Abstract
Cystic fibrosis (CF) is one of the most common, lethal autosomal recessive disease in the Caucasians with a life expectancy of 37-47 years. The CF transmembrane conductance regulator (CFTR) is a plasma membrane ion channel, confined to apical membrane of epithelia, and ensures transepithelial water and solute movement across secretory epithelia in several organs. Numerous CF mutations, including the most prevalent deletion of F508 (ΔF508) in the nucleotide binding domain 1 (NBD1) leads to CFTR global misfolding and premature intracellular degradation at the endoplasmic reticulum (ER). To better understand the misfolding mechanism caused by CF-causing point mutations in the NBD1, which is poorly understood, differential scanning fluorimetry (DSF) and hydrogen deuterium exchange coupled with mass spectrometry (HDX-MS) are the choice of techniques. These established methods can measure the conformational dynamics of the NBD1 globally and at peptide resolution level by monitoring backbone amide HDX, respectively, and will be instrumental to evaluate the mechanism of action of CF mutations and folding correctors that rescue CFTR folding defects via stabilizing the mutant NBD1.
Keywords: cystic fibrosis, conformational diseases, missense mutation, NBD1, unfolding kinetic, thermal stability, melting temperature
1. Introduction
Cystic fibrosis and CFTR folding
Cystic fibrosis (CF) is caused by the functional expression defect of the cystic fibrosis transmembrane conductance regulator (CFTR), a phosphorylation regulated anion selective channel that belongs to the ATP Binding Cassette transport family [1]. Impaired functional expression of CFTR at the apical plasma membrane of secretory epithelia causes CF, the most prevalent, lethal genetic disease in the Caucasian population [2]. While the channel functional expression defect can be attributed to mutations that impair CFTR transcription, splicing, translation, folding, function or stability, combinations of these defects have also been documented [3,4]. Channel misfolding represents the most frequent cause of CF, which renders the newly synthesized polypeptide susceptible to endoplasmic reticulum (ER) associated degradation [5]. Immunoblot analysis of 54 mutations revealed that steady-state expression of the mature, complex-glycosylated form of 44 CFTR variants were significantly reduced in Fisher Rat Thyroid (FRT) epithelia [6]. Impaired conformational maturation of newly synthesized channels at the ER similar to that described for the most common point mutation, deletion of F508 (ΔF508) in the nucleotide binding domain 1 (NBD1) accounts for the loss-of-expression phenotype.
CFTR contains two symmetrical halves, and each consists of a membrane spanning domain (MSD1 or MSD2) that forms the ion permeation pathway and a cytosolic NBD (NBD1 or NBD2). The dimerization of NBDs is coupled to CFTR channel opening, formed by the MSD1-2, via association of the NBDs to the cytosolic loops of MSDs [7]. The two symmetrical halves are connected with a largely unstructured regulatory (or R) region. The cAMP-dependent phosphorylation of the R region is a prerequisite for NBD1-2 dimerization and channel opening [7]. From the 44 processing mutants, 14 were confined to the NBD1 in FRT epithelia [6]. The underlying mechanism of CFTR misfolding, initiated by these point mutations in NBD1, however, remains enigmatic. Although the X-ray crystal structures of the human wild-type (WT) and ΔF508 NBD1, containing seven (F409L, F429S, F433L, G550E, R553Q, R555K and H667R) and two (F494N and Q637R) second site solubilizing or suppressor mutations, respectively, were very similar [8], more recent analysis showed substantial difference between the mutant and WT NBD1 local and global conformational stability [9,10].
The ΔF508 is located in the alpha subdomain of the NBD1 and interferes with both the thermodynamic and kinetic stability of NBD1, as well as uncouples the NBD1-CL4 interface that contributes to CFTR cooperative domain unfolding [11,12]. This paradigm may be relevant for elucidating the underlying cause of other CF mutations associated with the NBD1. While the isolated NBD1 with ΔF508 has been intensively studied using conventional structural biology techniques, such as X-ray crystallography and NMR, the dynamic perturbation of the NBD1 by other CF mutations remain elusive. This can be partly attributed to the low purification yield and the thermal instability of the mutant NBD1s, leading to increased unfolding and aggregation propensity. Here, we describe hydrogen deuterium exchange with mass spectrometry (HDX-MS) technique to establish the conformational dynamics of isolated NBD1 in the absence and presence of a CF mutation. As an initial screening assay to uncover the impact of CF mutation on the conformation stability of NBD1 differential scanning fluorimetry (DSF) is also described.
2. Materials
2.1. NBD1 preparation
pET26b-derived vectors with N-terminal His6–SUMO fusion tag containing the human WT-CFTR with the crystallization domain boundaries (389-678 amino acid residues) (see Note 1).
