Abstract
Cutaneous squamous cell carcinoma (CSCC) is a malignant tumor that originates from keratinocytes in the epidermis or appendage. Traditional Chinese medicine Huaier has anti-tumor activity in various malignancies. Little is known about the role of Huaier in CSCC. Here, we investigated the function of Huaier in CSCC. We treated CSCC cell line (SCL-1 and A431) with a series of concentration gradients of Huaier to examine the half maximal inhibitory concentration (IC50) of Huaier on SCL-1 and A431 cells. The IC50 of Huaier on growth of SCL-1 and A431 cells were 6.96 and 7.57 mg/mL, respectively. Moreover, Huaier reduced the methylation levels of CDKN2A and TP53, and enhanced the expression of CDKN2A and TP53 in SCL-1 and A431 cells in a dosage-dependent manner. The expression of DNA methyltransferase DNMT1 was severely repressed by Huaier treatment in SCL-1 and A431 cells. DNMT1 overexpression enhanced the methylation levels of CDKN2A and TP53, and suppressed the expression of CDKN2A and TP53 in Huaier-treated SCL-1 and A431 cells. Huaier treatment inhibited proliferation, migration, and invasion of SCL-1 and A431 cells. However, inhibition of CDKN2A or TP53 reversed the influence of Huaier treatment on proliferation, migration, and invasion of CSCC cells. In conclusion, our data demonstrate that Huaier inhibits proliferation, migration, and invasion of CSCC cells by regulating DNA methylation of CDKN2A and TP53, thereby attenuating the progression of CSCC. Thus, Huaier extract may act as a drug for treating CSCC.
Keywords: Huaier, CDKN2A, TP53, DNA methylation, cutaneous squamous cell carcinoma
Introduction
Cutaneous squamous cell carcinoma (CSCC) is a type of malignant tumor that originates from keratinocytes in the epidermis or appendage. CSCC usually follows certain skin diseases or precancerous diseases, or is formed by various precancerous lesions. Among non-melanoma skin cancers, the incidence of CSCC is second only to basal cell carcinoma. 1 However, little is known about the pathogenesis of CSCC. Previous study has confirmed that epigenetic changes play a crucial role in the occurrence, development, and prognosis of CSCC, especially DNA methylation. 2
DNA methylation is a hot spot in research on tumorigenesis. In recent years, many studies have reported that CSCC exhibits abnormal changes in DNA methylation.2,3 Toll et al 4 have found that the expression of miR-204 is down-regulated in CSCC, which contributes to the increased methylation levels of miR-204. DNA methylation also leads to down-regulation of TRIM29 in patients with CSCC, and TRIM29 knockdown promotes migration and invasion of CSCC cells. 5 The tumor tissue of CSCC patients exhibits a decrease of DSS1 expression and an increase in the methylation levels of DSS1. The low methylation levels of DSS1 are closely associated with the poor prognosis of CSCC patients. 6 Previously, we have found that the promoter regions of tumor suppressor genes, P14ARF and E-cad, are hypermethylated, while the promoter regions of proto-oncogenes, c-jun and c-fos, are hypomethylated. 7 Thus, hypermethylation of tumor suppressor genes and hypomethylation of proto-oncogenes may synergistically cause the occurrence of tumors. Cell cycle-related gene Cyclin-dependent kinase inhibitor 2A (CDKN2A) and p53 tumor suppressor gene (TP53) regulate cell proliferation and apoptosis by transcribing and synthesizing cell cycle-related proteins. CDKN2A and TP53 are involved in the regulation of tumorigenesis as tumor suppressor genes.8,9 Studies have shown that DNA methylation occurs in the promoter of CDKN2A and TP53, and their DNA methylation is closely related to the progression of tumors.10,11
The traditional Chinese medicine Huaier plays a crucial role in the adjuvant treatment of many tumors. The anti-tumor mechanism of Huaier may contribute to inhibit the growth, proliferation, migration and invasion of tumor cells, and induce apoptosis of tumor cells. Moreover, Huaier reduces the formation of blood vessels, reverses the drug resistance of tumor cells, and enhances the killing ability of immune cells; in combination with chemotherapy drugs it can enhance the anti-tumor effect.12-14 With the continuous in-depth study of DNA methylation theory, more and more Chinese medicine practitioners speculate that the anti-tumor mechanism of Chinese medicine is related to DNA methylation.15,16 Currently, the role of Huaier in CSCC has not been reported in the literature. In this article, in vitro experiments were conducted to study the effects of Huaier on proliferation, migration, and invasion of CSCC cells. We further explore whether Huaier can play a role by inhibiting the methylation levels of CDKN2A and TP53 and promoting the expression of CDKN2A and TP53.
