Abstract
The current pandemic of coronavirus disease 2019 (COVID-19) has affected >160 million individuals to date, and has caused millions of deaths worldwide, at least in part due to the unclarified pathophysiology of this disease. Identifying the underlying molecular mechanisms of COVID-19 is critical to overcome this pandemic. Metabolites mirror the disease progression of an individual and can provide extensive insights into their pathophysiological significance at each stage of disease. We provide a comprehensive view of metabolic characterisation of sera from COVID-19 patients at all stages using untargeted and targeted metabolomic analysis. As compared with the healthy controls, we observed different alteration patterns of circulating metabolites from the mild, severe and recovery stages, in both the discovery cohort and the validation cohort, which suggests that metabolic reprogramming of glucose metabolism and the urea cycle are potential pathological mechanisms for COVID-19 progression. Our findings suggest that targeting glucose metabolism and the urea cycle may be a viable approach to fight COVID-19 at various stages along the disease course.
Short abstract
This cross-sectional metabolomics study proved that serum metabolite profiles are significantly different in different COVID-19 stages. The TCA and urea pathways may participate in pathological processes associated with COVID-19 progression. https://bit.ly/3hyB9RK
Introduction
Coronavirus disease 2019 (COVID-19), with its devastating consequences for the patients and their families, remains a major public health concern that has imposed an increasing financial and social burden worldwide [1, 2]. Notably, infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, may be asymptomatic, or may result in a wide spectrum of clinical symptoms, ranging from mild acute febrile illness to moderate, clinically severe and critical life-threatening infections [3–5]. This disease progression of COVID-19 from mild to severe is supposed to be driven by multiorgan failure with specific metabolic alterations in patients. It is well known that viruses completely rely on their host's cell energy and metabolic resources to fuel the different stages of viral infection, such as entry, multiplication and exit for a new round of infection [6]. For certain respiratory tract viruses, the consequential development of hypoxia caused by lung function impairment may also alter metabolic profiles [7, 8]. The metabolite profiles of coronavirus infections have been investigated previously by several research groups and they showed that levels of a series of small-molecule metabolites, such as free fatty acids, kynurenine, sphingolipids, glucose and amino acids were altered as a result of virus infection, which were supposed to be involved in changes in organ function and immune responses [9, 10]. However, the genesis of COVID-19 is a multistep process, and samples collected from severely ill patients or the mixed groups (including both mild or moderate and severe types) were used in these investigations. Thus, the distinct stage-specific phenotypes of metabolite profiles caused by SARS-CoV-2 infection are still not fully understood.
In this study, a metabolomic analysis has been applied to screen metabolic variations, and to provide a comprehensive view of endogenous metabolites. Metabolomics approaches are generally classified as untargeted analysis or targeted analysis. The untargeted analysis is a relative-quantitative procedure that maximally extracts metabolite information from the complex biological matrix without bias, whereas targeted quantitative metabolomics focuses on the detection of specific metabolites associated with a detailed class or metabolic pathways through the standard curve. We first analysed the metabolic variations using an untargeted approach, and then confirmed our findings using a targeted approach. Further analysis showed that these altered metabolites are mainly involved in the tricarboxylic acid (TCA) cycle and the urea cycle pathway. More importantly, recovery groups were included in our investigation. We obtained more valuable information about metabolic variations by comparing the recovery group with other groups.
Materials and methods
Study design and participants
From January to March 2020, 63 adult patients with COVID-19 confirmed at the Guangzhou Center for Disease Control and Prevention (CDC) laboratory were recruited and referred to the designated treatment centre in Guangzhou for treatment. The diagnosis of COVID-19 was made following the protocol for novel coronavirus pneumonia diagnosis and treatment issued by the National Health Commission of the People's Republic of China [11]. According to Chinese clinical guidance for COVID-19 pneumonia diagnosis and treatment (supplementary table S1), patients were classified into three subgroups: 18 cases in the mild group, 12 cases in the severe group and 20 cases in the recovery group.
For the prospective validation study, a further 93 patients who had been diagnosed with COVID-19 were enrolled independently. Of these 93 peripheral blood samples, three had to be excluded due to haemolysis, leaving data from 90 patients for validation analysis.
During this same period, 13 and 28 healthy volunteers in the discovery and validation cohorts, respectively, including 22 males and 19 females with a mean age of 48 years (range 42–56 years), were recruited at the medical examination centre at No. 8 People's Hospital of Guangzhou.
Sample processing and clinical data collection
Blood samples from the mild and severe groups were collected at the time of diagnosis and when the patient's condition reached the severe standard. Samples from the recovery group were collected following two negative COVID-19 tests and release from isolation. Blood samples from healthy controls were collected from individuals who had an annual physical examination during that period of time. Peripheral blood samples were collected in vacuum negative-pressure blood collection vessels. All subsequent processes were handled under biosafety level 3 containment conditions following risk assessments and code of practice approved by Guangdong CDC. Blood samples in serum separator tubes were centrifuged at 4°C at 1500×g for 10 min and the serum aliquoted and stored at −80°C until analysis.
Microsoft Excel (2013, version 15.0) is used for data collection on demographics and clinical information. Data on demographics, clinical presentations on admission, chest computed tomography (CT) scans and laboratory tests were collected. Laboratory data collected on each patient included complete blood count, urine and stool analysis, blood biochemistry, coagulation function, biomarkers of infection, as well as viral testing through nasopharyngeal swabs. Clinical data were obtained by reviewing clinical charts, nursing records and results of laboratory testing and CT scans for all laboratory-confirmed patients. The accuracy of the clinical data was confirmed by two clinicians caring for the patients.
To explore whether SARS-CoV-2 infection could induce an intense infection response, we also tested three cytokines using samples from four groups by ELISA assay. The human tumour necrosis factor (TNF)-α (AD11069Hu), interleukin (IL)-1β (AD11042Hu) and IL-6 (AD10759Hu) ELISA kits were purchased from Beijing Andy Gene (Beijing, China). The concentrations of these three inflammatory biomarkers were determined according to the manufacturer's protocols and absorbance was measured at 450 nm using a microplate reader. All samples were analysed in triplicate and the average concentration for each patient was calculated.