BL21-CodonPlus (DE3)-RIL E. coli (Agilent).
Isopropyl β-D-1-thiogalactopyranoside (IPTG).
Lysis buffer: 100 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 5 mM ATP, 5 mM imidazole, 2 mM β-mercaptoethanol, 10% ethylene glycol, 10% glycerol.
Protease inhibitor cocktail P8849 (Sigma-Aldrich).
cOmplete, EDTA-free protease inhibitor cocktail (Roche).
IMAC Sepharose 6 Fast Flow (GE Healthcare).
Storage buffer: 50 mM NaPO4, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM ATP, 1 mM TCEP, 10% glycerol.
PD10 desalting column (BioRad).
Superdex 200 size-exclusion chromatography column (GE Healthcare).
Amicon Ultra 10 kDa cut-off (Millipore).
2.2. Differential scanning fluorimetry (DSF)
96-well plate with round or conical-bottom wells for preparing the stock solutions.
White, non-skirted, low-profile 96-well Real-Time (RT)-PCR plate (used to run on the machine).
Self-adhesive optical seal for PCR (BioRad).
Purified NBD1 protein at ~4 mg/ml concentration (see Methods 3.1).
SYPRO orange dye, 5000x concentration in DMSO (Thermo Fisher Scientific).
Screening buffer 3X: 300 mM HEPES, pH 7.5, 450 mM NaCl, 15 mM MgCl2.
250 mM ATP, pH 7.4.
Stratagene Mx3005P RT-PCR (Agilent).
Refrigerated centrifuge for 96 well plates.
2.3. HDX-MS
Deuterium oxide (D2O).
Deuterium chloride (DCl)
Sodium deuteroxide (NaOD).
Urea and urea-d4.
Water with 0.1% formic acid (FA) (LC/MS grade).
Acetonitrile (ACN) with 0.1% FA (LC/MS grade).
Quenching buffer: 300 mM glycine-HCl, pH 2.4, 8 M urea.
H2O-based buffer: 10 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM ATP, 2 mM MgCl2, 1 mM TCEP, 1% glycerol.
D2O-based buffer: same components as H2O-based buffer dissolved in D2O, pD 7.5 (see Note 2).
Pepsin column (see Note 3).
Pepsin column washing cocktails: a) 10 mM glycine-HCl, pH 2.5, 2 M guanidine HCl and b) 5% ACN, 5% isopropanol (IPA), 20% acetic acid (AcOH) [13].
Trap and analytical column washing solution: 80% IPA in ACN.
A 1260 Infinity II isocratic pump (Agilent) and a 1290 Infinity II binary pump (Agilent).
A 0.2 μm pre-column filter (Thermo Fisher Scientifc), a Hypersil GOLD C18 column, 1 mm × 50 mm, 1.9 μm (Thermo Fisher Scientifc) and an EXP C18 trap column (Optimize Technologies).
A LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) and Proteome Discoverer software (Thermo Fisher Scientific).
A model 8125 low dispersion sample injector with a 10 μL sample loop (Rheodyne).
A 1290 Infinity valve drive and 6-port valve head (Agilent).
HDExaminer software (Sierra Analytics).
PeakFit (Systat Software).
3. Methods
3.1. Isolated NBD1 expression and purification (see Note 4)
Revert two of the three solubilizing mutations (F429S and Q637R) in the pET26b-derived vectors to WT sequence by PCR mutagenesis.
Introduce the ΔF508 mutation using the same technique above.
Express the WT- and ΔF508-NBD1 containing the F494N second site solubilizing mutations (NBD1-1S) in BL21-CodonPlus (DE3)-RIL cells.
Scale up an overnight starter culture to 6-8 l at 30°C. At 0.2-0.4 optical density, cool cells to 12°C (2 h) and then induce with 0.1 mM IPTG.
After 16 h of IPTG-induction at 12°C, centrifuge cells (10 min, 6000 rpm) at 4°C. Resuspend cells in lysis buffer, supplemented with protease inhibitor cocktail, and sonicate for 12 × 10 s with 10 s intervals on ice.
Recover the soluble fraction after centrifugation (30 min, 15,000 rpm) at 4°C and apply the fraction onto an equilibrated 1 ml IMAC Sepharose 6 Fast Flow. Wash the IMAC column with eight column volumes (CV) of lysis buffer, including 30 mM imidazole. Elute with lysis buffer containing 500 mM imidazole.
Pool aliquots showing the highest protein concentration and perform buffer exchange with storage buffer using a PD10 desalting column.
Cleave His6-Sumo tag overnight at 4°C using 50 μg/ml of Ulp protease. His6-Sumo Tag and Ulp protease can be pulled down by incubating samples with 100 μl of nickel beads.
