Abstract
Sorafenib has been approved as a systemic drug for advanced liver cancer; however, the underlying mechanisms remain unclear. The present study aimed to investigate the effects of sorafenib on the proliferation, autophagy and apoptosis of HepG2 cells under hypoxia. Briefly, reverse transcription-quantitative PCR and western blotting was performed to quantify HIF-1, LC3II/I, mTOR and p70s6K expression levels. Cell proliferation was determined using the Cell Counting Kit-8 assay and the cell apoptosis rate was evaluated using flow cytometry. The results demonstrated that autophagy and apoptosis were induced by hypoxia, and that sorafenib further enhanced hypoxia-induced autophagy and apoptosis in HepG2 cells in a dose-dependent manner. Furthermore, the mechanism of sorafenib-mediated autophagy in liver cancer cell were investigated by using chloroquine (CQ). The results showed that CQ significantly inhibited autophagy by decreasing LC3II/LC3I ratio in HepG2 cells treated with sorafenib and/or hypoxia. By contrast, sorafenib could increase the expression of hypoxia-inducible factor-1 (HIF-1) and of the autophagy marker (LC3II/I) and decrease the expression of mammalian target of rapamycin and p70 ribosomal S6 kinase in HepG2 cells under normoxia and hypoxia conditions, suggesting that sorafenib could induce hypoxia and autophagy in liver cancer cells. In addition, sorafenib was demonstrated to prevent proliferation and induce apoptosis of HepG2 cells under normoxia and hypoxia. Sorafenib could also prevent the malignant behavior of HepG2 by inducing hypoxia and autophagy. In summary, the findings from the present study suggested that sorafenib may inhibit liver cancer progression by activating autophagy and HIF-1 signaling pathway.
Keywords: liver cancer, autophagy, hypoxia, sorafenib, hypoxia-inducible factor-1α
Introduction
Primary liver cancer is the second leading cause of cancer-associated mortality worldwide (1,2). Primary liver cancer is one of the few tumors with a steady increasing incidence and mortality. It is therefore a major public health challenge (3). Liver cancer normally comprises a heterogeneous group of malignant tumors, including hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (iCCA) and other rare tumors (4). The incidence of liver cancer is strongly correlated with cirrhosis, which is caused by alcohol abuse, hepatitis B and/or hepatitis C infection, aflatoxin exposure, smoking or other chronic liver injuries, such as metabolic liver diseases and autoimmune diseases (5,6). Sorafenib is a multiple kinase inhibitor targeting cell surface and intracellular kinases and that has been found to restrict tumor growth via inducing apoptosis and inhibiting angiogenesis (7-9). Furthermore, sorafenib activates liver cancer cell autophagy by regulating PI3K/AKT/mammalian target of rapamycin (mTOR) (10-12), Ras/RAF/MEK/ERK (13) and JAK-STAT (14) signaling pathways and induces the autophagic cell death (14). In clinical application, the efficacy of sorafenib in advanced liver cancer has been confirmed by two independent clinical trials (15,16) and sorafenib has been approved as a systemic agent for the treatment of the cancer. However, the underlying mechanisms of sorafenib in liver cancer are still being investigated.
Hypoxia commonly occurs in liver cancer due to intense oxygen metabolism (17). Tumor hypoxia has been linked to poor patient outcomes (18,19). Several techniques for assessing tumor hypoxia have allowed clinicians to determine the extent of hypoxia in order to adopt differential treatment strategies (20-22). In addition, various methods have been established to simulate hypoxic conditions in in vivo pre-clinical research, such as cell treatment with cobalt chloride or cell culture in hypoxic chamber (23,24). Hypoxia-inducible factors (HIFs) are transcription factors responding to hypoxia. They are composed of an α subunit and a β subunit and can be classified into the three isoforms HIF 1α, HIF 2α and HIF 3α (25,26). Degradation of HIFα by prolyl hydroxylase (PHD) can suppress the activity of HIF under normoxia. However, the HIF commonly remains active due to the deactivation of PHD under hypoxia (25). Upregulation of HIFs in liver cancer cells contributes to cell adaption to the hypoxic environment via regulation of proliferation, metabolism, inflammation and angiogenesis (27,28). Activation of HIF-1α is a marker for acute hypoxia since HIF-1α accumulation increases the expression of PHD by positive feedback regulation, which leads to HIF-1α degradation under chronic hypoxia (28). Multiple signaling pathways of hypoxia-induced autophagy, including the HIF-1/mTOR signaling pathway, have been reported (29). However, the effects of hypoxia in liver cancer have not yet been reported.
Autophagy is a highly conserved catabolic process designed to deliver cellular materials to autophagosome-liposome infused complex for the degradation of damaged organelles and energy recycling (30,31). In mammalian cells, autophagy is typically divided into the principal step-initiation, nucleation of the autophagosome, elongation of the autophagosome membrane, fusion with the lysosome and degradation (32). Multiple signaling pathways of autophagy have been reported, including the P53(33), PI3K (34), Ras (35,36) and JAK-STAT (32,37) signaling pathways. Chloroquine (CQ) is frequently used as an inhibitor of autophagy in both cell culture and in vivo (38). The direct effects of CQ on autophagy are related to the expression of HIF-1 and LC3II/I (39). Therefore, CQ is usually employed to study the mechanism of autophagy (40). It is hypothesized that autophagy might serve a complicated role in tumorigenesis, such as in liver tumorigenesis (41). However, whether sorafenib can influence the malignant behavior of liver cancer by regulating autophagy remains unknown.
