Abstract
Breast cancer is one of the most common malignant tumors in women. Although a number of homeobox (HOX) genes are known to serve an important role in breast cancer, the role of HOXD8 in breast cancer remains unclear. The aim of the present study was to investigate the role of HOXD8 in the physiological behaviors of breast cancer cells. The Gene Expression Profiling Interactive Analysis database was used to analyze the expression of HOXD8 in patients with breast cancer and in healthy subjects. Western blotting was performed to determine the expression levels of HOXD8 in several breast cancer cell lines; subsequently, HOXD8 expression was knocked down and overexpressed in MCF-7 cells. Cell Counting Kit-8, colony formation, wound healing and Transwell assays were used to evaluate the effects of HOXD8 on breast cancer cell viability, proliferation, migration and invasion, respectively. Chromatin immunoprecipitation and dual-luciferase reporter assays were conducted to identify the binding sites between HOXD8 and inhibitor of apoptosis-like protein-2 (ILP2). In addition, ILP2 expression levels were knocked down in MCF-7 cells. The results demonstrated that the expression levels of HOXD8 were significantly downregulated in breast cancer tissues and cell lines, and that the overexpression of HOXD8 inhibited the proliferation, invasion and migration of cancer cells. HOXD8 was shown to bind to the ILP2 promoter to regulate the expression of ILP2. Furthermore, ILP2 knockdown reversed the effects of HOXD8 knockdown on breast cancer cell proliferation, invasion and migration. In conclusion, the findings of the present study suggested that HOXD8 may inhibit the proliferation, migration and invasion of breast cancer cells by downregulating ILP2 expression.
Keywords: homeobox D8, inhibitor of apoptosis-like protein-2, breast cancer cells, proliferation, migration
Introduction
Breast cancer is the most commonly diagnosed type of cancer and the leading cause of cancer-related mortality in women worldwide (1). Breast cancer has been estimated to account for 24.2% of all new cancer diagnoses and 15.0% of all cancer-related deaths among women (1). Although the overall survival and prognosis of patients with breast cancer has significantly improved in recent years, metastasis remains the leading cause of mortality in patients with breast cancer (2). For example, patients with metastases have a 5-year survival rate of only 26% compared with a 5-year survival rate of 90% across all patients with breast cancer (3). Hence, it is important to further understand the mechanisms underlying the development and progression of breast cancer to identify novel targets for the treatment of this disease.
Baculoviral IAP repeat containing 8 (ILP2) is an inhibitor belonging to the inhibitor of apoptosis protein family (4), which can prevent pro-apoptotic stimulation and inhibit apoptosis. A previous study revealed that the expression levels of inhibitor of apoptosis-like protein-2 (ILP2) were upregulated in patients with breast cancer (5). In addition, the viability and migratory ability of human breast cancer cell lines (HCC-1937, MX-1 and MCF-7) were significantly inhibited following knockdown of ILP2 expression, and apoptosis was increased, compared with those in the control group (5). Collectively, these findings indicated that ILP2 may promote the proliferation, migration and invasion of breast cancer cells.
Transcription factors are proteins that bind to the DNA helix at specific regulatory sequences to activate or inhibit transcription through a transactivation or transrepression domain (6). Determining the activity of transcription factors is crucial for understanding the regulation of gene expression (7). A number of studies have reported that transcription factors serve an important role in the occurrence and development of numerous types of tumors. Therefore, the targeting of transcription factors may represent a novel method for tumor treatment (6,8). Homeobox (HOX) genes function as master regulatory transcription factors, and their expression levels have been found to be regularly altered in cancer (9). The expression levels of HOX genes were reported to be upregulated or downregulated in different tumor types, depending on the specific HOX gene involved and the type of cancer being investigated. HOXD8 is an important member of the HOX gene family, and was found to serve an important role in colorectal cancer, non-small cell lung cancer, advanced epithelial ovarian cancer, lung cancer and hepatocellular carcinoma (10-14). Therefore, the present study was undertaken to investigate the role of HOXD8 in the occurrence and development of breast cancer.
Bioinformatics software was used to examine the expression of HOXD8 in breast cancer tissues and predict its downstream genes. The aim of the present study was to examine the association between HOXD8 and its downstream genes, and to explore their roles and mechanism of action in breast cancer cells, in the hope that the results may uncover novel targets for the treatment of breast cancer.
Materials and methods
Cell lines and culture
Breast cancer cell lines (MCF-7, MDA-MB-231, SUM190PT and SK-BR-3) and a normal human breast epithelial cell line (MCF-10A) were purchased from Procell Life Science & Technology Co., Ltd. MCF-7 and MDA-MB-231 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.); SUM190PT cells were cultured in Ham's F-12 medium (Sigma-Aldrich; Merck KGaA); SK-BR-3 cells were cultured in McCoy's 5A medium (Sigma-Aldrich; Merck KGaA); and MCF-10A cells were cultured in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA) (15). All media aforementioned were all supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin/streptomycin (Thermo Fisher Scientific, Inc.). All cells were maintained in an incubator containing 5% CO2 at 37˚C.