Purify cleaved protein using Superdex 200 size-exclusion chromatography column pre-equilibrated in storage buffer.
Concentrate purified NBD1 to 3-5 mg/ml using Amicon Ultra 10kDa cutoff.
3.2. NBD1 melting temperature determination with DSF
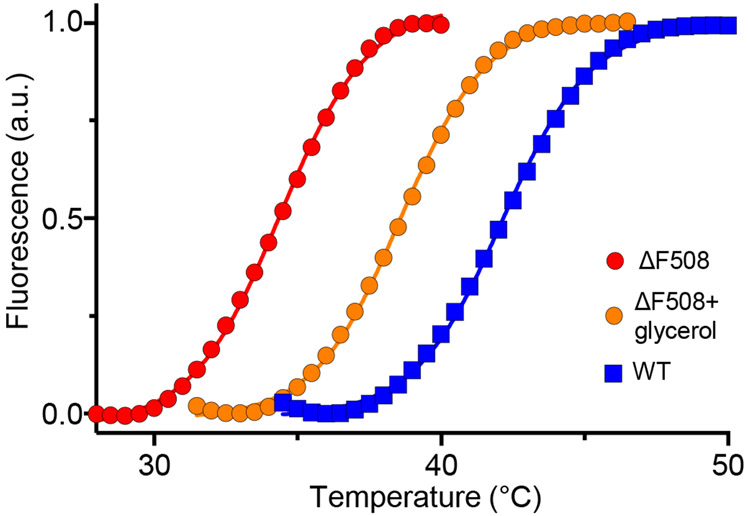
The advantage of monitoring NBD1 thermal unfolding by DSF over spectroscopic techniques (UV absorbance and circular dichroism) is that it requires considerable smaller amount of protein. DSF for NBD1 is performed in the presence of SYPRO orange [14], a fluorescent dye that has significantly increased quantum efficiency upon binding to the exposed hydrophobic patches of unfolded polypeptides as compared to it low binding affinity to the native polypeptides, largely exposed of hydrophilic residues. A polypeptide stability is related to its unfolding Gibbs free energy (ΔGu), which usually decreases with increasing temperature. At the melting temperature (Tm), the ΔGu value is zero and the solution contains equal amounts of folded and unfolded protein. Thus, monitoring augmented SYPRO orange fluorescence as a function of increasing temperature can reveal the impact of CF causing mutation on the isolated native-like NBD1 thermal stability using a RT-PCR machine (see Note 5). Introduction of the ΔF508 mutation into NBD1-1S causes a ~8°C reduction in the Tm as compared to that of the WT-NBD1-1S as illustrated by the DSF plots (Fig. 1). In contrast, 10% glycerol, a chemical chaperone, stabilized the ΔF508-NBD1-1S by ~4°C (Fig. 1). Similar measurements could identify the impact of CF mutations, second site suppressor mutations and specific ligands (or correctors) on the NBD1 domain stability as shown before [15-17].
Figure 1.
Thermal stability of the isolated human WT- and ΔF508-NBD1-1S. Unfolding of the domains was monitored by differential scanning fluorimetry (DSF) in the presence of SYPRO orange as described. The fluorescence intensity was expressed as the percentage of the maximum as a function of temperature. As indicated, ΔF508-NBD1-1S thermal melt was also determined in the presence of 10% glycerol.
3.2.1. Determining the melting temperature of NBD1 by DSF
Before starting, make sure that the RT-PCR machine is ready to go (see Note 6) and that centrifuge is at 4°C. Keep the 96-well plate on ice during preparation.
Prepare test compound(s) (e.g. glycerol) at 2X concentration in 1X Screening buffer (100 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2).
Pipette 5 μl of each condition to be tested into the 96-well plate at least in triplicate.
Prepare the protein mix, containing NBD1-1S at ~0.4 mg/ml, 8X SYPRO orange, 2 mM ATP in 1X screening buffer. Add NBD1 last, mix carefully but thoroughly.
Distribute 5 μl of the protein mix into the RT-PCR 96-well plate.
Cover plate with optical seal, and centrifuge it 2 min at 2.000 rpm at 4°C.
Insert plate into PCR machine and start the run.
3.2.2. Determining the thermal melt temperature (Tm)
To retrieve the fluorescence data from the RT-PCR machine, go to “Analysis” (top right) and choose the “Results” tab.
In the “Fluorescence” scroll down and choose “R (multicomponent view)”.
From “File” menu, choose “Export chart data”, then “Export data to excel” and finally “Format 2 - Horizontally grouped by plot”.