Untargeted liquid chromatography with tandem mass spectrometry analysis
Untargeted analysis of serum samples was similar to that described previously [12], with minor modifications. 100-µL serum samples were extracted by adding 300 µL of pre-cooled methanol and acetonitrile (2:1, v/v). Following vortex extraction for 1 min and incubation at −20°C for 2 h, samples were centrifuged for 20 min at 1610×g. The supernatant was transferred into Eppendorf tubes for vacuum freeze-drying. The metabolites were resuspended in 150 µL 50% methanol and centrifuged for 30 min at 1610×g, and the supernatants were transferred to autosampler vials for analysis. Quality control (QC) samples were prepared by pooling the same volume of each sample to evaluate the reproducibility of the analysis. The samples were performed on a system including Waters 2D Ultraperformance Liquid Chromatography (UPLC) (Waters, Milford, MA, USA), coupled to a Q-Exactive mass spectrometer (ThermoFisher Scientific, Waltham, MA, USA) with a heated electrospray ionisation source and controlled by the Xcalibur 2.3 software programme (ThermoFisher Scientific). Extracts were separated on a Waters ACQUITY UPLC BEH C18 column (1.7 μm, 2.1×100 mm; Waters). The column temperature was maintained at 45°C. The mobile phase consisted of 0.1% formic acid (A) and acetonitrile (B) in the positive mode, and consisted of 10 mM ammonium formate (A) and acetonitrile (B) in the negative mode. The separation was conducted through the following gradient: 0–1 min (2% B); 1–9 min (2–98% B); 9–12 min (98% B); 12–12.1 min (98–2% B); and 12.1–15 min (2% B). The flow rate was 0.35 mL·min−1 and the injection volume was 5 μL. The mass spectrometry parameters for positive/negative ionisation modes were set as following: spray voltage 3.8/−3.2 kV; sheath gas flow rate 40 arbitrary units (AU); auxiliary gas flow rate 10 AU; auxiliary gas heater temperature 350°C; capillary temperature 320°C.
The untargeted raw data (raw file) was imported into Compound Discoverer 3.0 (ThermoFisher Scientific) for data processing, including peak extraction, retention time correction within and between groups, additive ion pooling, missing value filling, background removal and metabolite identification. Metabolite identification was conducted against the BGI reference library (Shenzhen, China), mzCloud (ThermoFisher Scientific), ChemSpider, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Human Metabolome Database (HMDB). Statistical analysis was performed on the resulting normalised peak intensities using metaX software [13]. There are multiple steps in metaX processing, including data quality assessment, missing value imputation, data normalisation, univariate and multivariate statistics [14, 15].
Targeted liquid chromatography with tandem mass spectrometry verification of identified metabolites
Targeted liquid chromatography (LC) with tandem mass spectrometry (MS) analysis was performed using a UPLC-MS/MS system (ACQUITY UPLC-Xevo TQ-S). 306 standards were obtained from Sigma-Aldrich (St. Louis, MO, USA), Steraloids (Newport, RI, USA) and TRC Chemicals (Toronto, ON, Canada). All the standards were accurately weighed and prepared in an appropriate solution to obtain the individual stock solution at a concentration of 5.0 mg·mL−1. An appropriate amount of each stock solution was mixed to create stock calibration solutions. Serum samples were prepared and measured according to previously published methods [16–18]. Briefly, an aliquot of 25 μL of serum was added to a 96-well plate. 120 μL ice-cold methanol with internal standards was automatically added to each sample and vortexed for 5 min. Then the samples were centrifuged at 4000×g for 30 min. 30 μL supernatant was transferred to a clean 96-well plate, 20 μL of the derivative reagents were added to each well and the derivatisation was carried out at 30°C for 60 min. After derivatisation, 330 μL of 50% methanol solution was added and then the plate was stored at −20°C for 20 min. Then the samples were centrifuged at 4000×g at 4°C for 30 min. Finally, the supernatant was used for LC-MS analysis.
The mobile phases consisted of 0.1% formic acid in water (mobile phase A) and acetonitrile-methanol (70:30, mobile phase B). 5 µL of each sample was injected onto an ACQUITY BEH C18 column (1.7 µm, 100×2.1 mm) (Waters) maintained at 40°C. The flow rate was 0.40 mL·min−1 with the following mobile-phase gradient: 0.0–1.0 min (5% B); 1.0–11.0 min (5–78% B); 11.0–13.5 min (78−95% B); 13.5–14.0 min (95−100% B); 14.0–16.0 min (100% B); 16.0–16.1 min (100–5% B); and 16.1–18.0 min (5% B). The mass spectrometer was operated in the negative mode with a 2.0-kV capillary voltage and the positive mode with 1.5-kV capillary voltage. The source and desolvation temperatures were 150°C and 550°C, respectively.
The targeted raw data were processed through calibration curve of standards. The calibration curve was analysed by instrument response and standard concentration. Then, the targeted metabolites were analysed using the iMAP software (version 1.0; Metabo-Profile, Shanghai, China). Orthogonal partial least-square (OPLS) discriminant analysis (DA) was used to maximise identification of differences in metabolic profiles between groups. In addition to the multivariate statistical method, the test or Wilcoxon's rank-sum test was also applied to estimate the significance of each metabolite. The p-values across all metabolites within each comparison were adjusted by a false discovery rate method. Finally, the significantly differential metabolites need to meet the following criteria: 1) variable importance on projection (VIP) >1; 2) log2 fold change (FC) >0.25 or <−0.25; 3) false discovery rate (FDR) <0.05.
Statistical analysis
All data were analysed using R (version 3.5.0) and differential metabolomic analysis was performed using the MSstats R package, which includes log2 transformation, normalisation and p-value calculation on the Spectronaut and Skyline quantitative data. The clustering algorithm used hierarchical cluster analysis with the hcaMethods R package, and the distance calculation was performed in Euclidean distance with the heatmap package in R software to reveal the relationship between samples and metabolites. KEGG pathway enrichment analysis of differential metabolites was performed using the KEGG database (version 89.1) using Fisher's exact test by comparison with all the identified metabolites. Correlation analysis was performed using Pearson's correlation coefficient analysis with corrplot R package.
Categorical variables were summarised as percentages and compared between different groups using the Chi-squared test or Fisher's exact test. Continuous variables were expressed as mean (95% CI) or median (interquartile range), as deemed appropriate. For the multiple comparison of the four independent groups, the raw values of each metabolite were selected and normalised to the control group. Tukey's range test was conducted for all 11 metabolites, and the significance level for the test, α-value, was set to 0.5.
To verify whether the identified metabolites could be used as potential predictors for classification of different stage groups, a logistic regression model with a five-fold cross-validation approach was designed and performed to predict reliability and failure rate. All possible combinations containing n metabolites (n=1, 2, 3,…,9) were exhaustively tested by the model, and the predictive power of each combination was recorded and ranked according to the area-under-the-curve (AUC) value by receiver operating characteristic analysis. To ensure a large enough sample size, all corresponding AUCs were calculated for the discovery cohort and validation cohort as a whole, and presented as specific or average values.
Data and materials availability
This study did not generate new unique reagents. Untargeted and targeted metabolomics datasets are deposited into the public Mendeley database (https://data.mendeley.com/datasets/pcdgvmg9ws/draft?a=68f3ffb1–077e-47bc-945d-c9207cdc1c71).
Study approval
The study protocol for patient and healthy volunteer recruitment and sampling was approved by the ethical committee of Guangzhou CDC with reference number GZCDC-ECHR-2020A0002.
