Abstract
Background
The long noncoding RNA HOX transcript antisense RNA (HOTAIR) is highly expressed in breast cancer (BC) tissues and is associated with the recurrence and metastasis of BC. Until now, the results of studies on associations between several functional single nucleotide polymorphisms(SNPs) (rs920778, rs1899663, and rs4759314) in HOTAIR with BC susceptibility carried out in different regions of China are still inconsistent. There is no study on correlation between HOTAIR SNPs and prognosis of Chinese population. Therefore, we investigated the relationship between HOTAIR SNPs and susceptibility to and prognosis of BC.
Method
We conducted a population-based case-control study involving 828 BC cases and 905 healthy controls. Peripheral blood DNA was used for genotyping. The association between HOTAIR genotypes and BC risk were estimated by odds ratios (ORs) computed using the binary logistic regression model. The relationships between HOTAIR SNPs and clinicopathological features were tested by Pearson’s chi-square test or Fisher’s exact test. Survival was analyzed using the Kaplan-Meier method.
Results
The functional rs920778 genetic variant increased BC risk in the codominant model. Individuals with the rs920778 GG genotype had an OR of 2.426 (95% confidence interval [CI] = 1.491–3.947, P < 0.001) for developing BC compared to individuals with the AA genotype. Individuals with the AG genotype had an OR of 1.296 (95% CI = 1.040–1.614, P = 0.021) for developing BC compared to individuals with the AA genotype. Individuals with the rs4759314 GA genotype had a lower BC risk than individuals with the rs4759314 AA/GG genotype (OR = 0.566, 95% CI = 0.398–0.803, P = 0.001). The rs1899663 genotype had no correlation with BC susceptibility. Haplotypes composed of rs920778–rs1899663 and rs920778–rs1899663–rs4759314 could increase BC risk (all P < 0.001). There were no statistically significant associations between HOTAIR SNPs and clinicopathological characteristics. The rs920778 GG/AG genotypes were associated with worse disease-free survival (DFS) (p = 0.012), and the rs4759314 GA genotype was associated with worse DFS and overall survival (OS) (p = 0.011).
Conclusion
HOTAIR SNPs(rs920778 and rs4759314) are significantly related to BC susceptibility and prognosis in the northeastern Chinese population, indicating the significance in the occurrence and development of BC.
Keywords: HOTAIR, breast cancer, susceptibility, prognosis, single nucleotide polymorphisms
Introduction
Breast cancer (BC) is one of the most common cancers among women, and its morbidity and mortality have continued to increase worldwide in recent years, reflecting its strong invasive and metastatic characteristics (1, 2). In China, the incidence of BC is increasing annually and is currently the most common malignant tumor in women (3, 4).
Long noncoding RNAs are non-protein-coding transcripts longer than 200 nt and play important roles in the epigenetic regulation of gene expression. One such RNA, HOX transcript antisense RNA (HOTAIR), is transcribed from the antisense strand of the HOXC locus and mainly regulates HOXD genes. HOTAIR can guide the polycomb repressor complex 2/lysine-specific histone demethylase 1 complex to a specific target gene, where the complex then trimethylates lysine 27 of histone H3 and dimethylates lysine 4 of histone H3, causing chromatin remodeling (5–7). This can block some metastasis suppressor genes, such as junctional adhesion molecule 2, protocadherin beta 5, and protocadherin 10 (6).
HOTAIR is overexpressed in BC and is related to the occurrence, development, recurrence, and metastasis of BC. A large number of researches indicate that HOTAIR has oncogenic impacts. In the diagnosis of gastric cancer, pancreatic cancer, and colorectal cancer, the expression of HOTAIR is used to distinguish benign and malignant tissues, compared with benign tissues, the expression of HOTAIR in tumor tissues is higher. HOTAIR is a biomarker of therapeutic response and poor prognosis (8). In our previous studies, we identified several single nucleotide polymorphisms (SNPs) in HOTAIR (rs920778, rs4759314, and rs1899663). These SNPs are located in the intronic region of HOTAIR and can regulate its expression (9–11). Therefore, these SNPs are expected to be related to the occurrence, development, recurrence, and metastasis of BC. These SNPs may have the potential to be a new therapeutic target. Further research demonstrated that these sites are related to gastric cancer, esophageal cancer, and papillary thyroid cancer susceptibility. Several meta-analyses showed that these SNPs are associated with the susceptibility of gastrointestinal cancer and estrogen-dependent tumors (12–17), especially in Asian populations. However, these SNPs have different prevalences in different regions and races and are more common in Asian populations than in Caucasian populations. There are also different prevalences in different parts of Asia (12, 17). Few studies have reported a relationship between HOTAIR SNPs and BC susceptibility. The participants of the current study were mainly Chinese, Turkish, Iranian, and Indian. The results of the research on populations in different regions are inconsistent and controversial. There are obvious regional differences in the distribution of HOTAIR genetic polymorphisms in gastrointestinal cancer. The GG genotype of rs920778 in northeastern population is higher than in middle or southern population, the GG genotype of rs4759314 in southeastern population is higher than in middle and northern population, the GG genotype of rs1899663 in southeastern population is lower than in middle and northern population. Therefore, it is of great significance for us to study the role of HOTAIR gene polymorphisms in the occurrence, development, and prognosis of BC in the Northeast population for the first time. This can provide research basis for discovering new pathogenic targets of BC.Therefore, we retrospectively analyzed the relationship between HOTAIR SNPs (rs920778, rs1899663, and rs4759314) and BC clinicopathological features and prognosis in the northeastern Chinese population.
Materials and Methods
Ethics
This study was approved by the Institutional Ethics Committee of our hospital (ethical approval number 2014-031). Written informed consent was obtained from each participant at recruitment. The study methods were carried out in accordance with the relevant guidelines.
Study Design
Selection and Description of Participants
We investigated the relationship between HOTAIR SNPs (rs920778, rs1899663, and rs4759314) and the risk of BC in a case-control study. All of the participants were genetically unrelated Han Chinese individuals from northeast China. This study enrolled 828 BC patients and 905 age-matched healthy control individuals from The First Affiliated Hospital of Jilin University (Changchun, Jilin Province, China) between April 2013 and September 2016. The median follow-up time was 6.7 years. The participants’ clinical characteristics were collected through medical records. The inclusion criteria were female patients with early breast cancer diagnosed by pathology.
