was shown to inhibit the antiviral IFN expression [52], [53]. Mechanistically, with ectopic expression by transient transfection, a study by Zheng et al reported that SARS-CoV-2 M seems to be able to interact with RIG-I, MDA5, MAVS, and TBK1 to inhibit RIG-I–MAVS, MAVS–TBK1, and TRAF3–TBK1, but not MDA5–MAVS interactions, thus preventing phosphorylation and nuclear translocation of IRF3 [52], while Fu et al demonstrated that the viral protein specifically binds to MAVS but not RIG-I, MDA5, or TBK1, abating MAVS aggregation and recruitment of TRAF3, TBK1, and IRF3 to MAVS but having no noticeable effects on the RIG-I–MAVS or MDA5–MAVS interactions [53]. Coronavirus papain-like protease (PLpro, encoded in nsp3) is essential for the N-terminal cleavage of pp1a and pp1ab polyproteins, resulting in the release of nsp1, nsp2, and nsp3, while 3-chymotrypsin-like “main” protease (3CLpro or Mpro, harbored in nsp5) is responsible for the cleavage and hence mature of the other nsp proteins [54], [55], [56]. SARS-CoV-2 3CLpro was demonstrated to interact with RIG-I as well and thus obstruct K63-linked ubiquitination and activation of RIG-I by E3 ligase tripartite motif 25 (TRIM25) [57]. As for PLpro, besides the protease activity, it may also act as a deubiquitinase (DUB) and a deISGylase that directly remove ubiquitin or ubiquitin-like ISG15 modifications required for multiple signaling protein activation events, crippling innate immune signaling transduction [58], [59], [60]. SARS-CoV-2 PLpro shares ~ 83% sequence identity with its counterpart in SARS-CoV but show a different preference for substrates ubiquitin and ISG15 [61], [62], [63]. SARS-CoV PLpro disrupts ubiquitination but with a lesser effect on ISGylation, while SARS-CoV-2 PLpro exhibits notable preference for cleaving ISG15 from host protein substrates and in particular MDA5 and IRF3, therefore attenuating IFN induction [61], [64]. Additionally, SARS-CoV-2 PLpro may also directly cleave IRF3 to suppress IFN production [65]. Importantly, PLpro inhibitors can reverse PLpro-mediated suppression of type I IFN induction and attenuate SARS-CoV-2 replication [61], highlighting the dual intervention potential of these inhibitors against SAR-CoV-2 and COVID-19 that may not only inhibit viral nsp replicase mature but also rescue host antiviral immunity. SARS-CoV-2 ORF9b was shown to be localized to mitochondria and interact with translocase of outer mitochondrial membrane 70 (Tom70), a critical protein linking MAVS to TBK1 and IRF3, perhaps thus inhibiting type I IFN induction [66], [67]. Then, Wu et al. reported that ORF9b of SARS-CoV-2 appears to also target IKKγ, an essential regulator of NF-κB, and specifically interrupts IKKγ K63-linked polyubiquitination through its N-terminus, thereby inhibiting the IKKα/β/γ-NF-κB signaling and subsequent IFN promoter activation [68]. As stated above, SARS-CoV-2 nsp13 plays essential roles in viral replication as the helicase and 5’-triphosphatase; notably, this viral protein also exhibits significant inhibitory capacity against IFN induction (as well as IFN action that is discussed in the following section). Mechanistically, SARS-CoV-2 nsp13 may bind TBK1 and impedes the association of TBK1 with other signaling proteins including MAVS and TNF receptor-associated factors (TRAFs, which facilitate recruitment of TBK1 to MAVS), suppressing TBK1 and IRF3 activation and IFN expression [69], [70], [71]. Interestingly, SARS-CoV-2 nsp6 appears to interact with TBK1 to inhibit IRF3 activation as well; however, unlike nsp13, nsp6 expression does not affect TBK1 phosphorylation/activation [71]. Similar to SARS-CoV ORF6, SARS-CoV-2 ORF6 also robustly hampers IFN responses. However, SARS-CoV-2 ORF6 expression noticeably blocks IRF3 nuclear translocation but not phosphorylation [70], [72]; moreover, activation of type I IFN promoter by IRF3/5D (a constitutively active IRF3 mutant) overexpression also can be inhibited in the presence of SARS-CoV-2 ORF6 [7], suggesting that this viral protein likely acts at the level of IRF3 or downstream. Further, by reference to the studies on SARS-CoV, the interactions of SARS-CoV-2 ORF6 with karyopherin α proteins (KPNAs, nuclear import factors) were analyzed in settings of protein overexpression by transfection. Miorin et al. demonstrated SARS-CoV-2 ORF6 interactions with both KPNA1 and KPNA2 [73], while a specific interaction of the viral protein with KPNA2 but not KPNA1 was observed in another study by Xia et al. [71]. Although further validations are required, the potential targeting of these KPNAs by SARS-CoV-2 ORF6 may explain its specific blockade of IRF3 nuclear accumulation but not activation.