Results
Untargeted metabolomic profiling of sera from all stages of COVID-19 patients
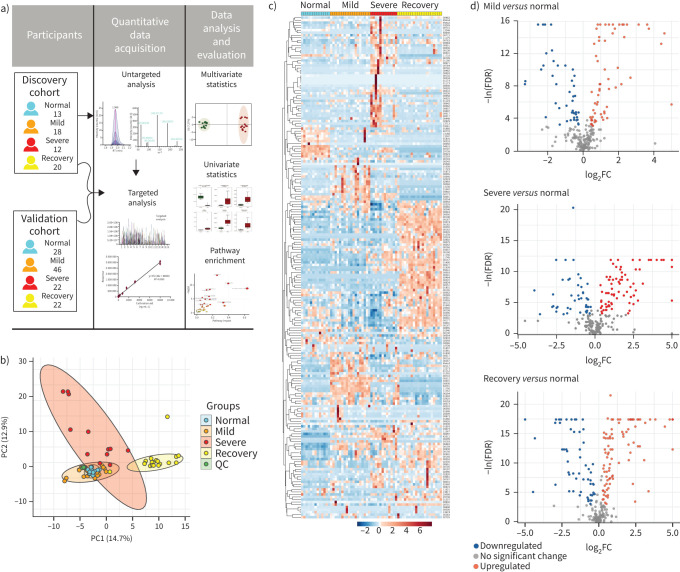
To determine the alterations in circulating metabolites from COVID-19 patients, we applied untargeted metabolomic analysis in the discovery cohort including 13 healthy subjects as normal group, 18 mild cases, 12 severe cases and 20 individuals in recovery from severe COVID-19. Clinical signs and symptoms, together with the laboratory parameters of patients infected with COVID-19 were collected and analysed (table 1). Compared to the mild group, patients in the severe group showed significant suppression of lymphocytes, but an increase of leukocytes and aspartate aminotransferase, indicating the disturbance of immune system and liver function.
TABLE 1.
Characteristics of discovery cohort patients infected with coronavirus disease 2019 at admission, according to disease severity
| Mild | Recovery | Severe | p-value | |
| Patients | 18 | 20 | 12 | |
| Age (years) mean (sd) | 50.67 (43.89–57.44) | 50.20 (42.05–58.35) | 65.33 (58.76–71.90) | 0.013 |
| Gender | ||||
| Male | 9 (50.0) | 12 (60.0) | 7 (58.3) | 0.811 |
| Female | 9 (50.0) | 8 (40.0) | 5 (41.7) | |
| Coexisting disorder # | 11 (61.1) | 7 (35.0) | 5 (41.6) | 0.257 |
| Clinical signs and symptoms | ||||
| Fever | 16 (88.9) | 12 (60.0) | 10 (83.3) | 0.091 |
| Cough | 14 (77.8) | 11 (55.0) | 7 (58.3) | 0.308 |
| Chills | 5 (27.8) | 4 (20.0) | 0 (0.0) | 0.146 |
| Myalgia | 7 (38.9) | 1 (5.0) | 0 (0.0) | 0.004 |
| Fatigue | 11 (61.1) | 0 (0.0) | 2 (16.7) | <0.001 |
| Rhinorrhoea | 4 (22.2) | 1 (5.0) | 0 (0.0) | 0.087 |
| Shortness of breath | 2 (11.1) | 0 (0.0) | 10 (83.3) | <0.001 |
| Sore throat | 5 (27.8) | 0 (0.0) | 1 (8.3) | 0.028 |
| Diarrhoea | 0 (0.0) | 1 (5.0) | 0 (0.0) | 0.465 |
| Chest CT findings | ||||
| Ground-glass opacity | 8 (44.4) | 15 (75.0) | 11 (100.0) | 0.005 |
| Local mottling shadow | 2 (11.1) | 2 (10.0) | 0 (0.0) | 0.499 |
| Bilateral mottling shadow | 9 (50.0) | 5 (25.0) | 3 (25.0) | 0.201 |
| Laboratory parameters mean (sd) | ||||
| Leukocytes (×109 cells·L−1; NR 3.5–9.5) | 5.50 (4.28–6.72) | 5.26 (4.54–5.98) | 9.32 (7.48–11.17) | <0.001 |
| Lymphocytes (×109 cells·L−1; NR 1.1–3.2) | 1.59 (1.13–2.06) | 1.39 (1.13–1.64) | 0.45 (0.30–0.60) | <0.001 |
| Neutrophils (×109 cells·L−1; NR 1.8–6.3) | 3.65 (2.51–4.80) | 3.35 (2.80–3.90) | NA | 0.605 |
| Platelets (×109 cells·L−1; NR 125.0–350.0) | 198.28 (169.52–227.04) | 184.30 (153.83–214.77) | 181.94 (137.44–226.43) | 0.723 |
| HGB (g·L−1; NR 130.0–175.0) | 127.48 (111.08–143.89) | 135.67 (120.24–151.10) | 108.24 (95.03–121.46) | 0.096 |
| APTT (s; NR 21.0–37.0) | 38.93 (36.80–41.05) | 39.73 (37.68–41.77) | NA | 0.574 |
| PT (s; NR 10.5–13.5) | 13.84 (13.41–14.27) | 14.26 (13.36–15.16) | NA | 0.399 |
| D-dimer (mg·L−1; NR 0–500) | 1833.9 (240.8–3427.0) | 1827.0 (976.4–2677.6) | NA | 0.993 |
| Albumin (g·L−1; NR 40.0–55.0) | 39.10 (36.17–41.99) | 39.51 (36.89–42.13) | 33.52 (28.93–38.10) | 0.033 |
| ALT (U·L−1; NR 9.0–50.0) | 30.93 (21.67–40.19) | 26.25 (17.74–34.76) | 95.75 (27.53–163.96) | <0.001 |
| AST (U·L−1; NR 15.0–40.0) | 26.82 (16.35–37.30) | 24.47 (20.36–28.58) | NA | 0.650 |
| Total bilirubin (μmol·L−1; NR 0.0–21.0) | 14.44 (9.87–19.02) | 11.01 (7.72–14.31) | 24.95 (9.40–40.49) | 0.014 |
| BUN (mmol·L−1; NR 3.6–9.5) | 4.43 (3.14–5.71) | 4.52 (3.48–5.56) | NA | 0.904 |
| Scr (μmol·L−1; NR 57.0–111.0) | 59.68 (53.15–66.22) | 78.68 (58.42–98.95) | NA | 0.075 |
| CK (U·L−1; NR 50.0–310.0) | 67.83 (47.14–88.53) | 81.95 (52.77–111.13) | NA | 0.423 |
| LDH (U·L−1; NR 120.0–250.0) | 217.89 (162.28–273.50) | 210.35 (176.99–243.71) | NA | 0.804 |
| CRP (mg·L−1; NR 0.0–5.0) | 19.10 (11.66–26.53) | 20.50 (12.45–28.55) | 9.25 (6.59–11.92) | 0.157 |
Data are presented as n, n (%), unless otherwise stated. CT: computed tomography; NR: normal reference value; HGB: haemoglobin; APTT: activated partial thromboplastin time; PT: prothrombin time; ALT: alanine aminotransferase; AST: aspartate aminotransferase; BUN: blood urea nitrogen; Scr: serum creatinine; CK: creatine kinase; LDH: lactate dehydrogenase; CRP: C-reactive protein; NA: not available. #: including hypertension, type 2 diabetes mellitus, cancer, hyperlipidaemia, liver disease and other chronic diseases.
