Abstract
Selective and sensitive detection of nucleic acid biomarkers is of great significance in early-stage diagnosis and targeted therapy. Therefore, the development of diagnostic methods capable of detecting diseases at the molecular level in biological fluids is vital to the emerging revolution in the early diagnosis of diseases. However, the vast majority of the currently available ultrasensitive detection strategies involve either target/signal amplification or involve complex designs. Here, using a p53 tumor suppressor gene whose mutation has been implicated in more than 50% of human cancers, we show a background-free ultrasensitive detection of this gene on a simple platform. The sensor exhibits a relatively static mid-FRET state in the absence of a target that can be attributed to the time-averaged fluorescence intensity of fast transitions among multiple states, but it undergoes continuous dynamic switching between a low- and a high-FRET state in the presence of a target, allowing a high-confidence detection. In addition to its simple design, the sensor has a detection limit down to low femtomolar (fM) concentration without the need for target amplification. We also show that this sensor is highly effective in discriminating against single-nucleotide polymorphisms (SNPs). Given the generic hybridization-based detection platform, the sensing strategy developed here can be used to detect a wide range of nucleic acid sequences enabling early diagnosis of diseases and screening genetic disorders.
Keywords: biomarkers, fluorescence resonance energy transfer (FRET), ultrasensitive sensor, hybridization sensor, single molecule, DNA/RNA detection, single-nucleotide polymorphism (SNP)
Graphical Abstract
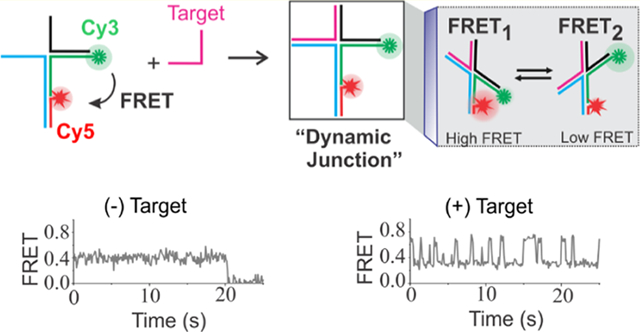
There are several technologies available for nucleic acid detection and analysis such as hybridization, strand displacement, and enzymatic and nonenzymatic amplification assays.1–6 These techniques employ either single-molecule or ensemble approaches including optical, electrochemical, and colorimetric assays.4,7–11 Although hybridization-based assays offer a simple and fast analysis of nucleic acid targets, they typically exhibit poor specificity between the target with perfect complementarity and the one with a point mutation.12,13 Therefore, any sensing approach that is sensitive down to single-nucleotide mismatch can be a very useful attribute to ensure specificity of diagnosis. Although enzymatic amplification approaches such as polymerase chain reaction (PCR), reverse transcriptase PCR (RT-PCR), and digital PCR are simple and highly sensitive, they rely on target amplification and are susceptible to false negatives/positives.14–16 This is problematic because false negatives run the risk of instilling a false sense of security and false positives may result in unnecessary panic.14–16 Therefore, novel single-molecule and ensemble techniques including DNA nanotechnology platforms with improved sensitivity and specificity are continuously emerging in recent years.17–21 Nonetheless, most of these methods are rather complicated and have limited applications.
For example, techniques including synthetic nanopores, barcodes, and force-based approaches are limited by the need for precise and sophisticated engineering.22–26 Also, most of these methods require targets to be labeled, modified, or amplified to enable detection.27–29 Over the years, DNA-based sensing using single-molecule fluorescence resonance energy transfer (smFRET) has gained significant popularity due to its several advantages. First, sensors made up of DNA can be used to detect any DNA or RNA sequences using a hybridization approach, which offers a great deal of flexibility. Second, the donor/acceptor fluorophores can be directly incorporated into sensors to enable FRET so that the target does not need to be labeled. In this case, the change in the FRET level after target binding can be used as a detection signal. Third, the smFRET approach provides quantitative information about the behavior of individual molecules, allowing simultaneous detection and quantitation.30–35 Additionally, the use of a total-internal reflection fluorescence (TIRF)-based FRET technique, as we have used in this study, enables high-throughput experiments by simultaneous imaging of several molecules in one movie.
In this manuscript, using a p53 tumor suppressor gene as our proof-of-concept target, whose mutation accounts for more than 50% of human cancers,36,37 we have demonstrated a simple background-free FRET-based sensor that enables an ultrasensitive detection of this biomarker. Unlike conventional fluorescence-based sensors, which require either a complex sensor design, signal amplification steps, or use of additional materials such as enzymes or nanocomposites, the sensor presented herein is simple and allows a single-step ultra-sensitive detection of DNA biomarkers without target/signal amplification. Further, in contrast to other bulk FRET-based assays where detection relies on either increase or decrease in the FRET signal or fluorescence life-time,38 the single-molecule approach used here takes advantage of the dynamic nature of a four-way DNA junction,39–41 which has several advantages. First, since the dynamic FRET is observed only in the presence of a target, this approach gives a zero background. In other words, there is no risk of a false signal. Second, due to direct binding of the target (no competition), our technique is ultrasensitive with a limit of detection of 50 fM (≈5 attomoles considering the sample volume of 100 μL) without the need for amplification and labeling. Third, the proposed method is direct because it does not require labeling of targets to achieve low fM detection. Fourth, unlike expensive enzymes or antibody-based sensors, this sensor can be readily prepared from short synthetic DNA strands and can be easily designed to detect “any” sequence of interest. Fifth, the sensor can discriminate targets even with single-nucleotide mutations. Therefore, the proposed approach is novel and has the potential to benefit clinical practices by allowing a high-confidence early diagnosis of diseases.
RESULTS AND DISCUSSION
Sensor Design and Working Principle.
The sensor design and working principle are outlined in Figure 1. The sensor molecules were custom designed using the nucleic acid hybridization principle. A retrospective design strategy was used to determine the single-stranded sequences that need to be incorporated into the binding regions of the sensor to enable target binding. In other words, when the target sequence is known, the target-binding region of the sensor can be created based on the principle of complementary base-pair hybridization. The rest of the sensor sequences were manually designed to obtain the desired sensor. A biotin was incorporated in one of the strands to allow surface immobilization of sensors on the streptavidin-modified microscope slide via biotin/streptavidin interaction (Figure 1).42,43 To enable FRET, a donor (Cy3) and an acceptor (Cy5) fluorophore were incorporated into the sensor molecules using fluorophore-modified oligonucleotides. The detailed sequence and fluorophore-labeling scheme of the sensor are shown in Figure S1. The formation of the sensor was characterized using native polyacrylamide gel electrophoresis (PAGE) (Figure S2).
Figure 1.