Materials and Methods
Cell Culture
Human cutaneous squamous cell carcinoma cell lines, SCL-1 and A431, were purchased from ATCC (Manassas, VA, USA). SCL-1 and A431 cells were cultured in RPMI 1640 medium (Solarbio, Beijing, China) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C and 5% CO2.
Drug Treatment
SCL-1 and A431 cells were incubated with various concentrations (0, 4, 8, 16 mg/mL) of Huaier. Huaier aqueous extract (Gaitianli Medicine Co. Ltd., Jiangsu, China) was dissolved in RPMI 1640 medium to obtain 100 mg/mL Huaier. The solution was sterilized with 0.22-µm filter and storage at −20°C.
Cell Transfection
Lentivirus harboring full-length DNMT1 expression vector (LV-DNMT1) was generated by GeneChem (Shanghai, China). The empty lentivirus vector (LV-NC) served as a negative control (NC). Small interfering RNA (siRNA) specifically targeting CDKN2A (si-CDKN2A) or TP53 (si-TP53) and the corresponding NC (si-NC) were purchased from GeneChem. Plasmids were transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as the protocol described.
Methylmion Specific PCR (MSP)
Genomic DNA was extracted from SCL-1 and A431 cells using Animal Tissues/Cells Genomic DNA Extraction Kit (Solarbio). Genomic DNA was then treated with bisulfite using BisulFlash DNA Modification Kit (Epigentek, Farmingdale, NY, USA). Bisulfite converted unmethylated cytosine residues in DNA to uracil, while methylated cytosine was without change. PCR was carried out in a 20 μL mixture containing Methylamp Master Mix (10 μL), primers (1 μL each 0.5 μM solution), Template (2 μL), and DNA/RNA-free water (6 μL). The cycling condition consisted of an initial denaturation at 95°C for 7 min, 40 cycles of denaturing at 95°C for 30 s, annealing temperature for 30 s, and extension at 72°C for 60 s, followed by final extension for 5 min at 72°C. The annealing temperatures were 63°C and 59°C for CDKN2A and TP53, respectively. The primer sequence (5′-3′) was shown as follows: CDKN2A (methylation): F-TTA TTA GAG GGT GGG GCG GAT CGC and R-GAC CCC GAA CCG CGA CCG TAA; CDKN2A (unmethylation): F-TTA TTA GAG GGT GGG GTG GAT TGT and R-CAA CCC CAA ACC ACA ACC ATA A; TP53 (methylation): F-TTC GGT AGG CGG ATT ATT TG and R-AAA TAT CCC CGA AAC CCA AC; TP53 (unmethylation): F-TTG GTA GGT GGA TTA TTT GTT T and R-CCA ATC CAA AAA AAC ATA TCA C. CpG universal methylated and unmethylated DNA (Zymo Research Corporation, USA) were similarly treated with bisulfite and were used as positive and negative controls. The products were examined by 1.5% agarose gel electrophoresis.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from SCL-1 and A431 cells using TaKaRa MiniBEST Universal RNA Extraction Kit (Takara, Dalian, China). Complementary DNA was generated using PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara). QRT-PCR was carried out using TB Green Premix Ex Taq II (Tli RNaseH Plus) (Takara) according to the instruction. PCR was carried out in a 25 μL mixture containing TB Green Premix Ex Taq II (12.5 μL), primers (1 μL each 10 μM solution), Template (2 μL), and DNA/RNA-free water (8.5 μL). The cycling condition consisted of an initial denaturation at 95°C for 5 min; 20 cycles of denaturation at 95°C for 40 s, annealing temperature plus 10°C for 40 s, and extension at 72°C for 1.5 min; 10 cycles of denaturation at 95°C for 40 s, annealing temperature for 40 s, and extension at 72°C for 90 s; followed by final extension at 72°C for 10 min. The annealing temperatures were 62°C and 58°C for CDKN2A and TP53, respectively. The primer sequence (5′-3′) was shown as follows: CDKN2A: F-AGC CTT CGG CTG ACT GGC TGG and R-GGC CCA TCA TCA TGA CCT GG; TP53: F-AGG GAT ACT ATT CAG CCC GAG GTG and R-ACT GCC ACT CCT TGC CCC ATT C; GAPDH (reference gene): F-TCC ACC ACC CTG TTG CTG TA and R-ACC ACA GTC CAT GCC ATC AC. The data were analyzed using the 2−∆∆CT method.