HOTAIR SNP Genotyping
DNA was extracted from peripheral blood samples. Genotypes were detected using the MassArray system (Agena, San Diego, CA, USA) by the matrix-assisted laser desorption ionization-time of flight mass spectrometry method. HOTAIR was selected and genotyped as described previously (9–11). SNP genotyping was performed without knowledge of case status. Reciprocal testing was performed in a random sample of 15%, and the reproducibility was 99.7%.
Statistics
SPSS 24.0 (IBM Corp., Armonk, NY, USA) and the online SNPStats program (https://www.snpstats.net/start.htm, developed by the Institut Català d’Oncologia) were used to analyze BC risk. Variables are characterized as percentages. The Hardy-Weinberg equilibrium test was conducted to test whether the allele frequency distribution of the case group and the control group is biased. Pearson’s chi-square test was used to examine differences in demographic variables and HOTAIR htSNP genotype distributions between BC cases and controls. Associations between HOTAIR genotypes and BC risk were estimated by odds ratios (ORs) and their 95% confidence intervals (CIs), which were computed using the binary logistic regression model. All ORs were adjusted by age whenever appropriate. Pearson’s chi-square test or Fisher’s exact test were used to evaluate the relationships between HOTAIR SNPs and clinicopathological features. The effects of the HOTAIR SNPs on disease-free survival (DFS) and overall survival (OS) were evaluated using the Kaplan-Meier method and the univariate Cox model. All statistical tests were two-sided. P values < 0.05 were considered statistically significant.
Results
Participant Characteristics
The control group was composed of healthy women who had undergone routine physical examination in our hospital who did not have a family history of cancer. The median age of the control group was 38 years (range 32–53 years). There were 678 premenopausal women and 226 postmenopausal women. The median age of the case group was 51 years (range 44–58 years), in which there were 398 premenopausal women and 430 postmenopausal women. Only 32 cases had a family history of cancer. Among 828 BC cases, 793 were of an invasive ductal carcinoma and 35 were of other types. Detailed information on the characteristics of the BC patients can be found in Table 1 .
Table 1.
Clinical characteristics of breast cancer patients.
| Characteristics | Cases No. (%) |
|---|---|
| Median age (years) | 51 (44-58) |
| Menstrual status | |
| Premenopause | 398 (48.07) |
| Postmenopause | 430 (51.93) |
| Family history | |
| Negative | 796 (96.14) |
| Positive | 32 (3.86) |
| Pathological type | |
| Invasive ductal carcinoma | 793 (95.77) |
| Other types | 35 (4.23) |
| Histological grade | |
| I | 31 (3.74) |
| II | 511 (61.71) |
| III | 286 (34.54) |
| Tumor size | |
| T1 | 422(50.97) |
| T2 | 365(44.08) |
| T3 | 27(3.26) |
| T4 | 14(1.69) |
| Lymph node | |
| N0 | 396 (47.83) |
| N1 | 285 (34.42) |
| N2 | 101 (12.20) |
| N3 | 46 (5.58) |
| Lymphovascular invasion | |
| Negative | 471 (56.88) |
| positive | 357 (43.12) |
| Total | 828 (100.00) |
Relationship Between HOTAIR SNPs and Risk of BC
The genotype distribution of cases and controls showed no deviation for different HOTAIR SNPs either in controls or in cases ( Table 2 ). The functional rs920778 genetic variant was associated with an increased risk of BC in three genetic models. We used the Akaike Information Criterion to select the optimal genetic model, and the lowest AIC was found in the codominant genetic model. We discovered that the rs920778 GG genotype had an OR for BC development of 2.426 (95% CI = 1.491–3.947, P < 0.001) compared to the AA genotype. The rs920778 AG genotype was also associated with an increased BC risk compared to the rs920778 AA genotype (OR = 1.296, 95% CI = 1.040–1.614, P = 0.021). The functional rs4759314 genetic variants had different associations with BC risk in different genetic models (i.e., the codominant model, dominant model, and overdominant model). The AIC was the lowest in the overdominant model; therefore, using that model, the rs4759314 GA genotype was associated with a lower risk of BC development (OR = 0.566, 95% CI = 0.398–0.803, P = 0.001) than the AA/GG genotype. The rs1899663 SNP did not show an association with BC risk ( Table 3 ).
Table 2.
Hardy-Weinberg equilibrium test for different HOTAIR SNPs.
| SNPs | Cases | Controls | ||||||
|---|---|---|---|---|---|---|---|---|
| 1H0 | 1He | χ2 | P | 1H0 | 1He | χ2 | P | |
| rs920778 | 0.3321 | 0.3545 | 3.2952 | 0.0695 | 0.3105 | 0.3072 | 0.1054 | 0.7455 |
| rs1899663 | 0.2923 | 0.3043 | 1.2960 | 0.2549 | 0.3127 | 0.2989 | 1.9426 | 0.1634 |
| rs4759314 | 0.0743 | 0.0870 | 17.5992 | <0.001 | 0.1149 | 0.1142 | 0.0398 | 0.8419 |
1H0:observed value of heterozygote frequency; 1He:expected value of heterozygote frequency.
Table 3.