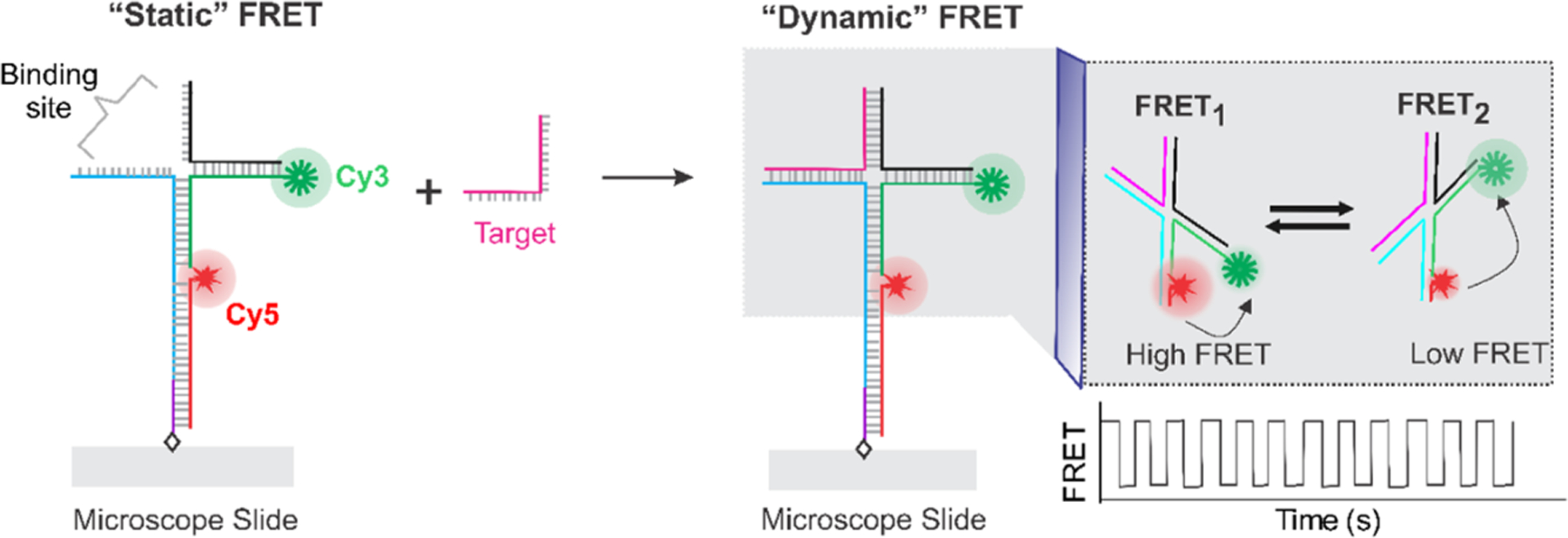
Working principle of the sensor. The sensor is composed of synthetic DNA strands, two of which are labeled with either a Cy3 or a Cy5 fluorophore. The DNA construct exhibits a relatively steady FRET efficiency in the absence of a target due to averaging of fast transitions among multiple states. However, binding of the target forms a four-way structure resulting in a dynamic switching between a high (FRET1) and a low (FRET2) FRET state. FRET represents FRET efficiency.
In this study, we took advantage of the unique nature of the four-way DNA junction (Figure 1) that spontaneously interconverts between two stacked (X) conformers.39–41,44 To leverage this nature of the four-way junction in sensing, the sensor was designed to have an incomplete junction so that it exhibits a medium but nondynamic FRET level at the experimental acquisition time (50–100 ms) in the absence of a target. However, binding of the target completes the four-way junction resulting in a dynamic FRET. Therefore, a characteristic FRET pattern exhibiting fluctuations between a low- and a high-FRET states was expected as a detection signal.
Single-Molecule Sensing.
First, the sensor molecules were immobilized on the microscope slide using the biotin/streptavidin interaction (Figure 1) as described in the Methods section. To remove unbound sensor molecules, an imaging buffer containing an oxygen scavenger system (OSS) was injected and incubated for ~2 min to let the OSS equilibrate. The OSS was used to retard fluorophore blinking and photobleaching upon laser illumination.45,46 The fluorescence movies were collected using a total-internal reflection fluorescence (TIRF) microscope as described in our previous publications and in the Methods section.41,47,48 Briefly, the flow cell was then irradiated with a 532 nm laser to excite the donor (Cy3) fluorophore of the surface-tethered molecules to enable fluorescence resonance energy transfer (FRET).30,32 The fluorescence emissions of both Cy3 and Cy5 fluorophores were recorded at 10 frames per second (100 ms time resolution and camera gain of 200). The presence of a Cy5 fluorophore was confirmed by direct excitation using a red laser (639 nm) toward the end of the movies. The movies were processed with IDL and MATLAB codes (see the Methods section) to create intensity–time traces.41,47,48 Only those single molecules showing evidence for the presence of both donor/acceptor fluorophores and single-step photobleaching of fluorophores were selected for further analysis.
We first examined the sensor alone in the absence of the target by analyzing intensity–time traces and the corresponding FRET state. Some typical molecules from this experiment are shown in Figure 2. As expected, the donor/acceptor emission traces were relatively flat without any definite dynamic pattern (Figure 2). When the raw intensity traces were converted to FRET traces (see the Methods section for detail), all of the molecules show relatively static FRET–time traces with a FRET value of ~0.5. These experiments suggested that the sensor molecules behave uniformly.
Figure 2.