In the present study, the effects of sorafenib and hypoxia on the proliferation, autophagy and apoptosis of liver cancer cells were investigated. Furthermore, the role of sorafenib on the expression of hypoxia and autophagy-related proteins in liver cancer cells was also evaluated.
Materials and methods
Materials
HepG2 cells and GFP-LC3 plasmid were kindly donated by the Department of Biochemistry and Molecular Biology of the Third Military Medical University. DMEM cell culture medium, FBS, penicillin G, streptomycin and trypsin were purchased from Gibco; Thermo Fisher Scientific, Inc. The transfection reagent Lipofectamine™ 2000 was purchased from Omega Bio-Tek, Inc. Cell Counting Kit-8 (CCK8-kit) was acquired from Abmole BioScience Inc. Developer kit for western blotting was obtained from Pierce; Thermo Fisher Scientific, Inc. The BCA protein concentration test kit Pierce BCA Protein Assay kit was purchased from Pierce; Thermo Fisher Scientific, Inc. Tris-base, glycine, SDS, acrylamide, DTT, Tween-20 and BIS-Acrylamide reagents for western blotting were obtained from Sangon Biotech Co., Ltd. PVDF membranes were purchased from EMD Millipore. X-ray films were purchased from Guangxi Yestar Medical System Co. Ltd. Methanol, glycerol, ethanol, isopropanol and concentrated hydrochloric acid used for western blotting were acquired from Guangzhou Chemical Reagent Factory. RNA extraction kit was purchased from Ambion; Thermo Fisher Scientific, Inc. CQ was provided by Sigma-Aldrich; Merck KGaA. Annexin V-FITC/propidium iodide (PI) apoptosis kit was purchased from eBioscience; Thermo Fisher Scientific, Inc. The following antibodies were used in the present study: i) primary antibodies against HIF-1 (cat. no. 3716S; 1:1,000; Cell Signaling Technology, Inc.), LC3-I (cat. no. 12741S; 1:1000; Cell Signaling Technology, Inc.), LC3-II (cat. no. 12741S; 1:1,000; Cell Signaling Technology, Inc.), mTOR (cat. no. 2972S; 1:1,000; Cell Signaling Technology, Inc.), p70 ribosomal S6 kinase (p70s6k; cat. no. 9202S; 1:1,000; Cell Signaling Technology, Inc.), GAPDH (cat. no. 2118S; 1:1,000; Cell Signaling Technology, Inc.); and ii) HRP-conjugated anti-mouse (1:1,000; Santa Cruz Biotechnology, Inc.) or anti-rabbit secondary antibody (1:1,000; Cell Signaling Technology, Inc.).
Cell culture and cell treatment
HepG2 cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin G and 100 mg/ml streptomycin placed at 37˚C in a humidified incubator containing 5% CO2. The cells (2.5x103) were transferred into a 6-well plate and were exposed to a hypoxic (0.1% oxygen) or a normoxic (~20% oxygen) environments for 24 h. The cells were subsequently treated with sorafenib at the specified concentrations and for the indicated durations. Cells in each group were then collected for further experiments. For CQ treatment group, cells exposed to 50 µM CQ and cultured at 37˚C with 5% CO2 for 24 h were subsequently treated according indicated conditions.
Autophagosome evaluation
The HepG2 cells (2.5x103) were seeded evenly in a 6-well plate and cultured at 37˚C with 5% CO2 for two days. Then cells were transfected with GFP-LC3 plasmid for 48 h at 37˚C (2 µg/well, GFP-LC3 was subcloned from pEGFPC1-LC3 into the SnaB1 and Sal1 sites of pBABEpuro) using Lipofectamine 2000 according to the manufacturers' instructions. After treatment with various concentration of sorafenib (10, 20, 30 and 40 µM) at 37˚C for 12 h, cells were fixed in 4% formaldehyde at 20 min in room temperature. The autophagosomes were detected and photographed under a fluorescence microscope (magnification, x100). The green fluorescence of cells were quantified by ImageJ version 2.1.0 (National Institutes of Health) Cells with more than 5 puncta were defined as autophagic cells (42).
CCK-8 assay
HepG2 cells (2x103 cells/100 µl) were transferred into a 96-well plate and were exposed to hypoxic (0.1% oxygen) or normoxic (~20% oxygen) environments for 24 h. Cells were then treated with 30 µΜ sorafenib for 0, 24, 48 and 72 h, and cells were incubated with 10 µl CCK-8 for 3h at 37℃. The absorbance at 450 nm was read using a microplate reader.
Flow cytometer
HepG2 cells were treated as described above. Cells were washed with PBS (Invitrogen; Thermo Fisher Scientific, Inc.) and adjusted to a density of 1x106 cells/ml. The cells were subsequently resuspended in 100 µl binding buffer, 5 µl Annexin V-FITC and 5 µl PI and incubated in the dark at 4˚C for 15 min. Early + late apoptotic cells were washed with PBS and then analyzed by BD LSR Fortessa flow cytometer (Becton, Dickinson and Company) and FlowJo software version 7.6.1 (FlowJo LLC).
