Key words: bioinformatics, gene expression, kinetoplastid, parasitology, single-cell transcriptomics
Abstract
Kinetoplastid parasites are responsible for both human and animal diseases across the globe where they have a great impact on health and economic well-being. Many species and life cycle stages are difficult to study due to limitations in isolation and culture, as well as to their existence as heterogeneous populations in hosts and vectors. Single-cell transcriptomics (scRNA-seq) has the capacity to overcome many of these difficulties, and can be leveraged to disentangle heterogeneous populations, highlight genes crucial for propagation through the life cycle, and enable detailed analysis of host–parasite interactions. Here, we provide a review of studies that have applied scRNA-seq to protozoan parasites so far. In addition, we provide an overview of sample preparation and technology choice considerations when planning scRNA-seq experiments, as well as challenges faced when analysing the large amounts of data generated. Finally, we highlight areas of kinetoplastid research that could benefit from scRNA-seq technologies.
Introduction
Parasites within the class Kinetoplastea (commonly referred to as kinetoplastids) comprise unicellular pathogens capable of parasitizing most plant and animal species and severely impacting global health and economic well-being (Jackson et al., 2016; Filardy et al., 2018). Three kinetoplastids infect humans, causing widespread disease: Trypanosoma brucei sp. (human African trypanosomiasis or sleeping sickness), Trypanosoma cruzi (Chagas disease) and Leishmania spp. (leishmaniasis). All three are recognized as neglected tropical diseases (NTDs) by the World Health Organization due to the substantial loss of life and disability caused to those living with the infection. Infection of livestock causes further socio-economic burden due to loss in farming production (Swallow, 1999; Muhanguzi et al., 2017). This is particularly evident in the ‘tsetse belt’ of sub-Saharan Africa, where there are extensive infections of agricultural animals by T. vivax, T. congolense and, to a lesser extent, T. brucei spp. (Giordani et al., 2016), causing wasting diseases. The evolution of mechanical transmission has allowed T. vivax and T. b. evansi to spread further into South America (Desquesnes et al., 2013), as well as Asia in the case of T. b. evansi (Payne et al., 1991; Lun et al., 1993). T. cruzi and Leishmania spp. are also frequently found in wild and domesticated animals providing major reservoirs for human disease (Mazloumi Gavgani et al., 2002; Kaszak et al., 2015; Jansen et al., 2018; Medkour et al., 2019). Study of these diverse pathogens has largely been dependent on research with relatively few culture-adapted strains and life cycle stages, mostly the replicative forms of T. b. brucei, many Leishmania spp. and T. cruzi. Methods allowing investigation of non-adapted strains, which may be more virulent and transmissible, will be highly valuable to assess the relevance of this research to disease. Additionally, kinetoplastids are frequently found as heterogeneous populations, such as differentiating life cycle phases (Kramer, 2012), including among diverse host cells (both intra and extracellularly) and various tissues. Some rare life cycle forms have also been reported, such as persister-like cells (Barrett et al., 2019) and gametes (Peacock et al., 2011; Gibson and Peacock, 2019), and these are often difficult to isolate and characterise. Thus, single-cell transcriptomics could be leveraged to provide important new insights into many aspects of kinetoplastid biology and disease.
Unusually, kinetoplastids lack gene expression control via regulation of transcription initiation for virtually all genes (Campbell et al., 2003; Kramer, 2012; Clayton, 2016) (with the notable exception of variant surface glycoprotein (VSG) expression by African trypanosomes). Genes are arranged in arrays with common promotor regions and are co-transcribed (Berriman, 2005; Kazemi, 2011; Berná et al., 2019). Coordinated trans-splicing of the 5′ RNA splice leader (SL) cap and polyadenylation generates mature individual transcripts. Thus, kinetoplastids primarily rely on mRNA degradation, storage, translation regulation and protein degradation to regulate gene expression. Nonetheless, population-based transcriptomic analyses have revealed extensive modulation of transcript levels across the cell cycle and life cycle, and during stress response regulation (Cohen-Freue et al., 2007; Geiger et al., 2011; Siegel et al., 2011; Cantacessi et al., 2015; Haydock et al., 2015; Patino and Ramírez, 2017).
Profiling gene expression of individual cells with scRNA-seq allows a more detailed analysis of heterogeneous populations, detection of rare cell types and dissection of developmental processes (Wen and Tang, 2018). Recently, scRNA-seq has been applied to the field of parasitology with exciting results. ‘Cell atlases’ have been complied of the complete Plasmodium berghei (Howick et al., 2019) and Toxoplasma gondii (Xue et al., 2020) life cycles, where the transcriptomes of parasites from every life cycle stage have been sequenced and compared. Constructing a computational model of the life cycle with these cells allows gene expression changes across the life cycle to be inferred dynamically and in greater detail than is obtainable by bulk-analysis of mixed populations. Additionally, complete atlases provide a resource that can be mined for cell sub-populations, such as male and female Plasmodium falciparum gametes (Real et al., 2020). These, and other, scRNA-seq data sets have allowed apicomplexan parasitologists to study the dynamic gene expression change during developmental processes, such as the sexual commitment of Plasmodium species and T. gondii tachyzoite to bradyzoite differentiation (Waldman et al., 2020; Xue et al., 2020), and identify putative novel regulators. Additionally, cells from a query scRNA-seq sample can be mapped to these reference atlases to assess their developmental stage and unique gene expression differences (Howick et al., 2019; Real et al., 2020). For example, Howick et al., mapped P. malariae and P. falciparum from clinical samples to the P. berghei life cycle atlas to clearly identify the parasite stages present in the infected patients (Howick et al., 2019).
In kinetoplastids, published scRNA-seq studies are currently limited to T. brucei and the whole life cycle has not yet been explored. Expression dynamics during differentiation of slender to stumpy bloodstream forms in the mammal (Briggs et al., 2020), and epimastigote to metacyclic development in the tsetse salivary glands (Vigneron et al., 2020), have both been dissected using scRNA-seq. We identified transient expression patterns of several genes unidentified by bulk-RNA-seq analysis and generated a bloodstream form T. brucei reference cell atlas, to which we could map perturbed cells for comparison (Briggs et al., 2020). Vigneron et al., were able to separate the mixed population of parasites present in the tsetse fly salivary glands and highlighted the expression of an invariant family of surface proteins specific to metacyclic stages (Vigneron et al., 2020). Expression switching of the monoallelic expressed VSGs in bloodstream form T. brucei parasites has also been investigated using scRNA-seq (Müller et al., 2018). We additionally found scRNA-seq a powerful method for detailing gene expression dynamics during the T. brucei cell cycle (Briggs et al., 2020), as have others for P. falciparum (Reid et al., 2018) and T. gondii (Xue et al., 2020).
Several reviews have detailed the choices in technology (Ziegenhain et al., 2017); Svensson et al., 2017; Hwang et al., 2018; Lafzi et al., 2018; Chen et al., 2019; Natarajan et al., 2019), associated costs (Ziegenhain et al., 2017); Natarajan et al., 2019) and data processing methods (Vallejos et al., 2017; Hwang et al., 2018; Chen et al., 2019; Luecken and Theis, 2019; Vieth et al., 2019) involved in single-cell transcriptomics, and more options are likely to become available as the field evolves. Here, we review methods reported to date with protozoan parasites and their potential application to kinetoplastid research.
Methodology and challenges
Method selection
Common steps required for scRNA-seq include the isolation of individual cells, lysis to release RNA, barcoding of transcripts with specialized library adaptors, reverse transcription to generate cDNA, and cDNA amplification (summarised in Fig. 1). The result is a library suitable for next-generation sequencing. However, technologies vary in how these steps are achieved. Importantly, the endpoint data captured will vary between technologies and so which is the most applicable will often be dictated by the biological question being addressed. Aspects to consider include the level of expression of the gene(s) of interest, the occurrence of rare cell types within a sample, the sequence similarity between members of the same gene families under investigation, and the effect of preparation methods on parasite viability.
Fig. 1.
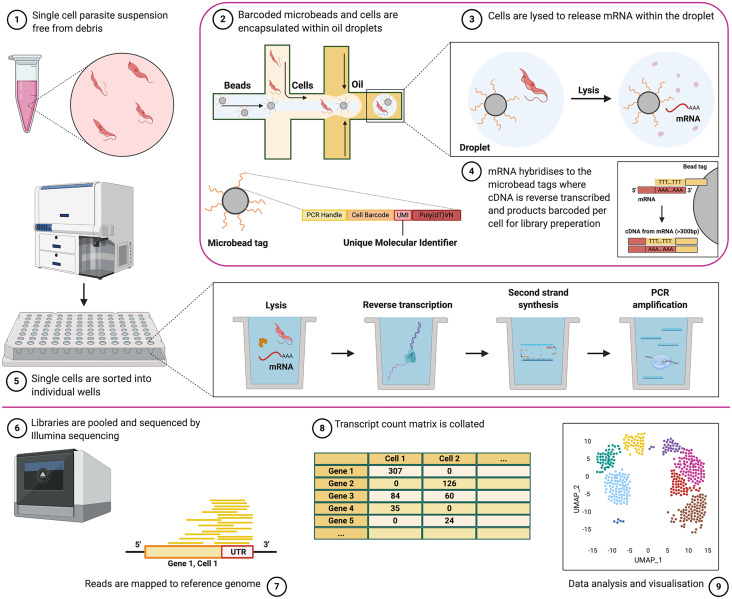
Overview of the scRNA-seq experimental approach. Single parasites, suspended free from debris and containing ideally >90% viable cells, (1) can be captured individually with barcoded library adaptors and lysis buffer either via droplet-based technology (2) or by sorting into individual plate-wells (5). Cells are then lysed (3) and polyadenylated RNA is reverse transcribed with barcode adaptors into cDNA and amplified (4). The resultant library is then sequenced by next-generation sequencing (6). Reads are mapped to the reference genome (7), and unique reads mapping to each gene are counted to generate raw transcript counts per gene, per cell (8). Quality control filtering, pre-processing and final analysis can then be performed (9). See Fig. 4 for an analysis overview. Note, Seq-well, which uses microarrays to capture cells, is not described in this figure but has been used to study T. gondii parasites (Waldman et al., 2020) (created with BioRender.com).
All scRNA-seq approaches aim to isolate the RNA of an individual cell and label transcripts with the same uniquely barcoded library adaptor. Each cell is isolated and once lysed, the released polyadenylated transcripts ligate to these adaptors and are labelled with the unique barcode, allowing the user to identify transcripts that originated from the same cell. The most widely used methods can be broadly grouped as droplet-based or plate-based, including droplet capture techniques Drop-seq (Poran et al., 2017) and Chromium (Howick et al., 2019; Sà et al., 2020; Vigneron et al., 2020), and the popular plate-based method, SMART-seq2 (Müller et al., 2018; Ngara et al., 2018; Reid et al., 2018; Howick et al., 2019; Xue et al., 2020). The former methods encapsulate each cell with an adaptor-coated bead using microfluidics, whereas the latter sort cells into individual wells of plates along with the unique barcoded adaptors. Alternative plate-based assays include SCRB-seq (Soumillon et al., 2014) and the NEBNext® Single Cell/Low Input RNA Library Prep Kit for Illumina® (New England Biolabs Inc. n.d.), both of which have been used to study P. falciparum (Brancucci et al., 2018; Real et al., 2020). Detailed summaries of the available methods can be found in reviews elsewhere (Haque et al., 2017; Hwang et al., 2018). The primary difference between these groups of methods is a trade-off between the higher throughput droplet-based technologies, which capture more cells per experiment, and the higher coverage of plate-based methods, which capture a greater number of transcripts per cell (Table 1). Even a greater number of cells can be captured with the microwell array-based method, Seq-well S3 (Hughes et al., 2020), which outperforms Chromium and Drop-seq in terms of the number of parasites recovered (Table 1).