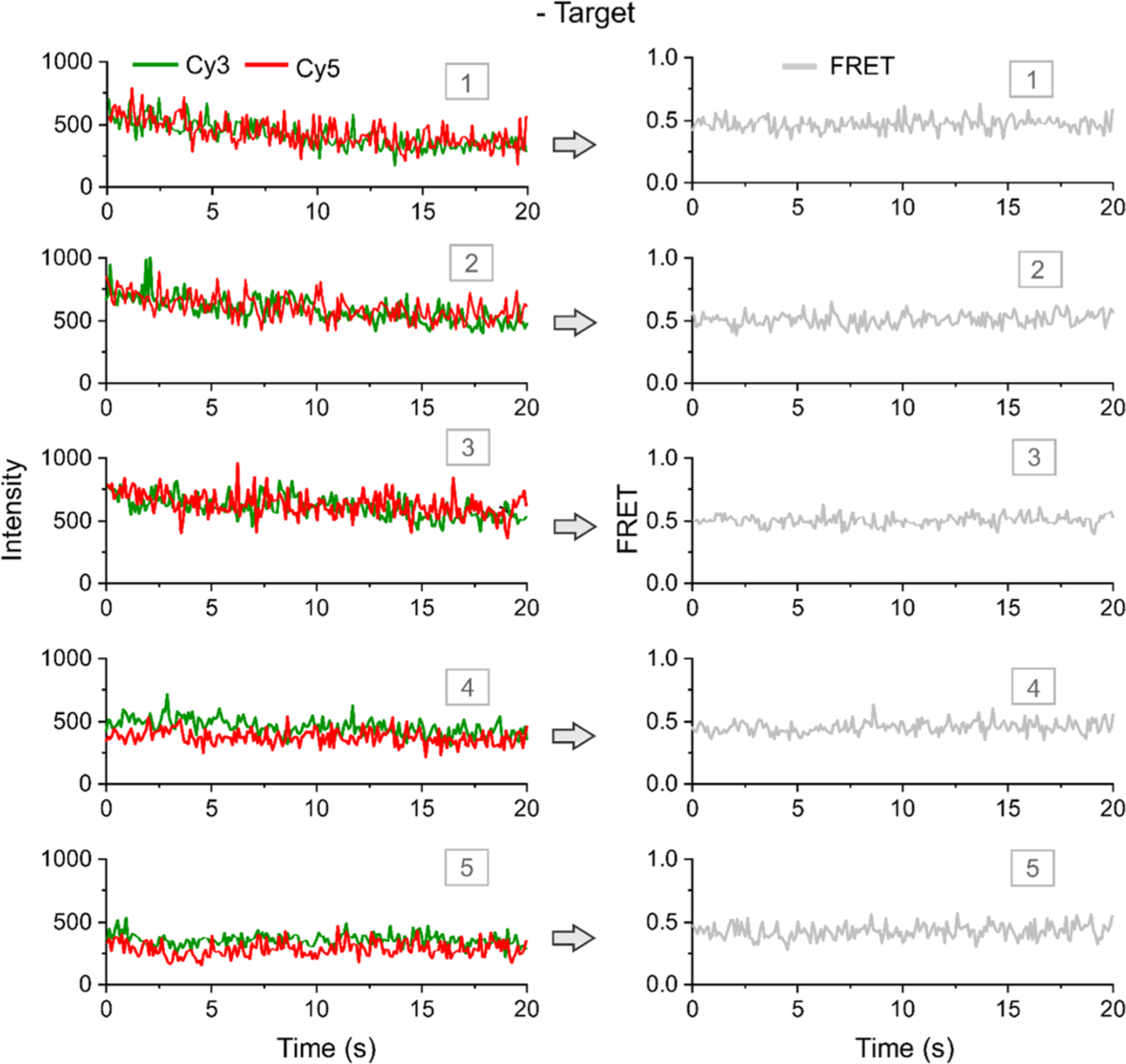
Typical single-molecule traces in the absence of a target. Typical intensity–time (left) and corresponding FRET–time traces (right). Five representative molecules are shown. The molecules exhibit static fluorescence intensities of Cy3 and Cy5 in the absence of a target, and a static FRET level of ~0.5 is observed in the absence of target DNA. All experiments are done at room temperature (23 °C). FRET represents FRET efficiency.
Interestingly, when the experiment in Figure 2 was repeated after injection of a target (p53 tumor suppressor gene: 5′-TTCCTCTGTGCGCCGGTCTCTCCT)37,49,50 and incubated for ~20 min, we observed a very different behavior of sensor molecules. In the presence of the target, sensor molecules showed very clear dynamics of Cy3 and Cy5 fluorescence intensities that were anticorrelated. The typical single-molecule traces from this experiment are shown in Figure 3. When the raw intensity–time traces were converted to FRET–time traces, all of the dynamic molecules showed a very clear switching pattern between ~0.3 and ~0.7 FRET states (Figure 3). Such dynamics is an inherent behavior of the fully formed four-way junction,39–41,44 and it is important to note that such dynamic switching was absent in the target-free experiments (Figure 2). Further, when we plotted FRET efficiency histograms for without and with target data, we observed a single population in the absence of the target but two populations with expected FRET levels in the presence of the target (Figure S3). Therefore, this sensing approach demonstrated a high-confidence detection of the target. Inspired by these results, we next performed experiments at various concentrations of the target to determine the analytical sensitivity of the sensor.
Figure 3.
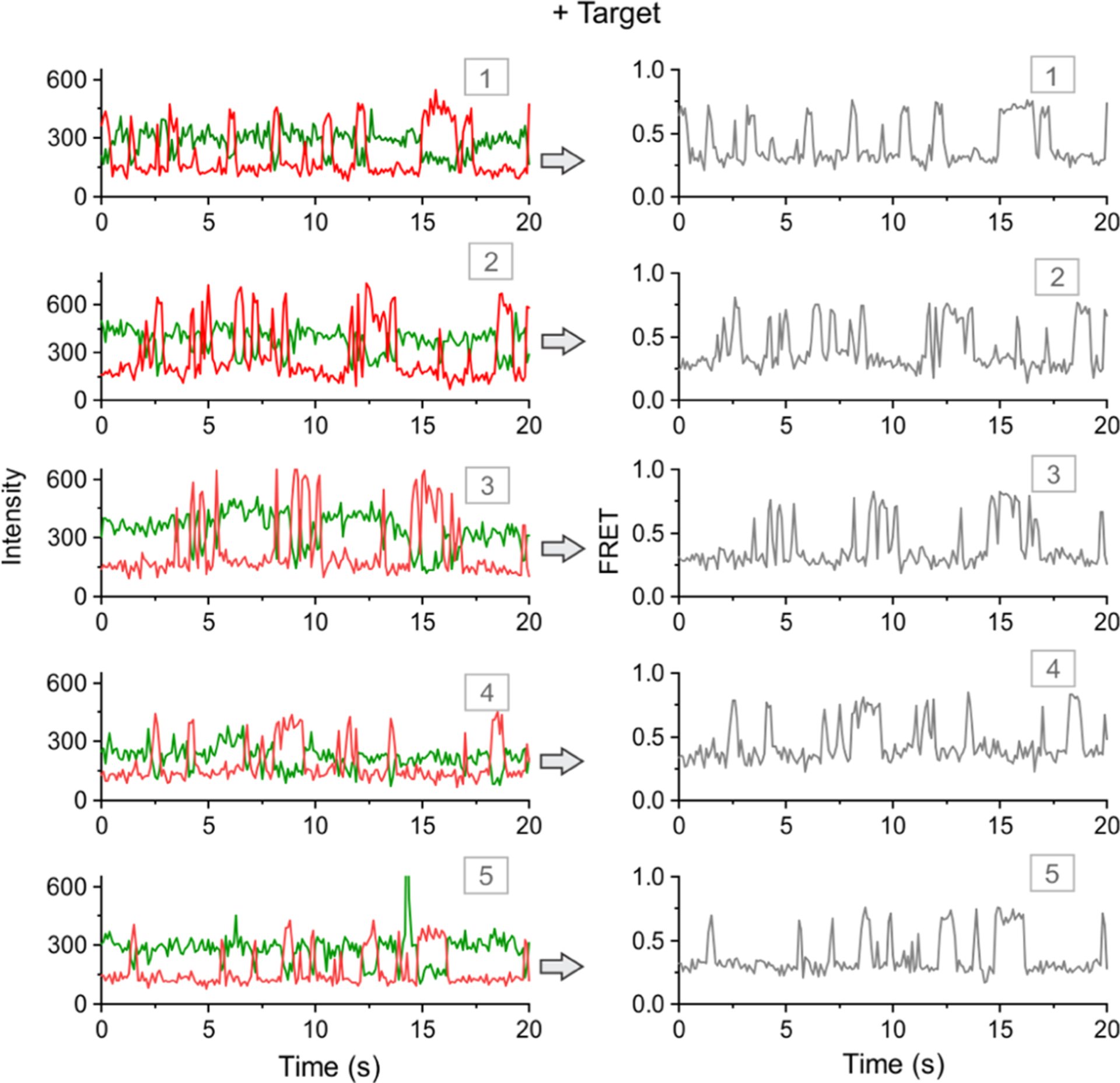
Detection of a target sequence (p53 tumor suppressor gene) using single-molecule FRET. Typical intensity–time (left) and corresponding FRET–time traces. Five representative molecules are shown. The molecules exhibit dynamic and anticorrelated fluorescence intensities of Cy3 and Cy5. Such dynamic FRET–time traces with FRET levels of ~0.3 and ~0.7 are obtained only in the presence of target DNA. All of the experiments are performed at room temperature (23 °C). FRET represents FRET efficiency. Note that a slightly revised design to improve the sensor stability will be discussed later.