This will generate a read only excel file, including two tables. The top table will include the fluorescence data from wells A1 to D12, and the bottom table the data for wells E1 to H12. In each table, the cycle number is indicated in column A, which corresponds to the plate temperature starting from 25°C in 0.5°C increment. The fluorescence intensity data of individual wells are shown in the following columns (e.g. fluorescence of A1 well will be in column B, A2 well in column C, and so on).
Change the cycle number in column A for the actual temperature (e.g. replace 1 by 25°C, 2 by 25.5°C and 3 by 26°C).
- Copy the raw data into GraphPad Prism and analyze them using “non-linear regression” from the “xy analysis” group, and choose “Boltzmann sigmoidal” equation for fitting according to
where Top and Bottom are the extrapolated maximum and minimum SYPRO orange fluorescence, respectively, X denotes the measured temperature, and V50 is the Tm.
3.3. HDX-MS
The principle of this technique is based on the impeded HDX kinetics of backbone amide hydrogens forming H-bonds or structurally buried in natively folded proteins, relative to flexible or unfolded segments that are more solvent accessible [18,19]. HDX can be initiated by diluting protein samples from stock solutions into high concentration D2O containing medium (Fig. 2a) and monitoring deuterium incorporation at backbone amides as a function of incubation time by mass determinations of proteolytic peptides. On line proteolysis is achieved by immobilized pepsin column, under conditions (pH 2.5, 0°C) that minimize deuterium back exchange. Considering that the back-exchange kinetics of hydrogen at side-chains is very rapid even under quenching conditions, HDX-MS only monitors the backbone amide hydrogen deuteration, which is considered as the primary determinant of the mass shift detected by HDX-MS. The difference in the centroid of isotopic envelopes between deuterated and non-deuterated samples indicates the deuterium incorporation at the peptide level.
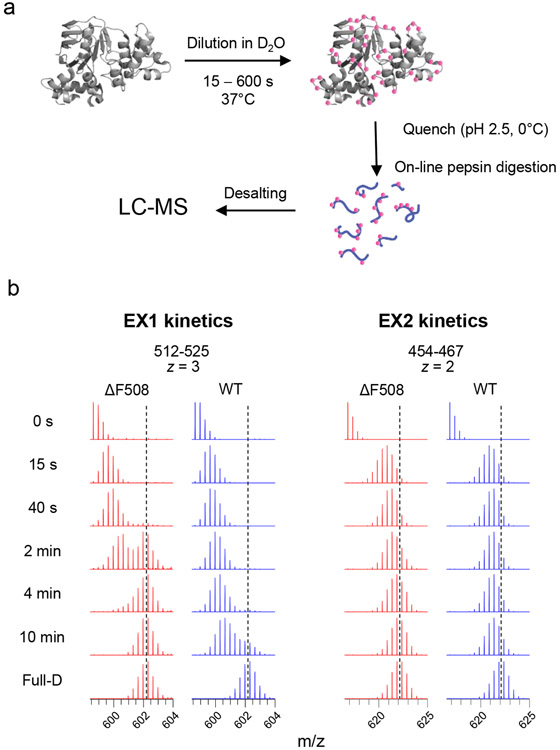
Figure 2.
a) The schematic work flow HDX-MS experiment to determine the NBD1 conformational dynamics. b) Mass spectra of peptides showing the bimodal peak distribution of the EX1 signature and EX2 signature unimodal peak distribution. The blue dotted lines indicate the centroids of isotopic envelopes from unfolded NBD1.
The advantage of HDX-MS for the NBD1 unfolding studies is the high sensitivity of the method that permits monitoring the domain conformation dynamics at relatively low NBD1 concentration (5-10 μM) that largely prevents domain aggregation during thermal unfolding. Unfolded and destabilized regions can be identified by comparing HDX kinetics between WT- and ΔF508-NBD1-1S and projected onto the X-ray crystal structures (e.g. PDB: 2BBT from Protein Data Bank). Backbone amides confined to secondary structural elements (i.e. α-helices and β-strands) during NBD1 unfolding are exposed to solvent display accelerated HDX. The hallmark of peptide/protein unfolding is the EX1 type kinetics (Fig. 2b). The characteristic bimodal peak of peptide mass distributions can be observed for the EX1 kinetics, as a consequence of the presence of both folded and unfolded peptide populations during the course of thermal unfolding. In contrast, the unimodal isotopic distributions of EX2 kinetics indicates a gradual increase of the deuteration of individual peptide population as a consequence of thermal conformational fluctuation of the polypeptide with regional differences (Fig. 2b). Using comparison of deuterium incorporation kinetics, the impact of CF-causing mutations on NBD1 dynamics can be studied in comparison to that of its WT counterpart (see Fig. 2b).