Other viral proteins of SARS-CoV-2, including nsp12, nsp14, nsp15, ORF3b and ORF8, may also repress IFN induction [7], [70], [74], [75], [76], [77], [78], [79]. However, the underlying mechanisms are more elusive. Like ORF6, nsp12 inhibits IRF3 nuclear translocation, but not phosphorylation, induced by Sendai virus (a model RNA virus triggering innate immune responses) [7], [74], suggesting that nsp12 may function at the level or downstream of IRF3. nsp14, nsp15, ORF3b and ORF8 were shown to impair nuclear translocation of IRF3 as well, but whether the activation of the transcription factor could be affected is unknown [70], [75], [79]. Moreover, the inhibitory capacity of ORF8 to interferon production is controversial [70], [75]. Suppression of IRF3 nuclear translocation is an experimental phenomenon which could be resulted by various interferences with the signaling events at the level of IRF3, or upstream or downstream. Thus, the exact cellular interaction targets of these SARS-CoV-2 proteins for IFN induction antagonism remain to be further determined. Despite the largely unclear mechanism, antagonistic activity of SARS-CoV-2 ORF3b to IFN induction likely depends on its C-terminal length [78], [79]. SARS-CoV-2 ORF3b is much shorter (22 amino acids) but appears to have greater capacity to antagonize Sendai virus infection-caused IFN promoter activation, in comparison with its SARS-CoV ortholog (153 amino acids). Moreover, interestingly, a natural SARS-CoV-2 ORF3b variant with extended C-terminus because of the loss of the first premature stop codon exhibited increased IFN-induction suppressive activity and was isolated from two severe COVID-19 cases [79].
In addition to inhibiting the classical RNA virus-triggered immune pathways, it has been recently reported that SARS-CoV-2 may also be able to antagonize the cGAS-STING signaling cascades [20]. ORF3a was shown to selectively inhibit cGAS-STING pathway but not RLR response. Mechanistically, ORF3a may interact with STING and block the nuclear accumulation of NF-κB, thus likely impeding IFN promoter activation in cGAS-STING signaling transduction. In comparison, 3CLpro seems to disturb both of the RLR (as described above) and cGAS-STING pathways. For the inhibition of cGAS-STING signaling, this viral protein was demonstrated to bind STING as well and abate its K63-ubiquitin modification, thus preventing STING functional complex assembly and downstream signalling [20].