Western Blot (WB)
Total protein was extracted from SCL-1 and A431 cells using Total Protein Extraction Kit (Solarbio). Protein samples were separated by polyacrylamide gel electrophoresis, and then electro-blotted onto polyvinylidene fluoride membranes (Merck Millipore, Billerica, MA, USA). The membranes were incubated with primary antibody anti-CDKN2A (1:1000, Proteintech, Wuhan, China), anti-TP53 (1:2000, Proteintech), anti-DNMT1 (1:1000, Proteintech), anti-DNMT3A (1:1000, Proteintech), anti-DNMT3B (1:1000, Proteintech), or anti-β-actin (1:5000, Proteintech) overnight at 4°C. After washed with Tris Buffered saline Tween, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (1:5000, Proteintech). WB signals were analyzed by Image J software.
Cell Counting Kit-8 (CCK-8) Assay
SCL-1 and A431 cells were seeded in 96-well plates at 37°C, respectively. The cell proliferation was tested at respective time points (24, 48, 72 h) using Cell Counting Kit-8 (Beyotime, Shanghai, China). To summarize, the cells (100 μL) were incubated with CCK-8 reagent (10 μL) at 37°C for 1 h. The absorbance of samples was measured at 450 nm using enzyme-labeled instrument (Thermo Fisher Scientific, Waltham, MA, USA). SCL-1 and A431 cells were treated with a series of concentration gradients of Huaier for 72 h. Then, the survival of cells was assessed by CCK-8 assay to calculate the half maximal inhibitory concentration (IC50) of Huaier on SCL-1 and A431 cells.
Wound-Healing Assay
Wound-healing assay was performed to examine cell migration of SCL-1 and A431 cells. SCL-1 and A431 cells were seeded in a 6-well plate, respectively. Then, the cells were scratched with a 20-μL pipette tip. The cells were washed with phosphate buffered saline for 3 times and cultured in serum-free RPMI 1640 medium at 37°C and 5% CO2. Wounds were observed by microscopy and photographed at 0, 24, 48, and 72 h after wounding. The data was analyzed using Image J software.
Transwell Invasion Assay
Transwell invasion assay was performed using a 24-well Transwell insert system (Corning, NY, USA). Matrigel was diluted to 1 mg/mL with serum-free medium and covered on the basolateral Transwell chambers. 0.2 mL of cell suspension (SCL-1 or A431 cells) (1.5 × 106 cells/mL) was added to the upper chamber at 37°C with 5% CO2. Fresh RPMI 1640 medium containing 10% FBS was added into the lower chamber. After 24 h of incubation, the invading cells on the bottom surface of the chamber were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. The number of invasive cells was counted and photographed under an inverted microscope (Olympus, Tokyo, Japan). Five random visual fields were selected to analyze the differences among groups.
Statistical Analysis
Each assay was performed at least 3 times. All data reported as mean ± standard deviation. SPSS 22.0 statistical software (IBM, Armonk, NY, USA) was used for statistical analysis. Two-tailed Student’s t, 1-way or 2-way ANOVA was used to analyze the statistical difference. P < .05 was considered as a significant difference.
