Abstract
Mutations in DYNC1H1 have been shown to cause spinal muscular atrophy lower extremity predominant type 1 (SMALED1), an autosomal dominant genetic neuromuscular disorder characterized by degeneration of spinal cord motor neurons resulting in muscle weakness. Here, we describe monozygotic twins, one with a more severe upper motor neuron phenotype as a result of a suspected perinatal hypoxic-ischemic event and the other presenting a typical lower motor neuron phenotype. Using exome sequencing, we identified the novel de novo variant c.752G>T; p.Arg251Leu in DYNC1H1. We thereby add this variant to the growing list of mutations in DYNC1H1 that cause SMALED1.
Keywords: DYNC1H1, Exome sequencing, Spinal muscular atrophy with lower extremity predominance type 1, SMALED1
Introduction
Spinal muscular atrophy lower extremity predominant type 1 (SMALED1) is an autosomal dominant genetic neuromuscular disorder characterized by congenital or very early onset static or slowly progressive muscle weakness primarily in the lower limbs without sensory abnormalities. 1 Mutations in the gene DYNC1H1 (MIM 600112) have been associated with SMALED1 (MIM 158600), as well as several other diseases including malformation of cortical development (MCD) (MIM 614563) and Charcot-Marie-Tooth disease, axonal type 2O (MIM 614228). 1,2 This gene has also been implicated in different neuropsychiatric disorders including autism spectrum disorder (ASD), intellectual disability (ID) and in a handful of cases of epileptic encephalopathy (EE). 3 DYNC1H1 (NM_001376) is located on chromosome 14q32 and encodes the heavy chain of cytoplasmic dynein-1, a motor protein complex which is critical for the transport of organelles, vesicles, and macromolecules towards the minus end of microtubules. 4 Neurons, as a result of their long processes, are extremely reliant on the functional operation of microtubules transport systems to traffic cellular components to and from their axons and dendrites. 4 Here, we present twin girls with a novel de novo mutation in the gene DYNC1H1 at position c.752G>T; p.Arg251Leu causing SMALED1.
Case
Informed consent was obtained from all participants. All experiments involving human participants or data were conducted in compliance with relevant ethical regulations. Approval for human subjects’ research was obtained from the Montreal Children's Hospital and the McGill University Health Centre Research Ethics Boards (project number 11-105-PED and 2019-4972).
Twin girls, now aged 11, were delivered at 35 weeks and 2 days of gestational age via an induced vaginal delivery due to weight discordance on follow-up ultrasound. Twin A weighed 2525 grams at birth with an Apgar score of 9-10-10. No cord pH was obtained. Her sister, Twin B, had a birth weight of 1800 grams with an Apgar score of 9-10-10.
Shortly after birth, Twin A developed respiratory distress with suspected transient tachypnea of the newborn and required continuous positive airway pressure ventilation (CPAP). On day of life 2, she developed lactic acidosis, became encephalopathic with generalized hypotonia and was later found to have neonatal seizures. Glucose and ammonia levels were both normal at that time. In the absence of a clear sentinel anoxic event, extensive metabolic investigations were done and are all negative to this day. Following 3 doses of intravenous bicarbonate, the pH normalized and there was no recurrence of acid-base imbalance. She later developed infantile spasms, spastic quadriparesis, severe global developmental delay, cortical blindness and acquired microcephaly. Twin B presents with a much milder phenotype with proximal weakness predominantly involving the lower limbs, mild appendicular hypotonia, and with advancement in age, development of areflexia. She has never had seizures, is able to walk by herself without support and attends a regular school. She is known for an attention deficit hyperactivity disorder (ADHD) and learning disability. Nerve conduction studies and electromyography were performed in both sisters and showed signs of a chronic denervating process suggestive of a lower motor neuron or root abnormality. Brain magnetic resonance imaging (MRI) of Twin A performed on day 4 of life demonstrated extensive changes in the bilateral hemispheres, basal ganglia, and brainstem with restricted diffusion consistent with ischemic changes. Brain MRI of Twin B at 3 years of age showed a terminal zone of myelination and no evidence of malformation of cortical development.
