Abstract
Interleukin (IL)-19, a designated IL-20 subfamily cytokine, has been implicated in inflammatory disorders including rheumatoid arthritis, psoriasis and, lately, asthma. Here, through the analysis of transcriptomic datasets of lung tissue of large asthma cohorts, we report that IL-19 expression is upregulated in asthma and correlates with disease severity. The gene expression of IL-19 was significantly higher in lung tissue from patients with severe and mild/moderate asthma compared to healthy controls. IL-19 protein level, however, was significantly higher in the blood and saliva of patients with severe asthma compared to mild/moderate subgroups as measured by ELISA assay. IL-19 protein level was not affected by corticosteroid treatment in plasma. Our data provide insights into the potential use of IL-19 as a saliva marker for asthma severity and a potential therapeutic target.
Short abstract
Plasma and saliva IL-19 are significantly upregulated in asthma and correlate with disease severity. Plasma IL-19 is not affected by steroid treatment. This suggests that IL-19 can be used as a biomarker of asthma severity. https://bit.ly/2QfYwpc
Introduction
A network of mutually interacting cytokines regulates the airway inflammation and bronchoconstriction that underlies asthma. Type 2 cytokines like interleukin (IL)-4, IL-5 and IL-13 play a crucial role in this process. However, the exact functional role of non-type 2 proinflammatory cytokines in asthma pathogenesis is not fully understood.
IL-19, a non-type 2 cytokine, belongs to the IL-10 family, which includes IL-10, IL-19, IL-20, IL-24 and IL-26. It is mainly produced by immune cells, such as macrophages and B-cells [1]. However, structural cells, e.g. airway epithelial cells, also secrete IL-19 in response to stimulation with IL-17 and type 2 cytokines like IL-4 and IL-13 [2]. IL-19 signals through IL-20R1 and IL-20R2 heterodimeric receptors and primarily activates signal transducer and activator of transcription 3 (STAT3) [1, 3], which is essential for differentiation of T-helper cell (Th)-17 and Th-2 cells, the key driving cells of allergic inflammation [4]. IL-19 and its receptors are also expressed by both synovial fibroblasts [5] and keratinocytes [6]. Pathogenic roles have, in fact, been attributed to IL-19 in rheumatoid arthritis and psoriasis based on its ability to increase the proliferation of these cells and enhance the production of chemokines and pro-inflammatory cytokines [5, 7, 8].
Little is known about the biological function and cellular source of IL-19 in asthma patients. IL-19 was found to be upregulated in the blood of asthma patients compared to healthy controls [9]. Higher IL-19 expression was also reported in airway epithelial cells from asthma patients compared to those from healthy controls [2], which may indicate its pathogenic role in the airways [10]. Moreover, blocking IL-20R1 attenuated lung inflammation in mice [11], which further supports its role in respiratory disease [1, 12]. However, the expression of IL-19 in the lung tissues of asthma patients relative to disease severity has never been investigated.
Here, we have assessed the gene expression level of IL-19 in datasets of bronchial biopsies from the Australian Unbiased Biomarkers for Prediction of Respiratory Disease Outcomes (U-BIOPRED) Project, bronchial brushing samples from the Severe Asthma Research Program (SARP) and sputum and peripheral blood mononuclear cells (PBMCs) from asthma cohorts. Furthermore, the level of IL-19 was evaluated in the plasma and saliva of asthma patients with different levels of disease severity to investigate the relationship between IL-19 expression and asthma severity.
Methods
Gene expression datasets
Publicly available gene expression datasets of adult asthmatic lung tissue available from the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) and the European Bioinformatics Institute (www.ebi.ac.uk) were used. The datasets used in this study are summarised in supplementary table S1.
The GSE147881 dataset comprises endobronchial biopsy tissues from patients with mild/moderate (n=18) and severe (n=42) asthma and from healthy controls (n=13), collected from the U-BIOPRED Project and Australian Newcastle severe asthma cohorts. Column-based extraction of total RNA was performed followed by gene expression array profiling. Patients’ characteristics are summarised in supplementary table S2.
The GSE43696 dataset comprises bronchial brushings obtained via bronchoscope-directed epithelial brushings from patients with moderate (n=50) and severe (n=38) asthma and from healthy adult controls (n=20), collected from the SARP cohort [13, 14]. Fresh, isolated bronchial brushings comprising >90% epithelial cells were analysed by gene expression array profiling. Demographic data for the cohort are summarised in supplementary table S3.
The GSE132006 dataset comprises PBMCs from patients with mild allergic asthma (n=5), defined as having positive skin test results to common aeroallergens and a forced expiratory volume in 1 s (FEV1) <80% predicted, and five healthy controls (negative skin test and FEV1 >80% predicted). Demographic data for the cohort are summarised in supplementary table S4.
The GSE137268 dataset comprises induced sputum samples from patients with controlled (n=21) and uncontrolled (n=21) asthma. RNA was extracted from sputum samples using column-based total RNA extraction followed by gene expression array profiling. Demographic data for the cohort are summarised in supplementary table S5.
The GSE83233 dataset comprises bronchial biopsy tissues from healthy male, non-smoker, non-allergic volunteers (age range 18–50 years) with normal lung function. Participants were randomised to receive either inhaled budesonide (1600 µg) or placebo. Bronchial biopsy samples were obtained via bronchoscopy 6 h after the placebo or budesonide inhalation.
The GSE50012 dataset comprises corticosteroid-treated PBMCs from healthy volunteers [15, 16]. The total RNA was obtained from aliquots of PBMCs treated with dexamethasone (500 nM) for 24 h.
