Abstract
Objective
Evidence supporting the utility of pharmacogenetic (PGX) tests in depression is scarce. The main objectives of this study were to summarize, update, and assess the quality of the available evidence regarding PGX testing in depression as well as estimating the impact of using PGX testing tools in depression outcomes in the Middle East/North Africa (MENA) region.
Methodology
Scientific databases were systematically searched from inception to June 30, 2020 for systematic reviews and randomized controlled trials (RCTs) assessing the clinical utility of PGX tests in the treatment of depression. Meta-analyses only and RCTs that were included in eligible systematic reviews were excluded. The quality of the eligible studies was assessed using the Crowe Critical Appraisal Tool (CCAT).
Results
Six systematic reviews and three RCTs met the inclusion criteria and were included in this study. The results of the systematic reviews provided weak evidence on the efficacy of PGX testing, especially in patients with moderate–severe depression at 8 weeks. In addition, there was a lack of evidence regarding safety outcomes. Newer RCTs with better quality showed clinical promise regarding efficacy outcomes, especially in patients with gene–drug interactions. No evidence was found regarding PGX testing impact in the MENA region.
Conclusion
This systematic review is an update and summary of the available literature on the clinical utility of PGX testing in depression. The findings of this study demonstrate that PGX testing prior to treatment initiation or during the course of therapy may improve efficacy outcomes. Further studies are warranted to assess the impact of PGX testing on safety outcomes.
Keywords: pharmacogenetic testing, major depressive disorder, clinical decision support, antidepressant treatment response
Introduction
Major depressive disorder (MDD) is one of the most prevalent mood disorders worldwide. The World Health Organization (WHO) reports that more than 300 million people from all age groups are affected by depression worldwide.1 Globally, depression is considered one of the major causes of disability, leading to high societal and economic burden.2,3 Since a variety of pharmacological options are available to treat depression, selection of an antidepressant is usually based on its safety profile and various individual clinical factors.4,5 Although a multicenter naturalistic study reported that up to 62% of patients with depression were able to achieve clinical remission after 1 year of treatment with antidepressants,6 evidence from randomized controlled studies, such as the landmark STAR*D trial, suggests that only up to one-third of patients with depression appear to achieve clinical remission over a period of up to 37 months with multiple treatments.7 The latter finding has also been supported in several effectiveness studies.8–10 In addition, an increase in the prevalence of treatment-resistant depression has been reported, with 20–30% of cases failing to demonstrate clinical improvement after the use of two medications at adequate dose and treatment duration.11,12 These findings represent a major concern since those individuals with poor outcomes are at high risk of developing complications that may eventually lead to declines in productivity and social functioning.13
Previous reports have demonstrated that genetic factors play an important role in antidepressant treatment response in terms of both safety and efficacy.14,15 These factors could be represented as genetic variants that can affect the pharmacokinetic and pharmacodynamic properties of antidepressants, related to changes in enzymes such as cytochrome P450 2D6 (CYP2D6) and CYP2C19 or in monoamine receptors such as serotonin (5-HT) and norepinephrine receptors. This has shed the light on the concept of implementing personalized medicine, which includes tailoring treatment and diagnostic approaches to individualized clinical factors along with genetic profiles.16 A wide variety of commercial pharmacogenetic (PGX)-based decision support tools are available worldwide. The concept behind these tools is to use patients’ biological material, such as saliva or plasma, to produce genetic information relevant to the genotype of each individual. This information is then integrated into algorithms and processed into data on patients’ phenotypes, which are finally interpreted to produce patient-specific recommendations for drugs, doses, and treatment duration.17 These tools have demonstrated promise in solving issues related to treatment response in various psychiatric disorders and related contexts. For example, a study by Ielmini et al reported on genetic testing-guided treatment versus treatment as usual in patients with bipolar disorder. The results showed that patients who had genetic testing-guided medication changes had a statistically significant improvement in their Clinical Global Impression Item Severity (CGI-S) score at 3-month follow-up compared to those not having changes consistent with the test.18 In another case study by Ielmini et al, PGX-guided treatment in two patients with bipolar disorder showed improvements in both psychopathology and tolerability, over 3 months of follow-up.19 In another PGX study involving subjects with depression, PGX testing-guided antidepressant treatment was associated with statistically significant positive changes in all depression outcomes (eg, depression rating scores, side effects, and response rate).20
Although the concept of genetic testing has been shown to be effective in mental health settings, it is not yet fully integrated into clinical practice, especially for depression. The reason behind this is the lack of robust clinical utility studies that confirm whether these tools bring real value to clinical practice.21,22 Clinical utility is defined as the extent to which a diagnostic tool improves health outcomes when compared to the current best alternative. Such studies need to involve an adequate sample representative of the population in order to examine the effect of these tools on real patients.23 Many systematic reviews of clinical utility studies with similar scope have been identified from the literature. However, some of the studies included had methodological gaps in terms of study design and samples included, and lacked an assessment of bias, and the majority were industry funded.23–28 In addition, newer randomized controlled trials (RCTs) with relevant important findings have become available in the literature since these reviews were published.29–31 Therefore, this study was conducted with the primary objective of updating the current evidence regarding the impact of PGX-based decision support tools on clinical (efficacy and safety) outcomes of depression treatment. Secondary objectives of this study included: 1) to compare the efficacy of different genetic testing strategies available in improving depression outcomes; and 2) to identify studies in the Middle East/North Africa (MENA) region which report on PGX-based decision support strategies, to guide prescribing for depression in countries that belong to this region, such as Qatar.
Methods
The protocol of this systematic review is registered with the International Prospective Register of Systematic Reviews (PROSPERO) database (registration ID: CRD42020182936, https://www.crd.york.ac.uk/prospero/#recordDetails),32 and followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) recommendations.33
Eligibility Criteria
Systematic reviews and meta-analyses that assessed the clinical efficacy (remission, response, change in depression severity, symptoms improvement, time to response, and quality of life), and safety (side effects, tolerability, suicide rate, and relapse) of using PGX testing tools in adult patients with depression of any severity as compared to treatment as usual (TAU) were included. In addition, recent RCTs with the same scope that were not included in previous systematic reviews or meta-analyses were included. Meta-analysis only, narrative reviews, observational, case–control studies, and reviews that assessed the predictive or theoretical abilities of PGX testing only without assessment of clinical outcomes were excluded. To avoid duplication, RCTs that were involved in the eligible systematic reviews were also excluded.
Search Strategy
PubMed, EMBASE, Cochrane Central Register of Controlled Trials, ISI Web of Science, Scopus, Database of Abstracts of Reviews of Effects (DARE), EBSCO (Arab world research source), EBSCO (Academic Search Ultimate), PsycINFO, PsyChJournal, Journal of Clinical Psychiatry, PROSPERO, and Google Scholar were systematically searched for relevant articles. Gray literature sources such as PharmGKB, ProQuest and conference registries were also searched to identify unpublished relevant articles. Reference lists of eligible studies were manually searched for additional unindexed pertinent references. The search was limited to human studies that were published from inception until June30, 2020. Various combinations of the following keywords were used to search databases: major depressive disorder, depression, mental illness, mood disorder, antidepressant, response, remission, outcome, pharmacogenetic, pharmacogenomics, pharmacodynamics, pharmacokinetic, genetic testing, pharmacogenetic testing tools, pharmacogenetic testing kits, clinical utility, safety, efficacy, using Medical Subject Headings (MESH) terms or an advanced search where relevant.
Data Selection and Extraction
All identified records followed two rounds of screening by two independent reviewers (HE and SA) after removal of duplicates. Round 1 consisted of screening based on titles and abstracts to identify potentially eligible articles; and round 2 consisted of screening based on full text to identify fully eligible articles. Data were extracted using a standardized Excel sheet and included the following domains: study title, authors, country, study design, aims, search strategy, eligibility criteria, population, intervention, comparator, outcomes, number of studies included, main findings, and limitations for systematic reviews and meta-analysis. For RCTs, the extraction process involved all the aforementioned data in addition to sample size, duration of study, and baseline characteristics. Conflicts between reviewers were resolved by discussion and consensus.