Cell transfection
Gene overexpression was performed with a pcDNA3.1 vector and gene knockdown with a pLVX-shRNA2 lentiviral vector. The full length coding sequence of HOXD8 (accession no. AH010089.2; https://www.ncbi.nlm.nih.gov/nuccore/AH010089.2/) was found from National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). Fragment with restriction sites was synthesized and inserted into the pcDNA3.1 vector (pcDNA-HOXD8) by Hunan Fenghui Biotechnology Co., Ltd., whereas the empty vector was the negative control (pcDNA-NC). Two types of short hairpin (sh)RNA-HOXD8 (shRNA-HOXD8-1 and shRNA-HOXD8-2), two types of shRNA-ILP2 (shRNA-ILP2-1 and shRNA-ILP2-2) and non-targeted shRNA as negative control (shRNA-NC) were purchased from Guangzhou RiboBio Co., Ltd. and transfected into MCF-7 cells. shRNA-1 and shRNA-2 correspond to different sequences being incorporated into the same vector. Briefly, cells (5x105 cells/well) were plated into six-well plates and cultured until they reached 50-70% confluence. Cells were transfected with 10 nM pcDNAs and shRNAs using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C, according to the manufacturer's protocol. Following 48 h of transfection, the expression levels of HOXD8 and ILP2 were analyzed, and the cells were used for subsequent experiments. The sequences of shRNA are as follows: shRNA-HOXD8-1 5'-CCGGAGCCGAAGGCCTGACAAATTACTCGAGTAATTTGTCAGGCCTTCGGCTTTTTTG-3'; shRNA-HOXD8-2 5'-CCGGGCCGAAGGCCTGACAAATTAACTCGAGTTAATTTGTCAGGCCTTCGGCTTTTTG-3'; shRNA-ILP2-1 5'-CCGGACGGTGGACAAGTCCTATATTCTCGAGAATATAGGACTTGTCCACCGTTTTTTG-3'; shRNA-ILP2-2 5'-CCGGTTTGGGCCACAACGTTAATATCTCGAGATATTAACGTTGTGGCCCAAATTTTTG-3'; shRNA-NC 5'-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG-3'.
Western blotting
All cells (5x105 cells/well) were seeded into a six-well plate separately and total protein was extracted from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology). Total protein concentration was quantified using the BCA method and 20 µg protein/lane was separated by 10% SDS-PAGE. The separated proteins were subsequently transferred onto PVDF membranes and blocked with 5% skimmed milk diluted in 5% BSA (Beijing Solarbio Science & Technology Co., Ltd.) for 2 h at room temperature. The membranes were then incubated with the following primary antibodies at a dilution of 1:1,000 overnight at 4˚C: Anti-HOXD8 (cat. no. sc-515357; Santa Cruz Biotechnology, Inc.), anti-MMP2 (cat. no. 40994; Cell Signaling Technology, Inc.), anti-MMP9 (cat. no. 13667; Cell Signaling Technology, Inc.), anti-ILP2 (cat. no. ab9664; Abcam) and anti-GAPDH (cat. no. 5174, Cell Signaling Technology, Inc.). Following primary antibody incubation, the membranes were incubated with an HRP-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibody (1:100,000; cat. nos. G-21234 and G-21040, respectively; Invitrogen; Thermo Fisher Scientific, Inc.) for 1.5 h at room temperature. GAPDH served as the internal reference control. Protein bands were visualized using an ECL kit (cat. no. 21342; Beyotime Institute of Biotechnology) and densitometric analysis was performed using ImageJ v1.51 software (National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
MCF-7 cells were seeded into 96-well plates at the density of 5x103 cells/well and cultured in an incubator containing 5% CO2 at 37˚C for 24, 48 or 72 h. In total, 10 µl CCK-8 solution (cat. no. ab228554; Abcam) was added to each well at each time point. Following incubation for another 2 h at 37˚C, the optical density was measured at a wavelength of 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation assay
MCF-7 cells (3x103 cells/well) were seeded into six-well plates and incubated with DMEM supplemented with 10% FBS at 37˚C. Following 14 days of incubation, the cells were fixed with 4% methanol for 15 min at room temperature and stained with 0.1% crystal violet dye solution for another 10 min at room temperature. The number of cell clone clusters containing >50 cells was counted under a light microscope (magnification, x100; Olympus Corporation). The assay was performed in triplicate to determine the number of colonies.
Wound healing assay
MCF-7 cells (5x105 cells/well) were plated into six-well plates and incubated at 37˚C until they reached 90% confluence. A scratch was subsequently created in the cell monolayer using a 200-µl pipette tip to generate an artificial wound. At 0 and 48 h of culture in serum-free medium at 37˚C, images of the wound area were captured in the same field using an inverted light microscope (Olympus Corporation; magnification, x100) and analyzed by the ImageJ v1.51 software (National Institutes of Health). Cell migration rate = wound area difference between 0 and 48 h/wound area at 0 h x 100%.
Transwell assay
Matrigel (cat. no. 356234; Corning, Inc.) was thawed overnight at 4˚C and diluted with serum-free medium (1:8) before 50 µl of this Matrigel was inoculated in the upper chamber of Transwell plates (cat. no. CLS3422; 8.0 µm; Corning, Inc.) at 37˚C. A total of 5x104 MCF-7 cells in 100 µl serum-free DMEM were seeded into the upper chamber of the Transwell plates and the lower chamber was filled with 500 µl DMEM supplemented with 10% FBS. Following 48 h of incubation at 37˚C, the cells in the lower chamber were fixed with 10% formaldehyde for 15 min at room temperature and stained with 0.1% crystal violet solution for 15 min at room temperature. The number of invasive cells was counted using a light microscope (magnification, x200) and analyzed by ImageJ v1.51 software (National Institutes of Health). Cell invasion rate=the number of invasive cells/number of inoculated cells x100%.