Cell culture
Primary bronchial fibroblasts were isolated from endobronchial tissue biopsies obtained from three non-smoking healthy donors (average age 43.7±12.5 years) and three patients with severe asthma (average age 43.4±8.3 years), as described previously [8, 9]. These primary bronchial fibroblasts were obtained from the biobank at the Quebec Respiratory Health Research Network. The original study was approved by the institutional review board (MUHC REB number BMB-02-039-t) and the subjects had provided written informed consent. The cells were cultured at confluence in six-well flat-bottom plates in complete media (DMEM (Sigma) supplemented with 10% FBS, 2 mM L-glutamine, 100 units·mL−1 penicillin and 100 ng·mL−1 streptomycin). The cells were then treated, or not, with 100 ng·mL−1 recombinant human IL-19 (9286-IL-19, R&D Systems) for 8 h. Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer's instructions. 5x HOT FIREPol EvaGreen qPCR SuperMix (Solis Biodyne) was used for cDNA amplification and reverse-transcriptase (RT) quantitative PCR (qPCR) was performed in QuantStudio 3 Real-Time PCR System (Applied Biosystems).
ELISA
Plasma IL-19, IL-6 and IL-17 cytokine concentrations in asthma patients (n=30) and healthy controls (n=6) were determined using commercially available human ELISA kits (IL-6, ab46042, Abcam; IL-19, ab231922, Abcam; IL-17, DY317-05, R&D Systems). Plasma used in our study was obtained from asthma patients recruited from Rashid Hospital, Dubai, United Arab Emirates. The patient characteristics are summarised in supplementary table S6. The study was approved by the Ethics Committee of Dubai Health Authority and Dubai Scientific Research Ethics Committee (DSREC-03/2019_10) and the subjects had provided written informed consent. Assays were performed strictly following the manufacturer's instructions. All samples were measured in duplicate.
Analysis procedures
The microarray data (CEL files) were normalised with the robust multi-array average procedure using the Bioconductor affy package [17–19]. The fold change of differentially expressed genes was calculated using the Bioconductor Limma package [20, 21], and genes with an adjusted p-value (false discovery rate) <0.05 were considered significantly differentially expressed.
Results of the gene expression analysis of the IL-19 heterodimeric receptors IL-20R1 and IL-20R2 in fibroblasts of patients with severe asthma treated, or not, with IL-19 are expressed as fold change relative to untreated bronchial fibroblasts. The following primers were used: human IL-20R1, forward 5′–3′: TGGGCTGAAAGTGGACGGTTCT and reverse 5′–3′: CCACTTCTCTGGAGCTGTCAGG; human IL-20R2, forward 5′–3′: ACAACCTTCGTGTCAGGGCCAC and reverse 5′–3′: TGATCTCCATCCCAGGTCGGGT; human 18s, forward 5′–3′: TGACTCAACACGGGAAACC and reverse 5′–3′: TCGCTCCACCAACTAAGAAC. Gene expression was analysed using the comparative Ct (ΔΔCt) method after normalisation to the housekeeping gene 18s rRNA. Analyses were performed using R software v 3.0.2 (R Foundation for Statistical Computing), SPSS Version 26 (IBM Corporation) and Graphpad Prism 8 (GraphPad Software Inc.). All t-tests were two-tailed and considered significant if the nominal p-value was <0.05.
Results
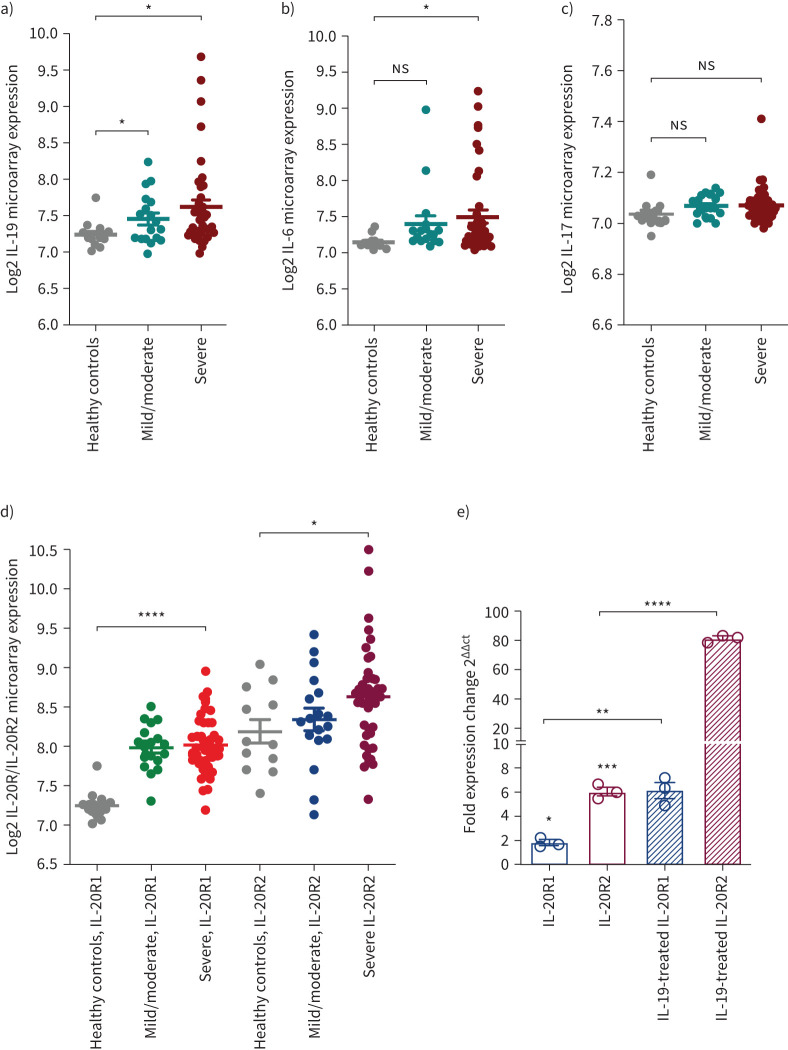
In this study we used gene expression datasets of bronchial biopsies of the Australian U-BIOPRED cohort consisting of 42 patients with severe asthma, 18 patients with mild/moderate asthma and 13 healthy subjects. This dataset was used to analyse the gene expression level of IL-19, which is known to correlate with asthma severity, along with other asthma-related cytokines such as IL-6 and IL-17 [22, 23]. A significant increase in the gene expression level of IL-19 was observed in the bronchial biopsies of patients with severe and mild/moderate asthma compared to healthy subjects (figure 1a). Although the expression levels of IL-6 and IL-17 cytokines were both elevated in patients with severe and mild/moderate asthma, the increase was not deemed statistically significant (figure 1b, c).