Association between HOTAIR SNPs and breast cancer risk.
| SNP | Genotype | Model | Cases No.(%) | Controls No.(%) | OR (95%CI) a | P value | AIC |
|---|---|---|---|---|---|---|---|
| rs920778 | AA | Codominant | 498(60.36) | 593(65.52) | 1.000 | <0.001 | 2174.4 |
| AG | 274(33.21) | 281(31.05) | 1.296(1.040-1.614) | 0.021 | |||
| GG | 53(6.43) | 31(3.43) | 2.426(1.491-3.947) | <0.001 | |||
| AA | Dominant | 498(60.36) | 593(65.52) | 1.000 | 0.001 | 2180.6 | |
| AG/GG | 327(39.64) | 312(34.48) | 1.406(1.140-1.735) | ||||
| AA/AG | Recessive | 772(93.58) | 874(96.57) | 1.000 | 0.001 | 2177.2 | |
| GG | 53(6.42) | 31(3.43) | 2.220(1.373-3.588) | ||||
| AA/GG | Overdominant | 551(66.79) | 624(68.95) | 1.000 | 0.078 | 2188.1 | |
| AG | 274(33.21) | 281(31.05) | 1.215(0.979-1.509) | ||||
| rs1899663 | CC | Codominant | 552(66.67) | 598(66.08) | 1.000 | 0.163 | 2190.6 |
| CA | 242(29.23) | 283(31.27) | 0.910(0.729-1.134) | 0.400 | |||
| AA | 34(4.10) | 24(2.65) | 1.586(0.900-2.793) | 0.111 | |||
| CC | Dominant | 552(66.67) | 598(66.08) | 1.000 | 0.721 | 2194.3 | |
| CA/AA | 276(33.33) | 307(33.92) | 0.962(0.778-1.190) | ||||
| CC/CA | Recessive | 794(95.89) | 881(97.35) | 1.000 | 0.087 | 2190.1 | |
| AA | 34(4.11) | 24(2.65) | 1.633(0.931-2.864) | ||||
| CC/AA | Overdominant | 586(70.77) | 622(68.73) | 1.000 | 0.296 | 2192.6 | |
| CA | 242(29.23) | 283(31.27) | 0.890(0.715-1.108) | ||||
| rs4759314 | AA | Codominant | 756(91.64) | 798(88.18) | 1.000 | 0.004 | 2182.8 |
| GA | 62(7.52) | 104(11.49) | 0.568(0.400-0.807) | 0.002 | |||
| GG | 7(0.84) | 3(0.33) | 1.930(0.459-8.119) | 0.370 | |||
| AA | Dominant | 756(91.64) | 798(88.18) | 1.000 | 0.004 | 2184 | |
| GA/GG | 69(8.36) | 107(11.82) | 0.609(0.434-0.855) | ||||
| AA/GA | Recessive | 818(99.15) | 902(99.67) | 1.000 | 0.331 | 2189.3 | |
| GG | 7(0.85) | 3(0.33) | 2.039(0.486-8.560) | ||||
| AA/GG | Overdominant | 763(92.48) | 801(88.51) | 1.000 | 0.001 | 2182 | |
| GA | 62(7.52) | 104(11.49) | 0.566(0.398-0.803) |
OR and 95%CI were analyzed by logistic regression and adjusted by age. Common genotype was taken as reference.
Haplotype Analysis
In order to analyze the influence of different haplotype systems composed of three HOTAIR SNP sites on the occurrence of BC, We explored the correlation between haplotypes and BC risk by comparing the distribution of each haplotype in the case group and the control group. There were significant differences between the case and control groups in the distributions of the following haplotypes: rs920778–rs1899663 and rs920778–rs1899663–rs4759314 (all P < 0.001). However, rs1899663–rs4759314 was not related to BC risk ( Table 4 ). Haplotype 1 is composed of wild-type genotypes of three SNPs. Haplotype 2 increased BC risk compared with haplotype 1 (OR=1.39, 95%CI=1.13-1.70, P=0.002). Haplotype 4,5, and 6 reduced BC risk compared with haplotype 1 (all P < 0.001) ( Table 5 ).
Table 4.
Association between haplotypes in HOTAIR and breast cancer risk.
| Haplotypes | df | Global P |
|---|---|---|
| rs920778-rs1899663 | 3 | <0.001 |
| rs1899663-rs4759314 | 3 | 0.100 |
| rs920778-rs1899663-rs4759314 | 7 | <0.001 |
adjusted by age.
Table 5.
Haplotype distribution analysis.
| Haplotype | rs920778 | rs1899663 | rs4759314 | Controls: Case frequency | OR(95%CI) | p | |
|---|---|---|---|---|---|---|---|
| 1 | 0.7353: | 0.7657 | 1.00 | ||||
| 2 | G | A | A | 0.1313: | 0.1854 | 1.39(1.13-1.70) | 0.002 |
| 3 | G | C | G | 0.0354: | 0.0453 | 1.32(0.91-1.92) | 0.140 |
| 4 | A | A | A | 0.0498: | 0.0018 | 0.02(1.01-0.07) | <0.001 |
| 5 | A | C | G | 0.0237: | 0.0006 | 0.01(0.00-0.10) | <0.001 |
| 6 | G | C | A | 0.0228: | 0.0012 | 0.03(0.01-0.12) | <0.001 |
| rare | A | A | G | 0.0017: | NA | 0.00(-) | – |
Relationship Between HOTAIR SNPs and Prognosis of BC
We did not find any significant associations between HOTAIR SNPs and clinicopathological characteristics of BC, including tumor size, lymph node metastasis, lymphovascular invasion, molecular type, histological grade, family history, menstrual status, and pathological type ( Table 6 ). We then assessed the correlation between HOTAIR SNPs and survival in Cox regression analysis. GA genotype of rs920778 and GA genotype of rs4759314 could predict poor prognosis both in univariate analysis and multivariate analysis ( Tables 7 and 8 ).
Table 6.
Association between HOTAIR SNPs and BC clinical characteristics.