Results
Huaier Inhibited the Methylation Levels of CDKN2A and TP53, and Promoted the Expression of CDKN2A and TP53 in CSCC Cells
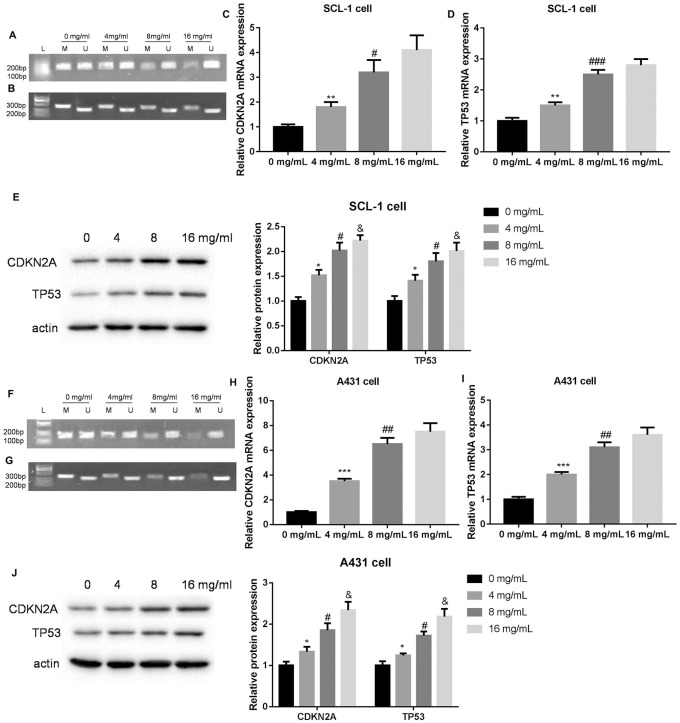
In order to explore the biological role of Huaier in CSCC, we first examined the IC50 of Huaier on SCL-1 and A431 cells by CCK-8 assay. The IC50 of Huaier on growth of SCL-1 and A431 cells were 6.96 and 7.57 mg/mL, respectively (Supplemental Figure 1). Subsequently, we examined the impact of Huaier on the methylation levels of CDKN2A and TP53 in SCL-1 and A431 cells by MSP assay. SCL-1 and A431 cells exhibited hypermethylation of CDKN2A and TP53. The methylation bands of CDKN2A and TP53 were weakened in SCL-1 and A431 cells in the presence of Huaier (8 and 16 mg/mL) (Figure 1A, B, F and G). Moreover, Huaier treatment enhanced the mRNA expression of CDKN2A and TP53 in SCL-1 and A431 cells. Compared with 4 mg/mL of Huaier treatment, Huaier at 8 mg/mL significantly enhanced the mRNA expression of CDKN2A and TP53 in SCL-1 and A431 cells. However, there was no obvious difference between 8 and 16 mg/mL of Huaier treatment (Figure 1C, D, H and I). We also found that Huaier enhanced the protein expression of CDKN2A and TP53 in SCL-1 and A431 cells in a dosage-dependent manner (Figure 1E and J). Thus, these data showed that Huaier inhibited the methylation levels of CDKN2A and TP53, and promoted the expression of CDKN2A and TP53 in CSCC cells.
Figure 1.
Huaier inhibits the methylation levels of CDKN2A and TP53, and promotes the expression of CDKN2A and TP53 in SCL-1 and A431 cells. SCL-1 cells were treated with 0, 4, 8, or 16 mg/mL of Huaier. (A and B) MSP was performed to assess the methylation levels of CDKN2A and TP53 in SCL-1 cells. (C-E) The gene and protein expression of CDKN2A and TP53 in SCL-1 cells were examined by qRT-PCR and WB. A431 cells were treated with 0, 4, 8, or 16 mg/mL of Huaier. (F and G) MSP was performed to assess the methylation levels of CDKN2A and TP53 in A431 cells. (H-J) The gene and protein expression of CDKN2A and TP53 in A431 cells were examined by qRT-PCR and WB.
Abbreviations: M, methylation; U, unmethylation.
*P < .05, **P < .01, ***P < .001, versus 0 mg/mL; #P < .05, ##P < .01, ###P < .001, versus 4 mg/mL; &P < .05, versus 8 mg/mL.
Huaier Reduced the Methylation Levels of CDKN2A and TP53 by Inhibiting DNMT1 Expression in CSCC Cells
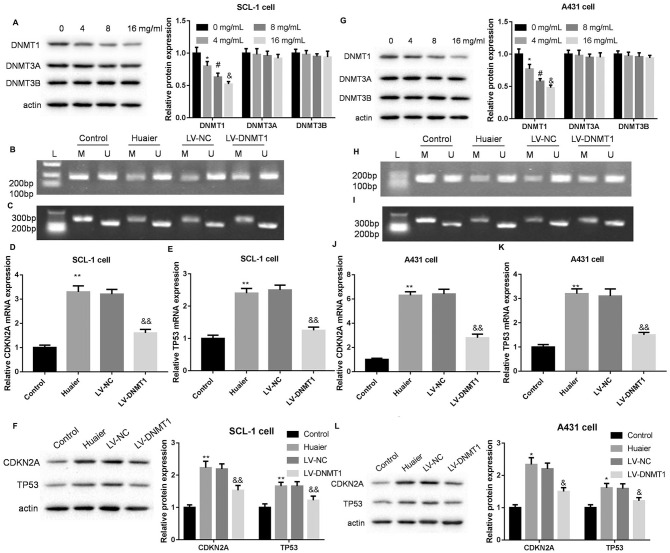
We estimated the effect of different concentrations of Huaier on the protein expression of DNA methyltransferase in SCL-1 and A431 cells. WB results showed that different concentrations of Huaier treatment significantly repressed the expression of DNMT1 in SCL-1 and A431 cells, although at different extents. However, Huaier treatment had no influence on the expression of DNMT3A and DNMT3B in SCL-1 and A431 cells (Figure 2A and G). Furthermore, SCL-1 and A431 cells were transfected with LV-DNMT1 to induce DNMT1 overexpression, followed with 8 mg/mL of Huaier treatment. The results of MSP assay revealed that the methylation bands of CDKN2A and TP53 were weakened in SCL-1 and A431 cells following Huaier treatment. Compared with LV-NC group, the methylation bands of CDKN2A and TP53 were elevated in the Huaier-treated SCL-1 and A431 cells in