FIGURE 1.
Gene expression of interleukin (IL)-19, IL-6 and IL-17 in bronchial biopsies from patients with asthma of different severity categories. The data are for bronchial biopsies from patients with severe asthma (n=42) and mild/moderate asthma (n=18) and healthy controls (n=13) from the U-BIOPRED Project and Australian Newcastle severe asthma cohorts (GSE147881). a) IL-19 gene expression was significantly increased in the lungs of patients with severe and mild/moderate asthma compared to healthy controls. b) IL-6 gene expression was significantly increased in the lungs of patients with severe asthma compared to healthy controls. c) There was a trend towards increased IL-17 gene expression in the lungs of patients with severe and mild/moderate asthma compared to healthy controls. d) IL-20R1 and IL-20R2 gene expression increased in the lungs of patients with severe and mild/moderate asthma compared to healthy controls. e) IL-20R1 and IL-20R2 gene expression increased in severe asthmatic bronchial fibroblasts compared to healthy bronchial fibroblasts. Treatment of these cells with 100 ng·mL−1 IL-19 increased IL-20R1 and IL-20R2 gene expression levels. Fold change presented relative to healthy controls. Two-way comparison was done using two-tailed t-test. ns: nonsignificant. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001.
IL-19 induces cellular responses via the IL-20R1/IL-20R2 receptor heterodimer and primarily activates STAT3 [3, 24]. When IL-20R1-deficient mice were challenged with house dust mite to induce airway hyperresponsiveness, IL-19 signalling and airway inflammation were diminished [11]. However, in a report that investigated the crystal structure of the 20R1/IL-20R2 receptor heterodimer, the affinity of IL-19 to IL-20R2 was shown to be higher than its affinity to IL-20R1 [25]. Thus, to better understand the expression level of these receptors in lung tissues of asthma patients, we evaluated the gene expression of IL-20R1 and IL-20R2 receptors in the bronchial biopsies of Australian U-BIOPRED participants. The expression of both of these receptors was elevated in asthmatic lung tissues compared to healthy controls (figure 1d). However, the increase in IL-20R2 lung expression was more pronounced than that of IL-20R1 in the lung tissues of healthy controls (close to 1-fold change increase in the mean expression levels of IL-20R1 (7.24) versus IL-20R2 (8.18); p<0.0001) as well as in the lung tissues of patients with mild/moderate (0.5-fold change increase in the mean expression levels of IL-20R1 (7.98) versus IL-20R2 (8.34); p=0.028) and severe (0.5-fold change increase in the mean expression levels of IL-20R1 (8.01) versus IL-20R2 (8.62); p<0.0001) asthma (figure 1d). Given the reported higher affinity of IL-19 to IL-20R2 [25], our finding suggests that IL-19 may exert its function more through the IL-20R2 receptor in asthma. To validate this observation, we used RT-PCR to assess the expression level of these two receptors in fibroblasts from patients with severe asthma. Similar to the Omics results, the IL-20R2 gene was expressed more highly than the IL-20R1 gene in severe asthma (figure 1e). In addition, upon treatment with recombinant IL-19, the gene expression level of both receptors was significantly increased. IL-19 induced a 3-fold increase in IL-20R1 expression, and over a 10-fold increase in IL-20R2 expression (figure 1e). Therefore, IL-19 and its receptors are upregulated in lung airways of asthma patients. Its role in asthma pathogenesis, however, requires further investigation. Proper understanding of the preferential signalling of IL-19 through IL-20R1 and/or IL-20R2 receptors may provide insight into the design of efficient inhibitors of IL-19 signalling.