Quality Assessment
A quality assessment of systematic reviews and RCTs was conducted using the Crowe Critical Appraisal Tool (CCAT).34 This tool was developed based on a number of previous critical appraisal tools and guidelines to account for various study designs. Therefore, it was chosen for this study owing to the heterogeneous nature of the included articles. This tool has undergone construct validity and reliability testing. A correlation coefficient of 0.89 was reported for consistency and 0.88 for absolute agreement.35–37 The CCAT is composed of eight domains; namely, preliminary, introduction, design, sampling, data collection, ethical matters, results, and discussion. Each domain is scored from 0 to 5, with 0 being the lowest and 5 the highest rank. As per the CCAT user guide,34 domain scores for each individual study should be reported along with the total score in order to obtain a clear insight on the actual quality and avoid making underestimated/overestimated judgments. The average score of the two reviewers (HE and SA) was calculated and divided by 40 to obtain a total score as a percentage. As the CCAT does not include an explanation of the percentages obtained, the quality of the included studies was classified as poor (score: 0–50%), moderate (score: 51–74%), and high (score: 75–100%), based on criteria used in previous studies.38–40
Results
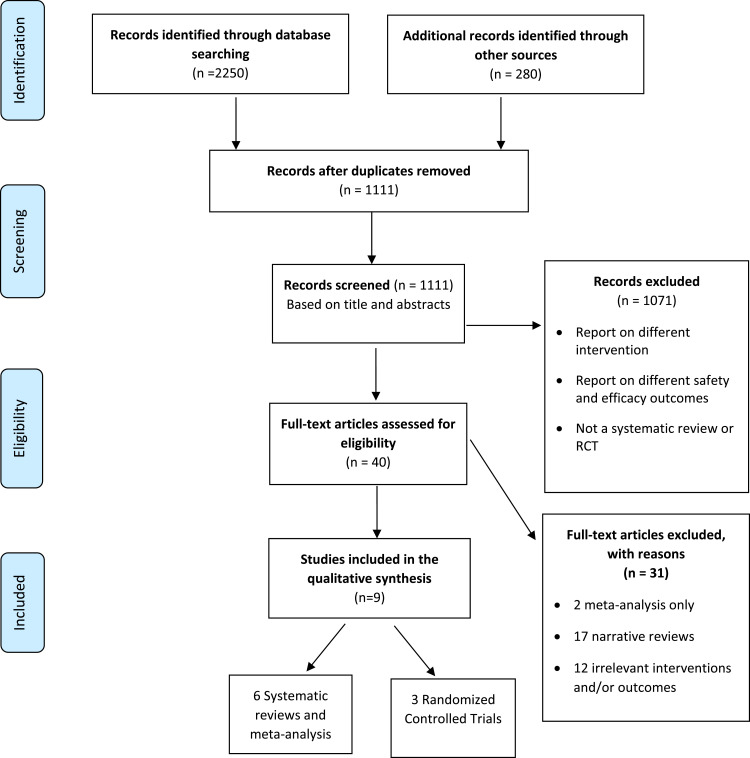
The systematic search yielded 2530 articles. As illustrated in Figure 1, a total of 1419 duplicates were removed, leaving 1111 studies. After reviewing titles and abstracts, 1071 studies were excluded because they assessed different outcomes or had different study designs. The remaining 40 potentially eligible studies were screened based on full text to assess their final eligibility. Of these, only nine articles fitted the full criteria for inclusion, and 31 were excluded. A variety of PGX tests are included in the eligible studies and a detailed description of each is presented in Table 1.
Figure 1.
Flow diagram of selection process.
Table 1.
Description of Pharmacogenetic Tests
| Test | Manufacturer | Eligibility | Genes | Medications |
|---|---|---|---|---|
| GeneSight Psychotropic57 | Assurex Health, Inc, Mason, OH, USA | Patients with depression |
56 alleles and variants across 12 genes: 8 Pharmacokinetic genes: CYP2D6, CYP2C19, CYP2C9, CYP3A4, CYP2B6, CYP1A2, UGT1A4, UGT2B15 4 Pharmacodynamic genes: SLC6A4, HTR2A, HLA-A*3101, HLA-B*1502 |
57 psychotropic medications |
| Genecept Assay58 | Dynacare, Inc, Brampton, Ontario, Canada | Patients with psychiatric and medical morbidities, patients with suboptimal response, patients with polypharmacy, and patients with medication adherence issues |
18 genes: 6 Pharmacokinetic genes: CYP2C19, CYP2C9, CYP2D6, CYP2B6, CYP1A2, CYP3A4/5 12 Pharmacodynamic genes: SLC6A4, CACNA1C, ANK3, 5HT2C, MC4R, DRD2, COMT, ADRA2A, MTHFR, BDNF, OPRM1, GRIK1 |
Not specified |
| CNSDose59 | CNSDose, Melbourne, Victoria, Australia | Patients with psychiatric disorders, patients with general medical conditions, patients with polypharmacy, and patients with depression and co-occurring conditions |
Liver and brain genes: ABCB1, ABCC1, CYP2C19, CYP2D6, UGT1A1 |
Antipsychotics, anxiolytics, mood stabilizers, hypnotics, ADHD, Alzheimer’s, cardiology, endocrinology, pain medicines, gastroenterology medicines |
| Neuropharmagen60 | AB-Biotics SA, Barcelona, Spain | Patients with psychiatric disorders |
25 genes: 10 Pharmacokinetic genes: CYP2C19, CYP2C9, CYP2D6, CYP2B6, CYP1A2, CYP3A4, ABCB1, CES1, EPHX1, UGT2B15 15 Pharmacodynamic genes: SLC6A4, COMT, BDNF, OPRM1, GRIK2, GRIK4, AKT1, AL157359, DDIT4, FCHSD1, HLA-A, HTR2A, HTR2C, LPHN3, RPTOR |
59 psychotropic medications |
| Amplichip CYP450 Test61 | Roche Molecular Systems, Basel, Switzerland | Patients receiving CYP2D6 and CYP2C19 substrates | 2 Pharmacokinetic genes: CYP2D6, CYP2C19 |
CYP2D6 and CYP2C19 substrates |
| NeuroIDgenetix62 | AltheaDx, Inc, San Diego, CA, USA | Patients with depression and anxiety |
10 genes: 6 Pharmacokinetic genes: CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5 4 Pharmacodynamic genes: SLC6A4, COMT, HTR2A, MTHFR |
Approx. 40 psychotropic medications |
| GeneLex63 | GeneLex, Seattle, WA, USA | Patients with depression, patients with epilepsy, patients with general medical conditions, patients with cancer, and patients with polypharmacy |
25 genes: 8 Pharmacokinetic genes: CYP2C9, CYP2C19, CYP2D6, CYP3A4/CYP3A5, CYP1A2, CYP2B6, CYP4F2 17 Pharmacodynamic genes: ADRA2A, COMT, DPYD, Factor II–Factor V, Leiden, GRIK4, HLA-A or HLA-B, HTR2A, HTR2C, IFNL3, MTHFR, NAT2, OPRM1, SLCO1B1, TPMT, UGT1A1 |
Not specified |
| HILOmet64 | Rennova Health, Inc, West Palm Beach, FL, USA | Patients with treatment resistance or drug intolerance to neuropsychiatric or cardiometabolic drugs |
3 Pharmacokinetic genes: CYP2D6, CYP2C9, CYP2C19 |
Not specified |
| Pillcheck65 | GeneYouIn Inc, North York, Canada | Patients with psychiatric disorders, patients with cardiovascular and gastroenterology medical conditions, patients taking analgesics, refractory patients, patients undergoing surgery and who will be prescribed analgesics during recovery, patients starting treatment for chronic conditions |
19 genes: 8 Pharmacokinetic genes: CYP2D6, CYP2C19, CYP2C9, CYP3A4/A5, CYP1A2, CYP2C8, CYP2B6 11 Pharmacodynamic genes: OPRM1, SLCO1B1, VKORC1, DPYD, F2, F5, IFNL3, TPMT, UGT1A1, UGT2B15, ADRB2 |
205 medications |
Features of the Systematic Reviews
Of the six reviews included, four were systematic reviews and meta-analyses,23,24,26,28 and two were systematic reviews only.25,27 All reviews were published between 2017 and 2020, with the latest published in April 2020. All studies had relatively broad inclusion criteria, with no or a very vague description for exclusion. Characteristics of the systematic reviews are summarized in Table 2.