All serum samples were processed and analysed using UPLC-MS/MS for untargeted metabolomics following the standardised protocol. Hydrophilic and hydrophobic molecules were analysed using both positive and negative ionisation to cover various endogenous biochemical classes. In total, 2466 metabolite peaks representing 631 and 1835 differential metabolites in negative and positive ion modes, respectively, were identified in the untargeted metabolomics analysis. With coefficients of relative standard deviation <0.30 across QC samples, 240 metabolites were identified based on MS/MS spectra through the BGI reference library, MzCloud, KEGG and HMDB (supplementary table S2). From the principal component analysis, the data from the QC set showed consistency and reproducibility (figure 1b). It was noteworthy that the metabolites of the mild group were partially coincident with the normal group, while the severe group and the recovery group were easily distinguished (figure 1b, supplementary figure S1). 193 out of 240 metabolites were significantly associated with COVID-19 (FDR <0.05; figure 1c). Compared to the normal group, the volcano plots showed that there were 48 significantly increased metabolites in the mild group and 33 decreased, while there were 59 increased metabolites and 29 that were decreased in the severe group; the number was even greater in the recovery group, with 64 increased and 34 decreased (figure 1d; FDR <0.05, log2FC >0.25 or <−0.25, VIP >1). Among these significant metabolites, numerous amino acids such as glutamine, glutamate, arginine, ornithine, kynurenine and tryptophan were correlated with disease severity. Some metabolites participating in the TCA cycle such as glucose, lactate and pyruvate showed disorder with the progression of COVID-19. These results suggest that the metabolites involved in the pathways such as nitrogen metabolism, the TCA cycle and fatty acid metabolism were imbalanced during progression of COVID-19 (supplementary figure S2).
FIGURE 1.
Untargeted metabolomic profiling of sera from coronavirus disease 2019 (COVID-19) patients at all stages in the discovery cohort. Untargeted metabolomic analyses were performed using sera from healthy control (normal) group and patients from the mild, severe and recovery groups. a) Schematic diagram of the study design; b) principal component (PC) analysis of untargeted metabolomics among the four groups; c) heatmap of 193 selected metabolites with false discovery rate (FDR) <0.05; d) volcano plots highlighted the serum metabolites that were increased (red) or decreased (blue) in the mild, severe and recovery groups, as compared to the normal group, with FDR <0.05, log2 fold change (FC) >0.25 or <−0.25, orthogonal partial least-square discriminant analysis variable importance on projection >1. QC: quality control.
Targeted metabolomic analysis revealed alterative metabolites in all stages of COVID-19 patients
To further quantify the changes of circulating metabolites between the four groups, we performed the targeted metabolomics analysis using the Q300 Kit (Metabo-Profile). The Q300 Kit has coverage of up to 306 metabolites and >12 biochemical classes; it includes the differential metabolites detected by untargeted screening such as glutamine, arginine, ornithine, glucose, lactate and pyruvate, but also contains other metabolites such as indoles, bile acids and short-chain fatty acids. These metabolites are involved in nitrogen metabolism, the TCA cycle, fatty acid metabolism, amino acid metabolism and bile acid metabolism.
In this study, we first quantified serum metabolites from 59 subjects (discovery cohort; four samples were excluded due to insufficient volume). Based on the concentrations presented in different groups, 199 metabolites were divided into 16 clusters (supplementary figure S3, supplementary table S3). We classified these clusters into three groups, according to whether the metabolites had returned to normal levels in the recovery group (supplementary figure S3a–c). Concentrations of metabolites in supplementary figure S3c returned to nearly normal levels, while those in supplementary figure S3a and b still showed substantial differences in contrast to normal levels. Concentrations of metabolites in supplementary figure S3a increased as the disease progressed, and were maintained at relatively higher level in the recovery group. Concentrations of metabolites shown in supplementary figure S3b were found to decrease as the disease developed, but did not return to normal levels.
We compared the metabolomic profiling of various stages of COVID-19 to the healthy group to identify and characterise specific metabolites and metabolic pathways involved in the progression of COVID-19. We focused on the difference between the various stages of COVID-19 with the normal group. A significance threshold of 0.5 was generally adopted for Q2 and R2 in the OPLS-DA model [19–21]. Clear differences were found for the following groups: normal versus mild, cumulative R2Y=0.941 and Q2Y=0.783; normal versus severe, R2Y=0.955 and Q2Y=0.79; normal versus recovery, R2Y=0.926 and Q2Y=0.815 (supplementary figure S4). Furthermore, we quantified serum metabolites from another 118 subjects as an independent validation (for demographic characteristics, see supplementary table S4). The same model was adopted, and the results showed high consistency, which were presented in supplementary figure S5.
Alteration of main metabolites across different stages
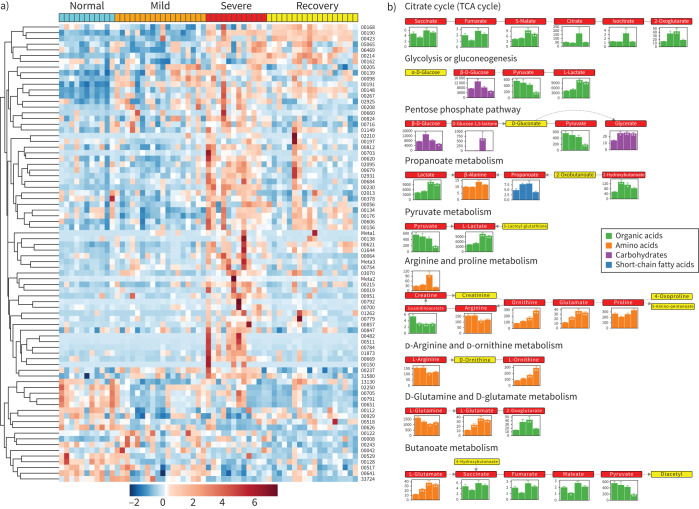
80 out of 199 metabolites quantified in the targeted metabolomics showed significant differences (FDR <0.05; supplementary table S5) in at least one of the stages compared to the normal group (figure 2a). Pathway analysis by the Small Molecule Pathway Database and KEGG showed that aspartate metabolism, the urea cycle, arginine and proline metabolism, glycine and serine metabolism and the TCA cycle were disordered (figure 2b, supplementary figure S6a and b). In particular, arginine and ornithine, both of which are crucial components in the urea cycle, presented with a remarkable decrease or increase (figure 2b, supplementary figure S6). The influences on TCA cycle, pyruvate metabolism and glycolysis merged to indicate the disturbance of energy metabolism, which was supported by the extreme high level of lactate in the severe group. This might be because patients still could not obtain oxygen effectively, and nutrition might also be affected, even with respiratory support, thus creating a huge obstacle for energy metabolism. This phenomenon could also be found in the patients who went through the severe scenario into the recovery group, indicating that a full recovery had not been achieved, even with a negative COVID-19 nucleic acid test. In other words, even if SARS-CoV-2 were completely removed, the damage still exists for some time, or a long time. Changes in the metabolite abundances in all four groups were shown in the pathway representation (figure 2b).