| Characteristic | Grouping | rs920778 genotype | P | rs4759314 genotype | P | rs1899663 genotype | P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA n(%) | GA n(%) | GG n(%) | AA n(%) | GA n(%) | GG n(%) | AA n(%) | GAn(%) | GG n(%) | |||||
| Molecular type | luminalA | 62 (12.45) | 37 (13.50) | 5 (9.44) | 0.650 | 95 (12.57) | 8 (12.90) | 1 (14.29) | 0.649 | 70 (12.68) | 30 (12.40) | 4 (11.76) | 0.527 |
| luminalB | 324 (65.06) | 161 (58.76) | 33 (62.26) | 473 (62.57) | 42 (67.74) | 3 (42.86) | 358 (64.86) | 142 (58.68) | 20 (58.82) | ||||
| HER2 | 58 (11.65) | 38 (13.87) | 7 (13.21) | 94 (12.43) | 7 (11.29) | 1 (14.29) | 65 (11.78) | 34 (14.05) | 4 (11.76) | ||||
| Triple negative | 54 (10.84) | 38 (13.87) | 8 (15.09) | 94 (12.43) | 5 (8.07) | 2 (28.56) | 5 9(10.68) | 36 (14.87) | 6 (17.66) | ||||
| Lymph node | N0 | 251 (50.40) | 120 (43.80) | 23 (43.40) | 0.258 | 363 (48.02) | 25 (40.32) | 6 (85.71) | 0.200 | 273 (49.46) | 111 (45.87) | 12 (35.29) | 0.407 |
| N1 | 168 (33.73) | 94 (34.31) | 22 (41.51) | 260 (34.39) | 24 (38.71) | 0 (0.00) | 187 (33.88) | 82 (33.88) | 16 (47.06) | ||||
| N2 | 53 (10.64) | 41 (14.96) | 7 (13.21) | 93 (12.30) | 7 (11.29) | 1 (14.29) | 60 (10.86) | 36 (14.88) | 5 (14.71) | ||||
| N3 | 26 (5.23) | 19 (6.93) | 1 (1.88) | 40 (5.29) | 6 (9.68) | 0 (0.00) | 32 (5.80) | 13 (5.37) | 1 (2.94) | ||||
| Tumor size | T1 | 264 (53.01) | 137 (50.00) | 19 (35.85) | 0.221 | 390 (51.59) | 30 (48.39) | 1 (14.29) | 0.397 | 288 (52.17) | 119 (49.17) | 15 (44.12) | 0.505 |
| T2 | 213 (42.77) | 121 (44.16) | 31 (58.49) | 327 (43.25) | 30 (48.39) | 6 (85.71) | 240 (43.48) | 109 (45.04) | 16 (47.06) | ||||
| T3 | 14 (2.81) | 11 (4.01) | 1 (1.89) | 26 (3.44) | 1 (1.61) | 0 (0.00) | 16 (2.90) | 10 (4.13) | 1 (2.94) | ||||
| T4 | 7 (1.41) | 5 (1.83) | 2 (3.77) | 13 (1.72) | 1 (1.61) | 0 (0.00) | 8 (1.45) | 4 (1.66) | 2 (5.88) | ||||
| Menstrual status | Premenopause | 244 (49.00) | 124 (45.26) | 29 (54.72) | 0.372 | 359 (47.49) | 34 (54.84) | 4 (57.14) | 0.475 | 276 (50.00) | 102 (42.15) | 20 (58.82) | 0.055 |
| Postmenopause | 254 (51.00) | 150 (54.74) | 24 (45.28) | 397 (52.51) | 28 (45.16) | 3 (42.86) | 276 (50.00) | 140 (57.85) | 14 (41.18) | ||||
| Family history | negative | 474 (95.18) | 266 (97.08) | 53 (100.00) | 0.136 | 726 (96.03) | 60 (96.77) | 7 b(100.00) | 1.000 | 526 (95.29) | 236 (97.52) | 34 (100.00) | 0.159 |
| positive | 24 (4.82) | 8 (2.92) | 0 (0.00) | 30 (3.97) | 2 (3.23) | 0 (0.00) | 26 (4.71) | 6 (2.48) | 0 (0.00) | ||||
| Histological grade | Ⅰ | 20 (4.02) | 8 (2.92) | 3 (5.66) | 0.877 | 28 (3.70) | 3 (4.84) | 0 (0.00) | 0.861 | 21 (3.80) | 9 (3.72) | 1 (2.94) | 0.992 |
| Ⅱ | 305 (61.24) | 172 (62.77) | 32 (60.38) | 464 (61.38) | 40 (64.52) | 5 (71.43) | 342 (61.96) | 149 (61.57) | 20 (58.82) | ||||
| Ⅲ | 173 (34.74) | 94 (34.31) | 18 (33.96) | 264 (34.92) | 19 (30.64) | 2 (28.57) | 189 (34.24) | 84 (34.71) | 13 (38.24) | ||||
| Total | 498 (100.00) | 274 (100.00) | 53 (100.00) | 756 (100.00) | 62 (100.00) | 7 (100.00) | 552 (100.00) | 242 (100.00) | 34 (100.00) | ||||
Table 7.
HR in different genotypes of HOTAIR SNPs in univariate Cox regression analysis.
| Genotype | DFS | OS | ||
|---|---|---|---|---|
| HR(95%CI) | P-value | HR(95%CI) | P-value | |
| rs920778 | ||||
| AA | 1.000 | – | 1.000 | – |
| GA | 1.646(1.141-2.375) | 0.007 | 1.786(1.008-3.163) | 0.047 |
| GG | 1.909(1.005-3.625) | 0.048 | 1.209(0.364-4.014) | 0.757 |
| rs1899663 | ||||
| AA | 1.000 | – | 1.000 | – |
| CA | 0.768(0.346-1.703) | 0.007 | 1.011(0.232-4.397) | 0.989 |
| CC | 0.537(0.247-1.166) | 0.062 | 0.892(0.214-3.724) | 0.876 |
| rs4759314 | ||||
| AA | 1.000 | – | 1.000 | – |
| GA | 1.850(1.078-3.173) | 0.026 | 2.792(1.357-5.745) | 0.007 |
| GG | 0.981(0.137-7.028) | 0.985 | <0.001(0–∞) | 0.996 |
Table 8.