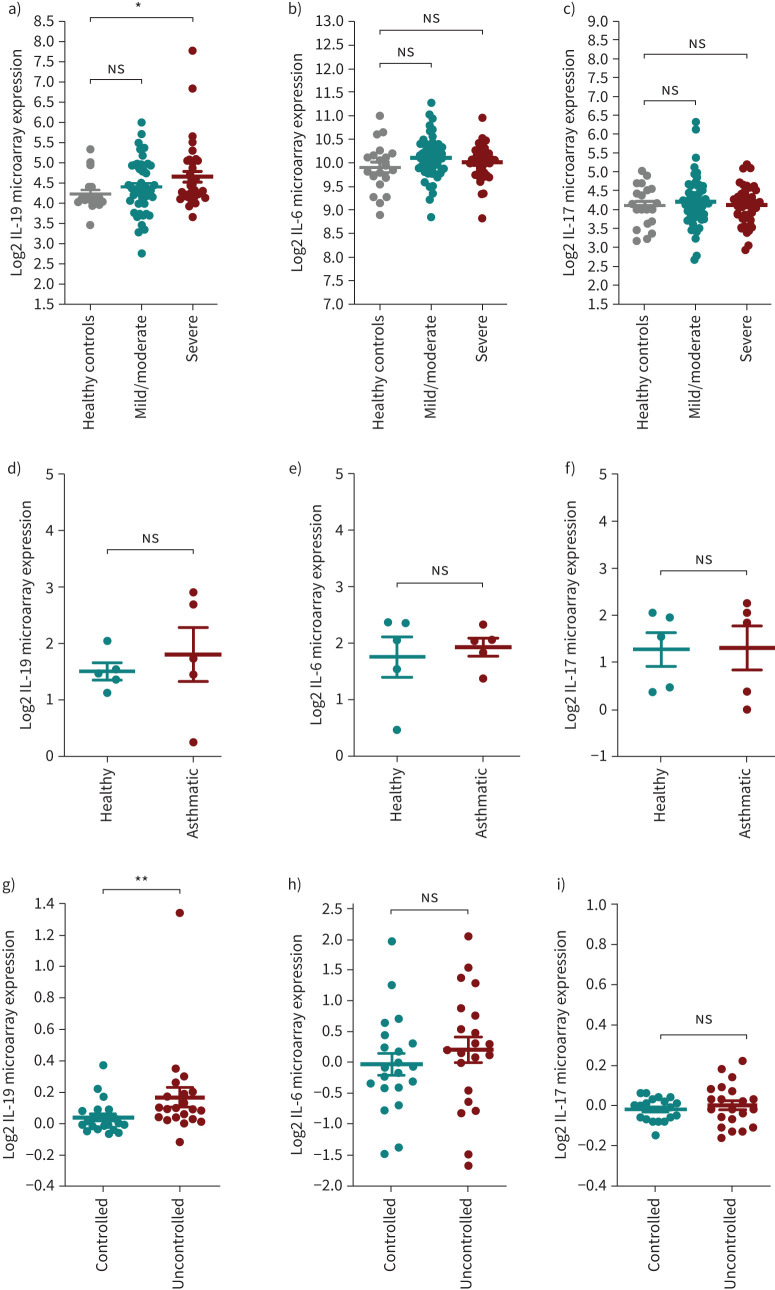
In addition, using datasets of bronchial brushings from the SARP cohort we observed that the IL-19 expression level was significantly increased in samples from patients with severe, but not mild/moderate, asthma compared to healthy subjects, although there was a trend for an increase in the mild/moderate group (figure 2a). There was no distinct pattern for IL-6 (figure 2b) and IL-17 (figure 2c) expression levels in asthmatic and healthy control subjects. Moreover, in PBMCs, no difference was observed in the levels of IL-6 (figure 2e) and IL-17 (figure 2f) between healthy and asthmatic subjects. There was an increase in IL-19 expression, but not to a significant level (figure 2d).
FIGURE 2.
Gene expression of interleukin (IL)-19, IL-6 and IL-17 in bronchial brushing, blood and sputum samples from patients with asthma of different severity categories. a–c) The data are for bronchial brushing from patients with severe asthma (n=38) and mild/moderate asthma (n=50) and healthy controls (n=20) from the SARP cohorts (GSE43696). a) IL-19 gene expression is significantly increased in the bronchial brushings from patients with severe asthma compared to healthy controls, and there is a trend towards increased IL-19 expression in bronchial brushings from patients with mild/moderate asthma compared to healthy controls. b) There was a trend towards increased IL-6 gene expression in bronchial brushing from patients with severe and mild/moderate asthma compared to healthy controls. c) No distinct pattern was found in IL-17 gene expression in bronchial brushings from patients with severe and mild/moderate asthma compared to healthy controls. d–f) The data are for peripheral blood mononuclear cells (PBMCs) (GSE132006) from patients with allergic asthma (n=5) and healthy controls (n=5). d) There was a trend towards increased IL-19 gene expression in PBMCs from patients with asthma compared to healthy controls. e) No distinct pattern was found in IL-6 gene expression in PBMCs from patients with asthma compared to healthy controls. f) No distinct pattern was found in IL-17 gene expression in PBMCs from patients with asthma compared to healthy controls. g–i) The data are for sputum (GSE137268) from patients with controlled (n=21) and uncontrolled (n=21) asthma. g) IL-19 gene expression was slightly increased in the sputum of patients with uncontrolled asthma compared those with controlled asthma. h) There was a trend towards increased IL-6 expression in the sputum of patients with uncontrolled asthma compared to those with controlled asthma. i) No distinct pattern was found for IL-17 expression in the sputum of patients with uncontrolled asthma compared to those with controlled asthma. Two-way comparison was done using two-tailed t-tests (GSE43696). Limma adjusted p<0.05 (GSE132006 and GSE137268). ns: nonsignificant. *: p<0.05; **: p<0.01.
We also investigated IL-19 expression levels in the sputum of asthmatic subjects. This dataset determined the differentially expressed genes in the sputum of patients with controlled and uncontrolled asthma (GSE137268). The level of IL-19 expression was slightly higher in the sputum of patients with uncontrolled asthma compared to controlled asthma (figure 2g). Similarly, IL-6 expression was higher in the sputum of patients with uncontrolled asthma, but not to a significant level (figure 2h). No difference in IL-17 levels was observed between patients with uncontrolled and controlled asthma (figure 2i).