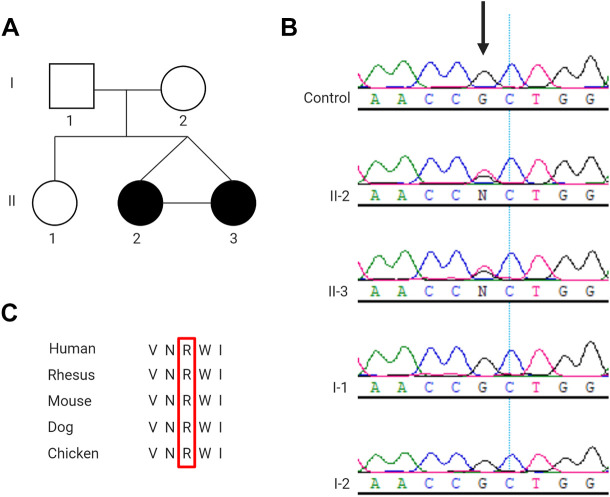
Exome sequencing was performed by Care4Rare according to previously published methods 5 using DNA extracted from whole blood of Twin A. Analyses were conducted in accordance with the American College of Medical Genetics (ACMG) guidelines 6 and revealed a heterozygous variant in DYNC1H1 at position c.752G>T; p.Arg251Leu. Sanger sequencing confirmed the presence of this variant in both Twin A, Twin B, and its absence in their healthy parents (Figure 1A & B). The ACMG guidelines predict this variant to be likely pathogenic as it is in a mutational hot spot (PM1), is absent from control populations (PM2), is predicted to be deleterious by multiple lines of computational evidence (PP3) (PolyPhen-2, Mutation Taster, CADD, Provean), and the amino acid residue is highly conserved in vertebrates (PP3) (Figure 1C). Most notably, this variant is located at an amino acid residue where a different missense change has already been found to be pathogenic (PM5). 7
Figure 1.
Detection of DYNC1H1 variant. (A) Pedigree of family, black circles represent the affected children, II-2 is Twin A and II-3 Twin B. (B) Sanger sequencing confirmation of novel de novo DYNC1H1 variant (NM_001376: c.752G>T; p.Arg251Leu) in Twin A and Twin B and absence from unaffected parents. (C) Arginine (R) at position 251 in DYNC1H1 is a highly conserved amino acid residue.
Discussion and Conclusion
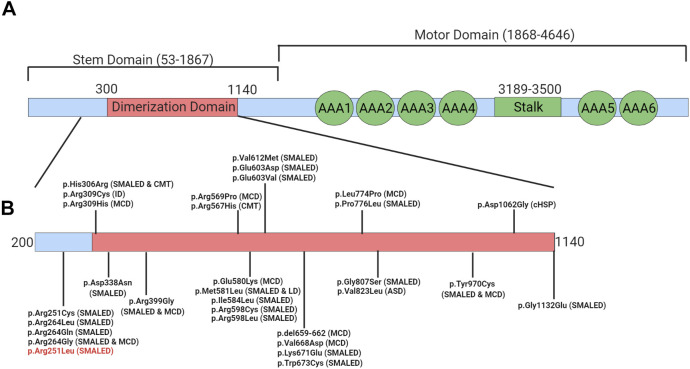
In this study, we report for the first time a de novo missense mutation in DYNC1H1 at position c.752G>T; p.Arg251Leu to cause SMALED1. The ACMG guidelines predict this variant to be likely pathogenic and its location on the DYNC1H1 protein is consistent with other mutations known to cause SMALED1. This is important as there appears to be a correlation between the location of the variant in the DYNC1H1 protein and the associated disease phenotype. 4,8,9 Variants in the stem domain of this protein, particularly those clustered in the important dimerization domain have been found to cause CMT and SMALED1 (Figure 2). 4,8,9 This novel variant is located just adjacent to the dimerization domain of this protein and is clustered amongst other variants reported to cause SMALED1 (Figure 2B). Notably, a distinct de novo variant at the same amino acid position (c.751C>T; p.Arg251Cys) has been reported as likely pathogenic in 4 unrelated patients with SMALED1. 7 This further supports the pathogenicity of our novel de novo variant and its association with the SMALED1 phenotype.
Figure 2.