3.2. Suppression of IFN action
The IFN action phase also can be circumvented by SARS-CoV-2. In SARS-CoV-2 infected cells, IFN-stimulated nuclear translocation of STAT1 and STAT2 can be generally diminished [73]. Several SARS-CoV-2 proteins may be involved here (Fig. 1 and Table 1). ORF6 ectopic expression was shown to abolish cellular STAT1 nuclear translocation but not phosphorylation driven by type I IFN treatment [7], [71], [73]. While the potential association of ORF6 with KPNAs as aforementioned appeared to be dispensable in the inhibition of IFN action, an interaction between ORF6 and nucleoporin 98 (Nup98) instead was shown to be important for the blockade of STAT1 nuclear import [67], [73]. Despite many controversial results existing in different reports, other proteins encoded by SARS-CoV-2 including ORF3a, ORF7a, ORF7b, nsp6, nsp13, M, and N may also dampen type I IFN action [7], [71], [80]. Therein, nsp6, nsp13, ORF7b, and N decrease both STAT1 and STAT2 phosphorylation [71], [80]; ORF3a and M may mainly reduce STAT1 phosphorylation, while ORF7a may mainly inhibit STAT2 activation [71]. In the study by Xia et al., it was further demonstrated that SARS-CoV-2 nsp6 has a stronger activity of inhibiting IFN action compared with MERS-CoV nsp6, whereas SARS-CoV nsp6 exhibits no such activity at all [71]. More interestingly, replacement of SARS-CoV-2 nsp6 by its orthologs from SARS-CoV or MERS-CoV in the context of a SARS-CoV-2 replicon system decreased the ability of resistance to type I IFN treatment [71]. The observation further supports the significance of IFN action antagonism for viral fitness to host cells in a context more like that of viral authentic infection. However, similarly, as impairments of the signaling components and events at the level or upstream of STATs all might result in the diminishment of STAT activation, the direct cellular targets and mechanisms employed by these IFN action antagonists (including nsp6) need to be further uncovered.
3.3. Global interference with production of host proteins including IFNs
As obligate intracellular parasites, viruses hijack host cell components to achieve their life cycles. In the course of infection, high-pathogenic coronaviruses likely have developed some strategies to globally interfere with production of host proteins including IFNs [81], [82], [83]. As for SARS-CoV-2, its nsp1, nsp8, nsp9, and nsp16 are likely involved in the disturbance of host protein synthesis or trafficking at various levels [84], [85], [86], [87] (Fig. 1 and Table 2). SARS-CoV nsp1 is notable as a host shutoff factor inhibiting cellular mRNA translation and this activity is likely also conservative in other pathogenic β- and α-coronaviruses, although the detailed mechanisms may differ [81], [88]. Recent studies have suggested that the nsp1 of SARS-CoV-2 is not exceptional. SARS-CoV-2 nsp1 interacts with the 40S ribosomal subunit by inserting its C-terminal domain containing two helices into the entrance region of the ribosomal mRNA channel, blocking mRNA translation [85], [86], [88], [89], [90]. However, how SARS-CoV-2 circumvents this translational blockage for the production of its own proteins is an open question. Here are two proposed models based on experimental data depicting the bipartite roles of SARS-CoV-2 nsp1 during infection: (1) nsp1 C-terminal locks the 40S in a conformation incompatible with host mRNA loading and disrupts initiation factor binding, while the 5′-UTR (especially the SL1 5′-UTR hairpin) of SARS-CoV-2 mRNA bypasses this inhibition by directly binding to nsp1 N-terminal and thus presumably resulting in a structural rearrangement of nsp1 and dissociation of nsp1 from the 40S ribosome during the initiation of viral translation [91], [92], [93]: (2) host genes harboring 5′ terminal oligo-pyrimidine (TOP) tracts can be spared, which makes sure the expression of the translation machinery components, RNA binding proteins, and other host factors important for viral propagation [94]. Additionally, SARS-CoV-2 nsp1 also interacts with the host mRNA export receptor heterodimer NXF1-NXT1 and prevents proper binding of NXF1 to mRNA export adaptors and NXF1 docking at the nuclear pore complex, rendering a significant number of cellular mRNAs retained in the nucleus and thus likely also contributing to the inhibition of host antiviral gene expression [84]. Further, SARS-CoV-2 nsp16 was shown to bind pre-mRNA recognition domains of U1/U2 snRNAs and disrupt global mRNA splicing and mature, while the nsp8 and nsp9 proteins bind to the 7SL RNA in the signal recognition particle (SRP) and interfere with protein integration into cell membrane and trafficking [87]. These activities leading to the global impairments of host protein production are all likely implicated in the viral subversion of the host antiviral responses to various extents.