FIGURE 2.
Alteration of main metabolites in sera from the discovery cohort at different stages. a) Heatmap of metabolites quantified in targeted metabolomics with false discovery rate <0.05. b) Pathway analysis identified significant differences in amino acid metabolism, especially regarding arginine, ornithine and glutamine, together with energy metabolism, including the tricarboxylic acid (TCA) cycle and glycolysis. Each box in a pathway represents a metabolite, and is marked in red if found in targeted metabolomics or in yellow for untargeted metabolomics. Bar plots show averaged absolute concentration of metabolites of normal, mild, severe and recovery stages (from left to right).
Dysregulated urea cycle and TCA cycle in disease progression
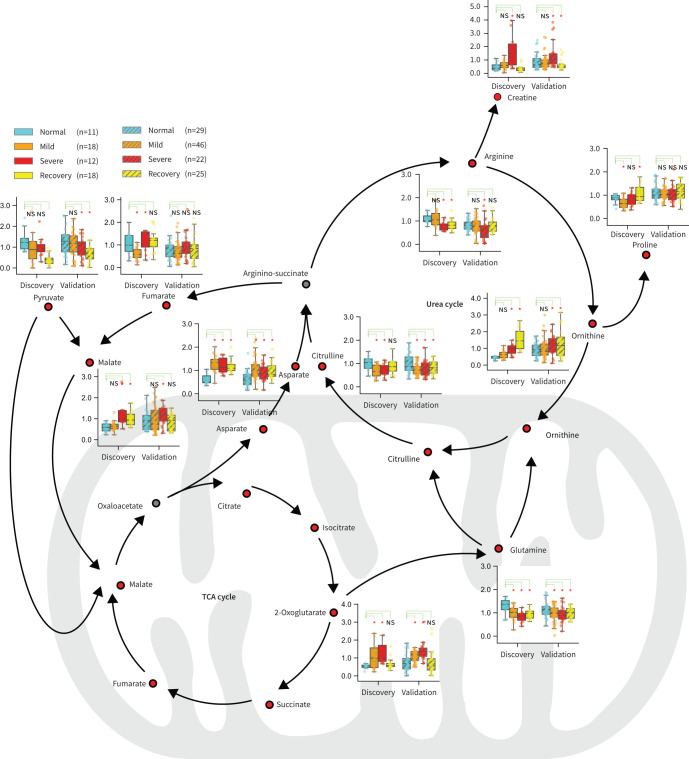
The urea cycle and TCA cycle responsible for amino acid metabolism and energy metabolism were ranked as the top two affected metabolic pathways according to the analysis (figure 3). 11 metabolites that belong to or are closely related to the two cycles were observed with significant changes among different groups (figure 3, supplementary table S6) in the discovery cohort. Furthermore, nine out of the 11 metabolites were found to have significant changes among different groups (figure 3, supplementary table S7). These nine metabolites were creatine, arginine, ornithine, asparate, pyruvate, malate, citrulline, glutamine and 2-oxoglutarate. Focused analysis highlighted significant increases of 2-oxoglutarate, asparate and ornithine levels, compared to the normal group. Malate and 2-oxoglutarate presented with the highest levels in the severe group, indicating that the TCA cycle was seriously affected. Ornithine, which plays the key role in the urea cycle, increased consistently and reached the highest level in the recovery group. In contrast, the level of arginine, another vital amino acid in the urea cycle, decreased significantly in the severe group compared to that in the normal group, suggesting the dysregulation of the urea cycle during the progression of COVID-19.
FIGURE 3.
Dysregulation of the urea cycle and the tricarboxylic acid (TCA) cycle might be involved in the pathological progression of coronavirus disease 2019 (COVID-19). An overview of metabolites closely related to the urea cycle and TCA cycle. Boxplots show levels of these metabolites in the discovery and validation samples at different stages. Metabolomics pathway analysis in both the discovery cohort and the validation cohort identified the urea cycle and the TCA cycle as the top two pathways affected in patients with COVID-19. ns: nonsignificant. *: p<0.05, **: p<0.01, ***: p<0.001.
Combination of sera metabolites may be a potential predictive biomarker
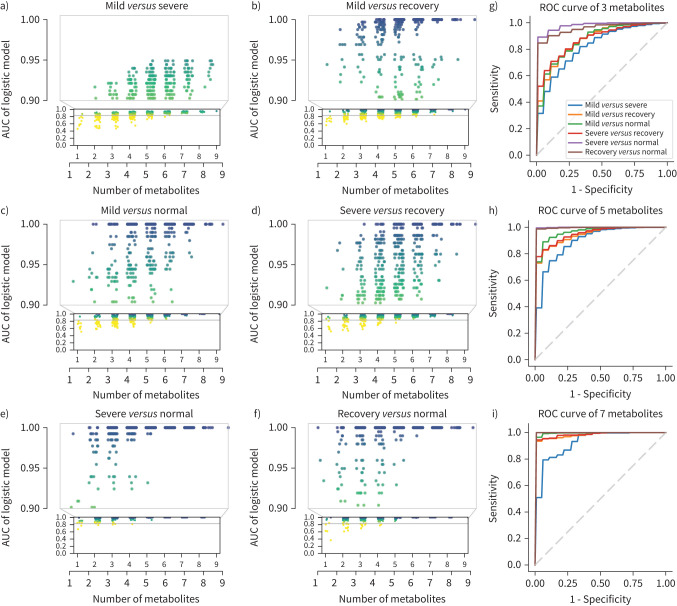
To study whether the nine metabolites could be used as biomarkers to risk-stratify COVID-19 patients, several classic models were trained with the discovery cohort and validated with the validation cohort including decision trees (supplementary figure S7), random forest (supplementary figure S8), support vector machine (supplementary figure S9) and logistic regression (supplementary figure S10). We adopted logistic regression for its better performance. The mean AUCs for logistic regression using three, five and seven metabolites in the validation cohort are presented in supplementary figure S11. Using different combinations of the above nine metabolites and whole-study participant data (discovery cohort+validation cohort), we found that the more metabolites involved, the higher the accuracy of classification distinguishing between any two groups (figure 4a–f). However, the alteration pattern of circulating metabolites was different during disease progression. For example, when we applied fewer metabolites, the model distinguished “severe” from “normal” with relatively higher accuracy, but not "mild" from “normal” (figure 4g). When the number of metabolites increased, the accuracy of the model distinguishing "mild" from "severe" was improved (figure 4g–i). Furthermore, the relatively higher AUC value needed to distinguish “recovery” from “normal” suggests that subjects who survived the severe scenario might still suffer from long-lasting collateral damage regarding to metabolic conditions. For further exploration, we performed logistic regression using nine metabolites individually in the discovery, validation and combined cohorts and found that creatine, 2-oxoglutarate and pyruvate maintained passing performance in three cohorts (supplementary figure S12), leading to the potential for the metabolites to serve as predictive biomarkers. To better validate our findings, we compared the nine metabolites with the results of Shen et al. [9] and found four metabolites that overlapped: malate, pyruvate, citrulline and glutamine. We then built a logistic regression model using these four metabolites as features and trained it with both the discovery and validation cohorts. The model was then validated utilising the published results from Shen et al. [9] and achieved good performance in distinguishing severe and normal (supplementary figure S13), proving that the model could hold up across studies. Such results indicate that malate, pyruvate, citrulline and glutamine might serve as potential biomarkers to risk-stratify COVID-19 severe patients.