Table 1.
Comparison of scRNA-seq experiments described with protozoan parasites
| Technique | Species | Cells recovered per experiment described | Average genes per cell | References |
|---|---|---|---|---|
| SMART-seq2 | P. falciparum | 161–191 | 1712–2090 | Reid et al., 2018 |
| P. berghii | 102–517* | 202–2995* | Howick et al., 2019 | |
| P. berghii | 144 | 1981 | Reid et al., 2018 | |
| T gondii | 849–2198 | 862–1290 | Xue et al., 2020 | |
| T. brucei | 418 | 1572 | Müller et al., 2018 | |
| P. falciparum (iRBCs) | 92 | 300–800* | Ngara et al., 2018 | |
| Chromium | P. falciparum | 6737 | 438 | Howick et al., 2019 |
| P. berghii | 4884 | 791 | Howick et al., 2019 | |
| P. knowlesi | 4237 | 557 | Howick et al., 2019 | |
| P. vivax | 22–2098 | 1019$ | Sà et al., 2020 | |
| T. brucei | 2045 | 298 | Vigneron et al., 2020 | |
| T. brucei | 2295–5321 | 1117–1494 | Briggs et al., 2020 | |
| Drop-seq | P. falciparum (iRBCs) | 436–2993 | 224–681 | Poran et al., 2017 |
| Seq-well | T. gondii | 26 560 | 685 | Waldman et al., 2020 |
| SCRB-Seq | P. falciparum | 191–364 | 212–503 | Brancucci et al., 2018 |
| NEBNext | P. falciparum | 88–451* | 108–1496.5* | Real et al., 2020 |
For each experiment, the method and species are given. iRBCs indicate the infected red blood cells were analysed without releasing parasites beforehand. The number of cells recovered after quality control filtering is given per experiment, as reported by the authors.
The asterisk indicates exceptions where the numbers of cell recovered are given for each life cycle stage studied. The average is given for the genes recovered per cell for each experiment or life cycle stage (asterisk), except where the dollar indicates the average of all experiments. NEBNext refers to NEBNext Single Cell/Low Input RNA Library Prep Kit for Illumina (New England Biolabs Inc.).
As only a small number of the total transcripts are recovered per cell with scRNA-seq, the lowest expressed genes are unlikely to be captured. If the aim is to characterize gene expression change for as many genes as possible, or target genes of interest that have known low expression, then a high-coverage approach may be necessary. Some cell types also have low RNA content [e.g. Plasmodium ring stages (Poran et al., 2017; Howick et al., 2019) and T. brucei metacyclics (Vigneron et al., 2020)] and may require more sensitive approaches. In the protozoan parasite studies discussed here, SMART-seq2 frequently recovered the most genes per cell (Table 1). The plate-based method SCRB-seq (Soumillon et al., 2014) has also been used to study protozoan parasites (P. falciparum) (Brancucci et al., 2018), and although the number of genes captured per cell was lower compared to the SMART-seq2 analysis of extracted P. falciparum (Reid et al., 2018), insight into sexual commitment was still gained. NEB also offers a single cell/low input RNA Illumina library preparation kit (New England Biolabs Inc.), which Real et al., used to perform scRNA-seq of P. falciparum and documented the parasite's journey through the mosquito in detail (Real et al., 2020). These methods all rely on assays being performed in 96- or 384-well plates, limiting the number of cells that can be analysed. If the aim is to characterize a cell type of interest that appears as a low or unknown proportion of the sampled population, then a higher throughput method may be preferable, such as Chromium, Drop-seq or Seq-well (Table 1). Only one available study has used Seq-well to study protozoan parasites; Waldman et al., profiled 26,560 T. gondii parasites, recovering an average of 685 genes comparable with other droplet-based methods, but fewer than the SMART-seq2 analysis of T. gondii in similar culture conditions (862–1290 average genes per cell; Table 1).
Preparation methods (see below) will also impact the number of transcripts recovered. For example, slender and stumpy bloodstream form T. brucei prepared in culture (Briggs et al., 2020) resulted in greater recovery of transcripts compared to T. brucei stages extracted from the tsetse salivary gland (Vigneron et al., 2020), despite both studies employing Chromium technology (Table 1). Notably, NEB reports more than three times greater recovery of transcripts compared to SMART-seq2 with the NEBNext® kit for low input RNA (New England Biolabs Inc. n.d.). The studies discussed here (Table 1) can aid users in selecting an appropriate method, considering the depth and cell number required to answer the biological question, in the context of the cell type and the necessary sample preparation steps.
Importantly, most methods (including Drop-seq, 3′ chemistry Chromium kits, Seq-well and SCRB-seq) only sequence the 3′ end of transcripts, whereas SMART-seq2 and NEBNext® sequence full-length transcripts. This consideration can be important, for instance, for studying highly similar paralog families where the 3′ end sequences may be indistinguishable, such as VSGs in T. brucei (Müller et al., 2018), var genes of Plasmodium species (Reid et al., 2018) and SAG1-related sequences (SRS) of T. gondii (Xue et al., 2020). Full-length transcripts are also key to studying splicing variation. However, kinetoplastids lack cis-splicing for all but a few genes (Campbell et al., 2003; Liang et al., 2003) and so this is not a concern for scRNA-seq studies with these parasites.
Cost and equipment availability will undoubtably also impact choice. Drop-seq can be compiled in-house, whereas Chromium technology requires the Chromium controller instrument to encapsulate single cells with barcoded beads. Although, Drop-seq may require more thorough optimization than the commercialized Chromium method. Seq-well, SMART-seq2 and SCRB-seq only need microarrays or plates to separate individual cells; although manual picking can be used, cell sorting machinery is often required to place cells into each well (Hu et al., 2016). The cost of scRNA-seq experiments is largely determined by the high price of library preparation consumables, reviewed here (Ziegenhain et al., 2017)). Notably, SCRB-seq (Soumillon et al., 2014) employed by Brancucci et al. was highly cost affective, at ~$30 per cell (Brancucci et al., 2018). In the case of high-throughput droplet-based methods, the cost can be reduced by mixing species with distinct transcriptomes that can be easily separated bioinformatically (Tung et al., 2017; Kang et al., 2018), as has been done with P. berghei and P. knowlesi (Reid et al., 2018; Howick et al., 2019), and T. brucei and L. mexicana (Briggs et al., 2020; Warren et al., unpublished). This approach also allows a doublet rate to be calculated (see below). Seq-well (Gierahn et al., 2017) and updated ‘Seq-well S3’ (Hughes et al., 2020) offer a comparable cost per read sequenced to Chromium v3, but has the advantage that it avoids the need to purchase Chromium instruments (Hughes et al., 2020). Sample labelling may also allow a further cost reduction in the future by multiplexing; for instance, methods such as CITE-seq (Stoeckius et al., 2017) or cell hashing (Stoeckius et al., 2018) rely on oligo-labelled antibodies that target expressed surface proteins and are captured with the individual cells they bind to. In the library preparation and sequencing steps, the sample barcode is captured along with the cell barcode, allowing transcripts to be traced to both the individual cell and the sample to which the cell belongs. In kinetoplastids, these approaches are more problematic, as commercial barcoded antibodies are not available, life cycle stages express unique surface proteins (Valente et al., 2019), and several prominent surface antigens are expressed from highly variable gene families, limiting options to generate in-house barcoded antibodies. MULTI-seq allows multiplexing of most cells by incorporating lipid-modified oligopeptides (LMOs) (McGinnis et al., 2019a) into the plasma membrane, and so will likely be highly useful for sample labelling and cost-cutting in kinetoplastid research.
Sample preparation
Irrespective of the method of scRNA-seq chosen, each sample should be a single cell suspension free from debris, where ideally at least 90% of the population are viable. Dead cells release cytoplasmic RNA into the sample, meaning a significant number of dying cells will result in both the poor capture of transcripts and the potential for contamination of all living cell transcriptomes with the free mRNA (Yang et al., 2020). Thus, the isolation of viable cells is critical. Cells that have clumped together will likely be labelled with the same barcode, meaning two distinct cell types could be considered as one during analysis. These limitations can be corrected downstream (see below), although cell clumps in the sample often prevent the formation of droplets, if using such methods, stopping the isolation of single cells.
Sample preparation is most easily achieved with extracellular parasites grown in culture, thus these conditions are most likely to result in the capture of high-quality transcriptomes, such as with bloodstream form T. brucei (Müller et al., 2018; Briggs et al., 2020). Here, the preparation may be as simple as washing and suspending cells in an appropriate buffer, such as PBS supplemented with glucose, to maintain viability. Isolation of extracellular forms from the host for in vivo studies will depend on the particular experiment (Vigneron et al., 2020). In this case, methods need to be optimized to ensure sufficient parasites are captured and cellular stress is minimized. Plate-based approaches will likely, additionally, require fluorescence-activated cell sorting (FACS) for cells to be sorted into individual wells, the impact of which on cellular stress and gene expression should be considered. Interestingly, FACS can also be leveraged to gain additional information of the cells before lysing; for instance, cell types can be quantified and enriched for if stained with specific fluorophore-conjugated antibodies. Furthermore, staining information and the plate position of positive cells can be recorded for subsequent association with the sequenced transcriptome (Paul et al., 2015).
In the case of intracellular life cycle stages (such as amastigote Leishmania and T. cruzi), scRNA-seq may be applied directly to the infected host cells. However, detection of the parasite-derived transcripts may be reduced due to the volume of host transcripts and individual parasites infecting the same host will not be distinguishable. The intracellular parasites could instead be extracted prior to scRNA-seq sample preparation. Single-cell analysis for kinetoplastids isolated from host cells has not been published to date, but has been used for apicomplexans: blood-stage Plasmodium spp (Reid et al., 2018; Howick et al., 2019) and T. gondii from human host cells (Waldman et al., 2020; Xue et al., 2020). Cellular stresses caused by these methods need to be considered and potentially tested. Interestingly, Poran et al., applied Drop-seq to P. falciparum infected red blood cells (iRBCs) without extracting parasites with encouraging results (224–681 average genes per cell). However, this method required each sample to be checked manually by microscopy for the number of parasites infecting each RBC to ensure >95% of iRBCs contained only one parasite (Poran et al., 2017). Ngara et al., used SMART-seq2 to profile iRBCs and recovered similar transcript levels (average of 300–800 per cell depending on P. falciparum life cycle stage). Here, the authors were able to confirm only one parasite was present in each iRBC by staining with MitoTracker dye and cell sorting (Ngara et al., 2018). Similar approaches will be useful for kinetoplastid research where extraction of parasites from host cells is difficult and therefore likely to significantly affect parasite viability and gene expression.