the presence of LV-DNMT1 (Figure 2B, C, H and I). In addition, Huaier-treated SCL-1 and A431 cells exhibited an increase the mRNA expression of CDKN2A and TP53. DNMT1 up-regulation repressed the mRNA expression of CDKN2A and TP53 in SCL-1 and A431 cells (Figure 2D, E, J and K). The protein expression of CDKN2A and TP53 in SCL-1 and A431 cells was enhanced by Huaier treatment. DNMT1 overexpression caused a down-regulation of CDKN2A and TP53 in Huaier-treated SCL-1 and A431 cells (Figure 2F and L). Therefore, these findings confirmed that Huaier reduced the methylation levels of CDKN2A and TP53 by inhibiting DNMT1 expression in CSCC cells.
Figure 2.
Huaier reduces the methylation levels of CDKN2A and TP53 by inhibiting DNMT1 expression in SCL-1 and A431 cells. SCL-1 cells were treated with 0, 4, 8, or 16 mg/mL of Huaier. (A) WB was performed to examine the expression of DNMT1, DNMT3A, and DNMT3B in SCL-1 cells. SCL-1 cells were transfected with LV-DNMT1 or LV-NC, and the modified SCL-1 cells were then treated with 8 mg/mL of Huaier. (B and C) MSP was performed to assess the methylation levels of CDKN2A and TP53 in the modified SCL-1 cells. (D-F) The gene and protein expression of CDKN2A and TP53 in modified SCL-1 cells were examined by qRT-PCR and WB. A431 cells were treated with 0, 4, 8, or 16 mg/mL of Huaier. (G) WB was performed to examine the expression of DNMT1, DNMT3A, and DNMT3B in A431 cells. A431 cells were transfected with LV-DNMT1 or LV-NC, and the modified A431 cells were then treated with 8 mg/mL of Huaier. (H and I) MSP was performed to assess the methylation levels of CDKN2A and TP53 in the modified A431 cells. (J-L) The gene and protein expression of CDKN2A and TP53 in modified A431 cells were examined by qRT-PCR and WB.
Abbreviations: M, methylation; U, unmethylation.
*P < .05, **P < .01, versus 0 mg/mL or control; #P < .05, versus 4 mg/mL; &P < .05, &&P < .01, versus 8 mg/mL or LV-NC.
Huaier Inhibited Proliferation, Migration, and Invasion of CSCC Cells by Promoting the Expression of CDKN2A and TP53
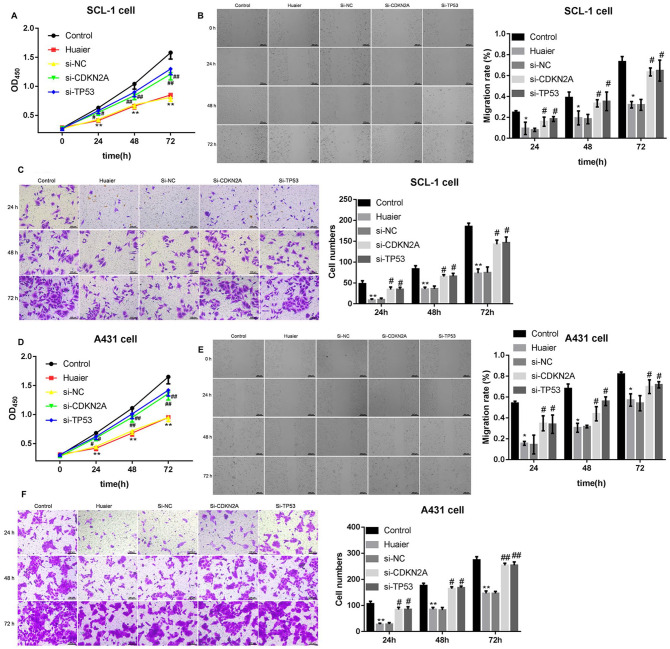
Finally, SCL-1 and A431 cells were transfected with si-CDKN2A or si-TP53 to induce CDKN2A or TP53 knockdown, following 8 mg/mL of Huaier treatment. The proliferation, migration, and invasion of SCL-1 and A431 cells were estimated by CCK-8, wound-healing, or transwell invasion assays. As shown in Figure 3A and D, Huaier treatment notably represses proliferation of SCL-1 and A431 cells. Both CDKN2A knockdown and TP53 deficiency significantly enhanced proliferation of Huaier-treated SCL-1 and A431 cells (Figure 3A and D). Moreover, Huaier treatment had an inhibiting effect on migration of SCL-1 and A431 cells. CDKN2A or TP53 knockdown reversed the inhibiting effect of Huaier on migration of SCL-1 and A431 cells (Figures 3B and E, Supplemental Figures 2 and 3). Furthermore, Huaier-treated SCL-1 and A431 cells displayed a decrease of invasion ability in SCL-1 and A431 cells. The invasion ability of SCL-1 and A431 cells was enhanced by CDKN2A silencing and TP53 knockdown (Figure 3C and F). Taken together, these data demonstrated that Huaier inhibited proliferation, migration, and invasion of CSCC cells by promoting the expression of CDKN2A and TP53.