Chromatin immunoprecipitation (ChIP) assay
To determine whether HOXD8 bound to the promoter of the ILP2 gene, a ChIP assay was performed in shRNA-NC- and shRNA-HOXD8-transfected cells using a SimpleChIP® enzymatic chromatin IP kit (Cell Signaling Technology, Inc.) according to the manufacturer's protocol. After MCF-7 cells were lysed, 50 µg protein G agarose beads was added to 1 ml supernatant and incubated at 4˚C for 1 h. The supernatant was then taken after centrifugation at 1,000 x g at 4˚C for 3 min before being divided into two groups. Afterwards, 3 µg IgG or HOXD8 antibodies were added to 500 µg protein samples and incubated overnight at 4˚C. The next day, 50 µg protein G agarose beads was added and the precipitate was collected after incubating at 4˚C for 6 h and centrifugated at 1,000 x g at 4˚C for 3 min. The precipitate was washed with 5X lysis buffer and resuspended in 150 µl 1X ChIP Elution Buffer. Chromatin from beads were eluted with gentle vortexing (1,200 rpm) at 65˚C for 30 min. DNA was purified using the DNA Purification kit (cat. no. D0033; Beyotime Institute of Biotechnology). Relative enrichment was performed by quantitative PCR (qPCR) analysis. The antibody against IgG (cat. no. 5415S; Cell Signaling Technology, Inc.) was diluted to the same concentration (2 µg/ml) as the HOXD8 antibody (cat. no. sc-515357; Santa Cruz Biotechnology, Inc.).
Dual-luciferase reporter assay
The full length (FL) was the fragment located at position 100-2,000 of ILP2 (Sequence ID, NC_000019.10, 53293434-53291335). Three deletion mutants were the fragments containing the predicted binding sites located at positions 420-427 (element 3, E3), 1601-1608 (E2), 1747-1754 (E1) separately mutated (Hunan Fenghui Biotechnology Co., Ltd.) and cloned into the pGL3 basic vector (BioVector NTCC Inc.). MCF-7 cells (1x105 cells/well) were seeded into 24-well plates and transfected with 100 ng luciferase vectors (FL; E3 Deletion, E3 Del; E2 Del or E1 Del) and 10 nM expression vectors (shRNA-NC or shRNA-HOXD8) using Lipofectamine® 2000 reagent (Thermo Fisher Scientific, Inc.) at room temperature. Following 5 h of incubation, the transfection solution was replaced by 500 µl DMEM and cells were incubated for another 24 h at 37˚C. The relative luciferase activity of cells was measured using a Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Database analysis
The Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn) database is a web server for cancer and normal gene expression profiling and interactive analyses (16). GEPIA analysis contains the expression analysis of RNA sequencing data from 9,736 tumors and 8,587 normal samples in The Cancer Genome Atlas (http://cancergenome.nih.gov) and Genotype-Tissue Expression (GTEx, http://commonfund.nih.gov/GTEx) projects. A total of 1,085 tumor samples and 291 normal samples were obtained from the breast invasive carcinoma (BRCA) data set of GEPIA, with |Log2FC|>1 and P<0.01 as the cutoff; where FC is fold-change.
JASPAR (http://jaspar.genereg.net) is a database of transcription factor binding profiles. There were a total of three profiles on HOXD8 found in the JASPAR database; the FASTA-formatted sequence of ILP2 was input to scan with the selected profile (Species: Homo sapiens; ID: MA0910.2), and three predicted sequences with high scores were selected as promising binding sites.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, Inc.) and the data are presented as the mean ± SD of triplicate experiments. The statistical significance of the differences between two groups were determined using an unpaired Student's t-test and among multiple groups using one-way ANOVA followed by a Dunnett's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
HOXD8 expression is downregulated in breast cancer tissues and cell lines
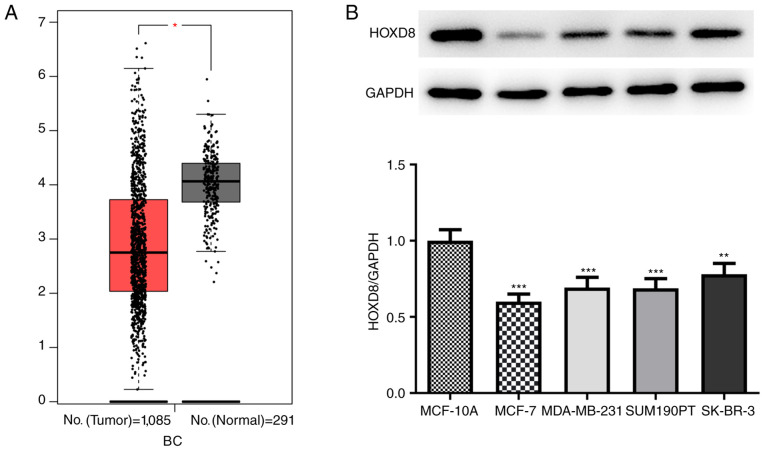
Data from 1,085 tumor samples and 291 normal samples were obtained from the BRCA dataset from the GEPIA database; it was revealed that the expression levels of HOXD8 were significantly downregulated in breast cancer tissues (Fig. 1A). In addition, the expression levels of HOXD8 were analyzed in four breast cancer cell lines (MCF-7, MDA-MB-231, SUM190PT and SK-BR-3), and the results revealed that HOXD8 protein expression was significantly lower in breast cancer cell lines compared with expression in the MCF-10A normal human breast epithelial cell line (Fig. 1B). HOXD8 expression was the lowest in MCF-7 cells; therefore, this cell line was selected for use in subsequent experiments.