Locations of reported disease-causing variants on DYNC1H1 protein. (A) Domains of DYNC1H1 protein. (B) Location of variants known to cause disease. Our novel variant p.Arg251Leu is indicated in red. LD = learning disability, MCD = malformations in cortical development, CMT = Charcot-Marie-Tooth disease, SMALED = Spinal muscular atrophy lower extremity predominant, cHSP = complex hereditary spastic paraplegia, ASD = autism spectrum disorder
To explain the brain MRI findings of Twin A, we suspect that a hypoxic-ischemic insult went unnoticed such as an in-utero insult or the post-natal respiratory distress leading to some degree of hypoxia. One can wonder whether Twin A was predisposed to such an event due to an underlying more severe neuromuscular disease. Indeed, a wide clinical variability within the same family has been previously described with another variant in the DYNC1H1 gene. 10 We hypothesize that mosaicism and/or epigenetic factors could have contributed to the potentially discordant phenotype in this set of monozygotic twins.
In conclusion, we report a novel de novo likely pathogenic missense variant (c.752G>T; p.Arg251Leu) in the stem domain of the DYNC1H1 gene in twin girls with SMALED1. This clinical case adds to the growing list of disease-causing variants in DYNC1H1. It also supports the correlation between variant location and disease phenotype, with variants in the stem domain most commonly being associated with SMALED1 or CMT.
Acknowledgements
The authors would like to thank the family for their participation in this report.
Footnotes
Author Contribution: Alexa Derksen and Amytice Mirchi are co-first authors as they were equally responsible for the work described in this paper.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was performed under the Care4Rare Canada Consortium funded by Genome Canada and the Ontario Genomics Institute (OGI-147), the Canadian Institutes of Health Research, Ontario Research Fund, Genome Alberta, Genome British Columbia, Génome Québec, and Children’s Hospital of Eastern Ontario Foundation. This research was enabled in part by support provided by Compute Canada (www.computecanada.ca). The authors also wish to acknowledge the McGill University and Genome Québec Innovation Center. A. Derksen is supported by the Canadian Institutes of Health Research (CIHR) Canadian Graduates Scholarships – Master’s, the Fondation du Grand Défi Pierre Lavoie Master’s Scholarship, and Heathy Brains for Healthy Lives Masters Fellowship. G. Bernard has received a Research Scholar Junior 1 award from the Fonds de Recherche du Québec – Santé (FRQS) (2012-2016), the New Investigator Salary Award from the Canadian Institutes of Health Research (2017-2022) and the Research Scholar Senior award from the FRQS (2022-2025).
ORCID iD: Alexa Derksen  https://orcid.org/0000-0001-7862-4890
https://orcid.org/0000-0001-7862-4890
References
- 1. Beecroft SJ, McLean CA, Delatycki MB, et al. Expanding the phenotypic spectrum associated with mutations of DYNC1H1. Neuromuscul Disord. 2017;27(7):607–615. [DOI] [PubMed] [Google Scholar]
- 2. Strickland AV, Schabhuttl M, Offenbacher H, et al. Mutation screen reveals novel variants and expands the phenotypes associated with DYNC1H1. J Neurol. 2015;262(9):2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin Z, Liu Z, Li X, et al. Whole-exome sequencing identifies a novel de novo mutation in DYNC1H1 in epileptic encephalopathies. Sci Rep. 2017;7(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoang HT, Schlager MA, Carter AP, Bullock SL. DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein-dynactin-cargo adaptor complexes. Proc Natl Acad Sci U S A. 2017;114(9):E1597–E1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamilton A, Tetreault M, Dyment DA, et al. Concordance between whole-exome sequencing and clinical Sanger sequencing: implications for patient care. Mol Genet Genomic Med. 2016;4(5):504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan SHS, van Alfen N, Thuestad IJ, et al. A recurrent de novo DYNC1H1 tail domain mutation causes spinal muscular atrophy with lower extremity predominance, learning difficulties and mild brain abnormality. Neuromuscul Disord. 2018;28(9):750–756. [DOI] [PubMed] [Google Scholar]
- 8. Tsurusaki Y, Saitoh S, Tomizawa K, et al. A DYNC1H1 mutation causes a dominant spinal muscular atrophy with lower extremity predominance. Neurogenetics. 2012;13(4):327–332. [DOI] [PubMed] [Google Scholar]
- 9. Poirier K, Lebrun N, Broix L, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45(6):639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viollet LM, Swoboda KJ, Mao R, et al. A novel pathogenic variant in DYNC1H1 causes various upper and lower motor neuron anomalies. Eur J Med Genet. 2020;63(12):104063. [DOI] [PubMed] [Google Scholar]