Western blotting
HepG2 cells were washed three times with PBS and diluted in Tris-HCl buffer. RIPA buffer (Beyotime Institute of Biotechnology) was added to HepG2 cells at 4˚C for 20 min. The samples were centrifuged at 13,500 x g at 4˚C for 10 min and the supernatants containing all proteins were collected and stored at -80˚C. Protein concentration was determined using Pierce BCA Protein assay kit. Proteins (20 µg) were separated by 10% SDS-PAGE and were transferred onto PVDF membranes. Membranes were blocked with 5% skimmed milk in TBS buffer overnight at 4˚C and washed three times with TBS buffer. Membranes were subsequently incubated in primary antibodies diluted into skimmed milk for 1 h at room temperature. After washing three times with TBS buffer, membranes were incubated with horseradish peroxidase-labeled secondary antibodies for 45 min at room temperature. Membranes were imaged using chemiluminescence (EMD Millipore). Relative expression levels were normalized to endogenous control GAPDH using software Image-Pro Plus 6.0 (National Institutes of Health).
Reverse transcription quantitative (RT-q) PCR
RNA extraction kit was utilized to extract total RNA from the treated HepG2 cells. RNA was reverse transcribed into cDNA using the cDNA Synthesis according to the manufacturers' instructions (PrimeScript RT reagent kit, Takara Bio, Inc.). RT-qPCR reactions were conducted using SYBR Green qPCR SuperMix (Invitrogen; Thermo Fisher Scientific, Inc.). The mRNA level of HIF was evaluated using RT-qPCR, GAPDH was used as the reference gene. The gene-specific primer sequences were: HIF-1-F: TATGAGCCAGAAGAACTTTTAGGC; HIF-1-R: CACCTCTTTTGGCAAGCATCCTG; GAPDH-F: CGGAGTCAACGGATTTGGTCGTAT; GAPDH-R: AGCCTTCTCCATGGTGGTGAAGAC. The conditions of PCR were: Pre-denaturation at 95˚C for 3 min, followed by 40 cycles of denaturation for 10 sec at 95˚C, annealing at 58˚C for 30 sec and then 95˚C for 10 sec. Melt Curve 65-95˚C with increments of 0.5˚C for 5 sec. Following this, the expression levels of HIF-1 was assessed. The relative quantification was performed using the comparative 2-∆∆Cq method (43). ∆∆Ct=[Ct(target gene)-Ct(internal gene)] experimental group-[Ct(target gene)-Ct(internal gene)] control group.
Statistical analysis
Each experiment was performed at least three times independently and data were presented as the means ± standard deviation. One-way ANOVA followed by Tukey's post-hoc test was used for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
Sorafenib induces the autophagy and apoptosis of HepG2 cells under hypoxia
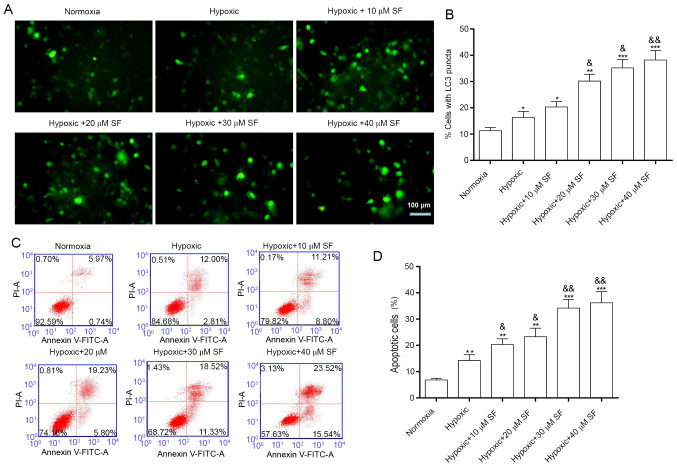
LC3 is a transmembrane protein that is abundant in the cytoplasm and located on the membrane of autophagosomes during autophagosome formation (44,45). The present study demonstrated that LC3 fluorescence intensity was significantly enhanced in HepG2 cells under hypoxia compared with cells under normoxia. Furthermore, treatment with sorafenib significantly enhanced LC3 fluorescence intensity in a concentration-dependent manner (P<0.05, P<0.01 and P<0.001; Fig. 1A and B). In addition, sorafenib and hypoxia-induced apoptosis was examined by flow cytometry. The results demonstrated that the percentage of apoptotic cells increased significantly in HepG2 cells under hypoxia compared with cells in the normoxia group (14.81 vs 6.71%). Furthermore, the percentage of apoptotic cells was significantly increased following treatment of HepG2 cell under hypoxia with sorafenib in a concentration-dependent manner (P<0.05, P<0.01 and P<0.001; Fig. 1C and D), up to 39.06%, for 40 µM sorafenib. Taken together, these findings demonstrated that sorafenib may enhance hypoxia-induced autophagy and apoptosis in HepG2 cells.
Figure 1.
SF induces autophagy and apoptosis of HepG2 cells under hypoxia. (A) GFP-LC3 plasmid was transfected into HepG2 cells and cells were incubated with 10, 20, 30 and 40 µM SF for 12 h. Fluorescence intensity of LC3 was determined using a fluorescence microscope. Magnification, x100. Scale bar=100 µm. (B) LC3 positive cells were quantified using results (A). (C) HepG2 cells were exposed to low oxygen levels for 24 h and were treated with various concentrations of SF. Cell apoptosis was estimated by Annexin V/PI staining. (D) Apoptotic rate was determined based on results from three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. normoxia group. &P<0.05 and &&P<0.01 vs. hypoxic group. SF, sorafenib.