Determining the Analytical Sensitivity.
To determine the analytical sensitivity of the sensor, a series of experiments were performed at various concentrations of the target. The percentage of dynamic molecules (≈detection) under each concentration of the target was then determined by dividing the number of dynamic molecules by the total number of single molecules (see the Methods section for details). Control experiments were performed in the absence of the target, which yielded no dynamic molecules. Through single-molecule counting, we determined that there were ~2% dynamic molecules (4 out of 204 total molecules) at 50 femtomolar (fM) target and no dynamic molecules were observed below this concentration. Therefore, the limit of detection (LOD) of this method was determined to be 50 fM under our experimental conditions. When the percentage of dynamic molecules was plotted against the target concentration (Figure 4), it showed a linearity up to around 10 pM target (Figure 4 inset), after which it was curved and eventually plateaued. It is important to notice that there were no dynamic molecules in the absence of the target (Figure 2), and thus this sensing approach exhibits a zero background. Further, given the flow cell volume of ~100 μL, the detection limit of 50 fM translates to 5 attomoles, which means that this sensing method is highly sensitive. In addition, it has a large dynamic range (~3 orders of magnitude) extending to ~100 pM.
Figure 4.
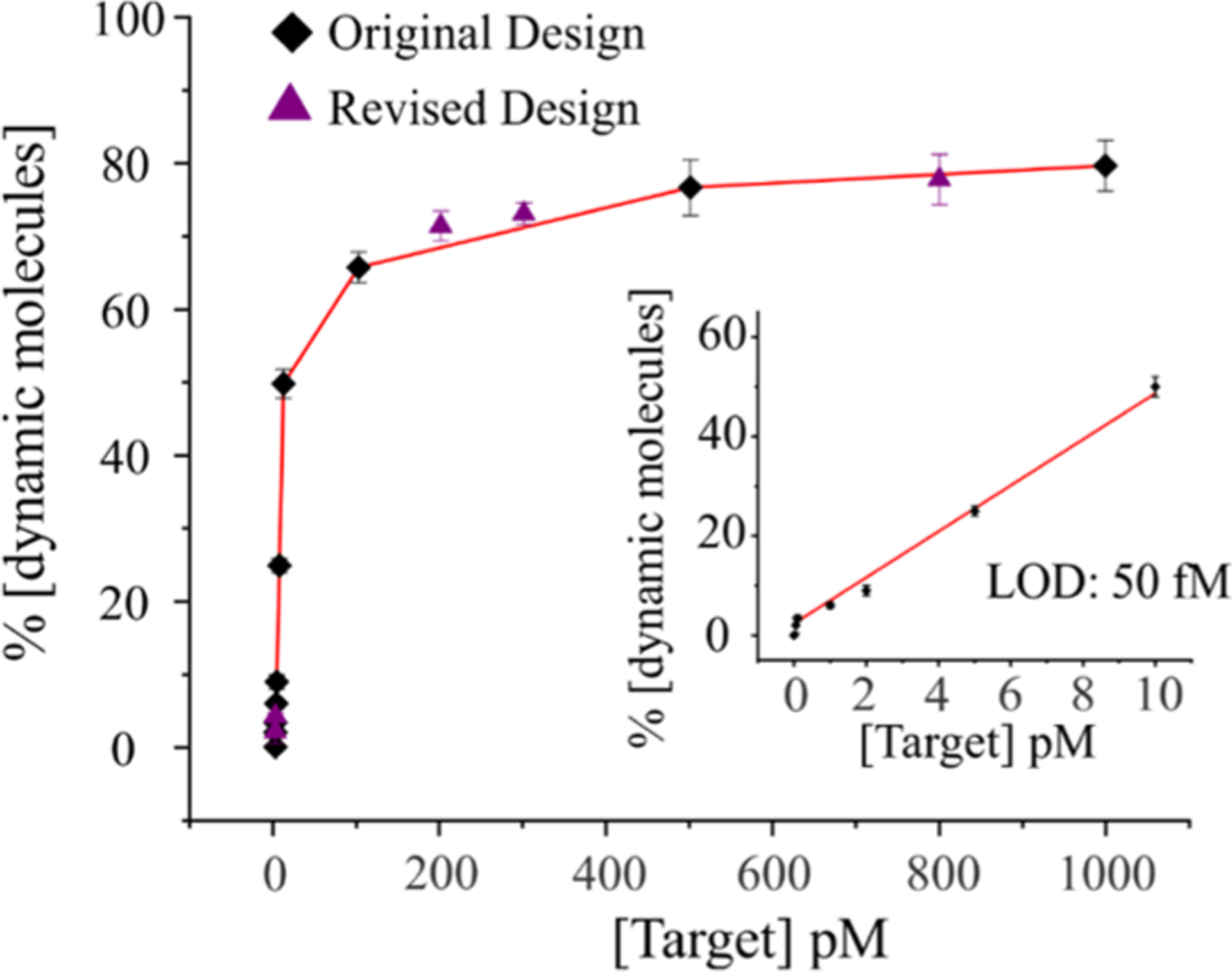
Determination of the limit of detection (LOD). A calibration curve is obtained by plotting the number of dynamic molecules (≈target-bound molecules) as a function of the target concentration. The inset shows the linear range of the calibration curve with a LOD of 50 fM. Considering our experimental volume of ~100 μL, this LOD is equivalent to 5 attomoles of the DNA target (1 attomoles = 1 × 10−18 moles). The results for both the original and revised designs are shown. The percentage of dynamic molecules is determined from more than 150 single molecules at each concentration. The error bars represent the standard deviation from three groups of independent movie files.
From the stability stand point, DNA-based sensors that are made up of short synthetic DNA strands can have a stability issue if they have to be stored for a long period of time (weeks). Since the sensor used in this study has short arms (11 bp each) and we observed a loss of Cy3 signal after about a week from sensor assembly, we sought to increase the arm length of the sensor and test it for dynamics and LOD. For this, we prepared a construct with slightly longer D/E and B/C arms (increased by 2 and 1 bp, respectively) and extended the Strand E by 4As to complement 4T in Strand B (Table S1 and Figure S1). Apart from these changes, the revised construct was identical to the original design. We tested this revised design for five different concentrations of targets (50 fM, 100 fM, 200 pM, 300 pM, and 800 pM) and obtained similar results as in the original construct in terms of the fraction of dynamic molecules. These results showed that the design can be tuned to enhance the sensor stability without compromising the sensitivity of the sensor. Given that the vast majority of sensing approaches available today require either amplification of target (enzymatic or nonenzymatic), labeling of target, or some sort of signal amplification such as the use of nanomaterials or nanocomposites to reach low nM to fM detection limits,51–62 the sensing approach that we demonstrated here offers an ultrasensitive detection of nucleic acids in a simpler format.