Table 2.
Characteristics of Eligible Systematic Reviews
| Study | Aim | Search Strategy | Population | Intervention | Comparator | Outcomes | Included Studies |
|---|---|---|---|---|---|---|---|
| Health Quality Ontario24 | To assess the effect of GeneSight test compared to TAU | Timeline: Until February 2016 Databases: >2 Search terms: Provided Flow diagram: Available Limits: English only Test: GeneSight |
Depression, GAD, psychiatric patients (n=13,377) |
PGX-guided treatment with GeneSight | Unguided treatment | Primary outcomes: Prevention of suicide, remission, response, depression score, quality of life, efficacy Secondary outcomes: Impact on therapeutic decisions, patient and clinician satisfaction |
2 Non-randomized open-label studies 1 RCT 1 propensity score cohort |
| Bousman et al23 | To examine the remission rates of 2nd generation PGX-guided treatment in MDD patients | Timeline: Until May 2018 Databases: >2 Search terms: Provided Limits: English only Flow diagram: Available Test: Not specified |
Adults with but not limited to severe or uncontrolled depression (n=1737) |
PGX-guided treatment | Unguided treatment | Remission | 5 RCTs: 2 GeneSight, 1 Neuropharmagen, 1 CNSDose, 1 NeuroIDgenetix |
| Fabbri et al25 | To provide an update on the currently available PGx tests for predicting treatment outcomes of drugs for depression with quality evaluation for each available study | Timeline: Until January 2018 Databases: >2 Search terms: Provided Limits: English only Flow diagram: Available Test: Not specified |
Not clearly defined, but patients with depression (n=not available) |
PGX-guided treatment | Unguided treatment | Safety and efficacy | 6 RCTs: GeneSight, CNSDose, Neuropharmagen, NeuroIDgenetix, Genelex, academic study based on FKBP5 SNPs 6 case–control non- RCTs: 3 GeneSight, Genecept, GeneLex, academic study based on ABCB1 SNPs 9 observational studies 2 AmpliChip, CNSDose GeneSight, Genecept, Genelex, HILOmet, Neuropharmagen, Pillcheck |
| Rosenblat et al26 | To determine the effect of PGX-guided treatment on response and remission rates in the acute treatment of MDD as compared to unguided treatment | Timeline: Until December 2017 Databases: 2 Search terms: Provided Limits: Human studies, English only Flow diagram: Available Test: Not specified |
Patients with MDD (n=not available) | PGX-guided treatment | Unguided treatment | Response and remission |
4 RCTs: GeneSight, CNSDOse, Neuropharmagen, NeuroIDgenetix 2 Unblinded, open-label cohort studies 2 GeneSight |
| Rosenblat et al27 | To determine the effect of PGX testing on clinical outcomes in MDD and assess its cost-effectiveness | Timeline: Until October 2015 Databases: 2 Search terms: Provided Limits: English only Flow diagram: Available Test: Not specified |
Patients with MDD (n=not available) | Guided treatment | Unguided treatment | Efficacy, cost effectiveness | 2 case–control non- RCTs: 2 GeneSight 2 RCTs: GeneSight, CNSDose 1 Observational: Genecept |
| Brown et al28 | To assess the clinical utility of the combinatorial test GeneSight in patients with MDD | Timeline: Not available. Database: 1 Search terms: Provided. Limits: N/A Flow diagram: Available Test: GeneSight |
Patients with MDD with prior medication failure (n=1556) | Guided treatment with GeneSight | Unguided treatment | Symptoms improvement, response, remission | 2 RCTs 2 Open- label trials 1 Pooled analysis |
Abbreviations: PGX, pharmacogenetic; TAU, treatment as usual; MDD, major depressive disorder; RCT, randomized controlled trial; GAD, generalized anxiety disorder.
Features of the Randomized Controlled Trials
Three RCTs that were not included in the systematic reviews and provided updates on the current knowledge were identified from the literature.29–31 The RCT by Greden et al30 assessed safety and efficacy outcomes and was included in one of the systematic reviews included in this study.28 However, the focus was only on efficacy outcomes; therefore, safety outcomes of this RCT were systematically reviewed and efficacy outcomes were briefly summarized. All studies were published between 2018 and 2019 and assessed the primary outcomes over a period of 8 weeks using the Hamilton Depression Rating Scale (HAMD). Detailed characteristics of the RCTs are summarized in Table 3.
Table 3.
Characteristics of Eligible Randomized Controlled Trials (RCTs)
| Study | Aim | Study Design | Sample Size | Eligibility Criteria | Population |
|---|---|---|---|---|---|
| Han et al29 | To evaluate the effectiveness and tolerability of commercial PGX test Neuropharmagen vs TAU in the Korean population |
Randomization: Stratified by study center with a 1:1 ratio Blinding: Single blinded Duration: 8 weeks Tool: Neuropharmagen Main outcomes: Primary outcomes: The mean change of scale (HAMD-17) from baseline to end of treatment. The change of total score of FIBSER from baseline to end of treatment was the co-primary endpoint Secondary outcomes: Response as reduction in HAMD-17 total score of at least 50% from baseline. Remission was defined as an absolute HAMD-17 total score of ≥7 at the end of treatment Other secondary endpoints: Changes in (PHQ-9/15), (CGI-S) score, (GAD-7) total score, and SDS |
100 |
Inclusion criteria 1) At least 20 years old 2) Diagnosis of MDD according to DSM-5 criteria 3) CGI-S score ≥3 4) Intolerance to current antidepressant therapy Exclusion criteria 1) Not currently on antidepressant treatment 2) Pregnancy or nursing 3) Substance abuse within the past 12 months 4) Diagnosed with unstable medical or neurological disorders 8) Had formal cognitive–behavioral therapy 9) Participated in a clinical trial within the past month 10) Hospitalized or had ECT within 8 weeks |
Groups: Guided (n=52), TAU (n=48) Age: 44 vs 43 years Sex: 77% female Ethnicity: Not reported Depression status: Moderate-to-severe MDD symptoms measured by HAMD-17 total scores (23.8±4.8). All the patients had a history of at least two previous failed antidepressant treatments for current MDD |
| Greden et al30 | To evaluate the utility of PGX testing in patients with MDD Data are presented from the Genomics Used to Improve DEpression Decisions (GUIDED) trial |
Randomization: 1:1 to guided or TAU Blinding: Patient- and rater-blinded Duration: Symptom improvement, response, and remission were monitored over 24 weeks with the primary endpoint at week 8 Tool: GeneSight Main outcomes: Assessed at baseline, week 4, week 8, week 12, and week 24 Primary outcome: Change in HAMD-17 Secondary outcome: Response and remission at week 8 according to HAMD-17, QIDS-C16, and PHQ-9, and side effects |
1541 patients were included in the final intent-to-treat cohort 1398 patients were included in per-protocol cohort |
Inclusion criteria 1) Over the age of 18 2) Diagnosed with MDD (≥11 on the QIDS-C16 and self-rated QIDS-SR16 at screening and baseline) 3) Had an inadequate response to at least one psychotropic treatment Exclusion criteria 1) Had significant suicidal risk 2) Severe co-occurring psychiatric or cognitive disorders, cognitive disorders and/or unstable or significant medical conditions |
Outpatients from 60 academic and community sites in the USA Groups: In the per-protocol cohort, 607 patients in TAU, 560 patients in the guided arm Age: Mean 47.5 years Sex: 71% female Ethnicity: 92.1% non-Hispanic non-Latino, 80.6% white Depression status: Moderate (392/1398, 28.0%), severe (493/1398, 35.3%), or very severe (513/1398, 36.7%) |
| Thase et al31 | To evaluate the extent to which PGX positively influenced antidepressant outcomes in patients entering the GUIDED trial taking medications subject to gene–drug interactions |
Randomization: 1:1 to guided treatment or TAU Blinding: Patient- and rater-blinded Duration: 8 weeks Tool: GeneSight Main outcomes: Assessed at baseline, week 4, week 8, week 12, and week 24. Outcomes assessed at week 8 included % change in HAMD-17 score from baseline. Response (≥50% decrease in HAMD-17 score). Remission (HAMD-17 score ≤7) Same outcomes were evaluated in patients whose medications were switched from baseline to week 8 |
912 patients, Guided (n= 439), TAU (n=473) | Same as Greden et al,30 but included mainly patients who were taking medications subject to gene–drug interactions at baseline (“use with caution” and “use with increased caution and with more frequent monitoring” report categories) | Same as Greden et al30 |
Abbreviations: PGX, pharmacogenetic; TAU, treatment as usual; MDD, major depressive disorder; DSM-5, Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition; GAD-7, Generalized Anxiety Disorder 7-item; CGI-S, Clinical Global Impression Scale; HAMD-17, 17-item Hamilton Depression Rating Scale; PHQ-9, Patient Health Questionnaire-9; QIDS-C16, Clinician-rated Quick Inventory of Depressive Symptomatology; QIDS-SR16, Self-Rated Quick Inventory of Depressive Symptomatology; FIBSER, Frequency, Intensity, and Burden of Side Effects Rating; SDS, Sheehan Disability Scale; ECT, electroconvulsive therapy.