Sorafenib significantly upregulates HIF-1 and LC3II/I and downregulated mTOR and p70s6K in HepG2 cells under hypoxia
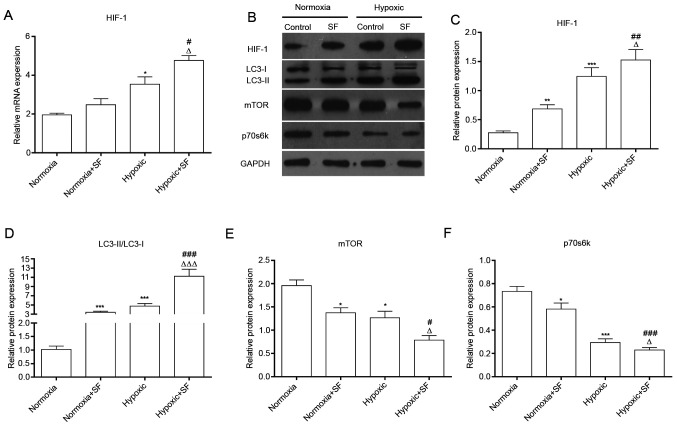
Due to the promotion of autophagy by Sorafenib, the signaling pathways that might be related to autophagy in hypoxic HepG2 cells were further investigated. The results demonstrated that hypoxia could upregulate HIF-1 at the mRNA and protein levels in HepG2 cells, and that hypoxia-induced HIF-1 overexpression was further enhanced by sorafenib (P<0.05; Fig. 2A and B). Subsequently, further investigation demonstrated that mTOR and p70s6K expression was significantly downregulated and that LC3II/LC3I ratio was significantly upregulated in HepG2 cells treated with SF under hypoxia. Furthermore, expression of mTOR and p70s6K was decreased in sorafenib-treated hypoxic HepG2 cells. These results showed that the combination of sorafenib and hypoxia resulted in a significantly higher HIF-1 expression and LC3II/LCI ratio and a significantly stronger downregulation of mTOR and p70s6K (P<0.05, P<0.01 and P<0.001; Fig. 2C and D). Taken together, these findings indicated that sorafenib could strengthen the autophagy of normal and hypoxic HepG2 cells.
Figure 2.
SF upregulates HIF-1, LC3II/I and downregulates mTOR and p70s6K in HepG2 cells under hypoxia. (A) After treatment with SF, total mRNA was collected and expression level of HIF-1 was measured by reverse transcription quantitative PCR in HepG2 cells under normoxia and hypoxia. (B) Proteins from treated HepG2 cells were collected and the expression of HIF-1, LC3, mTOR and p70s6K was determined using western blotting. (C-F) Relative protein expression of HIF-1, LC3II/I, mTOR and p70s6K was quantified. *P<0.05, **P<0.01 and ***P<0.001 vs. normoxia group. #P<0.05, ##P<0.01 and ###P<0.001 vs. normoxia + SF group. ∆P<0.05, ∆∆∆P<0.001 vs. hypoxic group. Data are presented as the means ± standard deviation (n=3). SF, sorafenib; mTOR, mammalian target of rapamycin; HIF-1, hypoxia-inducible factor-1; p70s6k, p70 ribosomal S6 kinase.
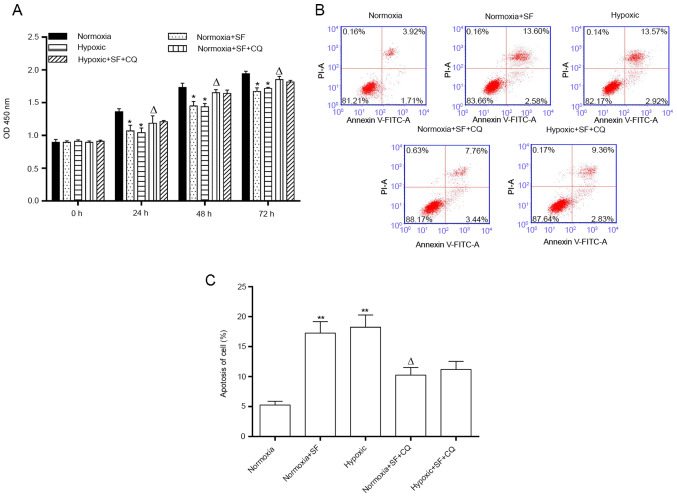
Sorafenib inhibits the proliferation and stimulates the apoptosis of HepG2 cells under normoxia and hypoxia
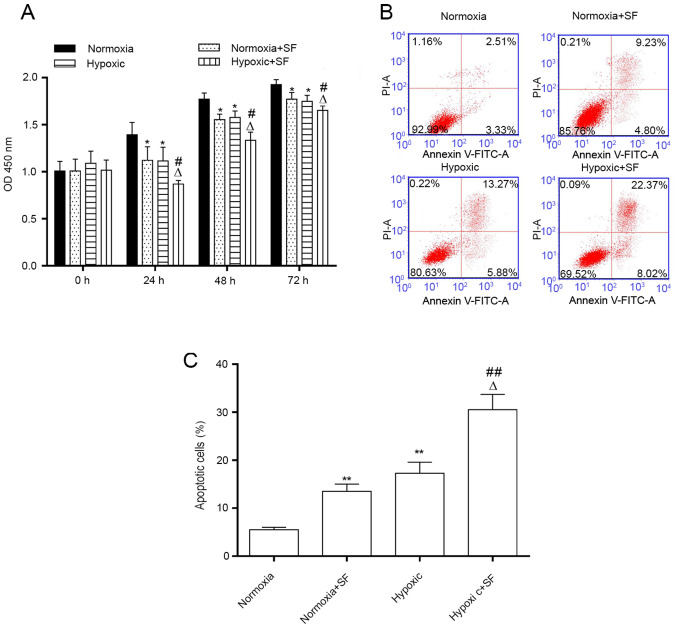
Consistently with the aforementioned findings, the results demonstrated that cell proliferation was significantly decreased in hypoxic HepG2 cells compared with cells in the normoxia group, and that sorafenib further inhibited the proliferation of HepG2 cells under hypoxia after 24, 48 and 72 h (P<0.05; Fig. 3A). Furthermore, the number of apoptotic cells was increased significantly in the hypoxic HepG2 cells compared with those in the normoxia group (19.15 vs 5.84%). This phenomenon was further enhanced following HepG2 cell treatment with 30 µM sorafenib for 24 h (30.39 vs. 19.12%; P<0.05 and P<0.01; Fig. 3B and C). The results demonstrated that sorafenib could inhibit the proliferation and promote the apoptosis of HepG2 cells under normoxia and hypoxia.