In addition to sensitivity, another requirement of a sensor is its specificity. Therefore, to test the specificity of the sensor, we designed three mutant sequences and compared the results with the original p53 target. As shown in Figure 5A, mutants 1 and 2 have their 12th nucleotide altered from the 5′-end. The rationale for this design was that, since the dynamic FRET is the result of an intact four-way junction, a single mismatch at the vicinity of the junction could result in loss in dynamics so that the signal in the presence of mutant will not overlap with the one from the specific target. Interestingly, both of the mutants showed less than 2% dynamic-like molecules even at nearly saturating concentration of mutants (100 pM). The typical single-molecule traces involving mutant 1 are shown in Figure S4. We also tested another mutant with two mutation sites (one on each arm) and obtained similar results as seen for single mutants 1 and 2. Overall, these experiments demonstrated that the four-way-junction based sensing can easily discriminate a fully matched target from its single-nucleotide mismatch mutants. In other words, these sensors can be retrospectively designed such that the mutation site directly falls at the junction to fully discriminate the mutant from the target.
Figure 5.
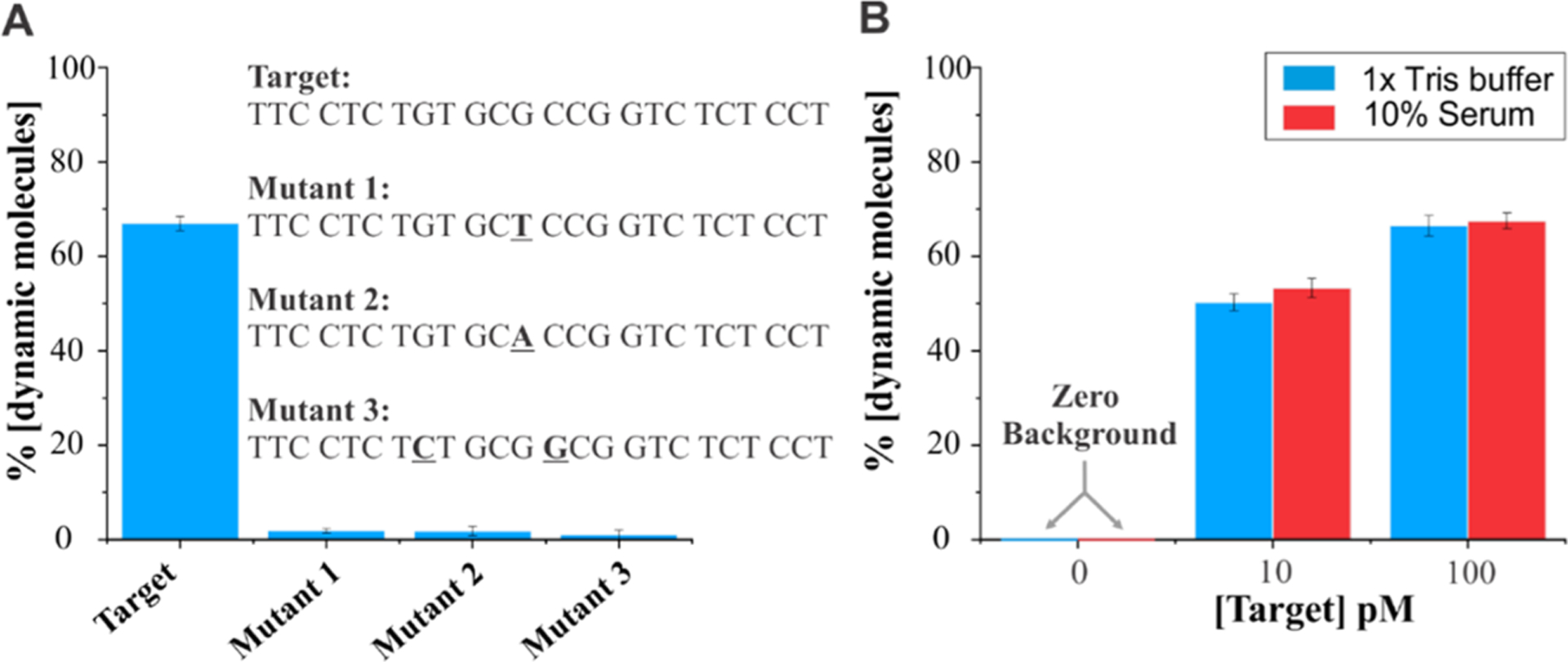
Specificity of sensors and their compatibility in serum. (A) Specificity test of the sensor using 100 pM target/mutants. While the target is perfectly complementary, mutants have one or two mismatched nucleotides (bolded and underlined). We found that 70% of molecules were dynamic in the presence of 100 pM target, while a negligible fraction of molecules showed dynamic behavior in the presence of 100 pM mutant, thus demonstrating a high specificity of our approach. (B) Sensors behave similarly in 1× Tris HCl and 10% human serum. Notice the zero background in the absence of the target.
Given that the body responds to the onset of diseases by the altered release of certain molecules such as miRNAs or hormones,63 sensors that are compatible with biological fluids warrant a wider range of applications. The serum is a suitable biological fluid for this purpose; therefore, we employed human serum and tested the performance of the sensor at 0, 10, and 100 pM of target and directly compared the results obtained in a regular 1× Tris buffer. Interestingly, the percentage of dynamic molecules determined in serum (10%) and regular buffer were the same within the error (Figure 5B). Further, similar to the regular buffer result, there were no dynamic molecules detected in the absence of the target in serum, demonstrating that the sensor offers a background-free detection in serum.
CONCLUSIONS
We have developed a novel single-step fluorescence-based sensor to detect DNA biomarkers, which we demonstrated using a p53 tumor suppressor gene as a proof-of-concept target. Since the detection relies on the target-induced formation of dynamic molecules, this sensing strategy enables a background-free, ultrasensitive, and high-confidence detection of DNA without the need for target/signal amplification. Further, a LOD of ~5 attomoles can be achieved without labeling the target. The sensor design is comprised of four-way junction with a 22-nucleotides binding site (11 nucleotides on each arm), which is a perfect size to implement for miRNA detection as the average mature miRNAs size falls between 20 and 23 nts.64–66 Further, the detection is based on the direct hybridization of sequences, which is a great advantage as the sensor can be easily designed to detect any sequence of interest by simply swapping the two unlabeled DNA strands. In addition, the LOD of 50 fM (~5 attomoles) is in the range of typical nucleic acid biomarkers including miRNAs, pathogenic DNA, and circulating tumor DNAs in biological samples and thus this sensor may offer direct applications in biotechnology to detect trace amounts of nucleic acid biomarkers or other pathogenic DNAs.
METHODS
Chemical Reagents and DNA Sequences.
