Abstract
Preeclampsia (PE) is a hypertensive disorder of pregnancy that is a major cause of maternal-fetal morbidity and mortality worldwide. Severe PE (sPE) is mediated by pathology of the placental villi resulting in repressed PlGF production and hyper-secretion of sFLT1, the net effect being widespread maternal endothelial dysfunction. Villous trophoblast differentiation is under control of the PPARγ and GCM1 axis which is dysregulated in sPE. We hypothesized that disruption of trophoblast differentiation via the PPARγ-GCM1 axis is a major contribution to excess production of sFLT1 and pharmacological activation of PPARγ in the sPE placenta could reduce sFLT1 to normal levels. sPE, age-matched control placentas and first trimester villous explants were used to investigate the molecular relationships between PPARγ-GCM1 and sFLT1. We modulated this pathway by pharmacologic activation/inhibition of PPARγ using Rosiglitazone and T0070907, respectively and through siRNA repression of GCM1. PPARγ and GCM1 protein expressions are reduced in the sPE placenta while FLT1 protein and sFLT1 secretion are increased. GCM1 reduction in the first trimester explants significantly increased sFLT1 secretion, suggesting GCM1 as a key player in this pathway. Activation of PPARγ restored GCM1 and significantly reduced sFLT1 expression and release in first trimester and sPE placental villi. Functional integrity of the PPARγ-GCM1 axis in the villous trophoblast is critical for normal pregnancy development and is disrupted in the sPE placenta to favor excessive production of sFLT1. Pharmacologic manipulation of PPARγ activity has the potential to rescue the anti-angiogenic state of sPE and thereby prolong pregnancy and deliver improved clinical outcomes.
Keywords: PPARγ, GCM1, Human placenta, FLT1, Preeclampsia, Pregnancy
Introduction
The placenta serves as a critical organ during pregnancy to support fetal growth and development [1–3]. Abnormal placental development is a hallmark of several pregnancy-related complications causing significant maternal and/or fetal morbidity and mortality, especially severe fetal forms of fetal growth restriction (sFGR) and preeclampsia (sPE) that result in stillbirth or early preterm delivery [3, 4]. sPE comprises new onset of maternal hypertension after 20 weeks of gestation with systemic endothelial dysfunction and critical end-organ injury involving the kidneys, liver, brain and coagulation system [1, 5]. In sPE, the placenta most commonly exhibits multiple histopathologic features, collectively described as maternal vascular malperfusion [4, 6]. The disease begins with reduced extravillous trophoblast invasion and transformation of the uteroplacental arteries, which results in chronic ischemia of the developing placental villi [6–8]. Patients at highest risk of sPE demonstrated bilateral abnormal uterine artery Doppler and low circulating levels of placenta growth factor (PIGF) [9] and subsequently begin to express very high levels of soluble fms-like tyrosine kinase 1 (sFLT1) [10, 11]. In combination, the high sFLT1/PlGF ratio is now an established diagnostic test for PE [12–14].
sFLT1 is a potent anti-angiogenic protein and major contributor to endothelial damage in PE [16]. sFLT1 is a splice variant of the vascular endothelial growth factor receptor 1 (VEGFR1) also known as FLT1. sFLT1 competitively binds to the receptor domains of vascular endothelial growth factor (VEGF) [17] and its dimeric partner PIGF [18], preventing their interaction with the endothelial cell surface receptors. Our group previously found that first trimester placentas secrete higher amounts of sFLT1 by tissue weight compared to healthy term placenta, suggesting that sFLT1 has important roles in early pregnancy [19]. Increased levels of sFLT1 throughout pregnancy combined with lowered levels of PlGF largely mediates the systemic endothelial dysfunction observed in PE [20–23].
The transcription factor, Glial cell missing 1 (GCM1) regulates villous trophoblast differentiation in human placental villi [24, 25] and analogous labyrinth formation in mice [26, 27]. GCM1 thereby modulates the expression of trophoblast-derived proteins involved in the maintenance of normal pregnancy and cardiovascular function, especially the promotion of PIGF following syncytial fusion [18]. Prior reports found reduced GCM1 expression in PE placentas [28] with similar repression of the downstream fusogenic partner, syncytin, that is required for syncytiotrophoblast fusion to grow the continuous outer layer covering the placental villi. GCM1 is a strong candidate to regulate sFLT1 production in human placental villi, since a previous study identified that heterozygous Gcm1 knockout murine placentas secrete significantly higher levels of sFLT1 [29]. In further support of this hypothesis, enhanced expression of GCM1 via anti-viral drugs reduced the expression of sFLT1 in murine placentas [30].