Table 2.
Global inhibition of the production of host proteins (including IFNs) by SARS-CoV-2.
| Antagonist | Function or mechanism | Cellular interaction target | Refs |
|---|---|---|---|
| nsp1 | Interacts with the 40S ribosomal subunit by inserting its C-terminal domain containing two helices into the entrance region of the ribosomal mRNA channel, blocking host mRNA translation; Interacts with the host mRNA export receptor NXF1-NXT1, leading to nuclear retention of cellular mRNAs. | 40S ribosomal subunit, NXF1-NXT1 | [89], [84], [85], [86] |
| nsp8, nsp9 | Bind to the 7SL RNA in the SRP and interfere with protein integration into cell membrane and trafficking. | 7SL RNA | [87] |
| nsp16 | Binds pre-mRNA recognition domains of U1/U2 snRNAs and disrupts mRNA splicing and mature. | U1/U2 snRNAs | [87] |
4. Conclusion and perspective
The world is currently suffering from the pandemic of COVID-19 caused by SARS-CoV-2. As a pivotal aspect of virus-host interactions and viral pathogenesis, the interplays of SARS-CoV-2 with host innate immune system have attracted many research interests that yield significant progress. However, we are still far from clearly understanding the complex immune regulation of SARS-CoV-2 including the IFN antagonism. In fact, which PRRs are involved in sensing of SARS-CoV-2 and how they cooperatively orchestrate the innate immune responses even remain to be clearly unraveled. Significantly, most of the potential IFN antagonists of SARS-CoV-2 were identified in the contexts of ectopic expression of individual viral proteins by transfection in cell lines and many controversial results exist in the functional screening studies. Further comparative analyses in the contexts of infections with specific mutated viruses generated by reverse genetics in both cultured cells and animal models are warranted to validate the functions and underlying mechanisms of the possible IFN antagonistic proteins summarized in this review. Identification of natural SARS-CoV-2 variants with the potential IFN antagonists mutated also could be valuable to assess the IFN-inhibiting activities and their corresponding significance to viral replication and pathogenicity, especially when combined with the clinical data (such as disease severity and viral loads in patients) or experimental analysis of animal model infections. Moreover, studies on mutant viruses with IFN antagonist(s) mutated or deleted might provide promising clues for attenuated vaccine development. Additionally, there is still debate as to whether some potential ORFs of SARS-CoV-2 encode viral proteins or not in the authentic viral infection. The map of SARS-CoV-2 coding capacity is generally based on computational predictions and homology analysis with other coronaviruses, yet the translation of some proteins like ORF3b, remains undetected [95]. Using a suite of ribosome-profiling techniques, 23 novel viral ORFs have been identified [95]. The function of these ORFs remains a mystery. Following the functional validations, mechanistic studies are then merited; however, even for the IFN antagonist candidates already identified, how they act in many aspects largely remains open questions.
Aside from attenuated vaccine development, drug design could also benefit from the knowledge of viral immune antagonists and their mechanisms. Intervention strategies against IFN-inhibiting factors encoded by SARS-CoV-2 might rescue host antiviral immunity and promote clearance of viral infection. Further, since many IFN-antagonistic proteins of SARS-CoV-2 (especially the viral structural proteins and the nonstructural replicase proteins) also play essential roles in viral replication, some drugs targeting these viral proteins may have dual antiviral activities by interfering with the viral immune evasion and by directly inhibiting virus replication, as suggested in the testing of PLpro inhibitors [61]. Additionally, identification of cellular targets and decipherment of molecular mechanisms underlying viral IFN antagonism may help inform the development of host-directed therapeutics that could have the potential of being broad-spectrum and more resistant to the emergence of escape mutation and drug-resistant strains. Then, combinatory usage of multiple drugs targeting viral IFN antagonists and host factors (including antiviral IFNs themselves) also might be considered in the study of anti-SARS-CoV-2 therapy.