3.3.1. Pepsin digestion pattern of NBD1 for HDX analysis
Detailed conformational dynamic analysis requires full coverage of the entire primary sequence of the NBD1 by its proteolytically digested peptides. To define the proteolytic coverage and facilitate the identification of the deuterated peptides, retention time and the mass-to-charge ratio (m/z) of peptic peptides generated from WT- and ΔF508-NBD1-1S on a pepsin column are identified by tandem MS (MS/MS) analysis.
Prepare 1 μM of NBD1 solution in quenching buffer. Aspirate 20 μl of the solution and inject onto the sample loop using a syringe (Fig. 3a, see Note 7). At this moment, the injector valve is set at “LOAD” position.
The flow rate of the isocratic pump is set to 30 μl/min. Change the injector valve to “INJECT” position. The NBD1 solution passes through an on-line pepsin column, resulting in peptide generation. Generated peptides are captured at the trap column.
At 1.5 min, a flow rate of the isocratic pump is changed to 200 μl/min to wash out any salts and urea.
At 3 min, load peptides onto a C18 analytical column by changing the position of the switching valve (Fig. 3b). Peptides are separated using a linear gradient of 5–40% can, containing 0.1% FA for 60 min at a 65 μl/min flow rate (see Note 8).
The analytical column is directly connected to an electrospray ionization (ESI) source of a mass spectrometer. Identify peptides by running MS/MS analysis in data-dependent acquisition and searching peptides with Proteome Discoverer. Save the results in Excel format (see Note 9).
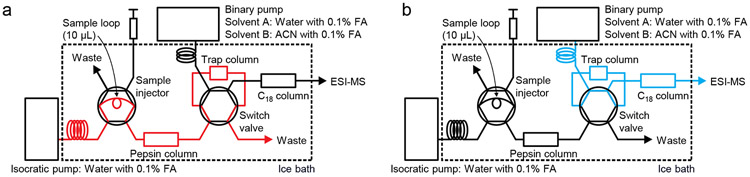
Figure 3.
The configuration of the liquid chromatography setup for HDX-MS experiments. a) Injected samples are loaded on the pepsin column and resulting peptic peptides are captured by a trap column. b) After washing out salts and urea, the captured peptides are eluted from the trap column and fractionated on a C18 analytical column. Separated peptides are directly injected to an ESI ion source.
3.3.2. Backbone amide HDX
Place ~50-100 μM of NBD1 stock solution on ice. Aliquot 27 μl of D2O-based buffer onto a 1.5 ml micro tube and pre-incubate at 37°C for 10 min. Meanwhile, prepare 5 micro tubes containing 16-19 μl of quenching solution (depending on the concentration of a NBD1 stock solution), and place them on ice.
HDX is initiated by mixing 3 μl of NBD1 stock solution with the pre-warmed D2O-based buffer. Incubate for 15, 40, 120, 240, and 300 s at 37°C, and then the HDX reaction is terminated by diluting 1-4 μl of the reaction mixture (pending on the NBD1 concentration) into quenching solutions 0°C. The final concentration of NBD1 should be ~1 μM. The quenched protein solution is flash frozen in either liquid nitrogen or methanol containing dry ice, and is stored at −80°C until use (see Note 10).
For non-deuterated controls, the initial dilution is made in H2O-based buffer. Quench, flash frozen and store as above.
Prepare fully deuterated samples by diluting NBD1 in D2O containing 8 M urea-d4 at room temperature for 3 h. Quench, flash frozen and store as above (see Note 11).
Prior to ultra-high performance liquid chromatography (UHPLC)-MS analysis, place a sample injection syringe on ice for 5 min. Deuterated samples are thawed and immediately loaded onto the injection valve.
Perform UHPLC-MS analysis. The LC settings are same as described in Subheading 3.2, except a 5–40% linear gradient of can, containing 0.1% formic acid for 10 min. Mass spectra of peptides are acquired in full-scan mode with m/z 200-2000.
After each LC-MS analysis, wash a pepsin column with pepsin column wash cocktails, as well as trap and analytical columns with 80% IPA in ACN in order to eliminate peptide carryover (see Note 12).
3.3.3. Data analysis
Prepare the NBD1 amino acid sequence in the FASTA file format. Import the NBD1 sequence in HDExaminer software.
Import a peptide source created in Subheading 3.2.1. Retention time should be adjusted at this stage (see Note 8).
Import mass spectrometry data files of undeuterated controls, followed by mass spectrometry data files of deuterated samples in each time point.
Export HDX results (the number of deuterium uptake and the percentage of deuteration in each time point) in Excel format (see Note 13).