Efficacy Summary
A summary of the results on efficacy as reported in the nine included studies is presented in Table 4. The efficacy studies reported several types of efficacy outcomes, including response, remission, symptom improvement, and quality of life. Response was assessed as a proportion of response, as change in depression scores, or as improved symptoms. The systematic review and meta-analysis conducted by the Ontario Health Technology team reported a greater proportion of patients with a significant response to depression therapy with GeneSight®-guided treatment than those who received usual care. For change in depression score, it was observed that more patients in the intervention group had a greater reduction in their scores for all outcome measures than those in the comparator group at 8 weeks, but this difference was not observed at 10 weeks.24 Fabbri et al highlighted that patients in the intervention arm had a borderline statistically significant improved response rate when compared to the comparator arm. These results were mainly observed in patients with moderate–severe depression in RCT studies.25 It was reported in RCTs included in the systematic review by Rosenblat et al in 2017 that 77% of participants using the Genecept® test showed improvement, and 39% demonstrated a treatment response as per Quick Inventory of Depressive Symptomatology Self-Report (QIDS-SR) scores. However, the study lacked a comparator arm, so the clinical efficacy could not be attributed solely to Genecept-guided treatment.27 The other meta-analysis study by Rosenblat et al, which had clearly defined outcomes, showed an overall pooled relative risk (RR) for response of 1.36 (95% [CI]: 1.14–1.62, p=0.0006; n=799), number needed to treat (NNT)=7, in favor of guided treatment.26 In the most recent systematic review and meta-analysis in the area of PGX testing in depression, conducted by Brown et al in April 2020, which focused on GeneSight and used HAMD-17 scores to measure outcomes, random effect model analysis showed that symptoms improvement was significantly better in the genetic-guided treatment arm relative to the unguided arm (Δ=10.08%, 95% CI: 1.67–18.50, p=0.019). For response outcome, the guided care arm had a 40% higher response rate compared with the unguided arm (RR=1.40, 95% CI: 1.17–1.67, p<0.001).28
Table 4.
Safety and Efficacy Results of Eligible Studies
| Study | Safety | Efficacy | Authors' Conclusion |
|---|---|---|---|
| Systematic reviews | |||
| Health Quality Ontario24 | N/A |
Depression score: Patients who received guided care had a greater reduction in depression at 8 weeks but not 10 weeks (RCT) on HAMD, QIDS-C16, and PHQ-9 Response: A greater proportion of patients had a response to depression therapy with GeneSight-guided care than those who received usual care Remission: There was a statistically significant benefit for the GeneSight guided group on QIDS-C16, but no significant difference on HAMD-17 or PHQ-9 |
Patients who received GeneSight-guided medication had improved response and greater improvement in measures of depression but no observed differences in rates of remission, based on low to very low confidence in the evidence |
| Bousman et al23 | N/A |
Remission: (RR=1.71, 95% CI: 1.17–2.48, p= 0.005) in favor of guided group Two trials, Singh and Perez et al, were identified as influential studies. Removal of the Singh trial resulted in a trend-level pooled RR of 1.46 (95% CI: 0.99–2.16, p=0.056), whereas removal of the Perez et al trial produced an increased pooled RR of 1.98 (95% CI: 1.47–2.66, p<0.001) |
This article serves as a “tipping point” of evidence in support of PGX-guided antidepressant treatment, in the context of moderate to severe depression |
| Fabbri et al25 | Better tolerability, lower rehospitalization and emergency room visits in the guided group |
RCT results: Higher response rates and remission in the guided group Case–control non-RCTs: Compared to TAU, patients had higher response and remission rates, and percent symptom improvement Meta-analysis of three GeneSight studies: No improvement in remission rate but better response rate and symptom improvement Observational studies: In one study all subjects were genotyped using Neuropharmagen, and patients whose treatment followed the PGX report showed a higher chance of symptom improvement |
Relatively weak clinical benefits were reported by RCTs and higher clinical benefits by non-RCTs, but with greater risk of bias |
| Rosenblat et al26 | N/A |
Response rate: Subgrouping by genetic test showed significant differences with some genetic tests (NeuroIDgenetix),(GeneSight, Neuropharmagen) showed no significant difference between PGX-guided and unguided treatment, and CNSDose was not included in this analysis. One study was influential, and did not appear to be an outlier: Perez et al, (2017) Remission rate: Subgrouping by genetic test showed significant differences with NeuroIDgenetix and GeneSight between PGX-guided versus unguided treatment, but not with Neuropharmagen. CNSDose was not included in this analysis. |
The current analysis provides preliminary support for improved response and remission rates in MDD when treatment is guided by pharmacogenomics |
| Rosenblat et al27 | N/A | There was an 18.2% reduction in HAMD-17 in the unguided group vs 30.8% reduction in the guided group (p=0.04) in a non-randomized, prospective cohort study 77% of participants showed improvement, with 39% showing a treatment response and 38% achieving full remission as per QIDS-SR scores, regardless of number of previous antidepressants, in a 3-month naturalistic, Genecept Assay (Genomind, King of Prussia, Pennsylvania). At the end of the 12-week trial, subjects receiving CNSDose guided prescribing (n=74) had a 72% remission rate, while the unguided group (n=74) had a remission rate of 28% with a 2.52-fold greater chance of remission (95% CI: 1.71–3.73, p<0.0001) with NNT=3 (95% CI: 1.7–3.5) in a 12-week prospective RCT with Caucasian adults (N=148) with moderate to severe MDD |
A limited number of studies have shown promise for the clinical utility of pharmacogenomic testing; Well-designed clinical trials with adequate sample sizes, randomization, and blinding are required |
| Brown et al28 |
Symptoms improvement: When either of the open-label trials were removed, there was no significant difference; Hall-Flavin et al, 2012 (Δ=9.54, 95% CI: −1.09–20.17, p=0.079) and Hall-Flavin et al, 2013 (Δ=5.76, 95% CI: −0.17–11.69, p=0.057) When the studies were grouped based on trial design, there was a statistically significant difference for the guided group for the RCTs and open-label studies, p<0.0045 and p<0.001, respectively Response: When each trial was removed, the analysis showed consistent results; significant response rate p<0.05 For analysis based on study type, guided group has shown statistical significance for both RCTs and open-label trials (RR=1.34, 95% CI: 1.10–1.62, p=0.004; and RR=1.72, 95% CI: 1.11–2.67, p=0.016), respectively Remission: When each trial was removed, the analysis showed no statistically significant difference except for Greden et al, p=0.075 When the studies were grouped based on trial design, there was no statistically significant difference between groups in the open-label trial, but there was significance in RCTs (RR=1.49, 95% CI: 1.14–1.96, p=0.004) |
This study has shown improved outcomes (response, remission, and symptoms improvement) for patients with MDD and prior medication failure who received treatment guided by GeneSight test as compared to the unguided group. This was visible across all studies and when analysis was conducted for RCTs only | |
| Randomized controlled trials | |||
| Han et al29 | FIBSER: frequency (p=0.0346), intensity (p=0.0001), and burden (p=0.0001) at the end of 8 weeks between the two groups favoring PGX-guided treatment by 18.1%, 69.0%, and 69.7% of differences |