HR in different genotypes of HOTAIR SNPs in multivariate Cox regression analysis.
| Genotype | DFS | OS | ||
|---|---|---|---|---|
| HR(95%CI) | P-value | HR(95%CI) | P-value | |
| rs920778 | ||||
| AA | 1.000 | – | 1.000 | – |
| GA | 1.480(1.024-2.139) | 0.037 | 1.547(0.871-2.746) | 0.138 |
| GG | 1.795(0.939-3.429) | 0.077 | 1.133(0.340-3.780) | 0.838 |
| rs1899663 | ||||
| AA | 1.000 | – | 1.000 | – |
| CA | 0.819(0.365-1.835) | 0.627 | 1.019(0.233-4.460) | 0.980 |
| CC | 0.648(0.296-1.416) | 0.276 | 1.091(0.260-4.570) | 0.905 |
| rs4759314 | ||||
| AA | 1.000 | – | 1.000 | – |
| GA | 2.076(1.206-3.571) | 0.008 | 3.472(1.675-7.197) | 0.001 |
| GG | 0.844(0.116-6.130) | 0.867 | <0.001(0–∞) | 0.968 |
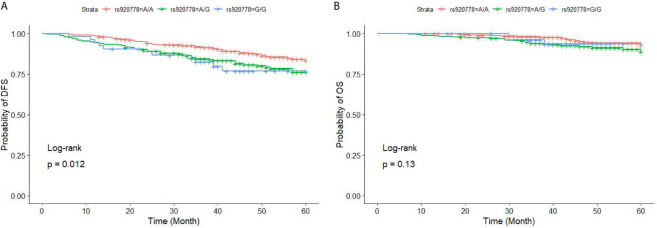
For the rs920778 SNP, there were many significant differences in DFS (P = 0.012) after comparing all three genotype of rs920778, the GG genotype was associated with the worst DFS of the three genotypes (GG, AG, and AA) in univariate analysis (HR = 1.909, P = 0.048). The AG genotype was associated with worse DFS than the AA genotype (HR = 1.48, P = 0.037). However, there was no significant difference in OS (P = 0.13). ( Figure 1 and Table 8 ).
Figure 1.
DFS (A) and OS (B) for BC patients with different genotypes of HOTAIR rs920778. BC, breast cancer; HOTAIR, HOX transcript antisense RNA; DFS, disease-free survival; OS, overall survival. The three curves of DFS are statistically significant (P = 0.012), subjects with GG genotype had a worst DFS (P = 0.048); the three curves of OS are not statistically different (P = 0.13), however, subjects with GA genotype had a worst OS than subjects with AA genotype (P = 0.047).
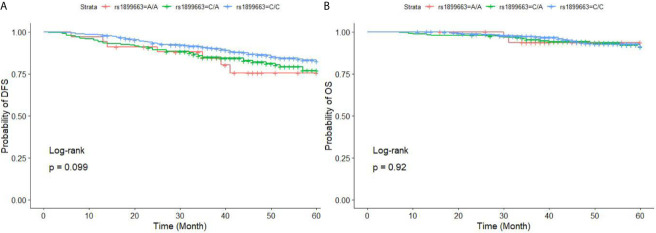
There was no difference in DFS or OS between individuals with the rs1899663 CC or CA genotypes and those with the AA genotype in multivariate analysis. ( Figure 2 and Table 8 ).
Figure 2.
DFS (A) and OS (B) for BC patients with different genotypes of HOTAIR rs1899663. BC, breast cancer; HOTAIR, HOX transcript antisense RNA; DFS, disease-free survival; OS, overall survival. The three curves of DFS/OS are not statistically different, the P value is 0.099 and 0.92 respectively. However, subjects with CA genotype had a worse DFS compared to subjects with AA genotype (P = 0.007).
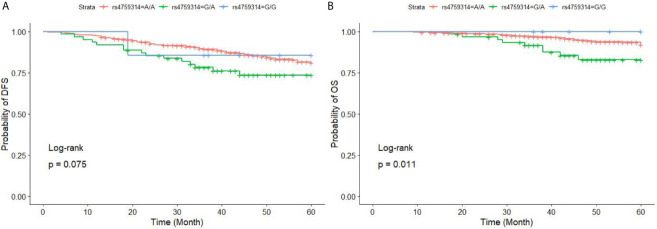
When comparing all three rs4759314 genotypes, the GA genotype had worse DFS and OS than those with the AA genotype (P = 0.008). The OS was significantly different when comparing all three genotypes (P = 0.011); individuals with the GA genotype had the worst OS(P=0.001). However, individuals with the GG genotype and those with the AA genotype had similar OS (P = 0.968) ( Figure 3 and Table 8 ).
Figure 3.
DFS (A) and OS (B) for BC patients with different genotypes of HOTAIR rs4759314. BC, breast cancer; HOTAIR, HOX transcript antisense RNA; DFS, disease-free survival; OS, overall survival. The overall three curves of DFS are insignificant different (P = 0.075), however, subjects with GA genotype had a worst DFS than subjects with AA genotype(P = 0.026); the overall three curves of OS are statistically significant, subjects with GA genotype had a worst OS in the three genotypes, however, subjects with GG genotype and AA genotype had a similar OS.
Discussion
HOTAIR is widely studied as an oncogene, and functional SNPs of HOTAIR have been related to cancer risk, including lung cancer, gastric cancer, esophageal cancer, cervical cancer and, prostate cancer, among others. Due to the difference in sample size and population characteristics, the relationship between the HOTAIR SNPs and BC risk is still contradictory. Our study may help to identify the significance of these three functional SNPs in BC susceptibility. Over-expression of HOTAIR is correlated with poor tumor prognosis, The expression of HOTAIR is regulated by multiple factors at the transcriptional and post-transcriptional levels, including estrogen receptors and estrogen receptor coregulators such as histone methylases MLL1 and MLL3 and CBP/p300 binding to the promoter of HOTAIR and regulating HOTAIR expression (18) and Pumilio homolog 1 regulating HOTAIR expression via a post-transcriptional mechanism (19). Three functional SNPs of HOTAIR can regulate HOTAIR expression (20–22), which may influence the BC prognosis. Our present study explored the relationship between the HOTAIR SNPs and BC prognosis.
The rs920778 SNP (G > A) is located in the intronic enhancer region of HOTAIR, and the AA genotype can increase the expression of HOTAIR. In our study, this SNP increased BC risk, which is consistent with the results of Bayram et al. (23), Rajagopal et al. (24), and Hassanzarei et al. (25) ( Table 9 ). However, Yan et al. (26) found that the A allele is the most common genotype in the central Chinese population and could increase BC risk, which is contrary to the findings for northeast Chinese, southeast Iranian, South Indian, and Turkish populations (the present study, Hassanzarei et al.’s study, Rajagopal et al.’s study, and Bayram et al.’s study, respectively). We found that the G allele is rare and can increase BC risk. The distributions of rs920778 genotypes in BC patients in these five BC studies differ slightly. However, in these five BC studies, the AA genotype is more common whereas the GG genotype is rare. They differ from the distributions observed in other tumor studies [Yan et al. (26) found that the GG genotype is more common, the AA genotype is rare, and the A allele carries disease risk]. One possible reason for this is differences in tumor type and gender. Further, the study by Yan et al. has limitations in terms of sample size, detection methods, research results, and population. Therefore, we think that the rs920778 GG/AG genotypes can increase BC risk.