Figure 1.
Expression levels of HOXD8 in breast cancer tissues and cell lines. (A) HOXD8 expression levels in breast cancer and normal tissues were obtained from the BRCA dataset from the GEPIA database. *P<0.05. (B) HOXD8 protein expression levels in different breast cancer cell lines and the normal human breast epithelial cell line MCF-10A was detected by western blotting. **P<0.01, ***P<0.001 vs. MCF-10A; n≥3. BRCA, breast invasive carcinoma; HOXD8, homeobox D8.
HOXD8 overexpression inhibits breast cancer cell proliferation, migration and invasion
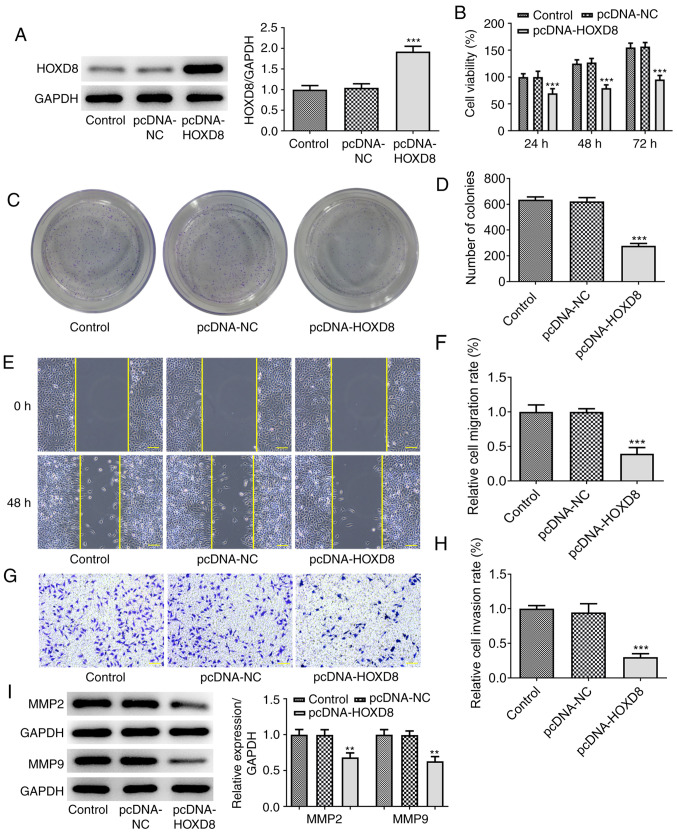
To further investigate the biological functions of HOXD8 in breast cancer, MCF-7 cells were transfected with pcDNA-HOXD8 or pcDNA-NC empty vector. Western blotting revealed that HOXD8 expression levels were significantly upregulated following transfection with pcDNA-HOXD8 compared with the untransfected MCF-7 cells (the control) and pcDNA-NC groups (Fig. 2A), indicating the successful transfection of pcDNA-HOXD8 into MCF-7 cells. The results of the CCK-8 assay demonstrated that the overexpression of HOXD8 significantly inhibited MCF-7 cell proliferation (Fig. 2B), and the number of cell clone clusters containing >50 cells was counted, the results of the colony formation assays were consistent with these findings (Fig. 2C-D). Moreover, the results obtained from the Matrigel and wound healing assays revealed that the overexpression of HOXD8 inhibited the invasive and migratory ability, respectively, of MCF-7 cells compared with the control and pcDNA-NC groups (Fig. 2E-H). In addition, western blotting demonstrated that the overexpression of HOXD8 significantly downregulated the expression levels of migration-related proteins, MMP2 and MMP9, compared with the control and pcDNA-NC groups (Fig. 2I). These results suggested that HOXD8 may act as a tumor suppressor in breast cancer cells.
Figure 2.
Overexpression of HOXD8 inhibits the proliferation, migration and invasion of MCF-7 breast cancer cells. (A) Transfection efficiency of pcDNA-HOXD8 was determined using western blotting. Cell proliferation was detected using (B) Cell Counting Kit-8 and (C and D) colony formation assays following overexpression of HOXD8. (E and F) Cell migration was analyzed using a wound healing assay. (G and H) Cell invasion was assessed using a Matrigel Transwell assay. (I) MMP2 and MMP9 protein expression was analyzed using western blotting. **P<0.01 and ***P<0.001 vs. pcDNA-NC; n≥3. Scale bar, 100 µm. HOXD8, homeobox D8; NC, negative control.