If it is not possible to use live cells, such as for clinical samples or time point preservation, cells may be cryopreserved with the cryoprotectant dimethyl-sulfoxide (DMSO) without affecting gene expression (Guillaumet-Adkins et al., 2017; Wohnhaas et al., 2019). Alternatively, cells can be fixed with methanol, stored and subsequently rehydrated for analysis (Chen et al., 2018), although comparisons with mammalian cells have found this method less favourable than cryofreezing (Wohnhaas et al., 2019). We ran a Chromium analysis of bloodstream from T. brucei fixed in methanol, following protocols from 10x Genomics, and sequenced ~30,000 reads per cell (Fig. 2). The percentage of cells recovered after quality control (QC) filtering (discussed below) was only 9.02% for fixed parasites vs 46.55% for live parasites. Although the percentage of kDNA and rRNA transcripts per cell (used for QC, discussed below; Fig. 2B and C) was similar in live and fixed samples, the number of genes per cell and unique transcripts (unique molecular identifiers; UMIs) per cell were dramatically reduced in the fixed sample (Fig. 2A). The effects of these differences meant that no clear grouping of slender and stumpy forms was detected in the fixed parasites, unlike in live parasites (Fig. 2D and E). Methanol preserved P. falciparum and P. malariae clinical samples have been profiled (Howick et al., 2019). Here, SMART-seq2 was used and resulted in more genes per cell being detected [630–1355 median genes per cell, dependant on life cycle stage (Howick et al., 2019)] compared with our Chromium analysis of fixed T. brucei (163 median cells per cell; Fig. 2A). However, relatively few P. falciparum and P. malariae transcriptomes were successfully profiled using fixed parasites, with just 2% and 45% of the starting cell number of early and late blood stages reaching quality control cut-offs, respectively (Howick et al., 2019). Comparison and optimization of fixed and cryostored samples will undoubtedly open new options for single-cell analysis of kinetoplastids in the future.
Fig. 2.
Comparison of methanol fixed and live T. brucei Chromium scRNA-seq data. Methanol fixed (20,000 cells; 1,804 recovered) and live (15,000 cells; 6,938 recovered) T. brucei bloodstream form parasites were previously subjected to Chromium scRNA-seq to a depth of ~30,000 reads per cell (Briggs et al., 2020). (A) UMI (unique molecular identifier;x-axis) and gene (y-axis) per cell counts for methanol fixed (above, red) and live (below, blue) cells. Each point is one cell. The red dashed line indicates the QC threshold used for filtering each sample. (B) As in a, where the percentage of transcripts aligning to the maxi circle kDNA sequence is used as a QC threshold (y-axis). (C) As in a, with a percentage of transcripts aligning to rRNA genes used as a QC threshold (y-axis). (D) UMAPs of each sample (methanol-fixed above, live below), generated as described previously (Briggs et al., 2020) using an identifical method and parameters, with the expections of the QC thresholds indicated in a, b and c. Each data point is one cell coloured by the cluster identity, where each cluster is a group of cells with similar transcriptomic profiles. Colours are not transferred between plots. (E) UMAP plots of cells coloured by expression of one slender marker gene (GAPDH; Tb927.6.4280) and one stumpy marker gene (PAD2; Tb927.7.5940). The scale shows the raw transcript count per cell.
Sequencing and read mapping
Sequencing depth also impacts the analysis of scRNA-seq, as increasing depth will increase the number of unique transcripts returned. Although increasing sequencing depth will therefore increase the information captured, the proportion of newly captured transcripts to sequencing reads will decrease with increasing depth (Fig. 3A). Thus, it is advisable to perform rounds of sequencing and analyse the return of unique transcripts with each round to balance the cost of extensive sequencing and data recovered. The cell ranger tool [10x Genomics (10x Genomics, 2020a)] estimates the unique transcripts to reads mapped ratio, termed sequencing saturation, to guide the user. As it is not possible to uniquely map short-read sequencing to repetitive genomic regions, a large proportion of reads sequenced may be unusable when working with highly repetitive genomes. Thus, calculations should consider confidently mapped reads and greater sequencing depth may be required in these cases. We tested the effect of sequencing depth on the identification of clusters and variable genes using bloodstream form T. brucei Chromium data (Briggs et al., 2020). The return of new transcripts for increased sequencing depth (sequencing saturation) is shown in Fig. 3A. Sequencing to a total depth of 52 ,971 mean reads per cell was performed (Briggs et al., 2020). After selecting different proportions (0.1, 0.25, 0.5 and 0.75) of these total mapped reads by downsampling, we found the mean genes per cell increased from 1,046 when using ~5,000 read per cells, to 1,121 with all reads (~50,000) per cell (Fig. 3A). The increase in mean UMIs per cell was greater, increasing from 1,324 to 1,680 (Fig. 3B). This increase translated into an increased number of clusters being identified (when using a clustering resolution of 0.35 in all cases; Fig. 3D) and revealed far more differentially expressed genes between clusters (Fig. 3C). Hence, sequencing depth can have a large impact on the biological conclusions that can be drawn from the data.
Fig. 3.
Evaluation of sequencing depth impact on cluster identification and differential gene expression analysis. T. brucei bloodstream form (Briggs et al., 2020) cells were previous subjected to Chromium scRNA-seq (Briggs et al., 2020) to a depth of 52,971 mean reads per cell. (A) Sequencing saturation [1−(number of unique reads/number of total mapped reads)] as calculated by cell ranger (10x Genomics, 2020a) for between 5,000 and 52,971 mean reads per cell. The dashed line is equal to 0.9 (90%) sequence saturation. (B) Median genes per cell for total sequencing (52,971 mean reads per cell) and four downsampled data sets. The shaded area shows standard deviation (SD.) from the mean for all cells after QC filtering to remove cells with <500 unique transcripts. (C) The median number of unique transcripts (UMIs) per cell for each data set, shaded area shows SD (D) Number of differentially expressed (DE) genes identified between clusters shown in e, using MAST (Finak et al., 2015). (E) UMAP plots of each data set. Each data point is one cell coloured by cluster identified with the same parameters (resolution = 0.35). Colours are not transferred between plots. Mean reads per cell for each data set are indicated above in bold. The analysis was performed as described previously (Briggs et al., 2020).
Like all genomics and transcriptomic studies, only uniquely mapping reads should be considered. Additionally, controls are needed to remove duplicate reads generated during the cDNA amplification steps of library preparation. Many technologies label individual transcripts with unique barcodes (in addition to cellular barcodes) before any amplification. Hence, replicate transcripts will be identical in the mapping transcript sequence and unique barcode, and can be removed to avoid polymerase chain reaction (PCR) bias. Chromium, Drop-seq, Seq-well and SCRB-seq incorporate these transcript barcodes into the adaptor sequences as UMIs, whereas duplicates are identified purely computationally with SMART-seq2. These computationally biased methods assume all identical reads result from PCR amplification and, thus, are unable to identify real duplicates originating from different transcript copies of the same gene, present before amplification steps (for details, see an investigation by Parekh et al., 2016).
Once mapped to the reference genome, the number of unique transcripts per gene is counted as a measure of expression level. A highly problematic issue when mapping to kinetoplastid genomes is the accuracy of untranslated region (UTR) annotations, as most methods capture only short sequences upstream of the polyadenylation site. Thus, most reads will map to the last 300 bp of the 3′ UTR (Zheng et al., 2017), which needs to be annotated for the aligning read to be included in transcript counts. Polyadenylation site usage, and therefore 3′ UTR sequences, are also variable between species and life cycle stage (Siegel et al., 2010; Kramer and Carrington, 2011; Sà et al., 2020), so may need to be assumed when no accurate reference is available. We overcame this issue when mapping reads from EATRO Antat 1.1 T. brucei to the TREU927 T. brucei genome (Berriman, 2005) by extending UTR sequences to a maximum of 2,500 bp (removing all overlaps with neighbouring genomic features). This dramatically improved the transcripts counted. Sà et al., noted different UTR usage between sexual and asexual stage P. vivax and looked for peaks of unassigned reads to attribute these to the most proximal genes (Sà et al., 2020). Care needs to be taken when interpreting results where UTRs are been assumed, as reads may, in fact, have originated from unannotated genes or non-coding RNA. Full-length transcripts from SMART-seq should show better assignment to genes, as documented in T. brucei when only 400 bp 3′ UTRs were required for mapping (Müller et al., 2018). Hence, full-length transcripts may be preferred when an accurate reference transcriptome is not available. Alternatively, 5′ end methods may be used, such as Chromium Single Cell 5′ Gene Expression from 10x, as part of the Single Cell V(D)J Protocol (10x Genomics, 2020b), although the 5′ UTR annotation may still need to be optimized.
After mapping is performed, read quality and mapping accuracy can be investigated with a genome viewer, and the number of cells captured can be established. As libraries can be stored for additional sequencing it may be cost-effective to the first sequence to a low depth (e.g. 5,000 reads per cell), and assess the number and quality of transcriptomes captured using the methods described below. Additional sequencing can then be performed in the knowledge that the library contains viable cells and with a better estimate of the cell number. Read length could also be customized to allow more gene sequence to be captured and improve mapping (Briggs et al., 2020). For example, for 3′ methods when performing paired-end sequencing, read 1 sequence contains the cell barcode and the UMI, so biasing sequencing to read 2 will increase the length of gene-specific sequence returned.
Data analysis
Methods for the analysis of scRNA-seq data are still evolving and different methods will likely perform better than others with specific data sets. Figure 4 summarizes the general data analysis steps, considerations and decision points where the user is advised to compare methods and/or parameters.
Fig. 4.