Change in HAMD-17 score was significantly different between the two groups, favoring guided group by −4.1 points of difference (p=0.010) Response rate based on HAMD total score was also significantly higher in PGX-guided arm vs TAU at the end of treatment by 28.1% difference (p=0.014) Remission rate was numerically higher (19.9% difference) in PGX-guided arm than in TAU groups, without statistical difference (p=0.071) The mean changes from baseline to endpoint in PHQ-9 total scores in the PGX-guided and TAU groups were significantly different between the two groups, favoring PGX-guided arm by −3.8 points of difference (p=0.011) The mean changes from baseline to endpoint in PHQ-15 total scores in the PGX-guided and TAU groups were not significantly different between the two groups, but numerically favored PGX-guided arm by −1.7 points of difference (p=0.155) |
The present study demonstrates that PGX testing may be a better option in the treatment of MDD in terms of effectiveness and tolerability; however, study shortcomings may limit a generalization. Adequately powered, well-designed studies are needed |
| Greden et al30 | For the mean number of side effects at week 8, p=0.855 For the proportion of patients who experienced side effects, p=0.881 |
There was 30% improvement in response, and 50% improvement in remission among difficult-to-treat patients in the guided-care arm compared to TAU. Sustained improvement was observed at 24 weeks |
These results from the GUIDED trial indicate that PGX testing is effective in improving response and remission rates among those with prior treatment resistance, particularly for patients who are treated with medications that are incongruent with their genetic profile |
| Thase et al31 | N/A |
Response rate (∆=8.0%, p=0.008) at week 8 favoring the guided arm Remission rate (∆=7.5%, p=0.003) at week 8 favoring the guided arm Durability of guided-care outcomes through week 24: 42.2% decrease in HDRS-17 scores at week 24 compared to baseline. This decrease in HDRS-17 score represents a 56% increase in symptom improvement from week 8 At week 24, the response rate was 44.3% and the remission rate was 33.2% Outcomes in patients who switched medications: HAMD-17 scores decreased by 30.0% from baseline to week 8 in the guided-care arm compared to 22.3% in TAU (∆=7.6%, p=0.011) The response rate at week 8 among patients who switched medications was 29.8% in the guided-care arm compared to 19.4% for TAU (∆=10.4%, p=0.011) The remission rate at week 8 for patients who switched medications in the guided-care arm was 20.3% compared to 11.1% in TAU (∆=9.2%, p=0.008) This represents a relative improvement of 54% for response and 83% for remission among patients whose medication switches were informed by pharmacogenomic testing |
All patient outcomes remained significantly improved for the guided-care arm vs TAU when the subset of patients who switched medications were evaluated separately. Patients with treatment-resistant depression have a low likelihood of reaching remission with standard treatment approaches |
Abbreviations: PGX, pharmacogenetic; TAU, treatment as usual; MDD, major depressive disorder; DSM-5, Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition; HAMD-17, 17-item Hamilton Depression Rating Scale; PHQ-9, Patient Health Questionnaire-9; QIDS-C16, Clinician-rated Quick Inventory of Depressive Symptomatology; QIDS-SR16, Self-Rated Quick Inventory of Depressive Symptomatology; FIBSER, Frequency, Intensity, and Burden of Side Effects Rating; RCT, randomized controlled trial; RR, risk ratio; NNT, number needed to treat.
Remission rates were assessed in all studies. Bousman et al defined remission as a HAMD score ≤7 and showed a significant association between PGX-guided care and remission (RR=1.71, 95% CI: 1.17–2.48, p=0.005).23 The systematic review and meta-analysis conducted by the Ontario Health Technology team assessed remission using the HAMD-17, Patient Health Questionnaire-9 (PHQ-9), and the Clinician-rated 16-item Quick Inventory of Depressive Symptomatology (QIDS-C16) and its patient self-rated component (QIDS-SR), although no clear definitions for the scores were provided. This study showed that there was a statistically significant change favoring patients in the GeneSight-guided arm when compared to the traditional arm in terms of QIDS-C16 scores, but no significant difference was observed when using HAMD-17 or PHQ-9. However, there was no clear reporting on the effect size of each outcome.24 In a meta-analysis of the GeneSight test reported in the study by Fabbri et al, no improvement in remission rates was found.25 In another RCT included in the systematic review by Rosenblat et al in 2017, which investigated the clinical utility of CNSDose®, subjects receiving PGX-guided treatment had a 72% remission rate versus 28% in the traditional arm, with a 2.5-fold greater chance of remission (95% CI: 1.71–3.73, p<0.0001).27 The other meta-analysis study by Rosenblat et al, which had clearly defined outcomes, showed an overall pooled RR for remission rates of 1.74 (95% CI: 1.09–2.77, p=0.02, n=735) in favor of guided treatment (NNT=7).26 In the 2020 study conducted by Brown et al, the pooled analysis RR showed that patients in the guided group had 49% increase in remission compared to the unguided group (RR=1.49, 95% CI: 1.17–1.89, p=0.001).28
Updates on Efficacy
An update of the efficacy data was retrieved from the three recent RCTs and is summarized in Table 4. The RCTs reported efficacy outcomes in a similar fashion to the systematic reviews and meta-analyses summarized earlier. For response outcomes, the RCT by Han et al in a Korean population showed a statistically significant difference in the mean change of HAMD-17 at 8 weeks, favoring patients on the Neuropharmagen®-guided arm, with a −4.1 point difference relative to TAU (p=0.010). Similarly, there was a statistically significant difference in response rate favoring the PGX-guided arm relative to TAU, of 28.1% (p=0.014).29 The RCT by Greden et al, however, did not show significant differences (p=0.107) between the GeneSight-guided arm and TAU in patients with inadequate treatment on previous antidepressant trials. The response rate among patients in the GeneSight arm was 26.0% at week 8, which was significantly higher than in TAU, at 19.9% (p=0.013).30 The RCT by Thase et al31 involved the same population and followed the same methodology as Greden et al;30 however, the analysis was conducted on a subgroup of patients who were resistant to treatment and found to have gene–drug interaction at baseline. The results showed a statistically significant decrease in HAMD scores by 27.1% from baseline to week 8 in the guided-care arm compared to 22.1% in TAU (∆=5.0%, p=0.029). Also, at week 8, the response rate was 27.0% in the guided-care arm compared to 19.0% in TAU (∆=8.0%, p=0.008).31
Remission rates were also assessed in all the RCTs. Han et al defined remission as a HAMD-17 total score of ≤7. At the end of the study period, the rate was higher in the PGX-guided group, but the difference did not reach statistical significance (p=0.071).29 The results of the RCT by Greden et al showed a statistically significant higher remission rate in the GeneSight-guided group (15.3%) compared to TAU (10.1%) (p=0.007).30 The remission rate in the RCT by Thase et al was 18.2% higher in the GeneSight-guided arm compared to 10.7% in TAU (∆=7.5%, p=0.003) in patients with previous antidepressant failures.31
Safety Summary
There was a general inconsistency and lack of data on safety outcomes in the included reviews. The systematic review by Fabbri et al showed that patients in the PGX group had a weak lower risk of medication tolerability problems, lower mean number of rehospitalizations, and borderline significant lower number of emergency room visits within 2-month follow-up based on RCT results.25 A summary of these results as reported in the individual reviews is presented in Table 4.