HOXD8 knockdown promotes breast cancer cell proliferation, migration and invasion
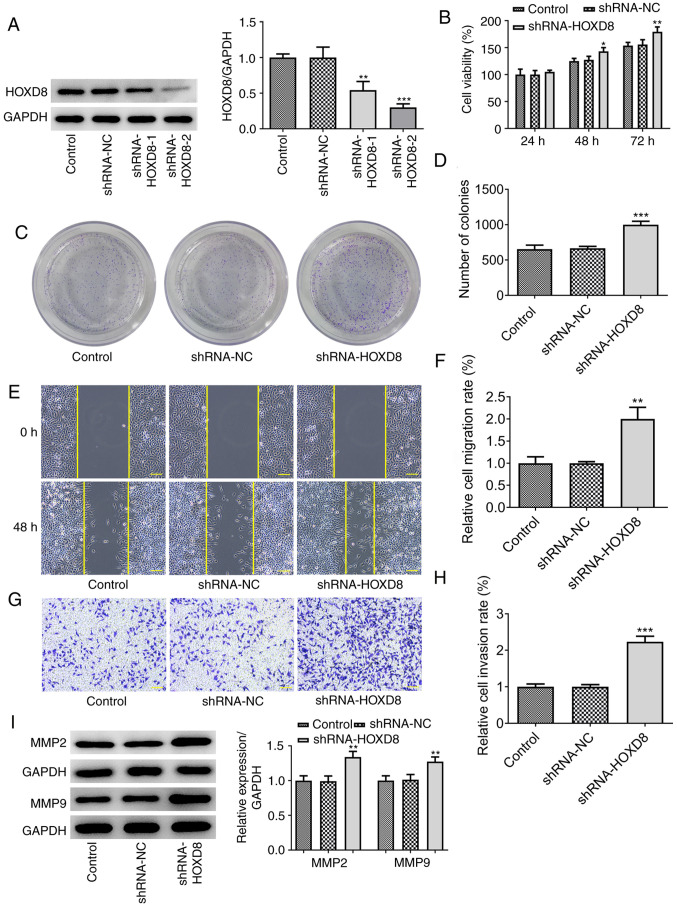
It was next investigated whether the knockdown of HOXD8 could promote the proliferation and colony forming ability of MCF-7 cells. Two types of shRNA-HOXD8 were separately transfected into MCF-7 cells to silence HOXD8 expression and shRNA-HOXD8-2 was selected for subsequent assays because it reduced the expression levels of HOXD8 to a greater extent compared with that of shRNA-HOXD8-1 (Fig. 3A). The results revealed that the proliferation (Fig. 3B), colony forming ability (Fig. 3C-D), invasion and migration (Fig. 3E-H) of MCF-7 cells were increased following HOXD8 knockdown. The protein expression levels of MMP2 and MMP9 were also upregulated following knockdown of HOXD8 compared with the control and shRNA-NC groups (Fig. 3I). These findings further suggested that HOXD8 may play a tumor-suppressive role in breast cancer.
Figure 3.
Knockdown of HOXD8 promotes the proliferation, migration and invasion of MCF-7 breast cancer cells. (A) Transfection efficiency of shRNA-HOXD8 was determined using western blotting. Cell proliferation was analyzed using (B) Cell Counting Kit-8 and (C and D) colony formation assays following knockdown of HOXD8. (E and F) Cell migration was determined using a wound healing assay. (G and H) Cell invasion was measured using a Matrigel Transwell assay. (I) MMP2 and MMP9 protein expression was analyzed using western blotting. *P<0.05, **P<0.01 and ***P<0.001 vs. shRNA-NC; n≥3. Scale bar, 100 µm. HOXD8, homeobox D8; NC, negative control; shRNA, short hairpin RNA.
ILP2 is a direct target of HOXD8 in MCF-7 breast cancer cells
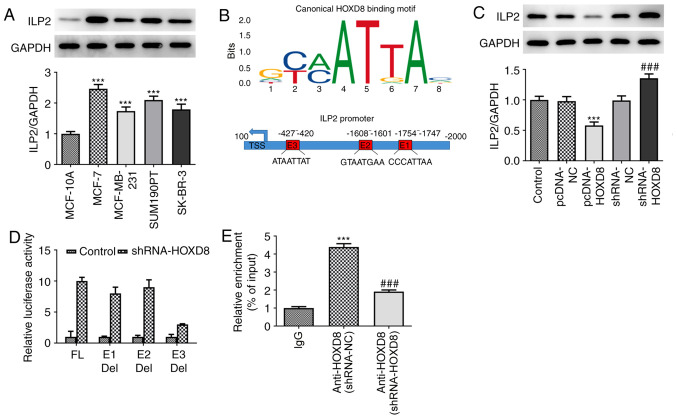
To determine the mechanism underlying the regulatory role of HOXD8 in breast cancer cells, the present study investigated potential targets using the JASPAR database; ILP2 was identified as a potential target and was investigated in further experiments. As shown in Fig. 4A, ILP2 expression levels were found to be significantly upregulated in breast cancer cell lines compared with MCF-10A cells, which indicated a potential role for ILP2 in breast cancer tumorigenesis and progression. Notably, in Fig. 4B, each column of HOXD8 sequence motif corresponds to a base position, and each base position is accumulated by the bases that will appear at that position. The larger the letter, the greater the total amount of information, and the greater the probability of the base appearing. Through combining the motif with the input sequence of ILP2, three putative HOXD8-binding elements (E1: CCCATTAA; E2: GTAATGAA; E3: ATAATTAT) were identified within the ILP2 promoter region using the JASPAR database. Subsequently, whether HOXD8 could regulate ILP2 expression was further determined. Western blotting demonstrated that HOXD8 overexpression downregulated ILP2 protein expression levels, whereas HOXD8 knockdown markedly upregulated ILP2 expression levels (Fig. 4C). To determine which binding site was required for HOXD8-mediated ILP2 expression, the three predicted HOXD8-binding sites were individually deleted and used in separate luciferase assays. The results revealed that HOXD8 almost failed to promote ILP2 transcriptional activity without the E3 element (Fig. 4D), which indicated that the E3 element may be essential for HOXD8 to activate ILP2 transcription. Notably, the enrichment of ILP2 from the ChIP assay and qPCR analysis demonstrated that HOXD8 bound to the promoter of the ILP2 gene (Fig. 4E).