The clinical course and outcome of COVID-19 are rather heterogeneous, ranging from asymptomatic and mild infections to severe infections and death. Impaired antiviral IFN responses have been suggested as a hallmark of severe COVID-19 patients [8], [12]. The virally encoded IFN antagonists, as potential virulence factors, could be significant determinants of the pathogenicity of different viral strains. As discussed above, IFN antagonist mutation in various viral strains is worth in-depth analysis and continuous monitoring. On the other hand, the disease heterogeneity also could be attributed to different individual patient susceptibility. Cell/tissue type variations and inter-individual differences should be precisely taken into consideration in further studies for elucidation of the viral immune antagonists and the evasion strategies, which is likely of critical importance for both the understanding of viral pathogenesis and the development of antivirals and vaccines. In addition to the mechanisms at the levels of viral RNAs and proteins, post-translation modification could be also involved in the viral immune antagonism. Particularly, the heavy glycosylation of SARS-CoV-2 S protein has been suggested to likely contribute to not only viral entry but also the escape from immune responses (including both antibody production and epitope recognition) [96], [97], [98], [99], [100], [101], although it is unclear whether the glycosylated viral protein is implicated in SARS-CoV-2 counteraction of IFN-based innate immunity which we here focus on. Additionally, palmitoylation of SARS-CoV-2 S protein seems to be involved in the viral protein-mediated, cholesterol-dependent syncytia formation which may facilitate viral cell-to-cell spread and hence escape from the extracellular neutralizing antibodies and immune recognition receptors [38], [102], [103]. Likewise, protein modification including glycosylation could differ among viral strains, cell types and individuals [97]. Therefore, it will be also interesting to address the effects of other viral and host factors, such as viral protein modification, cell types, and individual background, on the viral evasion from innate immunity and hence on the disease heterogeneity.
SARS-CoV-2 shares ~ 79.5% genomic sequence identity to its sister coronavirus SARS-CoV, while they exhibit notable differences in transmissibility and pathogenicity [104]. These differences could be attributed to various viral and host factors and their complex interactions, including the differentiation of IFN antagonists and the immune-suppressing strategies between the two viruses. Indeed, existing researches have shown both significant similarity and interesting variation in functions and mechanisms of IFN-antagonistic proteins encoded by SARS-CoV-2 and SARS-CoV (such as PLpro and nsp6, as discussed in this paper) [71], [61], [62], [63], [64]. Further comparative studies of the immune evasion strategies among SARS-CoV-2 and the related viruses (especially SARS-CoV and MERS-CoV) may provide new insights into the molecular basis of differential infectivity and pathogenicity of various coronaviruses and benefit the development of specific or broad-spectrum antiviral therapeutics.
With an extraordinarily large RNA genome, SARS-CoV-2 seems to be equipped with many IFN antagonists to interfere with multiple layers of the host antiviral IFN responses, which is likely a significant determinant for its well adaptation in human population as seen in the pandemic. Over the past year, a series of potential IFN antagonists with various possible mechanisms have been proposed as discussed here. However, greater efforts are undoubtedly needed to present a clearer and more comprehensive picture of the IFN antagonism of SARS-CoV-2. These efforts will help to not only understand the viral infection and pathogenesis but also develop novel antivirals and vaccines to combat the ongoing pandemic and to prepare better for dealing with the future outbreaks of emerging or re-emerging coronaviruses.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The research in our laboratory was funded by the National Natural Science Foundation of China (31870162 and 82161138003, to YJN), the National Key Research and Development Program of China (2018YFA0507202, to YJN and HLW), and the Youth Innovation Promotion Association of Chinese Academy of Sciences (to YJN). We thank the National Virus Resource Center for virus resources and the Core Facility and Technical Support of Wuhan Institute of Virology for technical assistance in the studies of our laboratory.