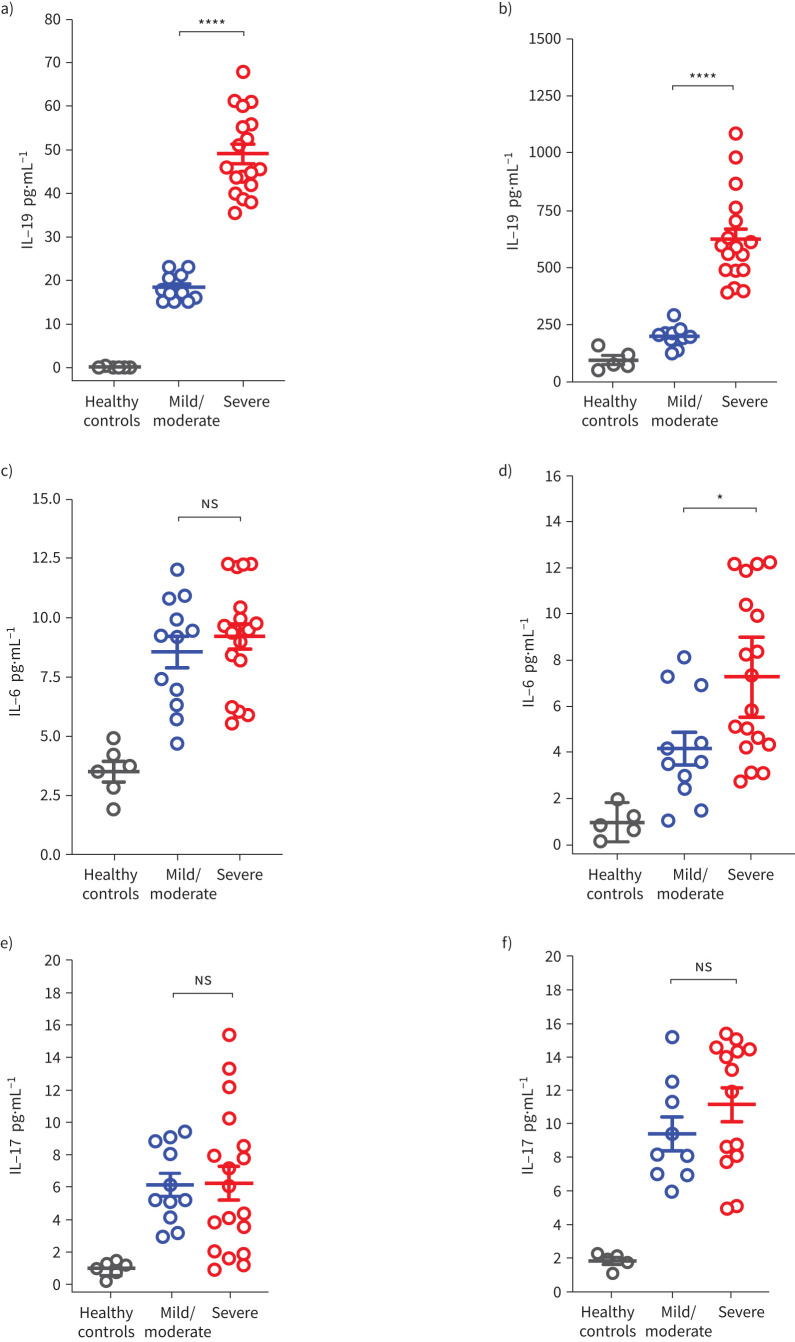
Using an ELISA assay, we evaluated the level of IL-19 in the plasma and saliva of asthmatic patients. IL-19 plasma (figure 3a) and saliva (figure 3b) protein levels were significantly increased in patients with severe asthma compared to patients with mild/moderate asthma and healthy controls. IL-6 levels in the saliva were significantly increased in patients with severe asthma compared to mild/moderate asthma and healthy controls (figure 3d). In the plasma, IL-6 level followed a similar trend; however, the increase in patients with severe asthma compared to mild/moderate asthma was not deemed statistically significant (figure 3c). No increase in IL-17 plasma level was observed (figure 3e). Although the level of IL-17 appeared higher in the saliva of patients with severe asthma, the increase was not statistically significant (figure 3f). The observed IL-19 upregulation in the plasma and saliva samples of patients with severe asthma in addition to the Omics results suggest that IL-19 could potentially be used as a biomarker for asthma severity; this, however, requires further investigation.
FIGURE 3.
Plasma and saliva interleukin (IL)-19, IL-6 and IL-17 levels in patients with asthma of different severity categories. The data are for samples from patients with severe asthma (n=18) and mild/moderate asthma (n=12) and healthy controls (n=6). a, b) IL-19 level was significantly increased in plasma (a) and saliva (b) of patients with severe asthma compared to mild/moderate asthma; the increase in IL-19 level was more pronounced in saliva than in plasma level. c, d) There was a trend towards increased plasma IL-6 level (c) in patients with severe asthma compared to mild/moderate asthma, and a significant increase in saliva IL-6 level (d) in patients with severe asthma compared to mild/moderate asthma. e, f) No distinct pattern was found in plasma IL-17 levels (e) in patients with severe asthma compared to mild/moderate asthma, but there was a trend towards increased saliva IL-17 levels (f) in patients with severe asthma compared to mild/moderate asthma. Two-way comparison was done using two-tailed t-test. ns: nonsignificant. *: p<0.05; ****: p<0.0001.