GCM1 and the nuclear steroid receptor, peroxisome proliferator activated receptor gamma (PPARγ), are critical proteins needed for placental development and pregnancy. PPARγ and GCM1 work as sequential partners to regulate proper villous trophoblast to syncytiotrophoblast differentiation [25, 29]. PPARγ functions as an upstream transcriptional regulator of GCM1 [27, 31] through binding at two PPARγ response elements in the GCM1 promoter [32]. Using the BeWo choriocarcinoma villous trophoblast cell line [32] and a first trimester placenta explant model [24], our group previously identified that pharmacological activation of PPARγ by Rosiglitazone led to an increase in GCM1 expression and villous trophoblast differentiation. These findings suggest that this pathway is important for normal trophoblast function and turnover.
In the past decade, PPARγ has emerged as an important player in placental development due to its regulatory roles in multiple cellular pathways including metabolism, nutrient balance, and anti-inflammatory response pathways [33–35]. Murine studies have shown that embryonic knockdown of PPARγ is lethal due to gross cardiovascular and placental abnormalities [36]. Prior studies have shown that hypoxia reduces PPARγ expression in the human placenta and murine trophoblast stem cells [37–39]. Therefore, the prolonged hypoxic/ischemic nature of the severe PE placenta is likely a cause for the reduced placenta expression of PPARγ observed in PE [40–43]. This further imposes an abnormal villous trophoblast structure and poorly-developed feto-placental vasculature observed in severely preeclamptic placentas [36]. In a rodent model of PE, decreased activity of PPARγ was found to correlate with increased sFLT1 levels [44] and re-introducing PPARγ in PPARγ−/− murine trophoblast stem cells rescued differentiation of the syncytiotrophoblast and labyrinthine trophoblast lineages by GCM1 upregulation [27].
While it is established that PPARγ and GCM1 are critical factors for normal placental development, the potential molecular connections between PPARγ, GCM1, and sFLT1 in the normal and diseased human placenta are unclear. In the current study, we hypothesized that maintenance of the PPARγ-GCM1 axis in healthy developing placental villi represses the expression and secretion of the anti-angiogenic sFLT1, whereas this axis is disrupted in favor of hyper-secretion of sFLT1. We tested this hypothesis in explanted human placental villi from first trimester or from healthy and severely-preeclamptic women to determine the expression of GCM1 and production of sFLT1, under conditions that pharmacologically activated or inhibited PPARγ via the drugs Rosiglitazone and T0070907, respectively.
Methods
The authors declare that all supporting data are available within the article and its online supplementary files.
Tissue collection
First trimester (10–12 weeks of gestation) placental tissues (n = 4) were obtained with written informed consent from healthy pregnant women undergoing elective termination of pregnancy. The Institutional Review Board (IRB) of Wayne State University approved all consent forms and protocols used in this study, which abide by the NIH research guidelines. Term placental samples were obtained either by the Research Centre for Women’s and Infants’ Health (RCWIH) BioBank program of Mount Sinai Hospital in Toronto, Canada, in accordance with the policies of the Mount Sinai Hospital Research Ethics Board or Women’s Health Center at Spectrum Hospital in Grand Rapids, MI. All placentas collected were approved by the IRB waiver of parental consent. Specimens were collected from age-matched idiopathic preterm without histological evidence of chorioamnionitis not complicated by PE (PTC) (n = 14; gestational age = 31–37 weeks), and pregnancies complicated by severe PE (n = 14; gestational age = 31–37 weeks) and were delivered either by Cesarean section or vaginal birth. Inclusion criteria for severe PE was in accordance with current guidelines including blood pressure > 160/110 mm Hg on two occasions longer than 6 hr apart, evidence of end-organ damage including proteinuria, with or without fetal growth restriction [45].