Figure 3.
SF inhibits the proliferation and induces the apoptosis of HepG2 cells under normoxia and hypoxia. HepG2 cells were exposed to hypoxic or normoxic conditions for 24 h and were treated with 30 µΜ SF for the indicated time. (A) Cell Counting Kit-8 assay was used to evaluate the proliferation of HepG2 cells treated with SF and under hypoxia at 0, 24, 48 and 72 h. (B) Flow cytometer was used to determine the effect of SF and hypoxia on HepG2 cell apoptosis. (C) Quantification of apoptotic rates from (B). *P<0.05 and **P<0.01 vs. normoxia group. #P<0.05 and ##P<0.01 vs. normoxia + SF group. ∆P<0.05, vs. hypoxic group. Data are presented as the means ± standard deviation (n=3). SF, sorafenib.
CQ reverses the sorafenib-induced increase in HIF-1 and LC3II/I expression in HepG2 cells under normoxia and hypoxia
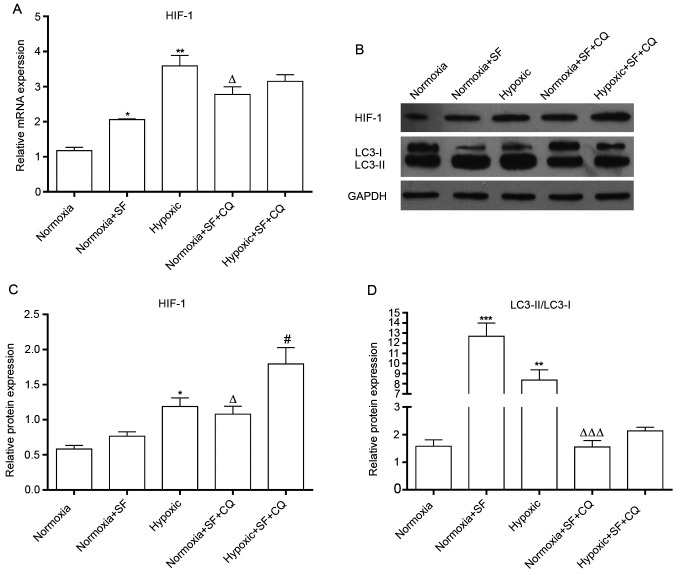
To further determine whether autophagy pathway might contribute to the apoptosis of liver cancer induced by sorafenib and hypoxia, the autophagy inhibitor CQ was used to evaluate the autophagy with or without sorafenib in HepG2 cells under normoxia and hypoxia. The results demonstrated that CQ could partially reverse the upregulation of HIF-1 induced by sorafenib and/or hypoxia in HepG2 cells (P<0.05 and P<0.01; Fig. 4A and B). In addition, CQ significantly inhibited autophagy by decreasing LC3II/LC3I ratio in HepG2 cells treated with sorafenib and/or hypoxia (P<0.05 and P<0.001; Fig. 4B-D). Taken together, these results suggested that hypoxia and sorafenib could upregulate HIF-1 expression via regulating autophagy.
Figure 4.
CQ reverses the increase in HIF-1 and LC3II/I expression induced by SF in HepG2 cells under normoxia and hypoxia. Under normoxia and hypoxia, HepG2 cells were treated with 30 µΜ SF or/and 50 µΜ CQ for 24 h. (A) Expression level of HIF-1 was measured by reverse transcription quantitative PCR in HepG2 cells. (B) Western blotting evaluated the expression of HIF-1 and LC3 in HepG2 cells under different conditions. (C and D) Relative protein expression of HIF-1 and LC3II/I was quantified. *P<0.05, **P<0.01 and ***P<0.001 vs. normoxia group. ∆P<0.05, ∆∆∆P<0.001 vs. normoxia + SF group. #P<0.05 vs. normoxia + SF + CQ group. Data are presented as the means ± standard deviation (n=3). SF, sorafenib; CQ, chloroquine.
CQ attenuates the inhibition of proliferation and induction of apoptosis mediated by sorafenib in HepG2 cells under normoxia and hypoxia
The present study demonstrated that the inhibition of HepG2 cell proliferation, which was mediated by sorafenib and/or hypoxia, could also be weakened by CQ (P<0.05; Fig. 5A). Furthermore, this study reported that the promoting effect of sorafenib and/or hypoxia on apoptosis could also be partially reversed by CQ in HepG2 cells (11.2 vs. 16.18% and 12.19 vs. 16.49%; P<0.05 and P<0.01; Fig. 5B). However, the percentage of necrotic cells was not significantly changed. These findings suggested that sorafenib and hypoxia may prevent the proliferation and accelerate the apoptosis of HepG2 cells by regulating the autophagy signaling pathway.