References
- 1.Wu F., Zhao S.u., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H., Lyu Y.Y., Hou F.J. SARS-CoV-2 infection and the antiviral innate immune response . J Mol Cell Biol. 2020;12:963–967. doi: 10.1093/jmcb/mjaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann H.-H., Schneider W.M., Rice C.M. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36(3):124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotenko S.V., Durbin J.E. Contribution of type III interferons to antiviral immunity: location, location, location. J Biol Chem. 2017;292(18):7295–7303. doi: 10.1074/jbc.R117.777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy D.E., Marié I.J., Durbin J.E. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol. 2011;1(6):476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acharya D., Liu GuanQun, Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. 2020;20(7):397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q., Bastard P., Liu Z.Y. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(eabd4570) doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meffre E., Iwasaki A. Interferon deficiency can lead to severe COVID. Nature. 2020;587(7834):374–376. doi: 10.1038/d41586-020-03070-1. [DOI] [PubMed] [Google Scholar]
- 12.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sampaio NG, Chauveau L, Hertzog J et al. The RNA sensor MDA5 detects SARS-CoV-2 infection. bioRxiv 2021. [DOI] [PMC free article] [PubMed]
- 14.Sa Ribero M., Jouvenet N., Dreux M., Nisole S., Stapleford K. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16(7):e1008737. doi: 10.1371/journal.ppat.100873710.1371/journal.ppat.1008737.g00110.1371/journal.ppat.1008737.g00210.1371/journal.ppat.1008737.g00310.1371/journal.ppat.1008737.t00110.1371/journal.ppat.1008737.t002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park A., Iwasaki A. Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 2020;27(6):870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazaleuskaya L., Veltrop R., Ikpeze N., Martin-Garcia J., Navas-Martin S. Protective Role of Toll-like Receptor 3-Induced Type I Interferon in Murine Coronavirus Infection of Macrophages. Viruses-Basel. 2012;4(5):901–923. doi: 10.3390/v4050901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akira S., Hemmi H. Recognition of pathogen-associated molecular patterns by TLR family. Immunol Lett. 2003;85(2):85–95. doi: 10.1016/s0165-2478(02)00228-6. [DOI] [PubMed] [Google Scholar]
- 18.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16(9):566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rui Y., Su J., Shen S.i., Hu Y., Huang D., Zheng W. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduct Tar. 2021;6(1) doi: 10.1038/s41392-021-00515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neufeldt C.J., Cerikan B., Cortese M. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. bioRxiv. 2020 doi: 10.1038/s42003-021-02983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J., Sun L., Chen X., Du F., Shi H., Chen C. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X.-D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science. 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnell J., Kerr I., Stark G. Jak-Stat Pathways and Transcriptional Activation in Response to Ifns and Other Extracellular Signaling Proteins. Science. 1994;264(5164):1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 26.Schindler C., Levy D.E., Decker T. JAK-STAT signaling: From interferons to cytokines. J Biol Chem. 2007;282(28):20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 27.Yang E., Li M.M.H. All About the RNA: Interferon-Stimulated Genes That Interfere With Viral RNA Processes. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.605024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu Rev Immunol. 2014;32(1):513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazewski C., Perez R.E., Fish E.N., Platanias L.C. Type I Interferon (IFN)-Regulated Activation of Canonical and Non-Canonical Signaling Pathways. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.606456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye L., Schnepf D., Staeheli P. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat Rev Immunol. 2019;19:614–625. doi: 10.1038/s41577-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 31.Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 32.Le Bon A., Tough D.F. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14(4):432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 33.Snijder E.J., Limpens R.W.A.L., de Wilde A.H. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. 2020;9 doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scutigliani E.M., Kikkert M. Interaction of the innate immune system with positive-strand RNA virus replication organelles. Cytokine Growth F R. 2017;37:17–27. doi: 10.1016/j.cytogfr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun. 2021;12(1) doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders D.W., Jumper C.C., Ackerman P.J. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. eLife. 2021:10. doi: 10.7554/eLife.65962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selvaraj C., Dinesh D.C., Panwar U., Abhirami R., Boura E., Singh S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. P Natl Acad Sci USA. 2009;106(9):3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan L., Ge J.i., Zheng L., Zhang Y., Gao Y., Wang T. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell. 2021;184(1):184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu T., Huang M., Wu D.i., Ren Y., Zhang X., Han Y. SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts. Virol Sin. 2020;35(3):321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with Severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78(11):5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J. 2 '-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. P Natl Acad Sci USA. 2020;117(14):8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13(2):e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. P Natl Acad Sci USA. 2017;114(21):E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh S.J., Shin O.S. SARS-CoV-2 Nucleocapsid Protein Targets RIG-I-Like Receptor Pathways to Inhibit the Induction of Interferon Response. Cells. 2021;10(3):530. doi: 10.3390/cells10030530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen K.L., Xiao F., Hu D.W. SARS-CoV-2 Nucleocapsid Protein Interacts with RIG-I and Represses RIG-Mediated IFN-beta Production. Viruses-Basel. 2021;13 doi: 10.3390/v13010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Y.i., Zhuang M.-W., Han L., Zhang J., Nan M.-L., Zhan P. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct Tar. 2020;5(1) doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Y.-Z., Wang S.-Y., Zheng Z.-Q., Yi Huang, Li W.-W., Xu Z.-S. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol Immunol. 2021;18(3):613–620. doi: 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiel V., Ivanov K.A., Putics A. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 56.Ziebuhr J., Siddell S.G. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J Virol. 1999;73(1):177–185. doi: 10.1128/jvi.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y., Ma L., Zhuang Z., Cai S., Zhao Z., Zhou L. Main protease of SARS-CoV-2 serves as a bifunctional molecule in restricting type I interferon antiviral signaling. Signal Transduct Tar. 2020;5(1) doi: 10.1038/s41392-020-00332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey-Elkin B.A., Knaap R.C.M., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B. Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression. J Biol Chem. 2014;289(50):34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe Acute Respiratory Syndrome Coronavirus Papain-Like Protease Ubiquitin-Like Domain and Catalytic Domain Regulate Antagonism of IRF3 and NF-kappa B Signaling. J Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282(44):32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587(7835):657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39(18) doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect Dis. 2020;6(8):2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 64.Liu GuanQun, Lee J.-H., Parker Z.M., Acharya D., Chiang J.J., van Gent M. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat Microbiol. 2021;6(4):467–478. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moustaqil M., Ollivier E., Chiu H.-P., Van Tol S., Rudolffi-Soto P., Stevens C. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg Microbes Infec. 2021;10(1):178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang H.-W., Zhang H.-N., Meng Q.-F., Xie J., Li Y., Chen H. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol Immunol. 2020;17(9):998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J., Shi Y., Pan X., Wu S., Hou R., Zhang Y. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34(7):108761. doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo G., Gao M., Gao X., Zhu B., Huang J., Luo K. SARS-CoV-2 non-structural protein 13 (nsp13) hijacks host deubiquitinase USP13 and counteracts host antiviral immune response. Signal Transduct Tar. 2021;6(1) doi: 10.1038/s41392-021-00509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infec. 2020;9(1):1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xia H., Cao Z., Xie X., Zhang X., Chen J.-C., Wang H. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33(1):108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kimura I., Konno Y., Uriu K., Hopfensperger K., Sauter D., Nakagawa S.o. Sarbecovirus ORF6 proteins hamper induction of interferon signaling. Cell Rep. 2021;34(13):108916. doi: 10.1016/j.celrep.2021.108916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K.e., Cohen P., Patel R.S. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. P Natl Acad Sci USA. 2020;117(45):28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang W., Zhou Z., Xiao X., Tian Z., Dong X., Wang C. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol Immunol. 2021;18(4):945–953. doi: 10.1038/s41423-020-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rashid F., Dzakah E.E., Wang H., Tang S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021;296:198350. doi: 10.1016/j.virusres.2021.198350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flower T.G., Buffalo C.Z., Hooy R.M., Allaire M., Ren X.F., Hurley J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. P Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y.e. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lam J.-Y., Yuen C.-K., Ip J.D., Wong W.-M., To K.-W., Yuen K.-Y. Loss of orf3b in the circulating SARS-CoV-2 strains. Emerg Microbes Infect. 2020;9(1):2685–2696. doi: 10.1080/22221751.2020.1852892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Konno Y., Kimura I., Uriu K. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mu J., Fang Y., Yang Q.i., Shu T., Wang A.n., Huang M. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Narayanan K., Ramirez S.I., Lokugamage K.G., Makino S. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lokugamage K.G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S.I., Tseng C.-T. Middle East Respiratory Syndrome Coronavirus nsp1 Inhibits Host Gene Expression by Selectively Targeting mRNAs Transcribed in the Nucleus while Sparing mRNAs of Cytoplasmic Origin. J Virol. 2015;89(21):10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci USA. 2006;103(34):12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang K.e., Miorin L., Makio T., Dehghan I., Gao S., Xie Y. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci Adv. 2021;7(6):eabe7386. doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27(10):959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 87.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell. 2020;183(5):1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Min Y.Q., Mo Q., Wang J., Deng F., Wang H., Ning Y.J. SARS-CoV-2 nsp1: Bioinformatics, Potential Structural and Functional Features, and Implications for Drug/Vaccine Designs. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.587317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lapointe C.P., Grosely R., Johnson A.G., Wang J., Fernandez I.S., Puglisi J.D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2017715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA. Mol Cell. 2020;80(6):1055–1066.e6. doi: 10.1016/j.molcel.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., Hammann P. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2021;27(3):253–264. doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakuraba S., Qilin X., Kasahara K., Iwakiri J., Kono H. Modeling the SARS-CoV-2 nsp1–5’-UTR complex via extended ensemble simulations. bioRxiv. 2021 [Google Scholar]
- 93.Shi M., Wang L., Fontana P. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv. 2020 [Google Scholar]
- 94.Rao S., Hoskins I., Tonn T. Genes with 5' terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 NSP1 protein. RNA. 2021 doi: 10.1261/rna.078661.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 96.Zhao X.H., Chen H., Wang H.L. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.629873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reis C.A., Tauber R., Blanchard V. Glycosylation is a key in SARS-CoV-2 infection. J Mol Med. 2021 doi: 10.1007/s00109-021-02092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramírez Hernández E., Hernández-Zimbrón L.F., Martínez Zúñiga N., Leal-García J.J., Ignacio Hernández V., Ucharima-Corona L.E. The Role of the SARS-CoV-2 S-Protein Glycosylation in the Interaction of SARS-CoV-2/ACE2 and Immunological Responses. Viral Immunol. 2021;34(3):165–173. doi: 10.1089/vim.2020.0174. [DOI] [PubMed] [Google Scholar]
- 99.Yang Q., Hughes T.A., Kelkar A.J. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. eLife. 2020;9 doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci Rep. 2020;10:14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Central Sci. 2020;6(10):1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leroy H., Han M., Woottum M., Bracq L., Bouchet J., Xie M. Virus-Mediated Cell-Cell Fusion. Int J Mol Sci. 2020;21(24):9644. doi: 10.3390/ijms21249644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sobocinska J., Roszczenko-Jasinska P., Ciesielska A., Kwiatkowska K. Protein Palmitoylation and its Role in Bacterial and viral infections. Front Immunol. 2018;9 doi: 10.3389/fimmu.2017.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]