Table 9.
Comparison of previous studies with our study in the association of HOTAIR SNPs and BC risk.
| HOTAIR SNPs | Ethnicity | Source of Control | No. of Cases | Assay methods | Genotype | Genetic model | OR (P value) | ||
|---|---|---|---|---|---|---|---|---|---|
| Author rs920778 | CC% | CT% | TT% | ||||||
| Bayram et al. (23) | Turkish | Hospitala | 123 | TaqManSNP Genotyping |
25.2 | 42.3 | 32.5 | Recessivea | 2.4 P=0.01 |
| Yan et al. (26) | Chinese | Population | 502 | PCR-RFLP CRS–RFLP | 2.4 | 30.1 | 67.5 | T Allele |
1.41 P=0.02 |
| Hassanzarei et al. (25) | Southeast Iranian | Population | 220 | PCR-RFLP | 15.0 | 54.1 | 30.9 | Dominanta | 2.64 P<0.0001 |
| Rajagopal et al. (24) | Indian | Population | 502 | PCR-RFLP | 17.3 | 50.2 | 32.5 | Over-dominanta | 1.31 P=0.031 |
| Present Study 2021 | Chinese | Population | 828 | MassAray system | 6.4 | 33.2 | 60.4 | Codominanta | 2.426 P<0.001 |
| rs1899663 | GG% | GT& | TT% | ||||||
| Yan et al. (26) | Chinese | Population | 502 | PCR-RFLP CRS–RFLP | 67.53 | 31.35 | 3.97 | T Allele |
0.88 P=0.25 |
| Hassanzarei et al. (25) | Southeast Iranian | Population | 220 | PCR-RFLP | 37.7 | 55.0 | 7.3 | Over-dominanta | 0.38 P<0.0001 |
| Khorshidi et al. (28) | Iranian | Population | 122 | ARMS-PCR | 30.0 | 52.0 | 18.0 | 1.433 P=0.118 |
|
| Lin et al. (21) | southeast Chinese | Population | 969 | PCR-RFLP | 82.7 | 16.2 | 0.01 | 2.08 P=0.027 |
|
| Rajagopal et al. (24) | Indian | Population | 502 | PCR-RFLP | 38.5 | 45.4 | 16.1 | dominanta | 1.32 P=0.03 |
| Present Study 2021 | Chinese | Population | 828 | MassAray system | 66.67 | 29.23 | 4.1 | Recessivea | 1.633 P=0.087 |
| rs4759314 | AA% | AG% | GG% | ||||||
| Yan et al. (26) | Chinese | Population | 502 | PCR-RFLP CRS–RFLP | 89.84 | 10.71 | 0.40 | G Allele |
0.9 P=0.57 |
| Hassanzarei et al. (25) | Southeast Iranian | Population | 220 | PCR-RFLP | 93.2 | 6.8 | 0 | Codominanta | 2.31 P=0.0808 |
| Khorshidi et al. (28) | Iranian | Population | 122 | ARMS-PCR | 79.0 | 21.0 | 1,0 | 0.755 P=0.316 |
|
| Lin et al. (21) | southeast Chinese | Population | 969 | PCR-RFLP | 82.7 | 16.2 | 0.01 | 1.12 P=0.52 |
|
| Present Study 2021 | Chinese | Population | 828 | MassAray system | 91.64 | 7.52 | 0.84 | overdominanta | 0.566 P=0.001 |
aoptimal model.
The rs1899663 SNP (C > A) is located in the intronic region of HOTAIR, and the AA genotype can increase the expression of HOTAIR by altering the binding affinity of various transcription factors, such as paired box 4, spermatogenic leucine zipper 1, and zinc finger protein 281 (ZFP281) (28) to HOTAIR. The results of studies on the relationship between the rs1899663 SNP and BC susceptibility remain controversial. It has been observed that rs1899663 polymorphism is associated with BC risk in the South Indian population (Rajagopal et al.’s study), the southeast Chinese population (Lin et al.’s study) (21), and the Southeast Iranian population (Hassanzarei et al.’s study) ( Table 9 ). However, no relationships were observed in the central Chinese population (Yan et al.’s study), northeastern Chinese population (the present study), or in the Iranian population (Khorshidi et al.’s study) (27). Two smaller studies from Iran (Hassanzarei et al. and Khorshidi et al.) have inconsistent results, as do three larger Chinese studies (Lin et al., Yan et al., and the present study). In Taheri et al.’s study, the relationship between rs1899663 SNP and prostate cancer susceptibility was not observed due to the sample size, however, they compared prostate hyperplasia tissues and prostate cancer tissues and identified that the risk of AA alleles in tumor tissues was higher than CC alleles, This result suggests that AA alleles might increase prostate cancer susceptibility (28).The P value of 0.087 in the present study is close to 0.05. Therefore, we think that SNP has a weak relationship with BC risk when increasing the sample size due to the weak effect of rs1899663 SNP on BC risk.
The rs4759314 SNP (A > G) is located in intronic region of HOTAIR, and the GG genotype can increase the expression of HOTAIR by enhancing the promoter activity of HOXC11. Of five studies examining the relationship between rs4759314 and BC susceptibility ( Table 9 ), only two Chinese studies [Yan et al. (26) and this study] have shown a significantly decreased risk of BC in individuals with at least one G allele (GA or GG) compared to individuals with homozygous A alleles. The other three studies did not show any association of rs4759314 with BC risk. Two studies in the Iranian population [Hassanzarei et al (25). and Khorshidi et al. (27)] are too small to draw such conclusions, and another Chinese study in southeast China (Lin et al.’s study) showed that rs4759314 has no correlation with the risk of BC (21). This may be because BC has a population bias, and the population in the other two studies are in middle and northeast China.