Optionally, export deuterium uptake plots (in PDF or EMF format), a deuteration heat map (EMF format for the primary structure, Pymol or Chimera format for the 3D structure, see Fig. 4a and 4b), and peptide spectra (Excel format) generated by HDExaminer.
Locate regions showing the difference in deuterium uptake by comparing different states (Fig. 4c).
Identify unfolded regions by inspecting EX1 kinetics in peptide mass spectra (Fig. 2b).
-
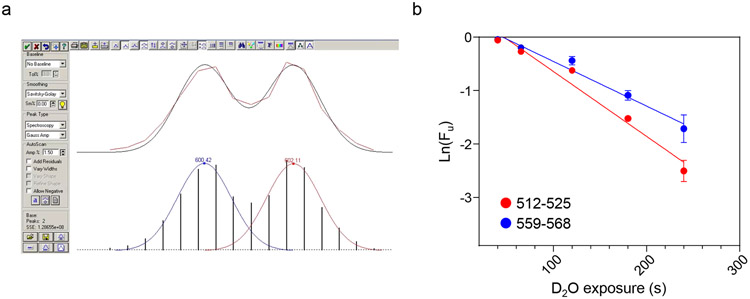
To determine unfolding kinetics, perform deconvolution of bimodal isotopic distributions to separate folded and unfolded populations using PeakFit software (Fig. 5a). The Gaussian distribution of folded and unfolded populations (Af and Au, respectively) are used to determine the fraction of unfolded population (Fu) according toThe natural logarithm of the Fu is plotted against D2O exposure time (Fig. 5b). Rate constant of unfolding is determined by fitting the data using an equation
where ku is the rate constant of unfolding, and t is the D2O exposure time.
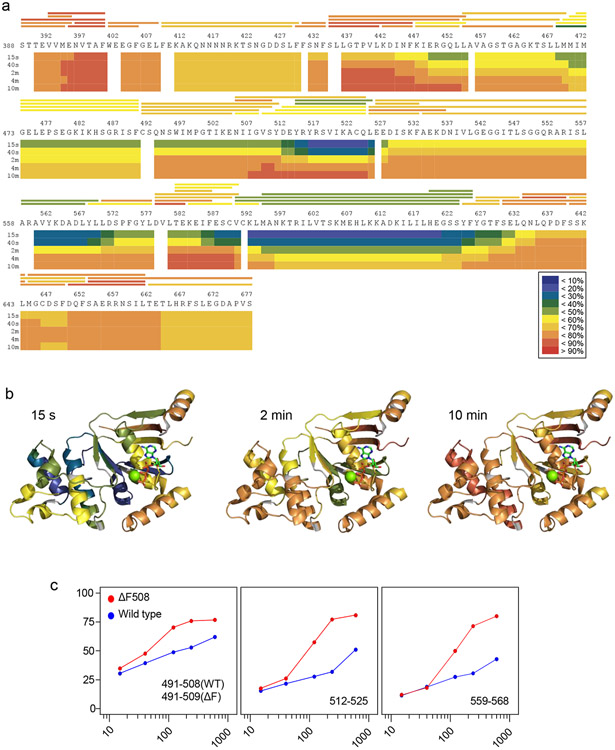
Figure 4.
Visualization of the HDX of the isolated NBD1s. a) Heat map of human NBD1-1S in the presence of 1% glycerol, representing the deuterium uptake over D2O exposure time. Blue (0%) to red (100%) indicates the extent of deuterium uptake expressed as % of the theoretical maximum. The horizontal bars above the sequence represent peptic peptides generated by on-line pepsin digestion. b) Heat map projection of ΔF508-NBD1-1S HDX after 15s, 2 min and 10 min incubation in D2O on the X-ray crystal structure of the mutant NBD1 (PDB: 2BBT). The 3D structure of NBD1 was created by the the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC (http://pymol.org). c) Deuteration kinetics of representative peptides from WT- and ΔF508-NBD1-1S. Percentage deuteration of peptides relative to the theoretical maximum are plotted for the ΔF508 (red) and WT-NBD1-1S (blue) as a function of D2O exposure time at 37°C.
Figure 5.
Unfolding rates (ku) of ΔF508-NBD1-1S peptides. a) Deconvolution of bimodal peak distribution was performed for each incubation time point using the PeakFit software and plotted in panel b. b) The ku of peptides 512-525 and 559-568 derived from ΔF508-NBD1-1S is determined by plotting the natural logarithm of unfolding fraction (Ln(Fu)) against the D2O exposure time at 37°C.
Acknowledgement
Experiments described were performed in GL lab and supported by CIHR, CFFT Inc., Cystic Fibrosis Canada, NIH-NIDDK and CFI. NS acknowledges partial financial support by Groupe de Recherche Axé sur la Structure des Protéines (GRASP). GL is the recipient of a Canada Research Chair.