Figure 5.
CQ attenuated the inhibition of proliferation and induction of apoptosis mediated by SF in HepG2 cells under normoxia and hypoxia. Normal and hypoxic HepG2 cells were treated with SF or/and CQ. (A) Cell proliferation was monitored using Cell Counting Kit-8 at 0, 24, 48 and 72 h. (B) Flow cytometer was used to determine HepG2 cell apoptosis. (C) Quantification of apoptotic rates from (B). *P<0.05 and **P<0.01 vs. normoxia group. ∆P<0.05 vs. normoxia + SF group. Data are presented as the means ± standard deviation (n=3). SF, sorafenib; CQ, chloroquine.
Discussion
The present study reported the underlying mechanism of sorafenib-induced liver cancer cell autophagy and apoptosis. The results demonstrated that sorafenib could promote cell autophagy and apoptosis under hypoxia. Furthermore, according to the effect of sorafenib on autophagy under hypoxia, we hypothesized that sorafenib may share a similar autophagic mechanism with hypoxia. The results demonstrated that both sorafenib and hypoxia induced cell autophagy via the HIF/mTOR-related signaling pathway. In addition, to address the crucial role of sorafenib-mediated autophagy in liver cancer cell, autophagy was inhibited using CQ. The results showed that CQ inhibited the apoptosis and promoted the proliferation of liver cancer cells, which were mediated by sorafenib or hypoxia. These findings strongly suggested that autophagy may contribute to the inhibition of proliferation in sorafenib-treated cells via the HIF/mTOR signaling pathway.
Although regorafenib and lenvatinib have been approved as second-line treatments to improve the clinical outcomes of patients with advanced liver cancer, these treatments only extend the median overall survival by ~3 months compared with patients treated with sorafenib (46). It is therefore crucial to determine the underlying mechanism of sorafenib in order to extend the overall survival of patients with advanced-stage liver cancer. Previous studies have reported that sorafenib combined with various autophagy-inducing drugs, such as metformin (13), capsaicin (47) and pemetrexed (48), can suppress the proliferation of liver cancer cells in vitro. Conversely to studies on autophagy-promoting cell death, sorafenib-induced autophagy can also be considered as a survival mechanism for liver cancer cells. It has been reported that liver cancer cells treated with sorafenib can be killed by inhibiting autophagy when combined with certain inhibitors, such as siRNA specific for ATG7(11) and CQ (49,50), compared with cells not treated with these inhibitors. The results from the present study demonstrated that autophagy induction via the HIF/mTOR signaling pathway may contribute to the suppression of the proliferation of cancer cells. Liver cancer cells are often in a state of moderate hypoxia (0.1% oxygen) due to impaired hepatic blood oxygen exchange and metabolic pressure. The present study also confirmed that chronic and moderate autophagy had a protective effect on cancer cells; however, severe hypoxia (<0.1% oxygen) could activate the HIF-1/mTOR signaling pathway and induce cell death (29). We performed the autophagy experiment in a normal state of hypoxia (0.1% oxygen).
Autophagy can be activated by mTOR dependent pathways and non-motor dependent pathways (51,52). Among the various autophagy pathways, the mTOR-related autophagy pathway is the most common one investigated (30). It has been demonstrated that LC3I can be converted into LC3II and transferred to the autophagosome membrane in the cytoplasm, which could also be induced by mTOR downregulation or dephosphorylation (53). p70 ribosomal S6 kinase (p70S6K) is a ribosomal protein downstream of mTOR that can regulate autophagy via AMPK activation mediated by the transforming growth factor-β-activated kinase 1(54). Furthermore, it has been reported that CQ can promote the apoptosis of liver cancer cells treated with sorafenib (49,50); however, in these studies, sorafenib at lower concentration (5-10 µΜ) might not activate the HIF/mTOR autophagy pathway and reversely leads to protective autophagy, which is inhibited by CQ. Of note, sorafenib has been previously confirmed to significantly downregulate mTOR expression in liver cancer, which is consistent with the findings from the present study (47). It has also been shown that mTOR phosphorylation could be markedly increased with sorafenib treatment (55). Summarized, the results from the present study might be explained by the fact that various signaling pathways of autophagy can be activated by different experimental conditions. The present study demonstrated that sorafenib may prevent the proliferation and induce the autophagy and apoptosis of liver cancer cells via the HIF-1/mTOR-related signaling pathway.