Biotinylated bovine serum albumin (bBSA) was purchased from Thermo Scientific. It was dissolved in filtered sterile water at 1 mg/mL and stored at −20 °C until needed. The reagents for the oxygen scavenging system including protocatechuate 3,4-dioxygenase (PCD), 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox), tris-(hydroxymethyl)-aminomethane (Tris), ethylenediaminetetraacetic acid disodium salt (EDTA), and acetic acid were also purchased from Fisher Scientific. Protocatechuic acid (PCA) and streptavidin were purchased from VWR. All DNA sequences were purchased from Integrated DNA Technologies (IDT Inc.) and stored at −20 °C until needed. Biotinylated and fluorophore-labeled sequences were purchased HPLC purified.
Sensor Design and Preparation.
The sensor was prepared by thermal annealing of five single-stranded DNA (ssDNA) oligonucleotides at 1 μM concentrations in 1× TAE-Mg buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA, and 100 mM Mg2+, pH 7.4). Among the five oligonucleotides, two were modified with either a Cy3 or Cy5 fluorophore to enable FRET and another strand was modified with a biotin so that sensors could be immobilized on the microscope slide via biotin/streptavidin interaction. The sample was heated at 95 °C for 5 min and then the temperature was gradually ramped down to 4 °C in the duration of <2 h.43,48,67 A donor (Cy3) and an acceptor (Cy5) fluorophore were introduced using labeled ssDNA to incorporate a FRET pair in each molecule.
Electrophoretic Mobility Shift Assay (EMSA).
The formation of the DNA sensor was characterized using native polyacrylamide gel electrophoresis (PAGE) (Figure S2). A 12% PAGE gel was prepared at 10 mL of total volume by mixing polyacrylamide, 1× TBE, 0.1% ammonium persulfate, 10 μL tetramethyethylenediamine (TEMED), and double sterile water. The gel was run at 75 V for 90 min in 1× TBE buffer and was stained with ethidium bromide (EtBr) for 20 min before imaging and visualized using an UV-transilluminator.
Single-Molecule Fluorescence Imaging.
The sensor molecules were immobilized on the surface of a quartz slide flow cell functionalized with biotin-BSA and streptavidin as described elsewhere.47,48 Briefly, after mounting the flow cell on the microscope stage, a 60–80 pM sensor solution prepared in 1× TAE buffer consisting of 100 mM MgCl2 and an oxygen scavenging system (OSS: 4 mM Trolox, 10 mM PCA, 100 nM PCD) was injected into the flow cell and incubated for ~30 s to allow surface immobilization of sensor molecules via biotin/streptavidin interaction. The unbound molecules were then removed by flushing the flow cell with 400 μL of imaging buffer (OSS containing 1× TAE buffer spiked with 100 mM Mg2+, pH 7.4).
The fluorescence imaging was carried out using a prism-based total-internal reflection fluorescence (pTIRF) microscope as described previously42,67 in a 1× TAE buffer spiked with 100 mM Mg2+ (pH 7.4). Using a 532 nm laser, the Cy3 fluorophores were continuously excited while emissions from Cy3 and Cy5 fluorophores were simultaneously recorded through green and red channels (512 × 256 pixels) on an EMCCD camera at a 100 ms time resolution. In all sensing experiments, a target DNA solution of a given concentration was injected into the flow cell and incubated for ~20 min before fluorescence imaging. The imaging was performed at room temperature (23 °C).
Single-Molecule Data Acquisition and Analysis.
The single-molecule movies were processed using IDL and MATLAB scripts and fluorescence-time trajectories of individual molecules were obtained as described previously.42,67 The presence of an active FRET pair was confirmed by turning on a 639 nm laser to directly excite the Cy5 fluorophore toward the end of each movie. Only those molecules that show the presence of both fluorophores and a single-step photobleaching were selected for further data analysis. The FRET efficiency value was calculated using the equation: IA/(ID + IA), where IA and ID represent the background-corrected fluorescence intensities of the acceptor and donor fluorophores, respectively.30,68 The dynamic vs static molecules were assigned by manual counting of two types of molecules as the dynamics was visually clear on the FRET–time traces exhibiting many transitions between the FRET levels of ~0.3 and ~0.7. A standard curve was prepared by plotting the percentage of dynamic molecules at different concentrations of target DNA. This calculation was performed by dividing the number of dynamic molecules by the total number of selected single molecules. The standard deviation of the percentage of dynamic molecules was determined using at least three groups of independent movie files at each concentration of the target.
Supplementary Material
ACKNOWLEDGMENTS
S.D. acknowledges the financial support from VCU Startup Grant and the VCU’s CTSA (UL1TR002649 from the National Center for Advancing Translational Sciences) and the CCTR Endowment Fund of Virginia Commonwealth University. The authors acknowledge Kumar Sapkota for his contribution to the project.
Footnotes
The authors declare the following competing financial interest(s): A patent application related to this work has been filed.
Complete contact information is available at: https://pubs.acs.org/10.1021/acssensors.1c00002
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.1c00002.