We also examined the haplotypes of these three SNPs. We found that the rs920778–rs1899663 and rs920778–rs1899663–rs4759314 haplotypes significantly increase BC risk (P < 0.001). We believe that the gene effect of rs920778 affects the gene effects of the other two SNPs, which leads to an increase in breast cancer susceptibility.
In Bayram’s study, researchers found an association between the rs920778 SNP and clinicopathological features in the Turkish population, including advanced TNM stage, larger tumor size, distant metastasis, perineural invasion, and poor histological grade (23). In Hassanzarei’s study, they found that the rs920778 SNP was only significantly associated with ER status (25). In Rajagopal’s study, they found that the rs920778 variant (AG + GG genotype) increased BC risk in premenopausal women (OR = 5.86, 95% CI = 3.87–8.88, P < 0.0001) (24). However, we did not find any relationship between the rs920778 SNP and any clinicopathological features. This may be because all of these studies were retrospective and there might be an inherent selection bias. Because of the low distribution frequency of the GG genotype (about 3–8% among common populations), a large sample size is needed to analyze the relationship between the GG genotype and clinical characteristics.
We initially found that the rs920778 SNP is associated with the prognosis of BC patients. Our study found that the DFS of patients with the AG/GG genotypes was much shorter than that of patients with the AA genotype (P = 0.012). However, we did not find similar results for OS. Our result is consistent with the result of Weng et al’s (29) study showing that subjects with GG genotype of rs920778 had a poor OS, however Xavier-Magalhhães et al’s study (30) had the opposite result that subjects with the AG genotype of rs920778 had a longer overall survival than GG subjects in glioma patients. The sample size and tumor type might result the inconsistent results. HOTAIR is regarded as an oncogene involved in both the initiation and progression of cancer. The rs920778 SNP is located in the intronic enhancer region of HOTAIR, and polymorphism of rs920778 could alter the activity of this enhancer and lead to overexpression of HOTAIR. Elevated expression of HOTAIR has been reported to be associated with reduced DFS and OS in cervical cancer patients (31). Therefore, we infer that the influence of the rs920778 SNP on BC prognosis is mediated by the resultant increased expression of HOTAIR. We need to prove this hypothesis further in BC tissue.
The rs1899663 SNP had no effect on DFS. However, in subgroup analysis, individuals with the CA genotype had worse DFS than those with the AA genotype (P = 0.007), which could provide references for future research. Individuals with the rs4759314 GA genotype had worse DFS and OS than patients with other genotypes(P=0.008 and P=0.001 respectively), which was also interesting and needed further study. Because of the low distribution frequency of the rare genotypes AA of rs1899663 and GG of rs4759314 (no more than 2.4%), a larger sample size is needed to assess their associations with prognosis. Because the rs1899663 and rs4759314 SNPs can increase the expression of HOTAIR, their effect on BC prognosis appears to be mediated by the increased expression of HOTAIR. However, we need to prove this hypothesis further in BC tissue. Although all the results of survival analysis have not been verified in multivariate analysis, our results suggest that some gene loci may play a role in the occurrence and development of BC.
In summary, this study demonstrates, for the first time, that functional HOTAIR SNPs rs920778 and rs4759314 are related to the risk and prognosis of BC in the northeastern Chinese population, suggesting that these two SNP sites may be involved in the occurrence, development, and metastasis of BC by regulating the expression of HOTAIR. This may have certain significance for future diagnosis, drug development, and prognostic judgment of BC. The distribution of gene frequency of the three functional HOTAIR SNP loci has a certain correlation with regions and populations. This study only examined the northeast Chinese population as its research object, and it therefore cannot explain why these three HOTAIR SNP loci are responsible for the occurrence and development of BC in the overall Chinese population. Therefore, we need a more large prospective multi-center, multi-regional, multi-ethnic population to analyze the significancy of HOTAIR SNP in BC development and find a target of treatment.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary files, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Ethics Committee of our hospital (ethical approval number 2014-031). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JC and MY conceived and designed the experiment. NC, LJ, and RB performed the experiment. CK analyzed the data. SX and GY collected clinical information. ZL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the Natural Science Foundation of Jilin Province (20200201474JC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
BC, breast cancer; HOTAIR, HOX transcript antisense RNA; SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; DFS, disease-free survival; OS, overall survival.
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136(5):E359–86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 3. Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al. Breast Cancer in China. Lancet Oncol (2014) 15(7):e279–89. 10.1016/S1470-2045(13)70567-9 [DOI] [PubMed] [Google Scholar]
- 4. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 5. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell (2007) 129(7):1311–23. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464(7291):1071–6. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science (2010) 329(5992):689–93. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghafouri-Fard S, Dashti S, Farsi M, Taheri M. HOX Transcript Antisense RNA: An Oncogenic lncRNA in Diverse Malignancies. Exp Mol Pathol (2021) 118:104578. 10.1016/j.yexmp.2020.104578 [DOI] [PubMed] [Google Scholar]