Updates on Safety
Similarly to the previous systematic reviews, there was a general inconsistency and lack of data on safety outcomes in the recent RCTs. The study conducted by Han et al showed that the proportion of patients with a score of two or less on the Frequency, Intensity, and Burden of Side Effects Rating (FIBSER) questionnaire at the end of 8 weeks was significantly different between the two groups, favoring the Neuropharmagen-guided arm: frequency (p=0.0346), intensity (p=0.0001), and burden (p=0.0001).29
Greden et al assessed patient-reported side effects in their study. The mean number of side effects and proportion of patients reporting side effects according to study arm were measured. Only side effects with a probability of being linked to medications administered (eg categorized as likely, probably, possibly, or definitely relating to medication) were included. The results showed that there is no statistically significant difference in the mean number of side effects at week 8 (0.243 vs 0.237, p=0.855) or the proportion of patients who experienced side effects (15.6% [88/560] vs 15.3% [93/607], p=0.881).30
Quality Assessment of the Available Systematic Reviews
Scores for each domain as well as total scores for individual articles are reported and presented in Table 5. The overall quality was found to be high (≥75%) for two of the included systematic reviews and the remaining four were of moderate quality (≥50%). All articles, including the high-quality ones, had the lowest scores in design, data collection, and discussion domains, as opposed to preliminaries and introduction domains. The quality of the included RCTs was also assessed, and is demonstrated in Table 6.
Table 5.
CCAT Quality Scores of Eligible Systematic Reviews
| Study | Domain Score | Overall Score | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Preliminaries | Introduction | Design | Sampling | Data Collection | Ethical Matters | Results | Discussion | ||
| Brown et al28 | 3 | 4 | 1 | 2 | 0 | 4 | 4 | 3 | 51% |
| Rosenblat et al27 | 3 | 5 | 2 | 2 | 0 | 2 | 3 | 3 | 53% |
| Health Quality Ontario 24 | 4 | 5 | 3 | 5 | 0 | 1 | 3 | 1 | 55% |
| Fabbri et al25 | 2 | 5 | 2 | 4 | 1 | 5 | 2 | 3 | 60% |
| Rosenblat et al26 | 5 | 5 | 4 | 3 | 5 | 4 | 3 | 4 | 83% |
| Bousman et al23 | 5 | 5 | 3 | 4 | 5 | 5 | 5 | 3 | 88% |
Notes: Poor quality (0–50%), moderate quality (51–74%), high quality (75–100%).
Table 6.
CCAT Quality Scores of Eligible Randomized Controlled Trials (RCTs)
| Study | Domain Score | Overall Score | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Preliminaries | Introduction | Design | Sampling | Data Collection | Ethical Matters | Results | Discussion | ||
| Han et al29 | 3 | 4 | 3 | 2 | 1 | 3 | 3 | 5 | 59% |
| Thase et al31 | 5 | 5 | 4 | 1 | 4 | 4 | 4 | 3 | 75% |
| Greden et al30 | 4 | 4 | 5 | 2 | 5 | 4 | 4 | 5 | 84% |
Notes: Poor quality (0–50%), moderate quality (51–74%), high quality (75–100%).
Other Results
Neither the systematic reviews nor the RCTs included in this review compared the efficacy or safety of different individual PGX tests; rather, they provided evidence on their effectiveness in clinical practice.
No studies were identified from the literature that assessed the clinical utility or effect of PGX-based treatment in any of the MENA region countries.
Discussion
This systematic review investigated the impact of PGX-guided treatment versus TAU on efficacy and safety outcomes in patients with depression. It provided a comprehensive summary and an update on the available literature on this topic. This study has highlighted important findings. First, despite the high level of heterogeneity and the methodological fallacies of the included studies, it showed that PGX-guided treatment had a positive effect on symptom remission and response rate. This effect was most prominent in patients with treatment-resistant depression who were found to have gene–drug interactions at baseline. Second, this review has demonstrated that there is a lack of studies assessing safety outcomes of PGX-guided treatment in depressed patients. The landmark RCT by Greden et al found significant differences between the two groups on the FIBSER scale; however, this finding was based on data extracted from a single cohort of patients and was specific to one genetic test, “GeneSight”.30 Newer RCTs, which assess the impact on safety, are underway and may provide a definitive answer to this important outcome.41–43
Furthermore, quality assessment conducted by authors of this study and the quality reported on the included systematic reviews showed that a high portion of the available literature on this area is of poor to moderate quality. This was found to be mainly due to major gaps in study design, including small sample size, inappropriate blinding of participants/clinicians, underestimation of confounding factors, and neglecting important measures needed to reduce the risk of bias in the design. In addition to gaps in the methodology, some of the included studies were found to be industry funded or their authors had financial relationships with the manufacturing companies, which may bias their results.23,25,28–31
The secondary objective of this study was make comparisons between the available tests in terms of safety and efficacy; however, none of the identified studies provided any clear comparisons. GeneSight by Assurex Health was the most commonly studied test, since it has demonstrated good clinical validity in a previous study.44 As a result, it is currently being covered by Medicaid and Medicare insurance plans in the USA, and is allowed to be ordered by psychiatrists only for patients with refractory response to antidepressants who continue to show moderate to severe symptoms.45
Moreover, this study aimed to identify literature related to PGX testing in patients with depression in the MENA region. However, no data with this scope were identified. This sheds light on a very important gap in the literature that should be addressed in future studies, especially considering that the population in the MENA region accounts for 18% of the global incidence of MDD.46–48
In addition to the study objectives, this study has allowed for the identification of many important gaps related to PGX testing. First, there is a general uncertainty regarding whether PGX testing should be recommended to patients before or after their initial treatment recommendation (pre-emptive versus reactive testing). Furthermore, the effect of these tests on patient-reported outcomes, such as patients’ satisfaction, adherence, quality of life improvement, and personal experience, is underinvestigated.24 In addition, in the available studies, the tests were used only by trained psychiatrists; thus, whether average clinicians will be able to incorporate testing into their clinical practice (eg primary care) remains unclear.24 This will depend on their PGX literacy, and since PGX and precision medicine are still emerging concepts, they may require extensive support from PGX experts and specialists. This is in line with findings from a previous survey study conducted on 217 chief psychiatry residents in New York City to assess their psychiatry training and preparedness to perform genetic testing. The results of the study showed that almost half of the respondents received 3 hours or less of training sessions in genetics, and only 14% out of 80 respondents indicated that they understood the role of genetic factor in mental health.49
The findings of this systematic review support the notion that the widespread use of PGX tests is still challenged by many factors; namely, lack of knowledge about research requirements to support their utility, heterogeneity of populations included in the current studies, and reproducibility of available results.50 To answer the question about research requirements, conducting pragmatic and group randomized trials with large sample sizes could be of value to provide relevant rigorous evidence. Moreover, a collaborative effort by many authors from various relevant disciplines dedicated to conducting a meta-analysis could be a promising strategy to answer the remaining questions. This will aid in accurately and collectively synthesizing reproducible evidence, avoiding duplication of data, and minimizing heterogeneity of results. The last of these could be achieved through the utilization of subgroup analysis on relevant data.51,52
The positive efficacy results demonstrated in this study are supported by results from complex case-reports of patients with psychiatric conditions including depression. The results of these cases have shown that genetic testing reports obtained from commercial tests not only were useful in guiding future medication selection but also explained the pattern of patients’ response to previous and current medications. Although these data support the clinical utility of PGX testing, it is important to consider that the majority of the current commercial tests contain more information/genes than is/are currently approved by the Food and Drug Administration (FDA) or recommended by the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines. Therefore, reports generated by these tools still cannot be fully relied upon.53,54
Aside from the technical limitations, clinicians’ perspectives, knowledge, and beliefs about routine PGX testing, especially in the psychiatric field, should also be considered, given that PGX test results may drive a complete change in their prescribing behavior.50 In addition, previous results have highlighted the presence of a gap in general practitioners’ trust in pharmacists’ clinical roles. Since clinical pharmacists, by virtue of their education and clinical skills, will most likely be responsible for interpreting test results and making medication/dosage recommendations, resistance in following tests recommendations by practitioners will most likely remain an unaddressed challenge in the incorporation of genetic tests into clinical practice.55
Strengths
This systematic review summarizes the findings, provides an update, and assesses the quality of the available systematic reviews on the clinical utility of PGX testing in depression. Furthermore, since data related to genetic testing in mental health have been accumulating recently, this study serves as a systematically synthesized reference for decision makers and experts when making clinically important decisions. In addition, by providing a summary of the gaps in the literature, this study paves the way for future, more robust PGX studies. Lastly, unlike the majority of the available systematic reviews, this study has been conducted by academic investigators with no financial conflicts of interest, which limits any risk of funding bias.