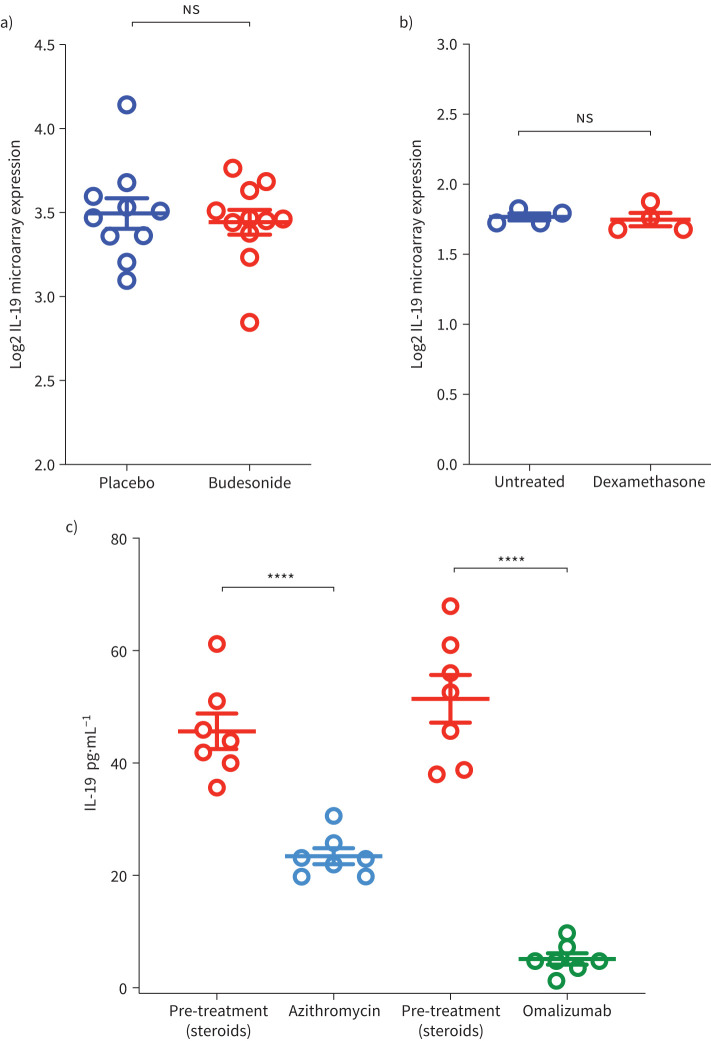
The plasma level of IL-6, a systemic asthma severity biomarker [26], could be affected by patients’ comorbidities as well as corticosteroid treatment. Here, we determined whether corticosteroid treatment affects IL-19 level. To test this hypothesis, we examined the expression level of IL-19 in relation to steroid treatment in two publicly available datasets: one for bronchial biopsies isolated from healthy subjects treated or not with inhaled corticosteroids, and one for PBMCs treated or not with dexamethasone. Treatment with corticosteroids did not affect IL-19 level in bronchial biopsies (figure 4a) or in PBMCs (figure 4b). However, in our cohort, treatment with omalizumab and with azithromycin suppressed IL-19 plasma level in patients with severe asthma (figure 4c).
FIGURE 4.
Plasma interleukin (IL)-19 levels in asthma patients on different treatment regimens. a) Gene expression level of IL-19 in bronchial biopsies from healthy, non-smoker, non-allergic male volunteers treated with placebo or inhaled budesonide (1600 μg). The data show no difference in IL-19 expression levels between the two groups. b) Gene expression level of IL-19 in the peripheral blood mononuclear cells (PBMCs) untreated or treated with dexamethasone (500 nM) for 24 h. The data show no difference in IL-19 expression level between the treated and untreated PBMCs. c) Azithromycin and omalizumab decreases IL-19 plasma level in severe asthma. The data show that treatment with these medications significantly decreased IL-19 plasma level in patients with severe asthma. Two-way comparison was done using two-tailed t-tests. ns: nonsignificant. ****: p<0.0001.
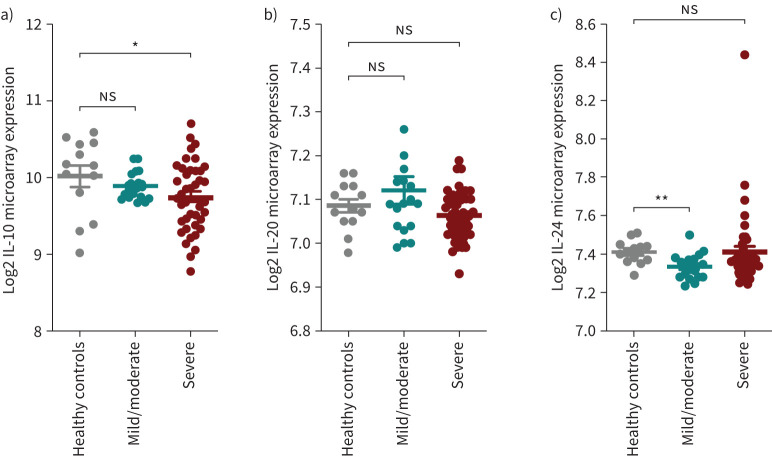
Because other IL-10 family cytokines, i.e. IL-10, IL-20 and IL-24, are in close proximity to the IL-19 gene on human chromosome 1q32, we further investigated their expression in the different types of samples from asthma patients and found no distinguishable pattern for their expression levels (supplementary figure S1). In fact, IL-10 expression level was significantly lower in the lung tissues of patients with severe asthma as compared to healthy controls (figure 5).