Figure 3.
Huaier inhibits proliferation, migration and invasion of SCL-1 and A431 cells by promoting the expression of CDKN2A and TP53. SCL-1 cells were transfected with si-CDKN2A or si-TP53 or si-NC, and the modified SCL-1 cells were then treated with 16 mg/mL of Huaier. (A) CCK-8 assay was performed to detect proliferation of SCL-1 cells. (B) Wound-healing assay was performed to examine migration of SCL-1 cells. (C) Transwell invasion assay was performed to assess invasion of SCL-1 cells. A431 cells were transfected with si-CDKN2A or si-TP53 or si-NC, and the modified A431 cells were then treated with 8 mg/mL of Huaier. (D) CCK-8 assay was performed to detect proliferation of A431 cells. (E) Wound-healing assay was performed to examine migration of A431 cells. (F) Transwell invasion assay was performed to estimate invasion of A431 cells.
*P < .05, **P < .01, versus control; #P < .05, ##P < .01, versus si-NC.
Discussion
Tumor suppressor genes, CDKN2A and TP53, participate in the development of various tumors. CDKN2A is down-regulated in recurrent oral squamous cell carcinoma patients, which is associated with poor prognosis and low survival rate of recurrent oral squamous cell carcinoma patients. 17 Bellmunt et al 18 have found that high grade T1 non-muscle invasive bladder cancer is rich in TP53 mutations, and the increased copy number of CDKN2A deletion is abundant in the recurrence or progression of high grade T1 non-muscle invasive bladder cancer. Thus, CDKN2A and TP53 may act as a predictor for high grade T1 non-muscle invasive bladder cancer. Lung adenocarcinoma and lung squamous cell carcinoma patients with longest survival display wild-type of TP53, and TP53 mutation status determines the survival time of lung cancer patients. 19 Existing studies have shown that CDKN2A and TP53 are frequently mutated genes in CSCC, showing that CDKN2A and TP53 participate in the occurrence of CSCC.20,21 CDKN2A mutations predict the poor outcome of CSCC. 22 In the present study, we investigated whether the traditional Chinese medicine Huaier can affect CSCC progression by regulating methylation levels of CDKN2A and TP53. The data showed that Huaier treatment notably enhanced the expression of CDKN2A and TP53, and suppressed the methylation levels of CDKN2A and TP53 in SCL-1 and A431 cells. We speculated that the mechanism of action of Huaier in CSCC may be associated with the methylation of CDKN2A and TP53.
DNA methylation participates in the progression of CSCC. The study of Li et al 23 has found that UVB irradiation represses the expression of ID4 via regulating DNA methylation, and ID4 overexpression suppresses proliferation, migration and invasion, and enhances apoptosis of CSCC cells. DNA methylation of miR-204 promoter accounts for miR-204 silencing in CSCC, and miR-204 affects the progression of CSCC by regulating MAPK pathway or STAT3 signaling pathways. 4 The present work determined the influence of Huaier on methylation levels of CDKN2A and TP53 in CSCC cells. Huaier treatment repressed the expression of DNMT1 in SCL-1 and A431 cells, but had no effect on the expression of DNMT3A and DNMT3B. DNMT1, DNMT3A, and DNMT3B are important DNA methyltransferases. DNMT1 maintains DNA methylation during DNA replication and repair. 24 DNMT3A and DNMT3B catalyze the de novo methylation of CpG. 25 Thus, Huaier may promote CDKN2A and TP53 expression by regulating DNMT1-mediated DNA methylation. Moreover, DNMT1 overexpression suppressed the expression of CDKN2A and TP53 in SCL-1 and A431 cells by promoting DNA methylation. Therefore, these data demonstrated that Huaier treatment promoted the expression of CDKN2A and TP53 in CSCC cells by repressing DNMT1-mediated DNA methylation.