Footnotes
The pET26b-derived vectors were kindly provided by H. Lewis (Structural GenomiX Inc. and CFFT Inc.) and C. Lima (Columbia University).
Adjust pD based on pD = pH read + 0.40 [20] using DCl or NaOD.
The on-line pepsin column provides fast and reproducible protein digestion, as well as less autolysis. Although immobilized pepsin resin can be prepared and packed in columns as described [21], they are also commercially available e.g. StyrosZyme (Orachrom Inc.), Poroszyme immobilized pepsin cartridge (Thermo Fisher) or Enzymate BEH pepsin column (Waters).
The expression and purification of recombinant NBD1 of human CFTR were previously described [12,22].
We assume that the mutant NBD1s can be isolated in their native-like conformation by inducing protein expression at permissive temperature. However, it is possible that mutations that cause severe structural defects in NBD1 cannot be rescued at reduced temperature and/or by chemical chaperons, and enhanced aggregation tendency will prevent the purification of native-like domain. This hurdle may be overcome by engineering sufficient number of second site stabilizing mutations in the NBD1 alone or in combination with chemical chaperones inclusion in the culture.
Storing the samples longer than 20-30 min before reading may negatively affect the experiment. The Stratagene Mx3005P RT-PCR can only start from 25°C. If lower temperature is required to prevent the unfolding of severely destabilized domain, one has to rely on an RT-PCR with cooling capacity e.g. QuantStudio™ 7 Flex System (Thermo Fisher Scientific). This RT-PCR can start the temperature ramp at 10°C. In preparation for running RT-PCR machine the UV-lamp has to warm up and plate holder cooled down for ~20 min.
The columns, solvent delivery lines, injector and other accessries should be placed in an ice bath to minimize back-exchange of deuterated analytes. Use stainless steel capillary tubing for solvent delivery lines in order to keep mobile phase chilled.
For LC-MS/MS analysis, slower gradient provides more peptide identification. Retention times of peptides should be adjusted to faster gradient for HDX analysis using the function “Use retention time adjustment” of HDExaminer.
At least three columns for peptide sequence, chage states and retention times must be included in Excel spreadsheets. A column for Xcorr values determined by Proteome Discoverer is optional.
Flash frozed samples can be stored at −80°C up to 3 weeks.
Fully deuterated sample is used to verify the deuteration level of fully unfolded states. By comparing the deuteration level, ΔF508-NBD1-1S was found to be completely unfolded at 37°C at 10 min.
Peptide carryover contributes to “false EX1” in mass specta and causes false-netagive in deuterium level [13,23]. Washing a pepsin column with washing cocktails (100 μL of each cocktail) as well as flashing trap and analytical columns using 80% IPA in ACN effectively eliminate peptide carry over.
Although HDExaminer generates deuterium uptake plots, the kintecs of deuterium uptake can be visualized by other software as well (e.g. Excel, GraphPad Prism or Sigmaplot).
References
- 1.Riordan JR (2008) CFTR function and prospects for therapy. Annu Rev Biochem 77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532 [DOI] [PubMed] [Google Scholar]
- 2.Rowe SM, Miller S, Sorscher EJ (2005) Cystic fibrosis. N Engl J Med 352 (19):1992–2001. doi:352/19/1992 [pii] 10.1056/NEJMra043184 [DOI] [PubMed] [Google Scholar]
- 3.Welsh MJ, Smith AE (1993) Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell 73 (7):1251–1254. doi:0092-8674(93)90353-R [pii] [DOI] [PubMed] [Google Scholar]
- 4.Veit G, Avramescu RG, Chiang AN, Houck SA, Cai Z, Peters KW, Hong JS, Pollard HB, Guggino WB, Balch WE, Skach WR, Cutting GR, Frizzell RA, Sheppard DN, Cyr DM, Sorscher EJ, Brodsky JL, Lukacs GL (2016) From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell 27 (3):424–433. doi: 10.1091/mbc.E14-04-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM (2006) Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126 (3):571–582. doi: 10.1016/j.cell.2006.06.041 [DOI] [PubMed] [Google Scholar]
- 6.Van Goor F, Yu H, Burton B, Hoffman BJ (2014) Effect of ivacaftor on CFTR forms with missense mutations associated with defects in protein processing or function. J Cyst Fibros 13 (1):29–36. doi: 10.1016/j.jcf.2013.06.008 [DOI] [PubMed] [Google Scholar]