Figure 4.
Association between HOXD8 and ILP2. (A) ILP2 protein expression in breast cancer cell lines was detected by western blotting. ***P<0.001 vs. MCF-10A; n≥3. (B) JASPAR analysis was used to identify putative HOXD8-binding elements within the ILP2 promoter region. The x-axis represents the position of the base, and the y-axis represents the probability of a base in the corresponding position. The larger the letter, the greater the probability of the base. (C) ILP2 protein expression in MCF-7 cells following HOXD8 overexpression or knockdown was detected by western blotting. ***P<0.001 vs. pcDNA-NC; ###P<0.001 vs. shRNA-NC; n≥3. (D) Relative luciferase activity was assessed using a dual-luciferase reporter assay. (E) Relative enrichment of ILP2 was evaluated using chromatin immunoprecipitation and quantitative PCR. ***P<0.001 vs. IgG; and ###P<0.001 vs. shRNA-NC; n≥3. Del, deletion; E1/E2/E3, promoter elements; FL, full length; HOXD8, homeobox D8; ILP2, inhibitor of apoptosis-like protein-2; NC, negative control; shRNA, short hairpin RNA.
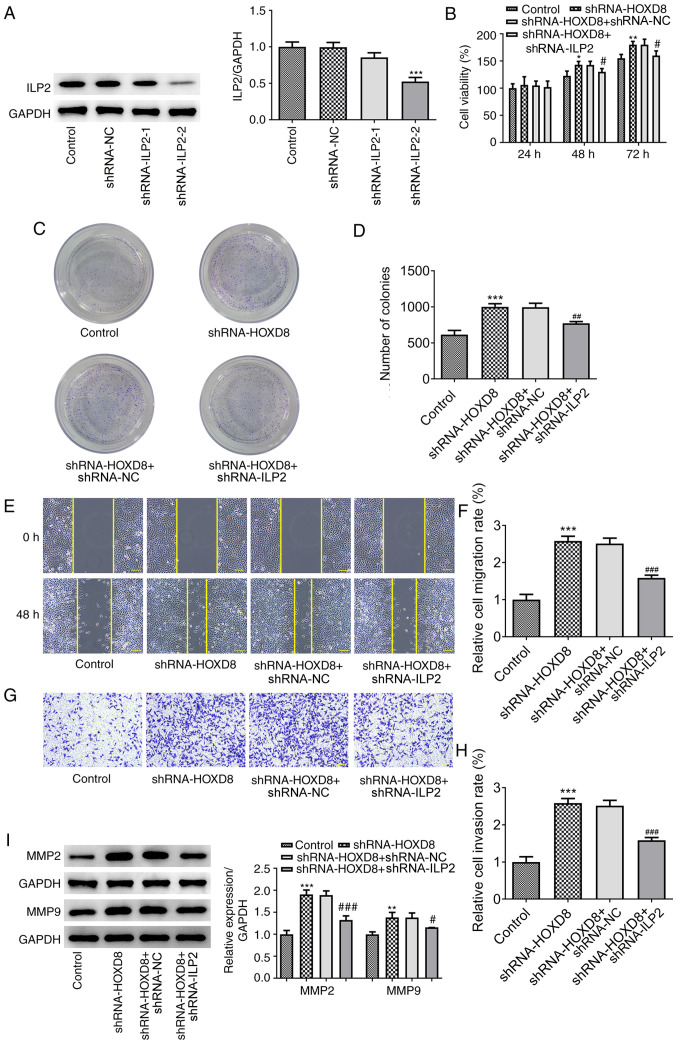
ILP2 knockdown reverses the promoting effects of HOXD8 knockdown on breast cancer cell proliferation, migration and invasion
To determine whether ILP2 influenced the effects of HOXD8 on the proliferation, migration and invasion, two types of shRNA-ILP2 were transfected into MCF-7 breast cancer cells to downregulate ILP2 expression, and shRNA-ILP2-2 was selected for subsequent experiments due to the higher potency of ILP2 knockdown compared with shRNA-ILP2-1 (Fig. 5A). As previously discussed, HOXD8 knockdown notably increased the proliferation (Fig. 5B), colony-forming ability (Fig. 5C-D), invasion and migration (Fig. 5E-H) of breast cancer cells, and these effects could be partially reversed by ILP2 knockdown. In addition, western blotting results demonstrated that transfection with shRNA-ILP2 downregulated the HOXD8 knockdown-induced upregulated expression levels of MMP2 and MMP9 (Fig. 5I). These results suggested that HOXD8 may regulate breast cancer cell progression by binding to the ILP2 promoter.
Figure 5.