Explant culture
For term tissues, a standardized random sampling protocol was applied dissecting random four 1cm3 cuboidal sections to avoid sampling bias. The collected tissues were washed and transported to the laboratory in ice cold HBSS (Hank’s Balanced Salt Solution) and processed within a maximum of 2 hr after delivery. On arrival, tissues were rinsed in chilled HBSS to remove residual blood and further dissected under a stereomicroscope to remove placental membranes and generate 20–30mg pieces of villous tissues for culture. First trimester explants were cultured according to our previously published floating villous explant protocol [46]. Individual clusters of villous trees were dissected under a stereomicroscope. Post dissection, the explants were cultured overnight in 500 uL of Dulbecco’s modified Eagle’s medium/Ham’s F-12 nutrient mixture (DMEM/F-12; 1:1; Life Technologies; Grand Island, NY) containing 10% fetal bovine serum (FBS; Life Technologies) and 1% Gibco™ antibiotic-antimycotic. Term explants were maintained overnight at 8% O2 with 5% CO2 at 37°C [47]. After an overnight culture, the tissues were treated with 10μM Rosiglitazone (Selleckchem) or 1μM T0070907 (R&D Systems) dissolved in dimethyl sulfoxide (DMSO, Sigma Life Sciences) for 18–24 hours. DMSO alone was used as a vehicle control. Comparison of DMSO to NT (not treated placental tissues) was performed to ensure there was not an effect from DMSO. After the culture period for each treatment, replicates were snap frozen for protein and RNA extraction and stored at −80°C. The media was also collected, snap frozen and stored at −80°C. In a few samples, an extra replicate was immediately fixed in 4% paraformaldehyde for immunohistochemistry. This was included in this study as a qualitative assessment to complement RNA/Protein expression findings.
Protein extraction and immunoblotting
Protein extraction from tissues (20–30 mg) was performed as previously described [48]. Protein concentration was determined with BCA™ protein assay reagent (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer’s instructions. Equal protein amounts (35 μg) were denatured (8 min, 95°C) in Laemmli sample buffer (Bio-Rad Laboratories; Hercules, CA) and separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, with subsequent semi-dry transfer (Trans-Blot®; Bio-Rad Laboratories) to a polyvinylidene difluoride membrane. The membranes were blocked with 5% nonfat dry milk in 1× Tris-buffered saline containing 0.05% Tween-20 and were incubated overnight at 4°C with anti-GCM1 (1:5,000; Aviva, San Diego, CA), anti-FLT1 (1:1000, Abcam), and anti-PPARγ (1:1,000; Cell Signaling Technology) primary antibodies. Subsequently, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hr at room temperature and were developed with Western Lightning® ECL Pro (PerkinElmer, Waltham, MA). Signals were visualized using a ChemiDoc™ Imaging System (Bio-Rad Laboratories) and Image Lab Version 5.1 software (Bio-Rad Laboratories). Densities of immunoreactive bands were measured as arbitrary units by the ImageJ software (NIH, Bethesda, MD). Protein levels were normalized to a housekeeping protein β-actin (1:4,000; Abcam). Protein expression values are reported as relative to β-actin.
ELISA
The media collected from PE, PTC, and first trimester placental explant cultures was assayed for levels of sFLT1 using the Human VEGFR1/Flt-1 DuoSet kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s instruction. Culture media was centrifuged at 4,500 × g for 10 minutes at 4°C to pellet all cell/tissue debris and the supernatant was used for ELISA analysis. The optical density of the final-colored reaction product was measured at 450 nm using a SoftMax Pro5 or a multispectral UV/VIS (Bio-Tek, VT) plate reader. A standard curve was used to calculate protein content, and this was normalized over wet weight of the explant to obtain the amount of protein secreted per milligram of explant tissue.
RNA extraction and qPCR analysis
The tissue was lysed in Qiazol and RNA was extracted using RNeasy Plus Universal Mini kit (Qiagen, Germany) as per the manufacturer’s protocol. The extracted RNA was quantified using Nanodrop and 1μg was reverse transcribed using iScript RT synthesis kit (Bio-Rad Laboratories, CA). Real-time PCR was performed on the Bio-Rad CFX384 real time system in triplicates in 10uL total reaction volume containing 10 ng of template cDNA, 5μL of SYBR-green master mix (LuminoCT, Sigma-Aldrich, MO) and 500nM of primers. The primers used for assessing the expression levels of target and housekeeping genes are outlined in Table 1. Data was analyzed using the delta-delta CT method as described in [49].