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was supported by the Youth Development Foundation of The First Affiliated Hospital of Hainan Medical University (grant no. HYFYPY201704).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
QZY and LHG designed all the experiments. LHG, XLH, JW and YKC performed the experiments. QZY and SBL analyzed the data. QSY collated the data, wrote and edited the manuscript. All authors confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cong WM, Bu H, Chen J, Dong H, Zhu YY, Feng LH, Chen J, Guideline Committee. Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J Gastroenterol. 2016;22:9279–9287. doi: 10.3748/wjg.v22.i42.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamashita T, Kaneko S. Liver cancer. Rinsho Byori. 2016;64:787–796. (In Japanese) [PubMed] [Google Scholar]
- 3.Murray CJL, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 4.Zhu XD, Sun HC. Emerging agents and regimens for hepatocellular carcinoma. J Hematol Oncol. 2019;12(110) doi: 10.1186/s13045-019-0794-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology. 2004;127 (Suppl 1):S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: From diagnosis to treatment. Surg Oncol. 2016;25:74–85. doi: 10.1016/j.suronc.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Keating GM. Sorafenib: A review in hepatocellular carcinoma. Target Oncol. 2017;12:243–253. doi: 10.1007/s11523-017-0484-7. [DOI] [PubMed] [Google Scholar]
- 8.Méndez-Blanco C, Fondevila F, García-Palomo A, González-Gallego J, Mauriz JL. Sorafenib resistance in hepatocarcinoma: Role of hypoxia-inducible factors. Exp Mol Med. 2018;50:1–9. doi: 10.1038/s12276-018-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haga Y, Kanda T, Nakamura M, Nakamoto S, Sasaki R, Takahashi K, Wu S, Yokosuka O. Overexpression of c-Jun contributes to sorafenib resistance in human hepatoma cell lines. PLoS One. 2017;12(e0174153) doi: 10.1371/journal.pone.0174153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gedaly R, Angulo P, Hundley J, Daily MF, Chen C, Evers BM. PKI-587 and sorafenib targeting PI3K/AKT/mTOR and Ras/Raf/MAPK pathways synergistically inhibit HCC cell proliferation. J Surg Res. 2012;176:542–548. doi: 10.1016/j.jss.2011.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, Hosui A, Ishida H, Tatsumi T, Kanto T, et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548–557. doi: 10.1002/ijc.26374. [DOI] [PubMed] [Google Scholar]
- 12.Zhang CZ, Wang XD, Wang HW, Cai Y, Chao LQ. Sorafenib inhibits liver cancer growth by decreasing mTOR, AKT, and PI3K expression. J BUON. 2015;20:218–222. [PubMed] [Google Scholar]
- 13.Ling S, Song L, Fan N, Feng T, Liu L, Yang X, Wang M, Li Y, Tian Y, Zhao F, et al. Combination of metformin and sorafenib suppresses proliferation and induces autophagy of hepatocellular carcinoma via targeting the mTOR pathway. Int J Oncol. 2017;50:297–309. doi: 10.3892/ijo.2016.3799. [DOI] [PubMed] [Google Scholar]
- 14.Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, Chen PJ, Chen KF. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 2013;4(e485) doi: 10.1038/cddis.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. SHARP Investigators Study Group: Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 16.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 17.Xiong XX, Qiu XY, Hu DX, Chen XQ. Advances in hypoxia-mediated mechanisms in hepatocellular carcinoma. Mol Pharmacol. 2017;92:246–255. doi: 10.1124/mol.116.107706. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Xiao Z, Yang L, Gao Y, Zhu Q, Hu L, Huang D, Xu Q. Hypoxia-inducible factors in hepatocellular carcinoma (Review) Oncol Rep. 2020;43:3–15. doi: 10.3892/or.2019.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaupel P, Mayer A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 20.Walsh JC, Lebedev A, Aten E, Madsen K, Marciano L, Kolb HC. The clinical importance of assessing tumor hypoxia: Relationship of tumor hypoxia to prognosis and therapeutic opportunities. Antioxid Redox Signal. 2014;21:1516–1554. doi: 10.1089/ars.2014.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch CJ, Evans SM. Optimizing hypoxia detection and treatment strategies. Semin Nucl Med. 2015;45:163–176. doi: 10.1053/j.semnuclmed.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Höckel M, Vaupel P. Tumor hypoxia: Definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 23.Rana NK, Singh P, Koch B. CoCl2 simulated hypoxia induce cell proliferation and alter the expression pattern of hypoxia associated genes involved in angiogenesis and apoptosis. Biol Res. 2019;52(12) doi: 10.1186/s40659-019-0221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen CB, Schneider BK, White CW. Limitations to oxygen diffusion and equilibration in in vitro cell exposure systems in hyperoxia and hypoxia. Am J Physiol Lung Cell Mol Physiol. 2001;281:L1021–L1027. doi: 10.1152/ajplung.2001.281.4.L1021. [DOI] [PubMed] [Google Scholar]
- 25.Chiu DK, Tse AP, Xu IM, Di Cui J, Lai RK, Li LL, Koh HY, Tsang FH, Wei LL, Wong CM, et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun. 2017;8(517) doi: 10.1038/s41467-017-00530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahraman DS, Diniz G, Sayhan S, Ayaz D, Yemen M, Karadeniz T, Sanci M. The clinicopathologic significance of hypoxia inducible factor 1 alpha expression in ovarian serous tumors. Eur J Gynaecol Oncol. 2019;40:242–245. [Google Scholar]
- 27.Lin C-A, Chang L-L, Zhu H, He Q-J, Yang B. Hypoxic microenvironment and hepatocellular carcinoma treatment. Hepatoma Res. 2018;4(26) [Google Scholar]