FIGURE 5.
Gene expression of interleukin (IL)-10, IL-20 and IL-24 in bronchial biopsies from patients with asthma of different severity categories. The data are from U-BIOPRED Project and Australian Newcastle severe asthma cohorts (GSE147881). a) IL-10 gene expression was significantly decreased in the lungs of patients with severe asthma compared to healthy controls. b) There was a trend towards increased lung IL-20 expression in patients with mild/moderate asthma compared to healthy controls. c) IL-24 gene expression was significantly decreased in the lungs of patients with mild/moderate asthma compared to healthy controls. Two-way comparison was done using two-tailed t-tests. ns: nonsignificant. *: p<0.05; **: p<0.01.
Discussion
In the era of personalised medicine, to deliver proper management for asthma it is important to be able to phenotype asthma and to define biomarkers that are able to predict the severity of disease and assess the response to treatment. Plasma IL-6 level has previously been associated with asthma severity and metabolic dysfunction [26]. However, there was a poor correlation between plasma and sputum IL-6 levels in the same cohort, indicating that IL-6 levels could not be used as a biomarker of severity in asthmatic sputum or saliva samples. In this study, IL-19 lung expression as well as its protein plasma and saliva levels were significantly increased in patients with asthma and correlated with disease severity. Its level was not affected by steroid treatment. This suggests that IL-19 can be used as a biomarker of asthma severity. However, further validation in larger asthma cohorts is needed.
Little is known about the cellular source of IL-19 in asthmatic patients. IL-19 was found to be upregulated in the blood of patients with asthma compared to healthy controls, and was highly expressed in the airway epithelia of patients with asthma compared to healthy controls [2]. Here, we have highlighted that, in asthmatic patients, besides airway epithelial cells, fibroblasts are also a cellular source of IL-19. Moreover, the expression of IL-20R1 and IL-20R2 was upregulated in lung fibroblasts of asthmatic patients as well as in response to IL-19 treatment.
In the current study, compared to IL-6 and IL-17 cytokines, IL-19 saliva level was highest in patients with severe asthma. This elevated IL-19 level could be related to the increased IL-17 level in airways of patients with severe asthma. The level of IL-17 in lung tissue is known to correlate with asthma severity [23]. IL-17 has also been shown to enhance IL-19 expression in airway epithelial cells [2]. Therefore, the elevated level of this cytokine in severe asthma could explain why IL-19 levels are highest in saliva from patients with severe asthma.
In summary, among the recruited patients, IL-19 protein levels were significantly higher in the plasma and saliva of patients with severe asthma compared with mild/moderate subgroups, and the increase in IL-19 level was more pronounced in saliva samples than in plasma samples. IL-19 protein level was not affected by corticosteroid treatment in plasma. Our data provide insights into the possible use of IL-19 as a saliva marker for asthma severity and as a potential therapeutic target.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary tables 00984-2020.SUPPLEMENT_TABLES (55.1KB, pdf)
Supplementary figures 00984-2020.SUPPLEMENT_FIGURES (164.2KB, pdf)
Footnotes
This article has supplementary material available from openres.ersjournals.com
Support statement: This work was supported by a Tissue Injury and Repair Group operational grant (code 150317), COVID-19 research grants (CoV19-0307) and a seed grant (code 2001090275); by a collaborative research grant (code 2001090278) to R. Halwani, University of Sharjah, UAE; by a Sandooq Al Watan Applied Research and Development Grant to R. Halwani (SWARD-S20-007); by an Al Jalila Foundation Seed Grant (AJF202019); and by the Prince Abdullah Ben Khalid Celiac Disease Research Chair, under the Vice Deanship of Research Chairs, King Saud University, Riyadh, Kingdom of Saudi Arabia. Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of interest: F. Saheb Sharif-Askari has nothing to disclose.
Conflict of interest: N. Saheb Sharif-Askari has nothing to disclose.
Conflict of interest: S. Goel has nothing to disclose.
Conflict of interest: B. Mahboub has nothing to disclose.
Conflict of interest: A.W. Ansari has nothing to disclose.
Conflict of interest: M-H. Temsah has nothing to disclose.
Conflict of interest: A.M. Zakri has nothing to disclose.
Conflict of interest: E. Ratemi has nothing to disclose.
Conflict of interest: R. Hamoudi has nothing to disclose.
Conflict of interest: Q. Hamid has nothing to disclose.
Conflict of interest: R. Halwani has nothing to disclose.