DNA oligonucleotides used in this study (Table S1); estimated interdye distances (R) as well as estimated and experimentally determined FRET efficiencies (E) (Table S2); the four-way sensor design with corresponding strand names (Figure S1); native gel characterization of the formation of the DNA sensor (Figure S2); smFRET analysis of the sensor (Figure S3); typical single-molecule traces in the presence of mutant 1 (Figure S4); and supporting references (PDF)
REFERENCES
- (1).Zhang M; Ye J; He JS; Zhang F; Ping J; Qian C; Wu J Visual detection for nucleic acid-based techniques as potential on-site detection methods. A review. Anal. Chim. Acta 2020, 1099, 1–15. [DOI] [PubMed] [Google Scholar]
- (2).Zhao Y; Chen F; Li Q; Wang L; Fan C Isothermal Amplification of Nucleic Acids. Chem. Rev 2015, 115, 12491–12545. [DOI] [PubMed] [Google Scholar]
- (3).Suea-Ngam A; Bezinge L; Mateescu B; Howes PD; deMello AJ; Richards DA Enzyme-Assisted Nucleic Acid Detection for Infectious Disease Diagnostics: Moving toward the Point-of-Care. ACS Sens. 2020, 5, 2701–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ho NRY; Lim GS; Sundah NR; Lim D; Loh TP; Shao H Visual and modular detection of pathogen nucleic acids with enzyme-DNA molecular complexes. Nat. Commun 2018, 9, No. 3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hill HD; Vega RA; Mirkin CA Nonenzymatic detection of bacterial genomic DNA using the bio bar code assay. Anal. Chem 2007, 79, 9218–9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Peng L; Yuan Y; Fu X; Fu A; Zhang P; Chai Y; Gan X; Yuan R Reversible and Distance-Controllable DNA Scissor: A Regenerated Electrochemiluminescence Biosensing Platform for Ultrasensitive Detection of MicroRNA. Anal. Chem 2019, 91, 3239–3245. [DOI] [PubMed] [Google Scholar]
- (7).Zhang J; Song S; Zhang L; Wang L; Wu H; Pan D; Fan C Sequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodes. J. Am. Chem. Soc 2006, 128, 8575–8580. [DOI] [PubMed] [Google Scholar]
- (8).Wei F; Lillehoj PB; Ho CM DNA diagnostics: nanotechnology-enhanced electrochemical detection of nucleic acids. Pediatr. Res 2010, 67, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ammanath G; Yeasmin S; Srinivasulu Y; Vats M; Cheema JA; Nabilah F; Srivastava R; Yildiz UH; Alagappan P; Liedberg B Flow-through colorimetric assay for detection of nucleic acids in plasma. Anal. Chim. Acta 2019, 1066, 102–111. [DOI] [PubMed] [Google Scholar]
- (10).Mayr R; Haider M; Thünauer R; Haselgrübler T; Schütz GJ; Sonnleitner A; Hesse J A microfluidic platform for transcription- and amplification-free detection of zepto-mole amounts of nucleic acid molecules. Biosens. Bioelectron 2016, 78, 1–6. [DOI] [PubMed] [Google Scholar]
- (11).Byun JY; Lee KH; Shin YB; Kim DM Cascading Amplification of Immunoassay Signal by Cell-Free Expression of Firefly Luciferase from Detection Antibody-Conjugated DNA in an Escherichia coli Extract. ACS Sens. 2019, 4, 93–99. [DOI] [PubMed] [Google Scholar]
- (12).Abi A; Safavi A Targeted Detection of Single-Nucleotide Variations: Progress and Promise. ACS Sens. 2019, 4, 792–807. [DOI] [PubMed] [Google Scholar]
- (13).Sachidanandam R; Weissman D; Schmidt SC; Kakol JM; Stein LD; Marth G; Sherry S; Mullikin JC; Mortimore BJ; Willey DL; Hunt SE; Cole CG; Coggill PC; Rice CM; Ning Z; Rogers J; Bentley DR; Kwok PY; Mardis ER; Yeh RT; Schultz B; Cook L; Davenport R; Dante M; Fulton L; Hillier L; Waterston RH; McPherson JD; Gilman B; Schaffner S; Van Etten WJ; Reich D; Higgins J; Daly MJ; Blumenstiel B; Baldwin J; Stange-Thomann N; Zody MC; Linton L; Lander ES; Altshuler D A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- (14).West CP; Montori VM; Sampathkumar P COVID-19 testing: The threat of false-negative results. Mayo Clin. Proc 2020, 1127–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chen Z; Li Y; Wu B; Hou Y; Bao J; Deng X A patient with COVID-19 presenting a false-negative reverse transcriptase polymerase chain reaction result. Korean J. Radiol 2020, 21, 623–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Feng H; Liu Y; Lv M; Zhong J A case report of COVID-19 with false negative RT-PCR test: necessity of chest CT. Jpn. J. Radiol 2020, 38, 409–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Johnson-Buck A; Li J; Tewari M; Walter NG A guide to nucleic acid detection by single-molecule kinetic fingerprinting. Methods 2019, 153, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cheng WC; Horn T; Zayats M; Rizk G; Major S; Zhu H; Russell J; Xu Z; Rothman RE; Celedon A Ultra-sensitive and rapid detection of nucleic acids and microorganisms in body fluids using single-molecule tethering. Nat. Commun 2020, 11, No. 4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cai S; Deng Y; Fu S; Li J; Yu C; Su X Single-molecule dynamic DNA junctions for engineering robust molecular switches. Chem. Sci 2019, 10, 9922–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Weng R; Lou S; Li L; Zhang Y; Qiu J; Su X; Qian Y; Walter NG Single-Molecule Kinetic Fingerprinting for the Ultrasensitive Detection of Small Molecules with Aptasensors. Anal. Chem 2019, 91, 1424–1431. [DOI] [PubMed] [Google Scholar]
- (21).Hayward SL; Lund PE; Kang Q; Johnson-Buck A; Tewari M; Walter NG Ultraspecific and Amplification-Free Quantification of Mutant DNA by Single-Molecule Kinetic Fingerprinting. J. Am. Chem. Soc 2018, 140, 11755–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kodadek T Protein microarrays: prospects and problems. Chem. Biol 2001, 8, 105–115. [DOI] [PubMed] [Google Scholar]
- (23).Wong KH; Jin Y; Moqtaderi Z Multiplex illumina sequencing using DNA barcoding. Curr. Protoc. Mol. Biol 2013, 101, 7.11.1–7.11.11. [DOI] [PubMed] [Google Scholar]
- (24).Sze JYY; Ivanov AP; Cass AEG; Edel JB Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun 2017, 8, No. 1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Li W; Jiang W; Dai S; Wang L Multiplexed detection of cytokines based on dual bar-code strategy and single-molecule counting. Anal. Chem 2016, 88, 1578–1584. [DOI] [PubMed] [Google Scholar]
- (26).Yang D; Ward A; Halvorsen K; Wong WP Multiplexed single-molecule force spectroscopy using a centrifuge. Nat. Commun 2016, 7, No. 11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Churchill GA Fundamentals of experimental design for cDNA microarrays. Nat. Genet 2002, 32, 490–495. [DOI] [PubMed] [Google Scholar]
- (28).Dunbar SA Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 2006, 363, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Shimron S; Wang F; Orbach R; Willner I Amplified detection of DNA through the enzyme-free autonomous assembly of hemin/G-quadruplex DNAzyme nanowires. Anal. Chem 2012, 84, 1042–1048. [DOI] [PubMed] [Google Scholar]