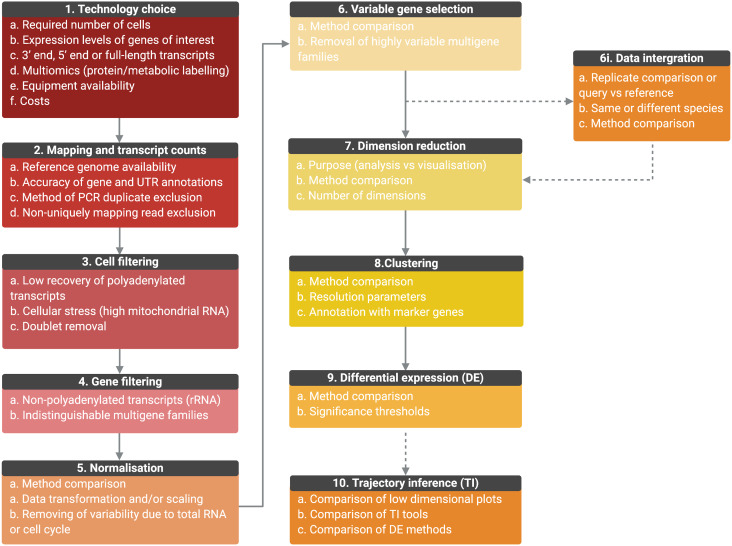
Outline of the general scRNA-seq analysis steps and user considerations. General analysis steps are indicate by numbers and points of consideration are listed below each. (1) The choice of technology will depend on the number of cells required, the expression level of genes, whether full-length transcripts are required, equipment availability and costs. Once scRNA-seq is performed, sequencing is mapped, and transcript counts per gene for each cell are calculated (2). Counts data will be affected by the accuracy of the genome, gene and UTR annotations, PCR duplicate removal and non-uniquely mapping reads. Data will then require filtering to remove cells of low quality or doublets (3) and genes for which transcript counts are likely to be inaccurate (4). Once filtered, data will require normalization, the best method for which will be data set-dependant (5). Data can also be scaled to remove variable gene expression due to total RNA per cell differences and cell cycle dependant gene expression variation. For further analysis, only the top variable genes should be selected to avoid introducing noise (6). Genes from multiple selection methods should be considered and some genes may require removal from variable gene lists, such as VSGs, if not under investigation. (6i) Replicate samples can be integrated, or query cells can be mapped to a control data set or cell ‘atlas’ of the same or different species. Methods should be compared and will depend on aims. As it is not possible to work in high-dimensional space, data should then be reduced (7) and the appropriate number of dimensions to include should be tested. The type of dimensional reduction performed will depend on aims (analysis or visualization) (8). Cells can be clustered by gene expression using reduced data and labelled by investigating the expression of marker genes. Cluster numbers will be dependent on parameters such as resolution. Differential expression (DE) analysis can be performed between clusters or between conditions if data is integrated (9). Tools are still under development to improve power and false discovery rates, and so methods should be compared. If investigating a biological progression between cellular states, trajectory inference (TI) can be performed (10). Over 70 tools exist and performance depends on the topology of the data in low-dimensional plots. Results should be compared and DE across trajectories investigated. (Created with BioRender.com)
Quality control and pre-processing
Quality control of kinetoplastid scRNA-seq data is broadly similar to other cells, whereby transcriptomes with higher than average levels of total RNA are removed as they are likely to include reads from two or more cells (doublets). Similarly, those with low RNA can be removed as they are likely to originate from prematurely lysed cells or where transcript capture has been inefficient. However, cut-offs need to be considered in the context of cell types. For example, metacyclic form T. brucei has substantially less total RNA than epimastigotes (Vigneron et al., 2020), and RNA levels in Plasmodium life cycle stages vary substantially (Poran et al., 2017; Reid et al., 2018; Howick et al., 2019; Real et al., 2020). In mammalian cells, it was found that mitochondrial-encoded genes are a good indicator of cell quality, as lysed cells preferentially lose cytoplasmic transcripts (since organelles are less likely to lyse), and the resulting transcriptomes have a greater percentage of mitochondrial transcripts (Ilicic et al., 2016). We found mapping of reads to the kDNA maxi circle of T. brucei provided an analogous method to remove low-quality parasites (Briggs et al., 2020). Additionally, we found some cells with an unusually high percentage of ribosomal RNA (rRNA) transcripts. As the Chromium adaptors should capture only polyadenylated transcripts, those cells with high rRNA were also considered low quality transcriptomes and were removed. Additional filtering to remove all rRNA transcript counts from further analysis can also be performed. Studies of Plasmodium have also noted contaminating rRNA, which required removal prior to analysis (Poran et al., 2017; Müller et al., 2018; Reid et al., 2018; Waldman et al., 2020; Xue et al., 2020).
Doublet rates vary between technologies and individual experiments. To assess the number of doublets when running the maximum number of cells (30,000) per Chromium sample, we combined T. brucei and L. mexicana in a 1:1 ratio. A total of 8.04% of cells were predicted to contain RNA from both parasites and guided us in removing these cells (Briggs et al., 2020). Similarly, Howick et al., mixed P. knowlesi and P. berghi 1:1 before subjecting 10,000 cells to Chromium scRNA-seq and found a doublet rate of 6.34% (Howick et al., 2019). As well as filtering transcriptomes with unusually high RNA levels, more sophisticated methods for removing doublets have been proposed, including, DoubletFinder (McGinnis, Murrow, and Gartner, 2019b), DoubletDecon (DePasquale et al., 2019) and souporcell, which also identifies free RNA released by damaged cells that have contaminated other transcriptomes (Heaton et al., 2020).
Spike-in control RNA can be included in various methods, though they are primarily used with SMART-seq, as done for analysis of T. brucei (Müller et al., 2018). The usefulness of spike-in RNA is debated, due to the difficulty with precisely adding equal amounts to each cell (Kolodziejczyk et al., 2015). However, this variability has been shown to cause a limited amount of technical noise (Lun et al., 2017) and spike-in RNA may be helpful for normalization, identification of empty wells and dying cells (Baran-Gale, Chandra, and Kirschner, 2018), and removing bias caused by contaminating free RNA (Kolodziejczyk et al., 2015; Marquina-Sanchez et al., 2020). Additionally, spike-in controls can help determine absolute RNA levels, although spike-ins are likely to be captured with different efficiencies to endogenous RNA (Svensson et al., 2017).
Post-filtering, the data requires normalization and can be log-transformed, the methods of which have been extensively reviewed elsewhere (Vallejos et al., 2017; Luecken and Theis, 2019; Lytal et al., 2020). It is worth noting that the total transcripts per cell can have a large effect on plots, and this can be regressed when scaling the data. The contribution of the cell cycle to gene expression variability can also be regressed from the data. However, this requires an assessment of known cell cycle-regulated genes. This was possible in T. brucei due to previous marker analysis (Archer et al., 2011), which is also available for T. cruzi (Chávez et al., 2017). Cell cycle markers are not currently published for Leishmania spp., although it may be possible to perform a similar analysis with orthologs of T. brucei and T. cruzi genes, with consideration of the caveats of inferring markers this way. However, it may be relevant to retain cell cycle variation in the data, for example when studying cell cycle exit.
Dimension reduction
Dimension reduction is performed, firstly, to separate biological variability in the data from technical noise. Many different approaches for dimension reduction have been developed (Luecken and Theis, 2019; Sun et al., 2019), including, but not limited to, variations of principal component (PC) analysis (PCA), diffusion maps (Angerer et al., 2016) and deep-learning methods for larger data sets (Sun et al., 2019). In most cases, the most variable genes must be selected first for use in data reductions to avoid introducing noise. Here, again, multiple methods are possible and the genes selected will affect downstream findings (Luecken and Theis, 2019; Townes et al., 2019). Yip et al., compared seven tools with the same data set and found only two genes were selected as significantly variable genes with all approaches (Yip et al., 2018). Thus, selecting genes with several methods and then comparing the results to select those in common may improve the robustness of analysis (Yip et al., 2018; Kiselev et al., 2019; Luecken and Theis, 2019). Differentially expressed gene families may need to be removed from the variable gene list to avoid grouping cells by predominately these factors, unless they are the subject of investigation. For example, when working with bloodstream form T. brucei it may be necessary to remove VSGs from the variable gene set as these show high variation between cells in some samples and may lead to grouping by the expressed VSG alone (Briggs et al., 2020). A second consideration is the number of PCs, or dimensions, to select for use in the next steps in the analysis. Selecting too few will ignore important variability, whereas selecting too many will introduce noise. Popular methods for selecting PCs include the JackStraw procedure (Macosko et al., 2015), and assessing the variability contributed by each PC with elbow plots. Both are implemented in the popular Seurat workflow (Satija et al., 2015).
The second aim of dimension reduction is visualization. PCA plots only allow limited dimensions (normally the top two) to be plotted and the distance between cells does not necessarily reflect the real distance between transcriptome similarities. Hence, the method of dimension reduction has to be considered to avoid overinterpretation of the data in the later analysis steps (see below). Non-linear dimensionality reduction methods, such as tSNE, UMAP and PHATE, allow further complexity to be incorporated and can conserve the distance between groups of cells. t-SNE plots focus on the local similarity of data points to group them together, at the expense of the global relationship between groups (Van Der Maaten and Hinton, 2008). UMAP (Uniform Manifold Approximation and Projection), however, is better able to conserve the global structure of data and so depicts the relationships between clustered cells as the distance (McInnes et al., 2018). PHATE projections capture local and global structure making it a preferred method for studying continuous data, such as a developmental progression (Moon et al., 2019). Therefore, UMAP is often better suited for scRNA-seq analysis to identify cell clusters and relationships between clusters (Luecken and Theis, 2019), whereas PHATE is well suited for trajectory analysis (see below). Again, the appropriate number of dimensions must be selected, as described above.
Clustering analysis
A common aim of scRNA-seq is to identify cell types present in the data by clustering and marker gene identification. Different clustering approaches have been applied to scRNA-seq data (Duò et al., 2018; Sun et al., 2019) and the choice of method will likely vary between experiments. Sun et al., compared 18 different dimension reduction methods, as well as 3 clustering approaches (k-means, Louvain and hierarchical), to provide a helpful guide (Sun et al., 2019). Each cluster can then be labelled by the cell type identified to begin understanding the biological significance of each population identified. With an existing cell atlas, it would be possible to transfer labels directly from these reference data sets to label cell clusters [a function now integrated into the Seurat pipeline (Stuart et al., 2019)]. Assessing the expression of known marker genes can aid cluster labelling (see differential expression below) in the absence of transferable labels. By exploring the clusters as such, the clustering parameters can be fine-tuned with various iterations to find the most appropriate number of clusters, in the context of the known biology.
Differential expression
As well as investigating known marker genes, novel differentially expressed genes can be identified to aid the interrogation of the identified cell populations. Methods for performing DE analysis between clusters (and across trajectories, see below) are still developing and will likely improve. Bulk RNA-seq DE methods have been applied to scRNA-seq data, although these do not consider the high dropout rate of scRNA-seq experiments, where only a proportion of genes are detected in each cell, or the high variability of most genes between cells (Vallejos et al., 2017). Gene expression of each cluster can be combined by ‘pseudo-bulking’ (Lun and Marioni, 2017; Kang et al., 2018; Crowell et al., 2020), before DE is performed with, for example, edgeR (Robinson et al., 2009) and DEseq2 (Love et al., 2014). scRNA-seq specialized methods have also been developed to tackle these problems (Finak et al., n.d.; Kharchenko et al., 2014), although Vieth et al., point out that the performance of DE tools was largely dependent on upstream analysis steps (Vieth et al., 2019). Bulk tools are improved when introducing gene weights, even out-performing specialist methods (Van den Berge et al., 2018), although this is computationally inefficient (Luecken and Theis, 2019). Notably, muscat has been developed to allow DE of single-cell data, including across multiple samples and multiple conditions, in what the authors’ term ‘differential state analysis’ (Crowell et al., 2020). Once performed, DE between clusters will reveal marker genes which can then be investigated to label known and novel cell types and reveal novel marker genes.
Trajectory inference
Another common aim in analysing scRNA-seq data is trajectory inference and pseudo-time analysis, which allows gene expression change across developmental processes to be investigated (Tanay and Regev, 2017), such as during differentiation of slender to stumpy T. brucei (Briggs et al., 2020), and tachyzoite to bradyzoite T. gondii differentiation (Waldman et al., 2020; Xue et al., 2020). First, an appropriate dimensionally reduced data plot needs to be generated. In general, PCA plots, diffusion maps (Haghverdi et al., 2015; Angerer et al., 2016) or PHATE maps (Moon et al., 2019) can capture simple linear or bifurcating processes that can then be inferred as a trajectory (Luecken and Theis, 2019; Saelens et al., 2019). UMAPs (Cao et al., 2019) or PAGA analysis (Wolf et al., 2019) are better suited to more complex data sets (Luecken and Theis, 2019). A huge number of trajectory inference tools now exist and so selecting one can be cumbersome. Saelens et al., helpfully compared and evaluated the performance of 45 of these tools (Saelens et al., 2019), concluding the choice is dependent on the topology of the trajectory: Slingshot (Street et al., 2018) and TSCAN (Ji and Ji, 2016) perform well when analysing linear processes; Slingshot (Street et al., 2018) also performs well in the detection of branches (Saelens et al., 2019); and PAGA (Wolf et al., 2019) was helpful for analysis of complex data with disconnected clusters of cells. Monocle v3 (Cao et al., 2019) was not available for inclusion in the comparison by Saelens et al., but has since proved helpful for analysing complex data using UMAP, as it can identify multiple branching and converging points in development. We inferred the circular trajectory of the T. brucei cell cycle (Briggs et al., 2020) by fitting a principle curve (Hastie and Stuetzle, 1989), although alternative approaches such as reCAT (Liu et al., 2017) are available (Saelens et al., 2019). Monocle v3 (Cao et al., 2019) provides a pipeline for differential gene expression across the trajectories it finds using graph-autocorrelation analysis. Alternatively, tradeSeq allows expression analysis across simple trajectories by fitting generalized additive models (Van den Berge et al., 2020).