Knockdown of ILP2 reverses the promoting effects of HOXD8 knockdown on the proliferation, migration and invasion of breast cancer cells. (A) Transfection efficiency of shRNA-ILP2 was determined using western blotting. Cell proliferation following ILP2 knockdown was analyzed using (B) Cell Counting Kit-8 and (C and D) colony formation assays. (E and F) Cell migration was measured using a wound healing assay. (G and H) Cell invasion was determined using a Matrigel Transwell assay. (I) MMP2 and MMP9 protein expression was analyzed using western blotting. *P<0.05, **P<0.01, ***P<0.001 vs. control; #P<0.05, ###P<0.001 vs. shRNA-HOXD8 + shRNA-NC; n≥3. Scale bar, 100 µm. HOXD8, homeobox D8; ILP2, inhibitor of apoptosis-like protein-2; NC, negative control; shRNA, short hairpin RNA.
Discussion
Numerous studies have demonstrated the important tumor-suppressive role of HOXD8. For example, HOXD8 expression levels were found to be downregulated in colorectal cancer tissues, which inhibited the proliferation, colony forming ability and invasion of colorectal cancer cells and upregulated the expression levels of apoptosis-associated proteins (10). A similar effect was also observed in hepatocellular carcinoma (14). However, to the best of our knowledge, the effects of HOXD8 in breast cancer have yet to be determined. Therefore, the present study aimed to investigate the role of HOXD8 in breast cancer. First, the expression levels of HOXD8 in multiple breast cancer cell lines were analyzed, and based on the results, the expression level of HOXD8 in the MCF-7 cell line was the lowest. In addition, since MCF-7 cell line is a representative in vitro model of HER2-negative luminal breast cancer (17), the MCF-7 cell line was selected for use in subsequent experiments. By constructing overexpression and knockdown vectors, the effects of HOXD8 overexpression or knockdown on cell proliferation, migration and invasion were determined. The results revealed that the overexpression of HOXD8 inhibited the proliferation, invasion and migration of breast cancer cells. The expression levels of MMP2 and MMP9 were also investigated to further determine the effects of HOXD8 on cell migration, and it was demonstrated that HOXD8 overexpression inhibited, and HOXD8 knockdown increased, the expression of MMPs. MMPs are a family of zinc and calcium ion-dependent proteases that can target and degrade numerous proteins in the extracellular matrix (ECM) (18), which is key to tumor invasion and metastasis, as the ECM constitutes the first barrier for tumor cells to overcome to effectively metastasize. The main component of the ECM is type IV collagen, which can be degraded by MMP2 and MMP9. Notably, the expression of MMP2 and MMP9 has been found to be implicated in the progression of several types of cancer (19-22). These findings suggested that HOXD8 may serve a role in breast cancer cell migration and invasion.
Apoptosis inhibitor proteins can inhibit cell apoptosis and promote cell proliferation, and have been shown to serve an important role in the occurrence and development of several types of cancer (23-25). In normal tissues, ILP2 is only expressed in the testis and lymphoblasts, where it inhibits cell apoptosis (4). Previous research on the role of ILP2 in breast cancer demonstrated that the expression levels of ILP2 were significantly upregulated in breast cancer tissues and cell lines, whereas knockdown of ILP2 expression significantly inhibited the proliferation, migration and invasion of breast cancer cells, and increased apoptosis (26). The results of our previous study revealed that ILP2 may serve as a serum biomarker for breast cancer, which is important for the diagnosis and treatment of this disease (5). In addition, a previous study reported that ILP2 overexpression exerted no inhibitory effects on TNF-mediated apoptosis, but effectively inhibited apoptosis mediated by Bax and apoptotic peptidase activating factor 1/caspase-9. In addition, an interaction was identified between ILP2 and caspase-9(4). Therefore, it was suggested that devising an inhibitor for the ILP2/caspase interaction may be an effective anticancer measure (27). The results of the present study revealed that HOXD8 exerted a tumor-suppressive effect on breast cancer by targeting the ILP2 promoter. Therefore, regulation of the HOXD8/ILP2 interaction may also represent a potential effective anticancer treatment.