- 9. Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, et al. The Identification of an ESCC Susceptibility SNP Rs920778 That Regulates the Expression of lncRNA HOTAIR via a Novel Intronic Enhancer. Carcinogenesis (2014) 35(9):2062–7. 10.1093/carcin/bgu103 [DOI] [PubMed] [Google Scholar]
- 10. Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, et al. A Functional lncRNA HOTAIR Genetic Variant Contributes to Gastric Cancer Susceptibility. Mol Carcinog (2016) 55(1):90–6. 10.1002/mc.22261 [DOI] [PubMed] [Google Scholar]
- 11. Zhu H, Lv Z, An C, Shi M, Pan W, Zhou L, et al. Onco-lncRNA HOTAIR and its Functional Genetic Variants in Papillary Thyroid Carcinoma. Sci Rep (2016) 6:31969. 10.1038/srep31969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tian T, Li C, Xiao J, Shen Y, Lu Y, Jiang L, et al. Quantitative Assessment of the Polymorphisms in the HOTAIR lncRNA and Cancer Risk: A Meta-Analysis of 8 Case-Control Studies. PloS One (2016) 11(3):e0152296. 10.1371/journal.pone.0152296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ge Y, Jiang R, Zhang M, Wang H, Zhang L, Tang J, et al. Analyzing 37,900 Samples Shows Significant Association Between HOTAIR Polymorphisms and Cancer Susceptibility: A Meta-Analysis. Int J Biol Markers (2017) 32(2):e231–42. 10.5301/jbm.5000235 [DOI] [PubMed] [Google Scholar]
- 14. Li J, Cui Z, Li H, Lv X, Gao M, Yang Z, et al. Long non-Coding RNA HOTAIR Polymorphism and Susceptibility to Cancer: An Updated Meta-Analysis. Environ Health Prev Med (2018) 23(1):8. 10.1186/s12199-018-0697-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X, Zhao Y, Li Y, Lin F, Zhang J. Association Between HOTAIR Genetic Polymorphisms and Cancer Susceptibility: A Meta-Analysis Involving 122,832 Subjects. Genomics (2020) 112(5):3036–55. 10.1016/j.ygeno.2020.05.018 [DOI] [PubMed] [Google Scholar]
- 16. Moazeni-Roodi A, Aftabi S, Sarabandi S, Karami S, Hashemi M, Ghavami S. Genetic Association Between HOTAIR Gene and the Risk of Cancer: An Updated Meta-Analysis. J Genet (2020) 99(1):48. 10.1007/s12041-020-01214-w [DOI] [PubMed] [Google Scholar]
- 17. Qi Q, Wang J, Huang B, Chen A, Li G, Li X, et al. Association of HOTAIR Polymorphisms Rs4759314 and Rs920778 With Cancer Susceptibility on the Basis of Ethnicity and Cancer Type. Oncotarget (2016) 7:38775–84. 10.18632/oncotarget.9608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J Mol Biol (2013) 425(19):3707–22. 10.1016/j.jmb.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, He XY, Qin S, Mo HQ, Li X, Wu F, et al. Upregulation of PUM1 Expression in Preeclampsia Impairs Trophoblast Invasion by Negatively Regulating the Expression of the lncRNA HOTAIR. Mol Ther (2020) 28(2):631–41. 10.1016/j.ymthe.2019.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hajjari M, Rahnama S. Association Between SNPs of Long Non-Coding RNA HOTAIR and Risk of Different Cancers. Front Genet (2019) 10:113. 10.3389/fgene.2019.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin Y, Guo W, Li N, Fu F, Lin S, Wang C. Polymorphisms of Long non-Coding RNA HOTAIR With Breast Cancer Susceptibility and Clinical Outcomes for a Southeast Chinese Han Population. Oncotarget (2018) 9(3):3677–89. 10.18632/oncotarget.23343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J, et al. The Association Analysis of lncRNA HOTAIR Genetic Variants and Gastric Cancer Risk in a Chinese Population. Oncotarget (2015) 6(31):31255–62. 10.18632/oncotarget.5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bayram S, Sumbul AT, Batmaci CY, Genc A. Effect of HOTAIR Rs920778 Polymorphism on Breast Cancer Susceptibility and Clinicopathologic Features in a Turkish Population. Tumour Biol (2015) 36(5):3863–70. 10.1007/s13277-014-3028-0 [DOI] [PubMed] [Google Scholar]
- 24. Rajagopal T, Seshachalam A, Akshaya RL, Rathnam KK, Talluri S, Jothi A, et al. Association of HOTAIR (Rs920778 and Rs1899663) and NME1 (Rs16949649 and Rs2302254) Gene Polymorphisms With Breast Cancer Risk in India. Gene (2020) 762:145033. 10.1016/j.gene.2020.145033 [DOI] [PubMed] [Google Scholar]
- 25. Hassanzarei S, Hashemi M, Sattarifard H, Hashemi SM, Bahari G, Ghavami S. Genetic Polymorphisms of HOTAIR Gene Are Associated With the Risk of Breast Cancer in a Sample of Southeast Iranian Population. Tumour Biol (2017) 39(10):1010428317727539. 10.1177/1010428317727539 [DOI] [PubMed] [Google Scholar]
- 26. Yan R, Cao J, Song C, Chen Y, Wu Z, Wang K, et al. Polymorphisms in lncRNA HOTAIR and Susceptibility to Breast Cancer in a Chinese Population. Cancer Epidemiol (2015) 39(6):978–85. 10.1016/j.canep.2015.10.025 [DOI] [PubMed] [Google Scholar]
- 27. Khorshidi HR, Taheri M, Noroozi R, Soudyab M, Sayad A, Ghafouri-Fard S. Investigation of the Association of HOTAIR Single Nucleotide Polymorphisms and Risk of Breast Cancer in an Iranian Population. Int J Cancer Manage (2017) 10(5):e7498. 10.5812/ijcm.7498 [DOI] [Google Scholar]
- 28. Taheri M, Habibi M, Noroozi R, Rakhshan A, Sarrafzadeh S, Sayad A, et al. HOTAIR Genetic Variants are Associated With Prostate Cancer and Benign Prostate Hyperplasia in an Iranian Population. Gene (2017) 613:20–4. 10.1016/j.gene.2017.02.031 [DOI] [PubMed] [Google Scholar]
- 29. Weng SL, Wu WJ, Hsiao YH, Yang SF, Hsu CF, Wang PH. Significant Association of Long non-Coding RNAs HOTAIR Genetic Polymorphisms With Cancer Recurrence and Patient Survival in Patients With Uterine Cervical Cancer. Int J Med Sci (2018) 15(12):1312–9. 10.7150/ijms.27505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xavier-Magalhaes A, Oliveira AI, de Castro JV, Pojo M, Goncalves CS, Lourenco T, et al. Effects of the Functional HOTAIR Rs920778 and Rs12826786 Genetic Variants in Glioma Susceptibility and Patient Prognosis. J Neurooncol (2017) 132(1):27–34. 10.1007/s11060-016-2345-0 [DOI] [PubMed] [Google Scholar]
- 31. Huang L, Liao LM, Liu AW, Wu JB, Cheng XL, Lin JX, et al. Overexpression of Long Noncoding RNA HOTAIR Predicts a Poor Prognosis in Patients With Cervical Cancer. Arch Gynecol Obstet (2014) 290(4):717–23. 10.1007/s00404-014-3236-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary files, further inquiries can be directed to the corresponding authors.