Data integration
Data integration allows the comparison of replicate experiments, but also the mapping of a query sample to an established ‘reference’ data set. For integrating replicate data sets and removing batch effects, methods include but are not limited to, Seurat (Stuart et al., 2019), BBKNN (Polański et al., 2020), Harmony (Korsunsky et al., 2019) and STACAS (Andreatta and Carmona, 2020). More complex data integration can also be performed to map cells onto a reference or ‘cell atlas’, even across different technologies, conditions, cells strains and species. We used STACAS (which is specialized for data sets that are expected to not fully overlap) to map ZC3H20 null T. brucei onto the trajectory of differentiating wild-type parasites to assess the regulation point of this factor (Briggs et al., 2020). Howick et al., employed scmap (Kiselev et al., 2018) to build a reference with P. berghei cells and then map P. falciparum and P. knowlesi transcriptomes to this complete life cycle atlas using orthologous genes (Howick et al., 2019). Thus, cells from different species could be directly compared. The authors were able to extend this approach to map transcriptomes of methanol-fixed P. malariae and P. falciparum from infected volunteers to the P. berghei atlas. Luecken et al., recently compared various combinations of pre-processing and integration methods, concluding Seurat v3 (Stuart et al., 2019) was better suited to simple data integration tasks, whereas BBKNN (Polański et al., 2020), Scanorama (Hie et al., 2019) and scVI (Lopez et al., 2018) successfully integrated more complex data sets (Luecken and Theis, 2019). Helpfully, the authors also provide their scripts so users can identify the optimal data integration method for new data sets (Luecken and Theis, 2019).
Applications
Kinetoplastids commonly exist as heterogeneous populations due to their diversity of life cycle stages. For example, infected tsetse salivary glands contain epimastigotes, early metacyclics and late metacyclics, as dissected by scRNA-seq (Vigneron et al., 2020), as well as gametes (Peacock et al., 2011; Gibson and Peacock, 2019). Several other highly varied populations exist: in culture Leishmania promastigotes exist as procyclic, nectomonad, leptomonad and metacyclic forms (Serafim et al., 2018); cell cycle arrested, or substantially slowed, ‘persister-like’ intracellular amastigote forms of Leishmania and T. cruzi exist as subpopulations with replicating amastigotes (Vickerman, 1985; Tarleton and Zhang, 1999; Fernandes and Andrews, 2012; Mandell and Beverley, 2017; Sánchez-Valdéz et al., 2018; Barrett et al., 2019; Ward et al., 2020); actively cycling and arrested bloodstream form African trypanosomes co-exist in the mammal blood and adipose tissue (Trindade et al., 2016; Rojas and Matthews, 2019); and T. cruzi trypomastigotes, spheromastigotes and epimastigotes are found in the triatomine bug intestinal tract (Chagas, 1909; Castro et al., 2007). Additionally, sexual life cycle stages have been noted for T. brucei, T. cruzi and Leishmania, yet the timing of their development, location within the host and integration into the respective life cycles are often poorly characterized. scRNA-seq is likely to aid our understanding of these diverse cell types, which can be difficult, or impossible, to separate from populations due to their small number or lack of marker genes. Additionally, scRNA-seq removes the need for large cell numbers, often required for bulk analyses. Thus, a detailed investigation of previously unreachable, rare populations which cannot be amplified in culture, should now be possible by profiling parasites immediately after extraction from the host.
Host-parasite interactions could also be analysed in tandem with scRNA-seq by, for instance, analysing infected host cells or capturing host cells along with infecting parasites in in vivo studies. scRNA-seq has been extensively used to analyse the immune response (Chattopadhyay et al., 2014; Chattopadhyay and Roederer, 2015; Buchholz et al., 2016; Buchholz and Flossdorf, 2018) and could be applied to study the host response to kinetoplastid infection. Comparison of parasites resident in differing host tissues, such as an invasion of the lymph and blood systems, skin (Capewell et al., 2016), adipose tissue (Trindade et al., 2016; Tanowitz et al., 2017) and brain (Grab and Kennedy, 2008; Kristensson et al., 2010) by African trypanosomes (Alfituri et al., 2020), should also be facilitated by scRNA-seq. In the case of T. cruzi, 10–30% of infected individuals will develop chronic Chagas (World Health Organization, 2002), where parasites invade the heart (Bellotti et al., 1996), and, in infected mice, parasites have been detected in the skeletal muscle, heart, bladder, peripheral nerve, liver, spleen, adrenal gland, brain and adipose tissue (Buckner et al., 1999). Understanding if and how T. cruzi adapts to these diverse host niches could be revealed by scRNA-seq. Leishmania promastigotes must make their way through the dermis extracellular matrix before infecting macrophages (Arango Duque and Descoteaux, 2015) and neutrophils (Ribeiro-Gomes et al., 2004) and then differentiating into amastigotes, and it remains possible scRNA-seq will uncover aspects of this journey. As well as isolating individual host cells and parasites for the scRNA-seq methods discussed, in situ analysis of single cells in the surrounding tissue can be performed. 10x Genomics now provides a spatial transcriptomic platform based on the method developed by Ståhl et al. (Ståhl et al., 2016). scRNA-seq can also be used to study cross-talk between physically interacting cells, such as T. congolense and the epithelial cells the parasites adhere to (Hemphill et al., 1994; Hemphill and Ross, 1995). PIC-seq combines cell sorting of physically interacting cells (PICs) with scRNA-seq, allowing analysis of the interacting cells and comparison to non-interacting single cells in the same sample to identify interaction-specific gene expression (Giladi et al., 2020). Thus, various iterations of scRNA-seq could improve understanding of the differences between tissue-specific parasites, as well as host cellular responses.
Complex life cycle stage differentiation can also be analysed in novel detail. We used trajectory inference to reconstruct the asynchronous differentiation of bloodstream form T. brucei from slender to stumpy form (Briggs et al., 2020). Epimastigote to early- and then late-stage metacyclic T. brucei has also been profiled to identify transitioning surface protein expression (Vigneron et al., 2020). Similar approaches should be capable of deconvolving the other life cycle stages of T. brucei, arguably the most tractable model of trypanosome differentiation. Different data sets from such studies can in the future be integrated (using the methods described above) to generate a T. brucei life cycle cell atlas. Cell atlases are valuable resources as they document gene expression changes across development processes (e.g. the life cycle or cell cycle) in greater detail than bulk analysis of selected populations.
Using a similar approach, complexities in the Leishmania life cycle could also be dissected. Bulk RNA-seq analysis showed extensive transcript regulation of promastigote, amastigote and axenic amastigote L. mexicana life cycle stages (Fiebig et al., 2015). Separating the various promastigote forms (procyclic, nectomonad and leptomonad) from sandfly tissues (Walters et al., 1989; Lawyer et al., 1990; Walters, 1993; Serafim et al., 2018; Coutinho-Abreu et al., 2020) is also difficult and so profiling these as a mixed population may aid understanding. The T. cruzi life cycle contains equal complexity, with epimastigotes, trypomastigotes and spheromastigotes, as well as many intermediate stages (Schaub, 1989), found together in the triatomine bug intestinal tract (Chagas, 1909; Onyekwelu, 2019). Both amasitogotes and infective trypomastigotes are additionally found in the mammalian host's blood (Onyekwelu, 2019). Hence, complete cell atlases for these life cycles would be equally valuable to understand these cell types and the development between stages via differentiation.
Importantly, these atlases would also provide a reference on which to map lower quality transcriptomes or those from experiments with low cell numbers. Howick et al., clearly demonstrated cross-species mapping of Plasmodium to characterize fixed clinical samples using a high-quality cell atlas (Howick et al., 2019). Clinical or field samples containing kinetoplastids will most likely also need to be stored before scRNA-seq can be applied. Thus, methods to improve the analysis of cryopreserved or chemically fixed parasites will be highly valuable and allow more flexibility when performing scRNA-seq in the future. With the methods discussed, scRNA-seq should allow parasitologists to reach beyond the most well-understood, laboratory-adapted species and cell types, and doing so will be valuable for the development of novel therapeutic approaches.
Perspectives
Despite their unusual approach to transcript regulation, single-cell transcriptomics is a valuable resource for kinetoplastid research. scRNA-seq should allow researchers to interrogate heterogeneous populations, study host–parasite interactions in greater detail, reconstruct maps of developmental processes, and study previously inaccessible species and rare cell types. As well as using the technology and methods discussed here, improved approaches [such as Seq-well S3 (Hughes et al., 2020)] and novel computational analysis [such as machine learning approaches to integration (Lotfollahi et al., 2020)] can be explored to allow larger kinetoplastid data sets to be leveraged. The generation of complete cell atlases for the most tractable species will greatly aid these aims, as will the improvement of protocols to analyse preserved samples. Equally, resources to make these large data sets accessible to the community and allow users to interrogate the data without specific bioinformatic knowledge will be valuable.
Acknowledgements
The authors thank J. Galbraith and P. Herzyk (Glasgow Polyomics, University of Glasgow) for their guidance, library preparation and sequencing. They also thank V. Howick (University of Glasgow) for providing data metrics for inclusion in this review.
Author contributions
E. M. B. and F. S. L. W. conducted literature review. All authors contributed to the discussion, writing and editing of this manuscript.
Ethical standards
Not applicable.
Financial support
This work was supported by the Wellcome Trust (218648/Z/19/Z to E.M.B., 104111/Z/14/ZR to T.D.O. and 103740/Z14/Z to K.R.M.), Wellcome Trust Institutional Strategic Support Fund (ISSF3) awards held at the University of Glasgow (204820/Z/16/Z awarded to E.M.B. and R.M.), the BBSRC-FAPESP (BB/N016165/1 to R.M), and the Edinburgh-Glasgow MRC Precision Medicine DTP (MR/N013166/1, F.S.L.W).
Conflict of interest
The authors declare there are no conflicts of interest.
References
- 10x Genomics (2020a) Cell ranger – Software overview. Available at: https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome (Accessed: 15 December 2020).
- 10x Genomics (2020b) Chromium Next GEM Single Cell 5′ v2. Available at: https://support.10xgenomics.com/single-cell-vdj/library-prep/doc/technical-note-chromium-next-gem-single-cell-5-v2-dual-index-reagent-workflow-and-performance-updates (Accessed: 15 December 2020).