- (30).Roy R; Hohng S; Ha T A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Shu D; Zhang H; Petrenko R; Meller J; Guo P Dual-channel single-molecule fluorescence resonance energy transfer to establish distance parameters for RNA nanoparticles. ACS Nano 2010, 4, 6843–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lerner E; Cordes T; Ingargiola A; Alhadid Y; Chung S; Michalet X; Weiss S Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 2018, 359, No. eaan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Phelps C; Israels B; Jose D; Marsh MC; von Hippel PH; Marcus AH Using microsecond single-molecule FRET to determine the assembly pathways of T4 ssDNA binding protein onto model DNA replication forks. Proc. Natl. Acad. Sci. U.S.A 2017, 114, E3612–E3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Christian TD; Romano LJ; Rueda D Single-molecule measurements of synthesis by DNA polymerase with base-pair resolution. Proc. Natl. Acad. Sci. U.S.A 2009, 106, 21109–21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kaur A; Dhakal S Recent applications of FRET-based multiplexed techniques. TrAC, Trends Anal. Chem 2020, 123, No. 115777. [Google Scholar]
- (36).Levine AJ p53, the cellular gatekeeper for growth and division. Cell 1997, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- (37).Mantovani F; Collavin L; Del Sal G Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).McGuinness CD; Nishimura MK; Keszenman-Pereyra D; Dickinson P; Campbell CJ; Bachmann TT; Ghazal P; Crain J Detection of single nucleotide polymorphisms using a DNA Holliday junction nanoswitch-a high-throughput fluorescence lifetime assay. Mol. BioSyst 2010, 6, 386–390. [DOI] [PubMed] [Google Scholar]
- (39).Hyeon C; Lee J; Yoon J; Hohng S; Thirumalai D Hidden complexity in the isomerization dynamics of Holliday junctions. Nat. Chem 2012, 4, 907–914. [DOI] [PubMed] [Google Scholar]
- (40).Joo C; McKinney SA; Lilley DM; Ha T Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J. Mol. Biol 2004, 341, 739–751. [DOI] [PubMed] [Google Scholar]
- (41).Gibbs DR; Dhakal S Single-molecule imaging reveals conformational manipulation of Holliday junction DNA by the junction processing protein RuvA. Biochemistry 2018, 57, 3616–3624. [DOI] [PubMed] [Google Scholar]
- (42).Kaur A; Sapkota K; Dhakal S Multiplexed Nucleic Acid Sensing with Single-Molecule FRET. ACS Sens. 2019, 4, 623–633. [DOI] [PubMed] [Google Scholar]
- (43).Sapkota K; Dhakal S FRET-based aptasensor for the selective and sensitive detection of lysozyme. Sensors 2020, 20, No. 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Okamoto K; Sako Y State transition analysis of spontaneous branch migration of the Holliday junction by photon-based single-molecule fluorescence resonance energy transfer. Biophys. Chem 2016, 209, 21–27. [DOI] [PubMed] [Google Scholar]
- (45).Aitken CE; Marshall RA; Puglisi JD An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J 2008, 94, 1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Fu J; Yang YR; Dhakal S; Zhao Z; Liu M; Zhang T; Walter NG; Yan H Assembly of multienzyme complexes on DNA nanostructures. Nat. Protoc 2016, 11, 2243–2273. [DOI] [PubMed] [Google Scholar]
- (47).Gibbs D; Kaur A; Megalathan A; Sapkota K; Dhakal S Build your own microscope: Step-by-step guide for building a prism-based TIRF microscope. Methods Protoc. 2018, 1, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Megalathan A; Cox BD; Wilkerson PD; Kaur A; Sapkota K; Reiner JE; Dhakal S Single-molecule analysis of i-motif within self-assembled DNA duplexes and nanocircles. Nucleic Acids Res. 2019, 47, 7199–7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Li F; Zhou YY; Peng T; Xu H; Zhang RB; Zhao H; Wang ZY; Lv JX; Wu ZS; Shen ZF Highly sensitive detection of cancer-related genes based on complete fluorescence restoration of a molecular beacon with a functional overhang. Analyst 2016, 141, 4417–4423. [DOI] [PubMed] [Google Scholar]
- (50).Yan Q; Duan Q; Huang Y; Guo J; Zhong L; Wang H; Yi G Symmetric exponential amplification reaction-based DNA nano-machine for the fluorescent detection of nucleic acids. RSC Adv. 2019, 9, 41305–41310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chang L; Li J; Wang L Immuno-PCR: An ultrasensitive immunoassay for biomolecular detection. Anal. Chim. Acta 2016, 910, 12–24. [DOI] [PubMed] [Google Scholar]
- (52).Guo R; Yin F; Sun Y; Mi L; Shi L; Tian Z; Li T Ultrasensitive Simultaneous Detection of Multiplex Disease-Related Nucleic Acids Using Double-Enhanced Surface-Enhanced Raman Scattering Nanosensors. ACS Appl. Mater. Interfaces 2018, 10, 25770– 25778. [DOI] [PubMed] [Google Scholar]
- (53).Jang H; Lee CY; Lee S; Park KS; Park HG Flap endonuclease-initiated enzymatic repairing amplification for ultra-sensitive detection of target nucleic acids. Nanoscale 2019, 11, 3633–3638. [DOI] [PubMed] [Google Scholar]
- (54).Kellner MJ; Koob JG; Gootenberg JS; Abudayyeh OO; Zhang F SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc 2019, 14, 2986–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Qin Y; Liao S; Huang Y; Zhao J; Zhao S Ultrasensitive fluorescent detection of nucleic acids based on label-free enzymatic-assisted cascade signal amplification. Anal. Chim. Acta 2018, 1039, 91–97. [DOI] [PubMed] [Google Scholar]
- (56).Sun Y; Peng P; Guo R; Wang H; Li T Exonuclease III-boosted cascade reactions for ultrasensitive SERS detection of nucleic acids. Biosens. Bioelectron 2018, 104, 32–38. [DOI] [PubMed] [Google Scholar]
- (57).Wang B; Ren D; You Z; Yalikun Y; Tanaka Y Ultrasensitive detection of nucleic acids based on dually enhanced fluorescence polarization. Analyst 2018, 143, 3560–3569. [DOI] [PubMed] [Google Scholar]
- (58).Westergaard Mulberg M; Taskova M; Thomsen RP; Okholm AH; Kjems J; Astakhova K New Fluorescent Nanoparticles for Ultrasensitive Detection of Nucleic Acids by Optical Methods. ChemBioChem 2017, 18, 1599–1603. [DOI] [PubMed] [Google Scholar]
- (59).Smith SJ; Nemr CR; Kelley SO Chemistry-Driven Approaches for Ultrasensitive Nucleic Acid Detection. J. Am. Chem. Soc 2017, 139, 1020–1028. [DOI] [PubMed] [Google Scholar]
- (60).Zhou W; Hu L; Ying L; Zhao Z; Chu PK; Yu XF A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun 2018, 9, No. 5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Peng XX; Guo T; Lu H; Yue L; Li Y; Jin D; Zhang GJ; Yang F Nanostructuring Synergetic Base-Stacking Effect: An Enhanced Versatile Sandwich Sensor Enables Ultrasensitive Detection of MicroRNAs in Blood. ACS Sens. 2020, 5, 2514–2522. [DOI] [PubMed] [Google Scholar]
- (62).Leonardi AA; Lo Faro MJ; Petralia S; Fazio B; Musumeci P; Conoci S; Irrera A; Priolo F Ultrasensitive Label-and PCR-Free Genome Detection Based on Cooperative Hybridization of Silicon Nanowires Optical Biosensors. ACS Sens. 2018, 3, 1690–1697. [DOI] [PubMed] [Google Scholar]
- (63).Steckl AJ; Ray P Stress Biomarkers in Biological Fluids and Their Point-of-Use Detection. ACS Sens. 2018, 3, 2025–2044. [DOI] [PubMed] [Google Scholar]
- (64).O’Brien J; Hayder H; Zayed Y; Peng C Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol 2018, 9, No. 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Farazi TA; Hoell JI; Morozov P; Tuschl T MicroRNAs in human cancer. Adv. Exp. Med. Biol 2013, 774, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Hagan JP; Croce CM MicroRNAs in carcinogenesis. Cytogenet. Genome Res 2007, 118, 252–259. [DOI] [PubMed] [Google Scholar]
- (67).Sapkota K; Kaur A; Megalathan A; Donkoh-Moore C; Dhakal S Single-Step FRET-Based Detection of Femtomoles DNA. Sensors 2019, 19, No. 3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Ha T Single-Molecule Fluorescence Resonance Energy Transfer. Methods 2001, 25, 78–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