FIGURE 4.
Area under the curve (AUC) of models by combination of nine significantly changed metabolites in the urea cycle and tricarboxylic acid cycle. Nine metabolites were used as features to build logistic regression models. a–f) The scatter plots of AUCs in distinguishing different stages using between one and nine metabolites. x-axes show the numbers of the metabolites used in the model. Models by all possible combinations were built, and each point shows the AUC of one single combination. The plots show that the models achieved better performance with increased numbers of metabolites. By combining three metabolites, most models’ AUC >0.90 for severe versus normal. By combining five metabolites, most models’ AUC >0.90 in all group pairs. By combining seven metabolites, all models’ AUC >0.90 in all group pairs. g–i) Mean AUC plots for models using three, five and seven metabolites. The mean AUC of models by all possible combinations with three, five and seven metabolites were calculated separately. The nine metabolites included creatine, arginine, ornithine, asparate, pyruvate, malate, citrulline, glutamine and 2-oxoglutarate.
Discussion
This study, for the first time, systematically provides a comprehensive view of metabolic characterisation of sera from COVID-19 patients at all stages, especially including the recovery stage of the disease course. Targeted and untargeted metabolomic profiling of sera from COVID-19 patients identified different alteration patterns of circulating metabolites from the mild, severe and recovery stages. Most individuals who acquire the SARS-CoV-2 infection experience mild disease, whereas some progress to severe or critical disease. These severe/critical and recovery cases are driven by the host response to the infection, and develop multisystem dysfunction and pathology. Comprehensive analysis of circulating metabolites from COVID-19 patients at all stages would help us develop a comprehensive strategy to better treat this disease.
We identified circulating metabolites, including malate, 2-oxoglutaratek and asparate, which were increased compared to normal levels, suggesting that the dysregulated glucose metabolism and TCA cycle could be crucial in susceptibility, severity and recovery during the COVID-19 disease course. Glucose either goes through aerobic metabolism and provides energy for diverse biological processes or is oxidised through the pentose phosphate pathway to yield NADPH to maintain redox homeostasis, which involves in host immune responses against pathogenic micro-organisms under aerobic conditions. While under anaerobic conditions, which is most common in COVID-19 patients, glucose goes through glycolysis and is fermented to lactate with a limited amount of ATP production, and causes the elevated blood lactate and lactate dehydrogenase levels [22, 23]. Our findings indicate that, in addition to anaerobic glycolysis, which usually occurs in most COVID-19 patients, the TCA cycle was also enhanced during all stages of the disease. We proposed that SARS-CoV-2 viruses hijack the host machinery, including glucose metabolism, to promote pathogenesis of COVID-19.
Infection of Caco-2 cells with SARS-CoV-2 was reported to upregulate glucose metabolism and blocking glycolysis with nontoxic concentrations of 2-deoxy-d-glucose prevented SARS-CoV-2 replication [24]. SARS-CoV-2 viruses in infected cells consumed large amount of cellular ATP to support the viral replication, as high ATP concentration promoted the translocation of SARS-CoV-2 in the unwinding of duplex RNA, which is required for viral replication [25].
Type I interferon (IFN-I) is a major component of the innate immune system and represents critical antiviral molecules [26, 27]. Viral infection causes the host to activate an antiviral response that, in part, is dependent on mitochondrial antiviral signalling protein (MAVS) to produce IFN-I. Cell and animal models of SARS-CoV-2 infection, in addition to transcriptional and serum profiling of COVID-19 patients, consistently revealed a unique and inappropriate inflammatory response. This response is defined by low levels of IFN-I and -III juxtaposed with elevated chemokines and high expression of IL-6. Reduced innate antiviral defences coupled with exuberant inflammatory cytokine production are the defining and driving features of COVID-19 [28]. In order to show the inflammation marker changes in our cohort, we selected 18 samples with sufficient volume from each group to examine IL-6, TNF-α and IL-1β levels, which were shown to be involved in COVID-19 infection. Our results showed that IL-1β level was higher in the mild stage, and was the highest in the severe stage, compared with controls. It was much lower in the recovery stage, although still higher than that in control group (supplementary figure S14a). IL-6 and TNF-α had the same trends (supplementary figure S14b and c). These results indicated that inflammation level was closely related with COVID-19 disease development, and might be involved in the production of metabolites. Therefore, we further conducted correlation analysis and found that the levels of asparate, creatine, malate and 2-oxoglutarate were positively correlated with the expression of cytokines (supplementary figure S15). A clinical study showed that patients with no IFN-α production presented poorer outcomes, and all of them required invasive ventilation and longer intensive care unit stays. The viral load tended to be higher in IFN-negative patients with COVID-19 at disease diagnosis [29].
In our study, a dysregulated urea cycle with decreased arginine and increased ornithine was identified in the sera of COVID-19 patients, suggesting that metabolic reprogramming may be involved in the COVID-19 disease progress. Another example showing metabolic reprogramming in the antiviral response came from a mouse model of virus infection. IFN-I was found to alter the expression and function of key enzymes of the urea cycle in hepatocytes, resulting in decreased arginine and increased ornithine concentrations in the circulation and suppressing virus-specific CD8+ T-cell responses [30–32]. However, whether IFN-I also regulates urea cycle in alveolar epithelial cells during SARS-CoV-2 infection is an important question, and remains to be investigated.
In our dataset, the level of arginine significantly decreased in sera of COVID-19 patients compared to that in healthy control group, which is consistent with the findings from other teams [33]. It is well known that arginine can either be converted to ornithine and urea using the enzyme arginase or to nitric oxide and citrulline through the enzyme nitric oxide synthase [34]. Nitric oxide or its derivatives was reported to affect the fusion between the S protein of SARS-CoV and its cognate receptor, angiotensin converting enzyme 2, and to reduce viral RNA production in the early steps of viral replication [35]. The effect of nitric oxide as a potential treatment in COVID-19 is well recognised, and a phase 3 clinical study (PULSE-CVD19–001) of inhaled nitric oxide therapy in up to 500 patients infected with COVID-19 was approved by the United States Food and Drug Administration in May 2020. Decreased levels of arginine with increased concentrations of ornithine but decreased production of ornithine in the sera of COVID-19 patients from our findings suggests that the conversion of arginine to ornithine in the urea cycle is dominant compared to that of arginine to ornithine in the nitric oxide cycle in COVID-19 patients. It would be interesting to investigate whether SARS-CoV-2 infection reduces nitric oxide production to maintain viral replication by switching the metabolic route to the urea cycle.