The traditional Chinese medicine Huaier has therapeutic effects on many diseases. For example, Huaier attenuates the progression of hepatoblastoma by repressing cell proliferation, and promoting cell apoptosis and cell cycle arrest via MEK-ERK signaling pathway. 26 Huaier restrains cholangiocarcinoma progression, which contributes to suppress cell proliferation, invasion, and metastasis of cholangiocarcinoma cells by modulating TP73-AS1 expression. 27 Huaier also exhibits antitumor and antimetastatic activities in malignant melanoma, and it inhibits the epithelial-mesenchymal transition by suppressing HIF-1/VEGF and AEG-1 signaling pathways. 28 In the present work, we revealed the function of Huaier in CSCC, showing that Huaier repressed the proliferation, migration, and invasion of CSCC cells. In addition, inhibition of CDKN2A or TP53 reversed the influence of Huaier treatment on proliferation, migration, and invasion of CSCC cells. Therefore, Huaier inhibited proliferation, migration, and invasion of CSCC cells by promoting the expression of CDKN2A and TP53.
In conclusion, our work suggests that Huaier attenuates CSCC progression, which contributes to inhibit proliferation, migration, and invasion of CSCC cells by regulating DNA methylation of CDKN2A and TP53. Thus, Huaier extract may act as a drug for treating CSCC.
Supplemental Material
Supplemental material, sj-pdf-1-ict-10.1177_15347354211031646 for Huaier Inhibits Proliferation, Migration, and Invasion of Cutaneous Squamous Cell Carcinoma Cells by Inhibiting the Methylation Levels of CDKN2A and TP53 by Liang Wang, Lei Xu and Yu Wang in Integrative Cancer Therapies
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Liang Wang  https://orcid.org/0000-0002-7261-7099
https://orcid.org/0000-0002-7261-7099
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. 2018;78:237-247. [DOI] [PubMed] [Google Scholar]
- 2. Hervás-Marín D, Higgins F, Sanmartín O, et al. Genome wide DNA methylation profiling identifies specific epigenetic features in high-risk cutaneous squamous cell carcinoma. PLoS One. 2019;14:e0223341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li L, Jiang M, Feng Q, et al. Aberrant methylation changes detected in cutaneous squamous cell carcinoma of immunocompetent individuals. Cell Biochem Biophys. 2015;72:599-604. [DOI] [PubMed] [Google Scholar]
- 4. Toll A, Salgado R, Espinet B, et al. MiR-204 silencing in intraepithelial to invasive cutaneous squamous cell carcinoma progression. Mol Cancer. 2016;15:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yanagi T, Watanabe M, Hata H, et al. Loss of TRIM29 alters keratin distribution to promote cell invasion in squamous cell carcinoma. Cancer Res. 2018;78:6795-6806. [DOI] [PubMed] [Google Scholar]
- 6. Venza M, Visalli M, Catalano T, Beninati C, Teti D, Venza I. DSS1 promoter hypomethylation and overexpression predict poor prognosis in melanoma and squamous cell carcinoma patients. Hum Pathol. 2017;60:137-146. [DOI] [PubMed] [Google Scholar]
- 7. Wang L. Role of oncogene and tumor suppressor gene methylation in skin squamous cell carcinoma. Master’s thesis. 王亮. 皮肤鳞状细胞癌与相关的原癌基因及抑癌基因甲基化关系的研究[D].吉林大学; 2012. [Google Scholar]
- 8. Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of genetic and epigenetic alterations of CDKN2A (p16(INK4a)) in cancer. EBioMedicine. 2016;8:30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aubrey BJ, Strasser A, Kelly GL. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb Perspect Med. 2016;6:a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiao Y, Feng Y, Wang X. Regulation of tumor suppressor gene CDKN2A and encoded p16-INK4a protein by covalent modifications. Biochemistry (Mosc). 2018;83:1289-1298. [DOI] [PubMed] [Google Scholar]
- 11. Saeed WH, Eissa AA, Al-Doski AA. Impact Of TP53 gene promoter methylation on chronic lymphocytic leukemia pathogenesis and progression. J Blood Med. 2019;10:399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Wu H, Wang X, et al. Huaier Granule extract inhibit the proliferation and metastasis of lung cancer cells through down-regulation of MTDH, JAK2/STAT3 and MAPK signaling pathways. Biomed Pharmacother. 2018;101:311-321. [DOI] [PubMed] [Google Scholar]