Limitations
This study possesses some limitations. First, the data presented here were synthesized utilizing a systematic review, as a meta-analysis was difficult to conduct considering the high heterogeneity of the studies included, the different study designs (RCTs, systematic reviews only, and systematic reviews and meta-analyses), the different safety and efficacy outcomes reported, the different sample sizes and population characteristics, as well as the different interventions reported. In addition, given the small number of eligible articles (n=9) and that each of them was found to have a risk of bias, conducting a meta-analysis would have most likely produced false results.
Second, conducting a systematic review that combines RCTs with a systematic review and meta-analysis may be regarded as not following best practice for conducting a systematic reviews of systematic reviews;56 however, this was the most appropriate design to answer the primary objectives of our study. Furthermore, the authors could not synthesize evidence related to efficacy outcomes since studies with this focus were sparse. While this is a significant limitation, it is also an important finding that needs to be addressed by future clinical trials.
Third, the use of a generic quality assessment tool for the eligible studies, instead of specific tools, may have affected the robustness of the quality results. Considering the heterogeneity of the study designs included in this study, the CCAT was a convenient option as it can be used across a broad range of quantitative and qualitative study designs, it has in-depth questions to fully assess research papers, and its validity and reliability have been well documented.
Finally, in order to search for studies reported on PGX-based decision support strategies to guide prescribing in depression in the MENA region, the names of all countries in the MENA region were used as keywords in the search strategy; it is acknowledged that countries participating in multicenter studies may have been missed using this search strategy. It is also acknowledged that the costs of PGX testing may limit their wider utilization in clinical practice. Because this systematic review only focused on the safety and efficacy of PGX testing, conducting future systematic reviews with this scope is recommended.
Conclusion
This systematic review has brought together all of the evidence related to the impact of PGX testing in depression in one place. In this review, we have portrayed the impact of PGX testing on patients’ clinically important outcomes, provided a quality assessment of the available literature, and highlighted the methodological limitations that need to be addressed in future studies. The findings of this study demonstrate that PGX testing could be an effective strategy in patients receiving antidepressants; however, owing to concerns over the quality of the available studies and the lack of safety data, further confirmatory studies are needed. This study lays a foundation for researchers to further investigate this topic to provide more robust evidence that could be incorporated into clinical practice, especially in the MENA region.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical Approval
None required.
Disclosure
The authors of this manuscript have no conflicts of interest to declare.
References
- 1.World Health Organization. Depression. Who.int. 2021. Available from: https://www.who.int/news-room/fact-sheets/detail/depression. Accessed October6, 2019.
- 2.Steel Z, Marnane C, Iranpour C, et al. The global prevalence of common mental disorders: a systematic review and meta-analysis 1980–2013. Int J Epidemiol. 2014;43(2):476–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.König H, König H, Konnopka A. The excess costs of depression: a systematic review and meta-analysis. Epidemiol Psychiatr Sci. 2019;29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Institute for Health and Care Excellence. Depression in Adults: Recognition and Management. London: NICE; 2009. Available fromhttps://www.nice.org.uk/guidance/cg90. Accessed October6, 2019. [PubMed] [Google Scholar]
- 5.Treatment Guidelines: Depression. CPNP. 2021. Available from: https://cpnp.org/guideline/external/depression. Accessed February21, 2021.
- 6.Seemüller F, Obermeier M, Schennach R, et al. Stability of remission rates in a 3-year follow-up of naturalistic treated depressed inpatients. BMC Psychiatry. 2016;16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warden D, Rush A, Trivedi M, Fava M, Wisniewski S. The STAR*D project results: a comprehensive review of findings. Curr Psychiatry Rep. 2007;9(6):449–459. [DOI] [PubMed] [Google Scholar]
- 8.Schulberg H. The ‘usual care’ of major depression in primary care practice. Arch Fam Med. 1997;6(4):334–339. [DOI] [PubMed] [Google Scholar]
- 9.Rost K, Nutting P, Smith J, Elliot C, Dickinson M. Managing depression as a chronic disease: a randomised trial of ongoing treatment in primary care. BMJ. 2002;325(7370):934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rush A, Trivedi M, Carmody T, et al. One-year clinical outcomes of depressed public sector outpatients: a benchmark for subsequent studies. Biol Psychiatry. 2004;56(1):46–53. [DOI] [PubMed] [Google Scholar]
- 11.Johnston K, Powell L, Anderson I, Szabo S, Cline S. The burden of treatment-resistant depression: a systematic review of the economic and quality of life literature. J Affect Disord. 2019;242:195–210. [DOI] [PubMed] [Google Scholar]
- 12.Tansey K, Guipponi M, Hu X, et al. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013;73(7):679–682. [DOI] [PubMed] [Google Scholar]
- 13.Trivedi M, Rush A, Wisniewski S, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163(1):28–40. [DOI] [PubMed] [Google Scholar]
- 14.Uher R, Tansey K, Henigsberg N, et al. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170(2):207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabbri C, Serretti A. Pharmacogenetics of major depressive disorder: top genes and pathways toward clinical applications. Curr Psychiatry Rep. 2015;17:7. [DOI] [PubMed] [Google Scholar]
- 16.Sanacora G. Is this where we stand after decades of research to develop more personalized treatments for depression? JAMA Psychiatry. 2020;77(6):E560. [DOI] [PubMed] [Google Scholar]
- 17.Abbott R, Chang D, Eyre H, Bousman C, Merrill D, Lavretsky H. Pharmacogenetic decision support tools: a new paradigm for late-life depression? Am J Geriatric Psychiatry. 2018;26(2):125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ielmini M, Poloni N, Caselli I, et al. The utility of pharmacogenetic testing to support the treatment of bipolar disorder. Pharmgenomics Pers Med. 2018;11:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ielmini M, Poloni N, Caselli I, Diurni M, Grecchi A, Callegari C. The role of pharmacogenetic testing in the treatment of bipolar disorder: preliminary results. Minerva Psichiatr. 2018;59(1):10–15. [Google Scholar]
- 20.Altar C, Carhart J, Allen J, Hall-Flavin D, Winner J, Dechairo B. Clinical utility of combinatorial pharmacogenomics-guided antidepressant therapy: evidence from three clinical studies. Mol Neuropsychiatry. 2015;1(3):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall-Flavin D, Winner J, Allen J, et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23(10):535–548. [DOI] [PubMed] [Google Scholar]
- 22.Bossuyt P, Reitsma J, Linnet K, Moons K. Beyond diagnostic accuracy: the clinical utility of diagnostic tests. Clin Chem. 2012;58(12):1636–1643. [DOI] [PubMed] [Google Scholar]
- 23.Bousman C, Arandjelovic K, Mancuso S, Eyre H, Dunlop B. Pharmacogenetic tests and depressive symptom remission: a meta-analysis of randomized controlled trials. Pharmacogenomics. 2019;20(1):37–47. [DOI] [PubMed] [Google Scholar]
- 24.Health Quality Ontario H. Pharmacogenomic Testing for Psychotropic Medication Selection: a Systematic Review of the Assurex GeneSight Psychotropic Test. PubMed Central (PMC). 2021. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5433545/. Accessed, 2019. [PMC free article] [PubMed] [Google Scholar]
- 25.Fabbri C, Zohar J, Serretti A. Pharmacogenetic tests to guide drug treatment in depression: comparison of the available testing kits and clinical trials. Prog Neuropsychopharmacol Biol Psychiatry. 2018;86:36–44. [DOI] [PubMed] [Google Scholar]
- 26.Rosenblat J, Lee Y, McIntyre R. The effect of pharmacogenomic testing on response and remission rates in the acute treatment of major depressive disorder: a meta-analysis. J Affect Disord. 2018;241:484–491. [DOI] [PubMed] [Google Scholar]
- 27.Rosenblat J, Lee Y, McIntyre R. Does pharmacogenomic testing improve clinical outcomes for major depressive disorder? J Clin Psychiatry. 2017;78(06):720–729. [DOI] [PubMed] [Google Scholar]
- 28.Brown L, Vranjkovic O, Li J, et al. The clinical utility of combinatorial pharmacogenomic testing for patients with depression: a meta-analysis. Pharmacogenomics. 2020;21(8):559–569. [DOI] [PubMed] [Google Scholar]
- 29.Han C, Wang S, Bahk W, et al. A pharmacogenomic-based antidepressant treatment for patients with major depressive disorder: results from an 8-week, randomized, single-blinded clinical trial. Clin Psychopharmacol Neurosci. 2018;16(4):469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greden J, Parikh S, Rothschild A, et al. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: a large, patient- and rater-blinded, randomized, controlled study. J Psychiatr Res. 2019;111:59–67. [DOI] [PubMed] [Google Scholar]
- 31.Thase M, Parikh S, Rothschild A, et al. Impact of pharmacogenomics on clinical outcomes for patients taking medications with gene-drug interactions in a randomized controlled trial. J Clin Psychiatry. 2019;80:6. [DOI] [PubMed] [Google Scholar]
- 32.PROSPERO. Crd.york.ac.uk. 2021. Available from: https://www.crd.york.ac.uk/prospero/#recordDetails. Accessed February21, 2021.