- Alfituri OA, Quintana JF, MacLeod A, Garside P, Benson RA, Brewer JM, Mabbott NA, Morrison LJ and Capewell P (2020) To the skin and beyond: the immune response to African trypanosomes as they enter and exit the vertebrate host. Frontiers in Immunology 11, e1250. doi: 10.3389/fimmu.2020.01250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M and Carmona SJ (2020) STACAS: sub-type anchor correction for alignment in Seurat to integrate single-cell RNA-seq data. Bioinformatics 1, 3. doi: 10.1093/bioinformatics/btaa755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C and Buettner F (2016) Destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243. doi: 10.1093/bioinformatics/btv715 [DOI] [PubMed] [Google Scholar]
- Arango Duque G and Descoteaux A (2015) Leishmania survival in the macrophage: where the ends justify the means. Current Opinion in Microbiology 26, 32–40. doi: 10.1016/j.mib.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Archer SK, Inchaustegui D, Queiroz R and Clayton C (2011) The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS ONE. Edited by N. Papavasiliou 6, e18425. doi: 10.1371/journal.pone.0018425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran-Gale J, Chandra T and Kirschner K (2018) Experimental design for single-cell RNA sequencing. Briefings in Functional Genomics 7, 233–239. doi: 10.1093/bfgp/elx035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett MP, Kyle DE, Sibley LD, Radke JB and Tarleton RL (2019) Protozoan persister-like cells and drug treatment failure. Nature Reviews Microbiology 17, 607–620. doi: 10.1038/s41579-019-0238-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellotti G, Bocchi EA, De Moraes AV, De Higuchi ML, Barbero-Marcial M, Sosa E, Esteves-Filho A, Kalil R, Weiss Jatene and Pileggi F (1996) In vivo detection of Trypanosoma cruzi antigens in hearts of patients with chronic Chagas’ heart disease. American Heart Journal 131, 301–307. doi: 10.1016/S0002-8703(96)90358-0 [DOI] [PubMed] [Google Scholar]
- Berná L, Pita S, Chiribao LM, Parodi-Talice A, Alvarez-Valin F and Robello C (2019) Biology of Trypanosoma cruzi. IntechOpen. 10.5772/intechopen.86144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M (2005) The genome of the African trypanosome Trypanosoma brucei. Science (New York, N.Y.) 309, 416–422. doi: 10.1126/science.1112642 [DOI] [PubMed] [Google Scholar]
- Brancucci NMB, De Niz M, Straub TJ, Ravel D, Sollelis L, Birren BW, Voss T, Neafsey DE and Marti M (2018) Probing Plasmodium falciparum sexual commitment at the single-cell level. Wellcome Open Research 3, 70. doi: 10.12688/wellcomeopenres.14645.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs EM, McCulloch R, Matthews KR and Otto TD (2020) ‘Single cell transcriptomic analysis of bloodstream form Trypanosoma brucei reconstructs cell cycle progression and differentiation via quorum sensing, bioRxiv, doi: 10.1101/2020.12.11.420976. [DOI] [PMC free article] [PubMed]
- Buchholz VR and Flossdorf M (2018) Advances in Immunology 10.1016/bs.ai.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Buchholz VR, Schumacher TNM and Busch DH (2016) T cell fate at the single-cell level. Annual Review of Immunology 34, 65–92. doi: 10.1146/annurev-immunol-032414-112014 [DOI] [PubMed] [Google Scholar]
- Buckner FS, Wilson AJ and Van Voorhis WC (1999) Detection of live Trypanosoma cruzi in tissues of infected mice by using histochemical stain for β-galactosidase. Infection and Immunity 67, 403–409. doi: 10.1128/iai.67.1.403-409.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DA, Thomas S and Sturm NR (2003) Transcription in kinetoplastid protozoa: why be normal? Microbes and infection, 5, 1231–1240. Doi 10.1016/j.micinf.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Dantas-Torres F, Nolan MJ and Otranto D (2015) The past, present, and future of Leishmania genomics and transcriptomics. Trends in Parasitology 31, 100–108. doi: 10.1016/j.pt.2014.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Speilmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C and Shendure J (2019) The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. doi: 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capewell P, Cren-Travaille C, Marchesi F, Johnston P, Clucas C, Benson RA, Gorman TA, Calvo-Alvarez E, Crouzols A, Jouvion G, Jamonneau V, Weir W, Stevenson ML, O'Neill K, Cooper A, Swar NK, Bucheton B, Ngoyi DM, Garside P and Macleod A (2016) The skin is a significant but overlooked anatomical reservoir for vector-borne African trypanosomes. eLife 5, e17716. doi: 10.7554/eLife.17716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DP, Seabra SH, Garcia ES, Souza W and Azembuja P (2007) Trypanosoma cruzi: ultrastructural studies of adhesion, lysis and biofilm formation by Serratia marcescens. Experimental Parasitology 117, 201–207. doi: 10.1016/j.exppara.2007.04.014 [DOI] [PubMed] [Google Scholar]
- Chagas C (1909) Nova Tripanozomiaze humana: estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homem. Memórias do Instituto Oswaldo Cruz 1 doi: 10.1590/s0074-02761909000200008 [DOI] [Google Scholar]
- Chattopadhyay PK, Gierahn TM, Roederer M and Love JC (2014) Single-cell technologies for monitoring immune systems. Nature Immunology 15, 128–135. doi: 10.1038/ni.2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay PK and Roederer M (2015) A mine is a terrible thing to waste: high content, single cell technologies for comprehensive immune analysis. American Journal of Transplantation 5, 1155–1161. doi: 10.1111/ajt.13193 [DOI] [PubMed] [Google Scholar]
- Chávez S, Chávez S, Eastman G, Smircich P, Becco LL, Oliveira-Rizzo C, Fort R, Potenza M, Garat B, Sotelo-Silveira JR and Duhagon MA. (2017) Transcriptome-wide analysis of the Trypanosoma cruzi proliferative cycle identifies the periodically expressed mRNAs and their multiple levels of control. PLOS ONE. 12, e0188441. doi: 10.1371/journal.pone.0188441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chen J, Cheung F, Shi R, Zhou H, Lu W and CHI Consortium (2018) PBMC fixation and processing for chromium single-cell RNA sequencing. Journal of Translational Medicine 16, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Ning B and Shi T (2019) Single-cell RNA-seq technologies and related computational data analysis. Frontiers in Genetics 10, e317. doi: 10.3389/fgene.2019.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C (2016) Gene expression in kinetoplastids. Current Opinion in Microbiology 32, 46–51. [DOI] [PubMed] [Google Scholar]
- Cohen-Freue G, Holzer TR, Forney JD and McMaster WR (2007) Global gene expression in Leishmania. International Journal for Parasitology 37, 1077–1086. doi: 10.1016/j.ijpara.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Coutinho-Abreu IV, Serafim TD, Meneses C, Kamhawi S, Oliveira F and Valenzuela JG (2020) Distinct gene expression patterns in vector-residing Leishmania infantum identify parasite stage-enriched markers. PLoS Neglected Tropical Diseases 14, e0008014. doi: 10.1371/journal.pntd.0008014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell HL, Soneson C, Germain P-L, Calini D, Collin L, Raposo C, Malhotra D and Robinson MD (2020) Muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nature Communications 11, 6077. doi: 10.1038/s41467-020-19894-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePasquale EAK, Schnell DJ, Van Camp PJ, Valiente-Alandí Í, Blaxall BC, Grimes HL, Singh H and Salomonis N (2019) DoubletDecon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Reports 26, 1718–1727. doi: 10.1016/j.celrep.2019.09.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desquesnes M, Holzmuller P, Lai DH, Dargantes A, Lun ZR and Jittaplapong S (2013) Trypanosoma evansi and surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. BioMed Research International 2013, e194176. doi: 10.1155/2013/194176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duò A, Robinson MD and Soneson C (2018) A systematic performance evaluation of clustering methods for single-cell RNA-seq data [version 3; peer review: 2 approved]. F1000 Research 7, e1141. doi: 10.12688/f1000research.15666.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes MC and Andrews NW (2012) Host cell invasion by Trypanosoma cruzi: a unique strategy that promotes persistence. FEMS Microbiology Reviews 36, 734–747. doi: 10.1111/j.1574-6976.2012.00333.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig M, Kelly S and Gluenz E (2015) Comparative life cycle transcriptomics revises Leishmania mexicana genome annotation and links a chromosome duplication with parasitism of vertebrates. PLoS Pathogens 11, e1005186. doi: 10.1371/journal.ppat.1005186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardy AA, Kelly S and Gluenz E (2018) Human kinetoplastid protozoan infections: where are we going next? Frontiers in Immunology 9, e1493. doi: 10.3389/fimmu.2018.01493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, Guimarães-Pinto K, Nunes MP, Zukeram K, Fliess L, Pereira L, Oliveira Nascimento D, Conde L and Morrot A (2015) MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biology 16, e278. doi: 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger A, Simo G, Grébaut P, Peltier JB, Cuny G and Holzmuller P (2011) Transcriptomics and proteomics in human African trypanosomiasis: current status and perspectives. Journal of Proteomics 74, 1625–1643. doi: 10.1016/j.jprot.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Gibson W and Peacock L (2019) Fluorescent proteins reveal what trypanosomes get up to inside the tsetse fly. Parasites and Vectors 12, e6. doi: 10.1186/s13071-018-3204-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierahn TM, Wadsworth MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love CJ and Shalek AK (2017) Seq-well: portable, low-cost RNA sequencing of single cells at high throughput. Nature Methods 14, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi A, Cohen M, Medaglia C, Baran Y, Li B, Zada M, Bost P, Blecher-Gonen R, Salame TM, Mayer JU, David E, Ronchese F, Tanay A and Amit I (2020) Dissecting cellular crosstalk by sequencing physically interacting cells. Nature Biotechnology 38, 629–637. doi: 10.1038/s41587-020-0442-2 [DOI] [PubMed] [Google Scholar]
- Giordani F, Morrison LJ, Rowan TG, DE Koning HP and Barrett MP (2016) The animal trypanosomiases and their chemotherapy: a review. Parasitology 143, 1862–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grab DJ and Kennedy PGE (2008) Traversal of human and animal trypanosomes across the blood-brain barrier. Journal of NeuroVirology 13, 344–351. doi: 10.1080/13550280802282934 [DOI] [PubMed] [Google Scholar]
- Guillaumet-Adkins A, Rodríguez-Esteban G, Mereu E, Mendez-Lago M, Jaitin DA, Villanueva A, Vidal A, Martinez-Marti A, Felip E, Vivancos A, Keren-Shaul H, Heath S, Gut M, Amit I, Gut I and Heyn H (2017) Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biology 18, e45. doi: 10.1186/s13059-017-1171-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Buettner F and Theis FJ (2015) Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics (Oxford, England) 31, 2989–2998. doi: 10.1093/bioinformatics/btv325 [DOI] [PubMed] [Google Scholar]
- Haque A, Engel J, Teichmann SA and Lönnberg T (2017) A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Medicine. BioMed Central Ltd, 1–12. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T and Stuetzle W (1989) Principal curves. Journal of the American Statistical Association 84, 502–516. doi: 10.1080/01621459.1989.10478797 [DOI] [Google Scholar]