References
- 1.Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines—from host defence to tissue homeostasis. Nat Rev Immunol 2014; 14: 783–795. doi: 10.1038/nri3766 [DOI] [PubMed] [Google Scholar]
- 2.Huang F, Wachi S, Thai P, et al. . Potentiation of IL-19 expression in airway epithelia by IL-17A and IL-4/IL-13: important implications in asthma. J Allergy Clin Immunol 2008; 121: 1415–1421. doi: 10.1016/j.jaci.2008.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumoutier L, Leemans C, Lejeune D, et al. . Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J Immunol 2001; 167: 3545–3549. doi: 10.4049/jimmunol.167.7.3545 [DOI] [PubMed] [Google Scholar]
- 4.Gavino AC, Nahmod K, Bharadwaj U, et al. . STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy 2016; 71: 1684–1692. doi: 10.1111/all.12937 [DOI] [PubMed] [Google Scholar]
- 5.Sakurai N, Kuroiwa T, Ikeuchi H, et al. . Expression of IL-19 and its receptors in RA: potential role for synovial hyperplasia formation. Rheumatology 2008; 47: 815–820. doi: 10.1093/rheumatology/ken061 [DOI] [PubMed] [Google Scholar]
- 6.Kunz S, Wolk K, Witte E, et al. . Interleukin (IL)-19, IL-20 and IL-24 are produced by and act on keratinocytes and are distinct from classical ILs. Exp Dermatol 2006; 15: 991–1004. doi: 10.1111/j.1600-0625.2006.00516.x [DOI] [PubMed] [Google Scholar]
- 7.Witte E, Kokolakis G, Witte K, et al. . IL-19 is a component of the pathogenetic IL-23/IL-17 cascade in psoriasis. J Investig Dermatol 2014; 134: 2757–2767. doi: 10.1038/jid.2014.308 [DOI] [PubMed] [Google Scholar]
- 8.Li HH, Lin YC, Chen PJ, et al. . Interleukin-19 upregulates keratinocyte growth factor and is associated with psoriasis. Br J Dermatol 2005; 153: 591–595. doi: 10.1111/j.1365-2133.2005.06665.x [DOI] [PubMed] [Google Scholar]
- 9.Liao S-C, Cheng Y-C, Wang Y-C, et al. . IL-19 induced Th2 cytokines and was up-regulated in asthma patients. J Immunol 2004; 173: 6712–6718. doi: 10.4049/jimmunol.173.11.6712 [DOI] [PubMed] [Google Scholar]
- 10.Kawaguchi M, Kokubu F, Kuga H, et al. . Modulation of bronchial epithelial cells by IL-17. J Allergy Clin Immunol 2001; 108: 804–809. doi: 10.1067/mai.2001.119027 [DOI] [PubMed] [Google Scholar]
- 11.Weng Y-H, Chen W-Y, Lin Y-L, et al. . Blocking IL-19 signaling ameliorates allergen-induced airway inflammation. Front Immunol 2019; 10: 968. doi: 10.3389/fimmu.2019.00968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blumberg H, Conklin D, Xu W, et al. . Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell 2001; 104: 9–19. doi: 10.1016/S0092-8674(01)00187-8 [DOI] [PubMed] [Google Scholar]
- 13.Voraphani N, Gladwin MT, Contreras AU, et al. . An airway epithelial iNOS–DUOX2–thyroid peroxidase metabolome drives Th1/Th2 nitrative stress in human severe asthma. Mucosal Immunol 2014; 7: 1175–1185. doi: 10.1038/mi.2014.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Hawkins GA, Moore WC, et al. . Expression of asthma susceptibility genes in bronchial epithelial cells and bronchial alveolar lavage in the Severe Asthma Research Program (SARP) cohort. J Asthma 2016; 53: 775–782. doi: 10.3109/02770903.2016.1158268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maranville JC, Baxter SS, Torres JM, et al. . Inter-ethnic differences in lymphocyte sensitivity to glucocorticoids reflect variation in transcriptional response. Pharmacogenomics J 2013; 13: 121–129. doi: 10.1038/tpj.2011.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kupfer SS, Maranville JC, Baxter SS, et al. . Comparison of cellular and transcriptional responses to 1, 25–dihydroxyvitamin d3 and glucocorticoids in peripheral blood mononuclear cells. PLoS One 2013; 8: e76643. doi: 10.1371/journal.pone.0076643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughey JJ, Butte AJ. Robust meta-analysis of gene expression using the elastic net. Nucleic Acids Res 2015; 43: e79. doi: 10.1093/nar/gkv229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier L, Cope L, Bolstad BM, et al. . affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307–315. doi: 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- 19.RMA codes. https://github.com/Bioma-85/Asthma-biomarkers/blob/main/RMA.txt Date last accessed: March 2021.
- 20.Dudoit S, Yang YH, Callow MJ, et al. . Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin 2002; 12: 111–139. [Google Scholar]
- 21.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: 1–25. [DOI] [PubMed] [Google Scholar]
- 22.Ilmarinen P, Tuomisto LE, Niemelä O, et al. . Comorbidities and elevated IL-6 associate with negative outcome in adult-onset asthma. Eur Respir J 2016; 48: 1052–1062. doi: 10.1183/13993003.02198-2015 [DOI] [PubMed] [Google Scholar]
- 23.Al-Ramli W, Préfontaine D, Chouiali F, et al. . TH17-associated cytokines (IL-17A and IL-17F) in severe asthma. J Allergy Clin Immunol 2009; 123: 1185–1187. doi: 10.1016/j.jaci.2009.02.024 [DOI] [PubMed] [Google Scholar]
- 24.Ouyang W, Rutz S, Crellin NK, et al. . Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 2011; 29: 71–109. doi: 10.1146/annurev-immunol-031210-101312 [DOI] [PubMed] [Google Scholar]
- 25.Logsdon NJ, Deshpande A, Harris BD, et al. . Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines. Proc Natl Acad Sci USA 2012; 109: 12704–12709. doi: 10.1073/pnas.1117551109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters MC, McGrath KW, Hawkins GA, et al. . Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: a cross-sectional analysis of two cohorts. Lancet Respir Med 2016; 4: 574–584. doi: 10.1016/S2213-2600(16)30048-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary tables 00984-2020.SUPPLEMENT_TABLES (55.1KB, pdf)
Supplementary figures 00984-2020.SUPPLEMENT_FIGURES (164.2KB, pdf)