In addition to our findings of changes in the urea cycle and glucose metabolism in COVID-19, Shen et al. [9] uncovered dysregulated metabolites in lipid metabolism, suggesting their significant regulation in signal transduction and immune activation process. Kimhofer et al. [31] also determined the metabolic effects of SARS-CoV-2 infection on human blood plasma using multiplatform metabolic phenotyping. They observed key discriminant metabolites including elevated α-1-acid glycoprotein and an increased kynurenine/tryptophan ratio in COVID-19 patients, indicating that liver dysfunction was one of the potential pathological mechanisms. In another study on plasma metabolomic and lipidomic alterations associated with COVID-19, carbamoyl phosphatase, which participates in the urea cycle, was shown to be reduced in patients [36]. This finding is particularly consistent with our finding that the level of citrulline, which is formed from carbamoyl phosphate, was significantly downregulated in mild and severe patients. Collectively, these data indicated that the dysregulated urea cycle was involved in the potential pathological mechanisms of COVID-19 disease progression. Bruzzone et al. [32] revealed abnormally high levels of ketone bodies and 2-hydroxybutyric acid, a readout of hepatic glutathione synthesis and marker of oxidative stress in COVID-19 patients, which suggested that SARS-CoV-2 infection induced liver damage associated with dyslipidaemia and oxidative stress. Bojkova et al. [24] established a human cell-culture model for infection with clinically isolated SARS-CoV-2 and determined the infection profiling of SARS-CoV-2 by translatome and proteome proteomics. They revealed the reshape of SARS-CoV-2 in central cellular pathways, including carbon metabolism and nucleic acid metabolism. Their findings further enabled the identification of drugs that inhibit viral replication by targeting these pathways. Shen et al. [9] compared proteomic and metabolomic profiling of sera from COVID-19 patients with that from control individuals. They identified molecular changes, which implicated dysregulation of macrophage, platelet degranulation, complement system pathways, and massive metabolic suppression and might be useful to screen the potential blood biomarkers for severity evaluation. Seriously ill COVID-19 patients are technically afflicted with sepsis, which involves life-threatening organ dysfunction caused by a person's dysregulated immune response to an infection. Our dysregulated metabolite data also showed considerable overlap with previous studies, suggesting an underlying relationship between dysregulated metabolic pathways and development of sepsis [37–40]. Generally speaking, elucidation of the full spectrum of pathophysiological mechanisms and how metabolic reprogramming can be targeted and providing a “proof of principle” for COVID-19 treatment will be important in further investigations.
Although we successfully recruited 63 and 118 patients for the discovery and validation cohorts, respectively, the cohorts were still relatively in small size to obtain the best reliability. To confirm our findings, metabolic analysis of serum samples from larger COVID-19 cohorts may be warranted. However, high consistency of the results from the validation cohort and the discovery cohort consolidated our results. In addition, the cohorts had some important parameters that were not matched. For example, severe cases often occur in the elderly. Although there were no significant differences between the control, mild and recovery groups, it did create a natural mismatch between the mild and recovery or severe group, which should be taken into account when interpreting results.
Taken together, our metabolomic data presented here provide a comprehensive view of circulating metabolite characterisation from COVID-19 patients at all stages and identify the metabolic reprogramming of glucose metabolism and the urea cycle as potential pathological mechanisms of COVID-19. Targeting host metabolism might be a viable approach to fight against COVID-19 at various stages along the disease course [41].
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures ERJ-00284-2021.Figures (2.6MB, pdf)
Supplementary tables ERJ-00284-2021.Tables (1.7MB, pdf)
Shareable PDF
Acknowledgements
We thank all patients and healthy individuals involved in this study, as well as the dedicated medical and research staff fighting against SARS-CoV-2 worldwide. We thank Jinling Tang (Clinical Data Center, Guangzhou Women and Children's Medical Center, Guangzhou, China) for editing the manuscript.
Footnotes
This article has an editorial commentary: https://doi.org/10.1183/13993003.02417-2021
Author contributions: H. Liang, J. Fan, H. Jia and B. Di conceived, designed and supervised the study. Y. Zhang, K. Li, Y. Xu, J. Lin, Q. Zeng and X. Deng collected clinical samples. Y. Wang, D. Zhou, J. Lin and C. Ke performed the experiments. C. Liu, D. Zhou, Y. Wang, Y. Tan, J. Hong, G. Chen and Haiqing Zhang conducted the data analysis. D. Li, C. Ye, Q. Huang, D. Liu, Hao Zhang, F. Lin and L. Zheng performed mapping. Y. Gao, J. Fan, H. Jia, D. Li and H. Liang wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest: H. Jia has nothing to disclose.
Conflict of interest: C. Liu has nothing to disclose.
Conflict of interest: D. Li has nothing to disclose.
Conflict of interest: Q. Huang has nothing to disclose.
Conflict of interest: D. Liu has nothing to disclose.
Conflict of interest: Y. Zhang has nothing to disclose.
Conflict of interest: C. Ye has nothing to disclose.
Conflict of interest: D. Zhou has nothing to disclose.
Conflict of interest: Y. Wang has nothing to disclose.
Conflict of interest: Y. Tan has nothing to disclose.
Conflict of interest: K. Li has nothing to disclose.
Conflict of interest: F. Lin has nothing to disclose.
Conflict of interest: H. Zhang has nothing to disclose.
Conflict of interest: J. Lin has nothing to disclose.
Conflict of interest: Y. Xu has nothing to disclose.
Conflict of interest: J. Liu has nothing to disclose.
Conflict of interest: Q. Zeng has nothing to disclose.
Conflict of interest: J. Hong has nothing to disclose.
Conflict of interest: G. Chen has nothing to disclose.
Conflict of interest: H. Zhang has nothing to disclose.
Conflict of interest: L. Zheng has nothing to disclose.
Conflict of interest: X. Deng has nothing to disclose.
Conflict of interest: C. Ke has nothing to disclose.
Conflict of interest: Y. Gao has nothing to disclose.
Conflict of interest: J. Fan has nothing to disclose.
Conflict of interest: B. Di has nothing to disclose.
Conflict of interest: H. Liang has nothing to disclose.