In conclusion, the results of the present study demonstrated that the expression levels of HOXD8 were downregulated in breast cancer tissues and cell lines. In addition, HOXD8 overexpression was found to inhibit the proliferation, migration and invasion of breast cancer cells by targeting ILP2. These findings provide a novel insight into potential therapeutic target for breast cancer; however, the exact mechanism underlying the role of ILP2 in breast cancer requires further investigation.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XW designed the study and wrote the manuscript; YC performed the experiments and analyzed the data; XF participated in the experiments, examined the data and critically revised the manuscript for important intellectual content. All authors confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 3.Peart O. Metastatic breast cancer. Radiol Technol. 2017;88:519M–539M. [PubMed] [Google Scholar]
- 4.Richter BW, Mir SS, Eiben LJ, Lewis J, Reffey SB, Frattini A, Tian L, Frank S, Youle RJ, Nelson DL, et al. Molecular cloning of ILP-2, a novel member of the inhibitor of apoptosis protein family. Mol Cell Biol. 2001;21:4292–4301. doi: 10.1128/MCB.21.13.4292-4301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang M, Zhou W, Gao D, Fang X, Liu Q. Inhibitor of apoptosis protein-like protein-2 as a novel serological biomarker for breast cancer. Int J Mol Sci. 2012;13:16737–16750. doi: 10.3390/ijms131216737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert M, Jambon S, Depauw S, David-Cordonnier MH. Targeting transcription factors for cancer treatment. Molecules. 2018;23(1479) doi: 10.3390/molecules23061479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bemer M, van Dijk ADJ, Immink RGH, Angenent GC. Cross-family transcription factor interactions: An additional layer of gene regulation. Trends Plant Sci. 2017;22:66–80. doi: 10.1016/j.tplants.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Bushweller JH. Targeting transcription factors in cancer-from undruggable to reality. Nat Rev Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brotto DB, Siena ADD, de B II, Carvalho SDCES, Muys BR, Goedert L, Cardoso C, Plaça JR, Ramão A, Squire JA, et al. Contributions of HOX genes to cancer hallmarks: Enrichment pathway analysis and review. Tumour Biol. 2020;42(1010428320918050) doi: 10.1177/1010428320918050. [DOI] [PubMed] [Google Scholar]
- 10.Mansour MA, Senga T. HOXD8 exerts a tumor-suppressing role in colorectal cancer as an apoptotic inducer. Int J Biochem Cell Biol. 2017;88:1–13. doi: 10.1016/j.biocel.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Sun P, Song Y, Liu D, Liu G, Mao X, Dong B, Braicu EI, Sehouli J. Potential role of the HOXD8 transcription factor in cisplatin resistance and tumour metastasis in advanced epithelial ovarian cancer. Sci Rep. 2018;8(13483) doi: 10.1038/s41598-018-31030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Miao L, Ni R, Zhang H, Li L, Wang X, Li X, Wang J. microRNA-520a-3p inhibits proliferation and cancer stem cell phenotype by targeting HOXD8 in non-small cell lung cancer. Oncol Rep. 2016;36:3529–3535. doi: 10.3892/or.2016.5149. [DOI] [PubMed] [Google Scholar]
- 13.Zhu W, Wang JP, Meng QZ, Zhu F, Hao XF. MiR-142-5p reverses the resistance to gefitinib through targeting HOXD8 in lung cancer cells. Eur Rev Med Pharmacol Sci. 2020;24:4306–4313. doi: 10.26355/eurrev_202004_21011. [DOI] [PubMed] [Google Scholar]
- 14.Sun S, Wang N, Sun Z, Wang X, Cui H. MiR-5692a promotes proliferation and inhibits apoptosis by targeting HOXD8 in hepatocellular carcinoma. J BUON. 2019;24:178–186. [PubMed] [Google Scholar]
- 15.Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei HC. Mathematical modeling of tumor growth: The MCF-7 breast cancer cell line. Math Biosci Eng. 2019;16:6512–6535. doi: 10.3934/mbe.2019325. [DOI] [PubMed] [Google Scholar]
- 18.Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31 (Suppl 1):177–183. doi: 10.3109/14756366.2016.1161620. [DOI] [PubMed] [Google Scholar]
- 19.Avădanei R, Căruntu ID, Amălinei C, Lozneanu L, Balan R, Grigoraş A, Ciobanu Apostol D, Giuşcă SE. High variability in MMP2/TIMP2 and MMP9/TIMP1 expression in secondary liver tumors. Rom J Morphol Embryol. 2013;54:479–485. [PubMed] [Google Scholar]
- 20.Kalhori V, Törnquist K. MMP2 and MMP9 participate in S1P-induced invasion of follicular ML-1 thyroid cancer cells. Mol Cell Endocrinol. 2015;404:113–122. doi: 10.1016/j.mce.2015.01.037. [DOI] [PubMed] [Google Scholar]
- 21.Sokołowska J, Urbańska K. Immunohistochemical assessment of metalloproteinases MMP2 and MMP9 expression in canine various subtypes of lymphomas in relation with proliferative and apoptotic markers. Pol J Vet Sci. 2019;22:203–211. doi: 10.24425/pjvs.2019.127087. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Yang B, She Y, Ye Y. The lncRNA TP73-AS1 promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. J Cell Biochem. 2018;119:7790–7799. doi: 10.1002/jcb.27158. [DOI] [PubMed] [Google Scholar]
- 23.Varfolomeev E, Vucic D. Inhibitor of apoptosis proteins: Fascinating biology leads to attractive tumor therapeutic targets. Future Oncol. 2011;7:633–648. doi: 10.2217/fon.11.40. [DOI] [PubMed] [Google Scholar]
- 24.Fulda S. Exploiting inhibitor of apoptosis proteins as therapeutic targets in hematological malignancies. Leukemia. 2012;26:1155–1165. doi: 10.1038/leu.2012.4. [DOI] [PubMed] [Google Scholar]
- 25.Fulda S. Inhibitor of apoptosis proteins as targets for anticancer therapy. Expert Rev Anticancer Ther. 2007;7:1255–1264. doi: 10.1586/14737140.7.9.1255. [DOI] [PubMed] [Google Scholar]
- 26.Zhu L, Zhou W, Zhu X, Xiang S, Wang S, Peng Y, Lu B, Tang P, Chen Q, Wu M, et al. Inhibitor of apoptosis protein-like protein-2: A novel growth accelerator for breast cancer cells. Oncol Rep. 2018;40:2047–2055. doi: 10.3892/or.2018.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalili S, Mohammadpour H, Shokrollahi Barough M, Kokhaei P. ILP-2 modeling and virtual screening of an FDA-approved library: A possible anticancer therapy. Turk J Med Sci. 2016;46:1135–1143. doi: 10.3906/sag-1503-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.