- 7.Hwang TC, Kirk KL (2013) The CFTR ion channel: gating, regulation, and anion permeation. Cold Spring Harbor perspectives in medicine 3 (1):a009498. doi: 10.1101/cshperspect.a009498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, Lu F, Sauder JM, Molnar KS, Coales SJ, Maloney PC, Guggino WB, Wetmore DR, Weber PC, Hunt JF (2010) Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol 396 (2):406–430. doi: 10.1016/j.jmb.2009.11.051 [DOI] [PubMed] [Google Scholar]
- 9.Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, Wetmore D, Hunt JF, Brouillette CG (2010) Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci 19 (10):1917–1931. doi: 10.1002/pro.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C, Protasevich I, Yang Z, Seehausen D, Skalak T, Zhao X, Atwell S, Spencer Emtage J, Wetmore DR, Brouillette CG, Hunt JF (2010) Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci 19 (10):1932–1947. doi: 10.1002/pro.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendoza JL, Schmidt A, Li Q, Nuvaga E, Barrett T, Bridges RJ, Feranchak AP, Brautigam CA, Thomas PJ (2012) Requirements for efficient correction of DeltaF508 CFTR revealed by analyses of evolved sequences. Cell 148 (1-2):164–174. doi: 10.1016/j.cell.2011.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabeh WM, Bossard F, Xu H, Okiyoneda T, Bagdany M, Mulvihill CM, Du K, di Bernardo S, Liu Y, Konermann L, Roldan A, Lukacs GL (2012) Correction of both NBD1 energetics and domain interface is required to restore DeltaF508 CFTR folding and function. Cell 148 (1-2):150–163. doi: 10.1016/j.cell.2011.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumdar R, Manikwar P, Hickey JM, Arora J, Middaugh CR, Volkin DB, Weis DD (2012) Minimizing carry-over in an online pepsin digestion system used for the H/D exchange mass spectrometric analysis of an IgG1 monoclonal antibody. J Am Soc Mass Spectrom 23 (12):2140–2148. doi: 10.1007/s13361-012-0485-9 [DOI] [PubMed] [Google Scholar]
- 14.Uniewicz KA, Ori A, Xu R, Ahmed Y, Wilkinson MC, Fernig DG, Yates EA (2010) Differential scanning fluorimetry measurement of protein stability changes upon binding to glycosaminoglycans: a screening test for binding specificity. Anal Chem 82 (9):3796–3802. doi: 10.1021/ac100188x [DOI] [PubMed] [Google Scholar]
- 15.Okiyoneda T, Veit G, Dekkers JF, Bagdany M, Soya N, Xu H, Roldan A, Verkman AS, Kurth M, Simon A, Hegedus T, Beekman JM, Lukacs GL (2013) Mechanism-based corrector combination restores DeltaF508-CFTR folding and function. Nat Chem Biol 9 (7):444–454. doi: 10.1038/nchembio.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Aleksandrov AA, An J, Cui L, Yang Z, Brouillette CG, Riordan JR (2015) Restoration of NBD1 Thermal Stability Is Necessary and Sufficient to Correct F508 CFTR Folding and Assembly. J Mol Biol 427 (1):106–120. doi: 10.1016/j.jmb.2014.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall JD, Wang H, Byrnes LJ, Shanker S, Wang K, Efremov IV, Chong PA, Forman-Kay JD, Aulabaugh AE (2016) Binding screen for cystic fibrosis transmembrane conductance regulator correctors finds new chemical matter and yields insights into cystic fibrosis therapeutic strategy. Protein Sci 25 (2):360–373. doi: 10.1002/pro.2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wales TE, Engen JR (2006) Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev 25 (1):158–170. doi: 10.1002/mas.20064 [DOI] [PubMed] [Google Scholar]
- 19.Percy AJ, Rey M, Burns KM, Schriemer DC (2012) Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry-a review. Anal Chim Acta 721:7–21. doi: 10.1016/j.aca.2012.01.037 [DOI] [PubMed] [Google Scholar]
- 20.Glasoe PK, Long FA (1960) Use of Glass Electrodes to Measure Acidities in Deuterium Oxide. Journal of Physical Chemistry 64 (1):188–190. doi:DOI 10.1021/j100830a521 [DOI] [Google Scholar]
- 21.Wang L, Pan H, Smith DL (2002) Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics 1 (2):132–138 [DOI] [PubMed] [Google Scholar]
- 22.Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, Maloney PC, Guggino WB, Hunt JF, Emtage S (2005) Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem 280 (2):1346–1353. doi: 10.1074/jbc.M410968200 [DOI] [PubMed] [Google Scholar]
- 23.Fang J, Rand KD, Beuning PJ, Engen JR (2011) False EX1 signatures caused by sample carryover during HX MS analyses. Int J Mass Spectrom 302 (1-3):19–25. doi: 10.1016/j.ijms.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]