Support statement: This work was supported by The Key Project of Medicine Discipline of Guangzhou (2021–2023-11), and the Basic Research Project of Key Laboratory of Guangzhou (202102100001). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Zhang X, Tan Y, Ling Y, et al. Viral and host factors related to the clinical outcome of COVID-19. Nature 2020; 583: 437–440. doi: 10.1038/s41586-020-2355-0 [DOI] [PubMed] [Google Scholar]
- 2.Tianbing W, Yanqiu W, Johnson L, et al. A four-compartment model for the COVID-19 infection – implications on infection kinetics, control measures, and lockdown exit strategies. Precis Clin Med 2020; 3: 104–112. doi: 10.1093/pcmedi/pbaa018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020; 395: 514–523. doi: 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murthy S, Gomersall CD, Fowler RA. Care for critically ill patients with COVID-19. JAMA 2020; 323: 1499–1500. doi: 10.1001/jama.2020.3633 [DOI] [PubMed] [Google Scholar]
- 5.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323: 1239–1242. doi: 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 6.Bley H, Schöbel A, Herker E. Whole lotta lipids – from HCV RNA replication to the mature viral particle. Int J Mol Sci 2020; 21: 2888. doi: 10.3390/ijms21082888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bavis RW, Millström AH, Kim SM, et al. Combined effects of intermittent hyperoxia and intermittent hypercapnic hypoxia on respiratory control in neonatal rats. Respir Physiol Neurobiol 2019; 260: 70–81. doi: 10.1016/j.resp.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maile MD, Standiford TJ, Engoren MC, et al. Associations of the plasma lipidome with mortality in the acute respiratory distress syndrome: a longitudinal cohort study. Respir Res 2018; 19: 60. doi: 10.1186/s12931-018-0758-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 2020; 182: 59–72. doi: 10.1016/j.cell.2020.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas T, Stefanoni D, Reisz JA, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020; 5: e140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020; 323: 1061–1069. doi: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning P, Zheng Y, Luo Q, et al. Metabolic profiles in community-acquired pneumonia: developing assessment tools for disease severity. Crit Care 2018; 22: 130. doi: 10.1186/s13054-018-2049-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen B, Mei Z, Zeng C, et al. metaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics 2017; 18: 183. doi: 10.1186/s12859-017-1579-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y, Wang Y, Chen T, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 2017; 355: eaaf4791. doi: 10.1126/science.aaf4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Hou G, Zeng C, et al. Metabolomic profiling reveals amino acid and carnitine alterations as metabolic signatures in psoriasis. Theranostics 2021; 11: 754–767. doi: 10.7150/thno.51154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gou W, Ling CW, He Y, et al. Interpretable machine learning framework reveals robust gut microbiome features associated with type 2 diabetes. Diabetes Care 2021; 44: 358–366. doi: 10.2337/dc20-1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Z, Sun TY, He Y, et al. Dietary fruit and vegetable intake, gut microbiota, and type 2 diabetes: results from two large human cohort studies. BMC Med 2020; 18: 371. doi: 10.1186/s12916-020-01842-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi HH, Zou S, Wu J, et al. EGF relays signals to COP1 and facilitates FOXO4 degradation to promote tumorigenesis. Adv Sci 2020; 7: 2000681. doi: 10.1002/advs.202000681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan X, Jin J, Su X, et al. Intestinal flora modulates blood pressure by regulating the synthesis of intestinal-derived corticosterone in high salt-induced hypertension. Circ Res 2020; 126: 839–853. doi: 10.1161/CIRCRESAHA.119.316394 [DOI] [PubMed] [Google Scholar]
- 20.Bharath LP, Agrawa M, McCambridge G, et al. Metformin enhances autophagy and normalizes mitochondrial function to alleviate aging-associated inflammation. Cell Metab 2020; 32: 44–55. doi: 10.1016/j.cmet.2020.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi M, Matsuo K, Tanabe K, et al. Comprehensive serum glycopeptide spectra analysis (CSGSA): a potential new tool for early detection of ovarian cancer. Cancers 2019; 11: 591. doi: 10.3390/cancers11050591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 2020; 11: 102. doi: 10.1038/s41467-019-13668-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, Liu X, Shen J, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell 2020; 182: 1360. doi: 10.1016/j.cell.2020.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bojkova D, Klann K, Koch B, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020; 583: 469–472. doi: 10.1038/s41586-020-2332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang KJ, Jeong S, Kang DY, et al. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci Rep 2020; 10: 4481. doi: 10.1038/s41598-020-61432-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theofilopoulos AN, Baccala R, Beutler B, et al. Type I interferons (α/β) in immunity and autoimmunity. Annu Rev Immunol 2005; 23: 307–336. doi: 10.1146/annurev.immunol.23.021704.115843 [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Wang W, Chen Z, et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020; 586: 572–577. doi: 10.1038/s41586-020-2599-8 [DOI] [PubMed] [Google Scholar]
- 28.Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020; 181: 1036–1045. doi: 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trouillet-Assant S, Viel S, Gaymard A, et al. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol 2020; 146: 206–208. doi: 10.1016/j.jaci.2020.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lercher A, Bhattacharya A, Popa AM, et al. Type I interferon signaling disrupts the hepatic urea cycle and alters systemic metabolism to suppress T cell function. Immunity 2019; 51: 1074–1087. doi: 10.1016/j.immuni.2019.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimhofer T, Lodge S, Whiley L, et al. Integrative modeling of quantitative plasma lipoprotein, metabolic, and amino acid data reveals a multiorgan pathological signature of SARS-CoV-2 infection. J Proteome Res 2020; 19: 4442–4454. doi: 10.1021/acs.jproteome.0c00519 [DOI] [PubMed] [Google Scholar]
- 32.Bruzzone C, Bizkarguenaga M, Gil-Redondo R, et al. SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. iScience 2020; 23: 101645. doi: 10.1016/j.isci.2020.101645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser DD, Slessarev M, Martin CM, et al. Metabolomics profiling of critically ill coronavirus disease 2019 patients: identification of diagnostic and prognostic biomarkers. Crit Care Explor 2020; 2: e0272. doi: 10.1097/CCE.0000000000000272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rath M, Müller I, Kropf P, et al. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front Immunol 2014; 5: 532. doi: 10.3389/fimmu.2014.00532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akerström S, Gunalan V, Keng CT, et al. Dual effect of nitric oxide on SARS-CoV replication: viral RNA production and palmitoylation of the S protein are affected. Virology 2009; 395: 1–9. doi: 10.1016/j.virol.2009.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu D, Shu T, Yang X, et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl Sci Rev 2020; 7: 1157–1168. doi: 10.1093/nsr/nwaa086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013; 5: 195ra195. doi: 10.1126/scitranslmed.3005893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seymour CW, Yende S, Scott MJ, et al. Metabolomics in pneumonia and sepsis: an analysis of the GenIMS cohort study. Intensive Care Med 2013; 39: 1423–1434. doi: 10.1007/s00134-013-2935-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Sun Y, Teng S, et al. Prediction of sepsis mortality using metabolite biomarkers in the blood: a meta-analysis of death-related pathways and prospective validation. BMC Med 2020; 18: 83. doi: 10.1186/s12916-020-01546-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, Zhang M, Han T, et al. Exploring the biomarkers of sepsis-associated encephalopathy (SAE): metabolomics evidence from gas chromatography-mass spectrometry. Biomed Res Int 2019; 2019: 2612849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayres JS. A metabolic handbook for the COVID-19 pandemic. Nat Metab 2020; 2: 572–585. doi: 10.1038/s42255-020-0237-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures ERJ-00284-2021.Figures (2.6MB, pdf)
Supplementary tables ERJ-00284-2021.Tables (1.7MB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00284-2021.Shareable (281KB, pdf)