- 28.Wilson GK, Tennant DA, McKeating JA. Hypoxia inducible factors in liver disease and hepatocellular carcinoma: Current understanding and future directions. J Hepatol. 2014;61:1397–1406. doi: 10.1016/j.jhep.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 29.Fang Y, Tan J, Zhang Q. Signaling pathways and mechanisms of hypoxia-induced autophagy in the animal cells. Cell Biol Int. 2015;39:891–898. doi: 10.1002/cbin.10463. [DOI] [PubMed] [Google Scholar]
- 30.Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM, Chen ZS. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36(52) doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White E. Autophagy and p53. Cold Spring Harb Perspect Med. 2016;6(a026120) doi: 10.1101/cshperspect.a026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Zhang JX, Zhang QL. PI3K/AKT/mTOR-mediated autophagy in the development of autism spectrum disorder. Brain Res Bull. 2016;125:152–158. doi: 10.1016/j.brainresbull.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25:620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Cheng Q, Yin H, Yang G. Regulation of autophagy and EMT by the interplay between p53 and RAS during cancer progression (Review) Int J Oncol. 2017;51:18–24. doi: 10.3892/ijo.2017.4025. [DOI] [PubMed] [Google Scholar]
- 37.Wu Q, Wu W, Fu B, Shi L, Wang X, Kuca K. JNK signaling in cancer cell survival. Med Res Rev. 2019;39:2082–2104. doi: 10.1002/med.21574. [DOI] [PubMed] [Google Scholar]
- 38.Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, Qiang L, Sample A, Shah P, He Y-Y. NF-κB signaling activation induced by chloroquine requires autophagosome, p62 protein, and c-Jun N-terminal kinase (JNK) signaling and promotes tumor cell resistance. J Biol Chem. 2017;292:3379–3388. doi: 10.1074/jbc.M116.756536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edelstein CL, Venkatachalam MA, Dong Z. Autophagy inhibition by chloroquine and hydroxychloroquine could adversely affect acute kidney injury and other organ injury in critically ill patients with COVID-19. Kidney Int. 2020;98:234–235. doi: 10.1016/j.kint.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiskirchen R, Tacke F. Relevance of autophagy in parenchymal and non-parenchymal liver cells for health and disease. Cells. 2019;8(8) doi: 10.3390/cells8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bravo-San Pedro JM, Pietrocola F, Sica V, Izzo V, Sauvat A, Kepp O, Maiuri MC, Kroemer G, Galluzzi L. High-throughput quantification of GFP-LC3+ dots by automated fluorescence microscopy. Methods Enzymol. 2017;587:71–86. doi: 10.1016/bs.mie.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Runwal G, Stamatakou E, Siddiqi FH, Puri C, Zhu Y, Rubinsztein DC. LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep. 2019;9(10147) doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee YK, Lee JA. Role of the mammalian ATG8/LC3 family in autophagy: Differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 2016;49:424–430. doi: 10.5483/bmbrep.2016.49.8.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasaki R, Kanda T, Fujisawa M, Matsumoto N, Masuzaki R, Ogawa M, Matsuoka S, Kuroda K, Moriyama M. Different mechanisms of action of regorafenib and lenvatinib on Toll-like receptor-signaling pathways in human hepatoma cell lines. Int J Mol Sci. 2020;21(3349) doi: 10.3390/ijms21093349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai N, Ye R, He Q, Guo P, Chen H, Zhang Q. Capsaicin and sorafenib combination treatment exerts synergistic anti- hepatocellular carcinoma activity by suppressing EGFR and PI3K/Akt/mTOR signaling. Oncol Rep. 2018;40:3235–3248. doi: 10.3892/or.2018.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bareford MD, Hamed HA, Tang Y, Cruickshanks N, Burow ME, Fisher PB, Moran RG, Nephew KP, Grant S, Dent P. Sorafenib enhances pemetrexed cytotoxicity through an autophagy-dependent mechanism in cancer cells. Autophagy. 2011;7:1261–1262. doi: 10.1158/0008-5472.CAN-11-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rong LW, Wang RX, Zheng XL, Feng XQ, Zhang L, Zhang L, Lin Y, Li ZP, Wang X. Combination of wogonin and sorafenib effectively kills human hepatocellular carcinoma cells through apoptosis potentiation and autophagy inhibition. Oncol Lett. 2017;13:5028–5034. doi: 10.3892/ol.2017.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi YH, Ding ZB, Zhou J, Hui B, Shi GM, Ke AW, Wang XY, Dai Z, Peng YF, Gu CY, et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159–1172. doi: 10.4161/auto.7.10.16818. [DOI] [PubMed] [Google Scholar]
- 51.Heras-Sandoval D, Pérez-Rojas JM, Hernández-Damián J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014;26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 52.Kim YC, Guan KL. mTOR: A pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fritzen AM, Frøsig C, Jeppesen J, Jensen TE, Lundsgaard AM, Serup AK, Schjerling P, Proud CG, Richter EA, Kiens B. Role of AMPK in regulation of LC3 lipidation as a marker of autophagy in skeletal muscle. Cell Signal. 2016;28:663–674. doi: 10.1016/j.cellsig.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Sun J, Mu Y, Jiang Y, Song R, Yi J, Zhou J, Sun J, Jiao X, Prinz RA, Li Y, et al. Inhibition of p70 S6 kinase activity by A77 1726 induces autophagy and enhances the degradation of superoxide dismutase 1 (SOD1) protein aggregates. Cell Death Dis. 2018;9(407) doi: 10.1038/s41419-018-0441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gedaly R, Angulo P, Hundley J, Daily MF, Chen C, Koch A, Evers BM. PI-103 and sorafenib inhibit hepatocellular carcinoma cell proliferation by blocking Ras/Raf/MAPK and PI3K/AKT/mTOR pathways. Anticancer Res. 2010;30:4951–4958. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.