- 33.Liberati A, Altman D, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339(jul211):b2700–b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crowe Critical Appraisal Tool (v1.4). Conchra. Conchra.com.au. 2020. Available from: https://conchra.com.au/2015/12/08/crowe-critical-appraisal-tool-v1-4/. Accessed March17, 2020.
- 35.Crowe M, Sheppard L. A general critical appraisal tool: an evaluation of construct validity. Int J Nurs Stud. 2011;48(12):1505–1516. [DOI] [PubMed] [Google Scholar]
- 36.Crowe M, Sheppard L, Campbell A. Reliability analysis for a proposed critical appraisal tool demonstrated value for diverse research designs. J Clin Epidemiol. 2012;65(4):375. [DOI] [PubMed] [Google Scholar]
- 37.Crowe M, Sheppard L, Campbell A. Comparison of the effects of using the Crowe Critical Appraisal Tool versus informal appraisal in assessing health research: a randomised trial. Int J Evid Based Healthc. 2011;9(4):444–449. [DOI] [PubMed] [Google Scholar]
- 38.Ismail A, Munro K, Armitage C, Dawes P. What do hearing healthcare professionals do to promote hearing aid use and benefit among adults? A systematic review. Int J Audiol. 2019;58(2):63–76. [DOI] [PubMed] [Google Scholar]
- 39.Corrigan F, Broome H, Dorris L. A systematic review of psychosocial interventions for children and young people with epilepsy. Epilepsy Behav. 2016;56:99–112. [DOI] [PubMed] [Google Scholar]
- 40.Sznitman S, Taubman D. Drug use normalization: a systematic and critical mixed-methods review. J Stud Alcohol Drugs. 2016;77(5):700–709. [DOI] [PubMed] [Google Scholar]
- 41.Genecept Assay™ vs. Treatment-as-Usual to Evaluate Efficacy of Assay-Guided Treatment in Adults With MDD - Full Text View - ClinicalTrials.gov. Clinicaltrials.gov. 2021. Available from: https://clinicaltrials.gov/ct2/show/NCT02634177. Accessed February21, 2021.
- 42.Clinical Study to Evaluate Patient Outcomes Following Pharmacogenetic Testing of Subjects Exhibiting Neuropsychiatric Disorders - Full Text View - ClinicalTrials.gov. Clinicaltrials.gov. 2021. Available from: https://clinicaltrials.gov/ct2/show/NCT02411123. Accessed February21, 2021.
- 43.Drug & Gene Interaction Risk Analysis With & Without Genetic Testing Among Patients Undergoing MTM - Full Text View - ClinicalTrials.gov. Clinicaltrials.gov. 2021. Available from: https://clinicaltrials.gov/ct2/show/NCT02428660. Accessed February21, 2021.
- 44.Benitez J, Jablonski M, Allen J, Winner J. The clinical validity and utility of combinatorial pharmacogenomics: enhancing patient outcomes. Appl Trans Genomics. 2015;5:47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.RETIRED Local Coverage Determination for MolDX: geneSight Assay for Refractory Depression (L36323). Cms.gov. 2021. Available from: https://www.cms.gov/medicare-coverage-database/details/lcd-details.aspx?LCDId=36323&ContrId=364. Accessed February21, 2020.
- 46.Eloul L, Ambusaidi A, Al-Adawi S. Silent Epidemic of Depression in Women in the Middle East and North Africa Region: emerging tribulation or fallacy? PubMed. 2021. Available from: https://www.ncbi.nlm.nih.gov/pubmed/21509269. Accessed March17, 2020. [PMC free article] [PubMed]
- 47.Ghubash R, Hamdi E, Bebbington P. The Dubai Community Psychiatric Survey: I prevalence and socio-demographic correlates. Soc Psychiatry Psychiatric Epidemiol. 1992;27(2):53–61. [DOI] [PubMed] [Google Scholar]
- 48.Okasha A, Kamel M, Sadek A, Lotaif F, Bishry Z. Psychiatric Morbidity Among University Students in Egypt. Br J Psychiatry. 1977;131(2):149–154. [DOI] [PubMed] [Google Scholar]
- 49.Winner J, Goebert D, Matsu C, Mrazek D. Training in psychiatric genomics during residency: a new challenge. Acad Psychiatry. 2010;34(2):115–118. [DOI] [PubMed] [Google Scholar]
- 50.Dias M, Sorich M, Rowland A, Wiese M, McKinnon R. The routine clinical use of pharmacogenetic tests: what it will require? Pharm Res. 2017;34(8):1544–1550. [DOI] [PubMed] [Google Scholar]
- 51.Teutsch S, Bradley L, Palomaki G, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genetics Med. 2009;11(1):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sorich M, Polasek T, Wiese M Challenges and Limitations in the Interpretation of Systematic Reviews: Making Sense of Clopidogrel and CYP2C19 Pharmacogenetics. 2021. [DOI] [PubMed]
- 53.Brown J, Bishop J, Schneiderhan M. Using pharmacogenomics and therapeutic drug monitoring to guide drug selection and dosing in outpatient mental health comprehensive medication management. Mental Health Clinician. 2020;10(4):254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hicks J, Bishop J, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline forCYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerlach N, Michiels-Corsten M, Viniol A, et al. Professional roles of general practitioners, community pharmacists and specialist providers in collaborative medication deprescribing - a qualitative study. BMC Fam Pract. 2020;21:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasanpoor E, Hallajzadeh J, Siraneh Y, Hasanzadeh E, Haghgoshayie E. Using the Methodology of Systematic Review of Reviews for Evidence-Based Medicine. 2021. [DOI] [PMC free article] [PubMed]
- 57.DNA Medical Testing. Genetic Testing | geneSight. GeneSight. 2020. Available from: https://genesight.com/. Accessed October21, 2020.
- 58.Dynacare - GENECEPT ASSAY (English - Canada). Dynacare.ca. 2020. Available from: https://www.dynacare.ca/specialpages/secondarynav/find-a-test/nat/genecept%C2%A0assay.aspx?sr=ont&st=&. Accessed October21, 2020.
- 59.CNSdose | Genetic Testing | Home. Cnsdose.com. 2021. Available from: https://cnsdose.com/. Accessed October21, 2020.
- 60.Precision medicine especially developed for psychiatry | AB-Biotics. AB-Biotics. 2021. Available from: https://www.ab-biotics.com/c/precision-medicine/. Accessed March21, 2021.
- 61.Jain K Applications of AmpliChip™ CYP450. 2020.
- 62.AltheaDx. Depression and Anxiety. 2020. Available from: https://www.altheadx.com/clinical-trial-portfolio/idgenetix-tests/. Accessed October21, 2020.
- 63.Genelex . Precision Medicine Made Possible with Pharmacogenetics. Genelex.com. 2020. Available from: https://www.genelex.com/. Accessed October21, 2020.
- 64.Genomas:: Rennova Health, Inc. (RNVA). Rennovahealth.com. 2021. Available from: https://www.rennovahealth.com/brands/genomas. Accessed October21, 2020.
- 65.Pillcheck. Tailor your medications with Pillcheck DNA analysis. Pillcheck. 2020. Available from: https://www.pillcheck.ca/. Accessed October21, 2020.