- 13. Yang A, Zhao Y, Wang Y, et al. Huaier suppresses proliferative and metastatic potential of prostate cancer PC3 cells via downregulation of Lamin B1 and induction of autophagy. Oncol Rep. 2018;39:3055-3063. [DOI] [PubMed] [Google Scholar]
- 14. Gao S, Li X, Ding X, Jiang L, Yang Q. Huaier extract restrains the proliferative potential of endocrine-resistant breast cancer cells through increased ATM by suppressing miR-203. Sci Rep. 2017;7:7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qing Y, Hu H, Liu Y, et al. Berberine induces apoptosis in human multiple myeloma cell line U266 through hypomethylation of p53 promoter. Cell Biol Int. 2014;38:563-570. [DOI] [PubMed] [Google Scholar]
- 16. De Oliveira DT, Savio AL, Marcondes JP, et al. Cytotoxic and toxicogenomic effects of silibinin in bladder cancer cells with different TP53 status. J Biosci. 2017;42:91-101. [DOI] [PubMed] [Google Scholar]
- 17. Padhi SS, Roy S, Kar M, et al. Role of CDKN2A/p16 expression in the prognostication of oral squamous cell carcinoma. Oral Oncol. 2017;73:27-35. [DOI] [PubMed] [Google Scholar]
- 18. Bellmunt J, Kim J, Reardon B, et al. Genomic predictors of good outcome, recurrence or progression in high grade T1 non-muscle invasive bladder cancer. Cancer Res. 2020;80:4476-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Freudenstein D, Litchfield C, Caramia F, et al. TP53 status, patient sex, and the immune response as determinants of lung cancer patient survival. Cancers. 2020;12:1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Al-Rohil RN, Tarasen AJ, Carlson JA, et al. Evaluation of 122 advanced-stage cutaneous squamous cell carcinomas by comprehensive genomic profiling opens the door for new routes to targeted therapies. Cancer. 2016;122:249-257. [DOI] [PubMed] [Google Scholar]
- 21. Inman GJ, Wang J, Nagano A, et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat Commun. 2018;9:3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Küsters-Vandevelde HV, Van Leeuwen A, Verdijk MA, et al. CDKN2A but not TP53 mutations nor HPV presence predict poor outcome in metastatic squamous cell carcinoma of the skin. Int Cancer. 2010;126:2123-2132. [DOI] [PubMed] [Google Scholar]
- 23. Li L, Li F, Xia Y, et al. UVB induces cutaneous squamous cell carcinoma progression by de novo ID4 methylation via methylation regulating enzymes. EBioMedicine. 2020;57:102835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li J, Wang R, Jin J, et al. USP7 negatively controls global DNA methylation by attenuating ubiquitinated histone-dependent DNMT1 recruitment. Cell Discov. 2020;6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yagi M, Kabata M, Tanaka A, et al. Identification of distinct loci for de novo DNA methylation by DNMT3A and DNMT3B during mammalian development. Nat Commun. 2020;11:3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu DQ, Yuan XJ, Hirayama M, Toyoda H. Via Huaier extract induces apoptosis in hepatoblastoma cells the MEK/ERK signaling pathway. In Vivo. 2020;34:2381-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ji D, Zheng W, Huang P, et al. Huaier restrains cholangiocarcinoma progression in vitro and in vivo through modulating lncRNA TP73-AS1 and inducing oxidative stress. Onco Targets Ther. 2020;13:7819-7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su D, Jiang B, Yang Y, Miao Y, Fu Q, Zhang F. Effect of Huaier on melanoma invasion, metastasis, and angiogenesis. BioMed Res Int. 2020;2020:8163839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ict-10.1177_15347354211031646 for Huaier Inhibits Proliferation, Migration, and Invasion of Cutaneous Squamous Cell Carcinoma Cells by Inhibiting the Methylation Levels of CDKN2A and TP53 by Liang Wang, Lei Xu and Yu Wang in Integrative Cancer Therapies