- Haydock A, Terrao M, Sekar A, Ramasamy G, Baugh L and Myler PJ (2015) RNA-Seq approaches for determining mRNA abundance in Leishmania. Methods in Molecular Biology. Humana Press Inc 1201, 207–219. doi: 10.1007/978-1-4939-1438-8_12 [DOI] [PubMed] [Google Scholar]
- Heaton H, Talman AM, Knights A, Imaz M, Gaffney DJ, Durbin R, Hemberg M and Lawniczak MKN (2020) Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nature Methods 17, 615–620. doi: 10.1038/s41592-020-0820-1 [DOI] [PubMed] [Google Scholar]
- Hemphill A and Ross CA (1995) Flagellum-mediated adhesion of Trypanosoma congolense to bovine aorta endothelial cells. Parasitology Research 81, 412–420. doi: 10.1007/BF00931503 [DOI] [PubMed] [Google Scholar]
- Hemphill A, Frame I and Ross CA (1994) The interaction of Trypanosoma congolense with endothelial cells. Parasitology 109, 631–641. doi: 10.1017/S0031182000076514 [DOI] [PubMed] [Google Scholar]
- Hie B, Bryson B and Berger B (2019) Efficient integration of heterogeneous single-cell transcriptomes using scanorama. Nature Biotechnology 37, 685–691. doi: 10.1038/s41587-019-0113-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howick VM, Russell AJC, Andrews T, Heaton H, Reid AJ, Natarajan K, Butungi H, Metcalf T, Verzier LH, Rayner JC, Berriman M, Herren JK, Billker O, Hemberg M, Talman AM and Lawniczak MKN (2019) The malaria cell atlas: single parasite transcriptomes across the complete Plasmodium life cycle. Science (New York, NY) 365, eaaw2619. doi: 10.1126/science.aaw2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Zhang W, Xin H and Deng G (2016) Single cell isolation and analysis. Frontiers in Cell and Developmental Biology 4, 116. doi: 10.3389/fcell.2016.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TK, Wadsworth MH, Gierahn TM, Do T, Weiss D, Andrade PR, Ma F, de Andrade Silva BJ, Shao S, Tsoi LC, Ordovas-Montanes J, Gudjonsson JE, Modlin RL, Love JC and Shalek AK (2020) Second-strand synthesis-based massively parallel scRNA-Seq reveals cellular states and molecular features of human inflammatory skin pathologies. Immunity 53, 878–894.e7. doi: 10.1016/j.immuni.2020.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B, Lee JH and Bang D (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental and Molecular Medicine 50, 1–14. doi: 10.1038/s12276-018-0071-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC and Teichmann SA (2016) Classification of low quality cells from single-cell RNA-seq data. Genome Biology 17, 29. doi: 10.1186/s13059-016-0888-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Otto TD, Aslett M, Armstrong SD, Bringaud F, Schlacht A, Hartley C, Sanders M, Wastling JM, Dacks JB, Acosta-Serrano A, Field MC, Ginger ML and Berriman M (2016) Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Current Biology 26, 161–172. doi: 10.1016/j.cub.2015.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AM, Xavier SCDC and Roque ALR (2018) Trypanosoma cruzi transmission in the wild and its most important reservoir hosts in Brazil. Parasites and Vectors 11, 502. doi: 10.1186/s13071-018-3067-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z and Ji H (2016) TSCAN: pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Research 44, e117–e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA and Ye CJ (2018) Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nature Biotechnology 36, 89–94. doi: 10.1038/nbt.4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszak I, Planellas M and Dworecka-Kaszak B (2015) Canine leishmaniosis – an emerging disease. Annals of Parasitology 24, 182–188. doi: 10.1053/j.tcam.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Kazemi B (2011) Genomic organization of Leishmania species. Iranian Journal of Parasitology 6, 1–18. [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L and Scadden DT (2014) Bayesian approach to single-cell differential expression analysis. Nature Methods 11, 740–742. doi: 10.1038/nmeth.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Yiu A and Hemberg M (2018) Scmap: projection of single-cell RNA-seq data across data sets. Nature Methods 15, 359–362. doi: 10.1038/nmeth.4644 [DOI] [PubMed] [Google Scholar]
- Kiselev VY, Andrews TS and Hemberg M (2019) Challenges in unsupervised clustering of single-cell RNA-seq data. Nature Reviews Genetics 20, 273–282. doi: 10.1038/s41576-018-0088-9 [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC and Teichmann S (2015) The technology and biology of single-cell RNA sequencing. Molecular Cell 58, 610–620. doi: 10.1016/j.molcel.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P ru and Raychaudhuri S (2019) Fast, sensitive and accurate integration of single-cell data with harmony. Nature Methods 16, 1289–1296. doi: 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S (2012) Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Molecular and Biochemical Parasitology 181, 61–72. doi: 10.1016/j.molbiopara.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Kramer S and Carrington M (2011) Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends in Parasitology 27, 23–30. doi: 10.1016/j.pt.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensson K, Nygård M, Bertini G and Bentivoglio M (2010) African trypanosome infections of the nervous system: parasite entry and effects on sleep and synaptic functions. Progress in Neurobiology 91, 152–171. doi: 10.1016/j.pneurobio.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Lafzi A, Moutinho C, Picelli S and Heyn H (2018) Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nature Protocols 13, 2741–2757. doi: 10.1038/s41596-018-0073-y [DOI] [PubMed] [Google Scholar]
- Lawyer PG, Ngumbi PM, Anjili CO, Odongo SO, Mebrahtu YB, Githure JI, Koech DK and Roberts CR (1990) Development of Leishmania major in Phlebotomus duboscqi and Sergentomyia schwetzi (Diptera: psychodidae). American Journal of Tropical Medicine and Hygiene 43, 31–43. doi: 10.4269/ajtmh.1990.43.31 [DOI] [PubMed] [Google Scholar]
- Liang X-H, Haritan A, Uliel S and Michaeli S (2003) Trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryotic Cell 2, 830–840. doi: 10.1128/EC.2.5.830-840.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lou H, Xie K, Wang H, Chen N, Aparicio OM, Zhang MQ, Jiang R and Chen T (2017) Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nature Communications 8, 22. doi: 10.1038/s41467-017-00039-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R, Regier J, Cole MB, Jordan MI and Yosef N (2018) Deep generative modeling for single-cell transcriptomics. Nature Methods 15, 1053–1058. doi: 10.1038/s41592-018-0229-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfollahi M Naghipourfar M, Luecken M, Khajavi M, Büttner M, Avsec Z, Misharin A and Theis F (2020) ‘Query to reference single-cell integration with transfer learning’, bioRxiv, p. doi: 10.1101/2020.07.16.205997. [DOI]
- Love MI, Huber W and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken MD and Theis FJ (2019) Current best practices in single-cell RNA-seq analysis: a tutorial. Molecular Systems Biology 52, e8746. doi: 10.15252/msb.20188746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ZR, Fang Y, Wang CJ and Brun R (1993) Trypanosomiasis of domestic animals in China. Parasitology Today 9, 41–45. doi: 10.1016/0169-4758(93)90029-F [DOI] [PubMed] [Google Scholar]
- Lun ATL, Calero-Nieto FJ, Haim-Vilmovsky L, Göttgens B and Marioni JC (2017) Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data. Genome Research 27, 1795–1806. doi: 10.1101/gr.222877.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL and Marioni JC (2017) Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostatistics (Oxford, England) 18, 451–464. doi: 10.1093/biostatistics/kxw055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytal N, Ran D and An L (2020) Normalization methods on single-cell RNA-seq data: an empirical survey. Frontiers in Genetics 11, 41. doi: 10.3389/fgene.2020.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A and McCarroll SA (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. doi: 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell MA and Beverley SM (2017) Continual renewal and replication of persistent Leishmania major parasites in concomitantly immune hosts.. Proc Natl Acad Sci U S A 114, e801–e810. doi: 10.1073/pnas.1619265114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquina-Sanchez B, Fortelny N, Farlik M, Vieira A, Collombat P, Bock C and Kubicek S (2020) Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets. Genome Biology 21, 106. doi: 10.1186/s13059-020-02006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazloumi Gavgani AS, Hodjati MH, Mohite H and Davies CR (2002) Effect of insecticide-impregnated dog collars on incidence of zoonotic visceral leishmaniasis in Iranian children: a matched-cluster randomised trial. Lancet 360, 374–379. doi: 10.1016/S0140-6736(02)09609-5 [DOI] [PubMed] [Google Scholar]
- McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS, Werb Z, Chow ED and Gartner ZJ (2019a) MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nature Methods 16, 619–626. doi: 10.1038/s41592-019-0433-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM and Gartner ZJ (2019b) DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Systems 8, 329–337. doi: 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J and Melville J (2018) ‘UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction’. Available at: http://arxiv.org/abs/1802.03426 (Accessed: 28 October 2020).
- Medkour H, Davoust B, Dulieu F, Maurizi L, Lamour T, Marié J Lou and Mediannikov O (2019) Potential animal reservoirs (dogs and bats) of human visceral leishmaniasis due to Leishmania infantum in French Guiana. PLoS Neglected Tropical Diseases 13, e0007456. doi: 10.1371/journal.pntd.0007456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, Yim K, Elzen A van den, Hirn MJ, Coifman RR, Ivanova NB, Wolf G and Krishnaswamy S (2019) Visualizing structure and transitions in high-dimensional biological data. Nature Biotechnology 37, 1482–1492. doi: 10.1038/s41587-019-0336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhanguzi D, Mugenyi A, Bigirwa G, Kamusiime M, Kitibwa A, Akurut GG, Ochwo S, Amanyire W, Okech SG, Hattendorf J and Tweyongyere R (2017) African animal trypanosomiasis as a constraint to livestock health and production in Karamoja region: a detailed qualitative and quantitative assessment. BMC Veterinary Research 13, 355. doi: 10.1186/s12917-017-1285-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller LSM, Cosentino RO, Förstner KU, Guizetti J, Wedel C, Kaplan N, Janzen CJ, Arampatzi P, Vogel J, Steinbiss S, Otto TD, Saliba A-E, Sebra RP and Siegel TN (2018) Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature 563, 121–125. doi: 10.1038/s41586-018-0619-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan KN, Miao Z, Jiang M, Huang X, Zhou H, Xie J, Wang C, Qin S, Zhao Z, Wu L, Yang N, Li B, Hou Y, Liu S and Teichmann SA (2019) Comparative analysis of sequencing technologies for single-cell transcriptomics. Genome Biology 20, 70. doi: 10.1186/s13059-019-1676-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New England Biolabs Inc (no date) NEBNext® Single Cell/Low Input RNA Library Prep Kit for Illumina®. Available at: https://international.neb.com/products/e6420-nebnext-single-cell-low-input-rna-library-prep-kit-for-illumina#Product Information (Accessed: 15 December 2020).
- Ngara M, Palmkvist M, Sagasser S, Hjelmqvist D, Björklund ÅK, Wahlgren M, Ankarklev J and Sandberg R (2018) Exploring parasite heterogeneity using single-cell RNA-seq reveals a gene signature among sexual stage Plasmodium falciparum parasites. Experimental Cell Research 371, 130–138. doi: 10.1016/j.yexcr.2018.08.003 [DOI] [PubMed] [Google Scholar]
- Onyekwelu K (2019) Biology of Trypanosoma cruzi: Life cycle of Trypanosoma cruzi in the invertebrate and the vertebrate hosts [Google Scholar]
- Parekh S, Ziegenhain C, Vieth B, Enard W and Hellmann I (2016) The impact of amplification on differential expression analyses by RNA-seq. Scientific Reports 6, 25533. doi: 10.1038/srep25533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino LH and Ramírez JD (2017) RNA-seq in kinetoplastids: a powerful tool for the understanding of the biology and host-pathogen interactions. Infection, Genetics and Evolution 49, 273–282. [DOI] [PubMed] [Google Scholar]
- Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, David E, Cohen N, Lauridsen FKB, Haas S, Schlitzer A, Mildner A, Ginhoux F, Jung S, Trumpp A, et al. (2015) Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677. doi: 10.1016/j.cell.2015.11.013 [DOI] [PubMed] [Google Scholar]
- Payne RC, Sukanto IP, Djauhari D, Partoutomo S, Wilson AJ, Jones TW, Boid R and Luckins AG (1991) Trypanosoma evansi infection in cattle, buffaloes and horses in Indonesia. Veterinary Parasitology 38, 109–199. doi: 10.1016/0304-4017(91)90121-B [DOI] [PubMed] [Google Scholar]
- Peacock L, Ferris V, Sharma R, Sunter J, Bailey M, Carrington M and Gibson W (2011) Identification of the meiotic life cycle stage of Trypanosoma brucei in the tsetse fly. Proceedings of the National Academy of Sciences of the United States of America 108, 3671–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA and Park JE (2020) BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics (Oxford, England) 36, 964–965. doi: 10.1093/bioinformatics/btz625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poran A, Nötzel C, Aly O, Mencia-Trinchant N, Harris CT, Guzman ML, Hassane DC, Elemento O and Kafsack BFC (2017) Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551, 95. doi: 10.1038/nature24280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real E, Howick VM, Dahalan F, Witmer K, Cudini J, Andradi-Brown C, Blight J, Davidson MS, Kumar Dogga S, Reid AJ, Baum J and Lawniczak MKN (2020) ‘A single-cell atlas of Plasmodium falciparum transmission through the mosquito Short Title Plasmodium falciparum transmission atlas’, bioRxiv, doi: 10.1101/2020.10.11.333179. [DOI] [PMC free article] [PubMed]
- Reid AJ, Talman AM, Bennett HM, Gomes AR, Sanders MJ, Illingworth CJR, Billker O, Berriman M and Lawniczak MKN (2018) Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. eLife 7, e33105. doi: 10.7554/eLife.33105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Gomes FL, Otero AC, Gomes NA, Moniz-de-Souza MCA, Cysne-Finkelstein L, Arnholdt AC, Calich VL, Coutinho SG, Lopes MF and DosReis GA (2004) Macrophage interactions with neutrophils regulate Leishmania major infection. The Journal of Immunology 172, 4454–4462. doi: 10.4049/jimmunol.172.7.4454 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ and Smyth GK (2009) EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 26, 139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas F and Matthews KR (2019) Quorum sensing in African trypanosomes. Current Opinion in Microbiology 52, 124–129. [DOI] [PubMed] [Google Scholar]
- Sà JM, Cannon M V., Caleon RL, Wellems TE and Serre D (2020) Single-cell transcription analysis of Plasmodium vivax blood-stage parasites identifies stage- and species-specific profiles of expression. PLoS Biology 18, e3000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W, Cannoodt R, Todorov H and Saeys Y (2019) A comparison of single-cell trajectory inference methods. Nature Biotechnology 37, 547–554. [DOI] [PubMed] [Google Scholar]
- Sánchez-Valdéz FJ, A Padilla, W Wang, D Orr and RL Tarleton (2018) Spontaneous dormancy protects Trypanosoma cruzi during extended drug exposure. eLife 7, e34039. doi: 10.7554/eLife.34039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF and Regev A (2015) Spatial reconstruction of single-cell gene expression data. Nature Biotechnology 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub GA (1989) Trypanosoma cruzi: quantitative studies of development of two strains in small intestine and rectum of the vector Triatoma infestans. Experimental Parasitology 3, 260–273. doi: 10.1016/0014-4894(89)90108-2 [DOI] [PubMed] [Google Scholar]
- Serafim TD, Coutinho-Abreu I V., Oliveira F, Meneses C, Kamhawi S and Valenzuela JG (2018) Sequential blood meals promote Leishmania replication and reverse metacyclogenesis augmenting vector infectivity. Nature Microbiology 3, 548–555. doi: 10.1038/s41564-018-0125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S and Cross GAM (2010) Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Research 38, 4946–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Gunasekera K, Cross GAM and Ochsenreiter T (2011) Gene expression in Trypanosoma brucei: lessons from high-throughput RNA sequencing. Trends in parasitology 27, 434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A and Mikkelsen T (2014) ‘Characterization of directed differentiation by high-throughput single-cell RNA-Seq’, bioRxiv. doi: 10.1101/003236. [DOI]
- Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, et a (2016) Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82. doi: 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R and Smibert P (2017) Simultaneous epitope and transcriptome measurement in single cells. Nature Methods 14, 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM, Smibert P and Satija R (2018) Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biology 19, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E and Dudoit S. (2018) Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P and Satija R (2019) Comprehensive integration of single-cell data. Cell 17, 1888–1902. doi: 10.1016/J.CELL.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhu J, Ma Y and Zhou X (2019) Accuracy, robustness and scalability of dimensionality reduction methods for single-cell RNA-seq analysis. Genome Biology 20, 269. doi: 10.1186/s13059-019-1898-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V, Natarajan KN, Ly LH, Miragaia RJ, Labalette C, Macaulay IC, Cvejic A and Teichmann SA (2017) Power analysis of single-cell RNA-sequencing experiments. Nature Methods 14, 381–387. doi: 10.1038/nmeth.4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow BM (1999) ‘Impacts of trypanosomiasis on African agriculture’. Available at: http://www.fao.org/ag/againfo/programmes/en/paat/documents/papers/Paper_1999.pdf (Accessed: 13 March 2018).
- Tanay A and Regev A (2017) Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. doi: 10.1038/nature21350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanowitz HB, Scherer PE, Mota MM and Figueiredo LM (2017) Adipose tissue: a safe haven for parasites? Trends in Parasitology 33, 276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton RL and Zhang L (1999) Chagas disease etiology: autoimmunity or parasite persistence? Parasitology Today 15, 94–99. doi: 10.1016/S0169-4758(99)01398-8 [DOI] [PubMed] [Google Scholar]
- Townes FW, Hicks SC, Aryee MJ and Irizarry RA (2019) Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model. Genome Biology 20, 295. doi: 10.1186/s13059-019-1861-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade S, Rijo-Ferreira F, Carvalho T, Pinto-Neves D, Guegan F, Aresta-Branco F, Bento F, Young SA, Pinto A, Van Den Abbeele J, Ribeiro RM, Dias S, Smith TK and Figueiredo LM (2016) Trypanosoma brucei parasites occupy and functionally adapt to the adipose tissue in mice. Cell Host & Microbe 19, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung PY, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK and Gilad Y (2017) Batch effects and the effective design of single-cell gene expression studies. Scientific Reports 7, 39921. doi: 10.1038/srep39921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente M, Castillo-Acosta VM, Vidal AE and González-Pacanowska D (2019) Overview of the role of kinetoplastid surface carbohydrates in infection and host cell invasion: prospects for therapeutic intervention. Parasitology 146, 1743–1754. doi: 10.1017/S0031182019001355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos CA, Risso D, Scialdone A, Dudoit S and Marioni JC (2017) Normalizing single-cell RNA sequencing data: challenges and opportunities. Nature Methods 14, 565–571. doi: 10.1038/nmeth.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge K, Perraudeau F, Soneson C, Love MI, Risso D, Vert JP, Robinson MD, Dudoit S and Clement L (2018) Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biology 19, 24. doi: 10.1186/s13059-018-1406-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge K, Roux de Bézieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S and Clement L (2020) Trajectory-based differential expression analysis for single-cell sequencing data. Nature Communications 11, 1201. doi: 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L and Hinton G (2008) Visualizing data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Vickerman K (1985) Developmental cycles and biology of pathogenic trypanosomes. British medical bulletin, 41, pp. 105–114. Doi 10.1093/oxfordjournals.bmb.a072036 [DOI] [PubMed] [Google Scholar]
- Vieth B, Parekh S, Ziegenhain C, Enard W and Hellmann I (2019) A systematic evaluation of single cell RNA-seq analysis pipelines. Nature Communications 10, 4667. doi: 10.1038/s41467-019-12266-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron A, O’Neill MB, Weiss BL, Savage AF, Campbell OC, Kamhawi S, Valenzuela JG and Aksoy S (2020) Single-cell RNA sequencing of Trypanosoma brucei from tsetse salivary glands unveils metacyclogenesis and identifies potential transmission blocking antigens. Proceedings of the National Academy of Sciences of the United States of America 117, 2613–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman BS, Schwarz D, Wadsworth MH, Saeij JP, Shalek AK and Lourido S (2020) Identification of a master regulator of differentiation in toxoplasma. Cell 180, 359–372.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters LL, Modi GB, Chaplin GL and Tesh RB (1989) Ultrastructural development of Leishmania chagasi in its vector, Lutzomyia longipalpis (Diptera: psychodidae). American Journal of Tropical Medicine and Hygiene 41, 295–317. doi: 10.4269/ajtmh.1989.41.295 [DOI] [PubMed] [Google Scholar]
- Walters LL (1993) Leishmania differentiation in natural and unnatural sand fly hosts. Journal of Eukaryotic Microbiology 40, 196–206. doi: 10.1111/j.1550-7408.1993.tb04904.x [DOI] [PubMed] [Google Scholar]
- Ward AI, Olmo F, Atherton R and Taylor MC (2020) ‘Trypanosoma cruzi amastigotes have a reduced replication rate during chronic stage infections’, bioRxiv, p. doi: 10.1101/2020.08.04.236240. [DOI] [PMC free article] [PubMed]
- Wen L and Tang F (2018) Boosting the power of single-cell analysis. Nature Biotechnology 36, 408–409. doi: 10.1038/nbt.4131 [DOI] [PubMed] [Google Scholar]
- Wohnhaas CT, Leparc GG, Fernandez-Albert F, Kind D, Gantner F, Viollet C, Hildebrandt T and Baum P (2019) DMSO cryopreservation is the method of choice to preserve cells for droplet-based single-cell RNA sequencing. Scientific Reports 9, 10699. doi: 10.1038/s41598-019-46932-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L and Theis FJ (2019) PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biology 20, 59. doi: 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2002) Control of Chagas Disease: Second report of the WHO Expert Committee, World Health Organization – Technical Report Series.
- Xue Y, Theisen TC, Rastogi S, Ferrel A, Quake SR and Boothroyd JC (2020) A single-parasite transcriptional atlas of Toxoplasma gondii reveals novel control of antigen expression. eLife 9, e54129. doi: 10.7554/eLife.54129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Corbett SE, Koga Y, Wang Z, Johnson WE, Yajima M and Campbell JD (2020) Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biology 21, 57. doi: 10.1186/s13059-020-1950-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip SH, Sham PC and Wang J (2018) Evaluation of tools for highly variable gene discovery from single-cell RNA-seq data. Briefings in Bioinformatics 20, 1583–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, et al. (2017) Massively parallel digital transcriptional profiling of single cells. Nature Communications 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I and Enard W (2017) Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell 65(4), 631–643. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]