Table 1.
qPCR Primer Sequences
| Gene Name | Gene Symbol | Sequence |
|---|---|---|
| Cytochrome - C 1 | Cyc1 | 5’-CAT CAT CAA CAT CTT GAG CC-3’ |
| 5’-CAG ATA GCC AAG GAT GTG TG-3’ | ||
| Tyrosine 3-monooxygenase | Ywhah | 5’- CCG CCA GGA CAA ACC AGT AT -3’ |
| 5’- ACT TTT GGT ACA TTG TGG CTT CAA -3’ | ||
| TATA Box Binding Protein | Tbp | 5’-CAC ATC ACA GCT CCC CAC CA-3’ |
| 5’-TGC ACA GGA GCC AAG AGT GAA-3’ | ||
| Glial cell missing 1 | Gcm1 | 5’-TGA ACA CAG CAC CTT CCT C-3’ |
| 5’-CCA CTG TAA CTA CCA GGC AAT-3’ | ||
| Soluble fms-like tyrosine kinase 1 | sFlt1 | 5‘- CCT CAA ATG ATC CAC CTG CCT-3‘ |
| 5‘- CAG GAA GCA CCA TAC CTC CTG -3‘ |
PPARγ Transcription Factor Assay
Nuclear proteins were isolated from first trimester tissue after treatment with Rosiglitazone (10μM), T0070907 (1nM) and DMSO using a Nuclear Extract Kit (ActiveMotif, Carlsbad, California). The assay was performed using PPARγ binding assay (TransAM, ActiveMotif, Carlsbad, California) following the manufacturer’s protocol. Briefly, 3μg of nuclear proteins from treatment and control groups were used. The proteins from each group were added to the provided 96 well plate (in triplicates) and volumes were adjusted to 10uL using the complete lysis buffer from the kit. 5ug of given positive control and complete binding buffer containing 40pmol of the consensus site (from the kit) was then added to each well. The plate was incubated for 1 hour followed by 3 washes with the 200uL of 1X wash buffer. 100uL of the supplied PPARγ antibody was then added to all wells and the plate was incubated again for 1 hour at RT. After the incubation, the wells were washed again 4 times and 100uL of developing solution was then added to each well and incubated for 5 mins. The reaction was stopped using 100uL of Stop solution and the absorbance was read at 665nm. The absorbance for the blank wells was subtracted from all the readings and then values from Rosiglitazone and T0070907 samples were normalized to the values from DMSO vehicle for comparison between the treatments.
Immunohistochemistry
Immunostainings of placental villi were performed as described in [50]. Briefly, the sections were deparaffinized and rehydrated, followed by antigen retrieval using Dako Target retrieval solution (Agilent-DAKO, USA). The intrinsic peroxidase activity was then quenched by incubating the sections with 3% Hydrogen peroxide (Fisher Scientific, MA) for 30 mins at RT, followed by a wash with 1X PBS. The sections were then incubated overnight at 4°C with anti-FLT1 (Santa Cruz, TX) or 10μg/ml nonimmune Rabbit IgG (Jackson Immunoresearch, PA) (used as a negative control). The following day, the slides were washed 3 times (5 minutes/wash) with 1X PBS containing 0.1% Tween 20. The samples were then incubated for 30 min with a peroxidase-conjugated polymer coupled to anti-rabbit IgG (EnVision Systems Peroxidase, Agilent-DAKO, USA). The peroxidase was visualized with 3,3-diaminobenzidine (DAB, Agilent-DAKO, USA) and hydrogen peroxide for 5 min. Tissues were counterstained with hematoxylin, dehydrated and were cover slipped. The staining was visualized using Nikon Eclipse 90i epifluorescence microscope (Nikon Inc., Japan) and the images were analyzed using ImageJ software.
siRNA-mediated GCM1 suppression
For silencing, Silencer™ Select Pre-Designed siRNA assays (Thermo Fischer) were used. For GCM1 specific knockdown, assay ID s16199 was used and a Cy™3-labeled scramble sequence was used a negative control (AM4621, ThermoFisher). A non-silencing control was additionally incorporated as a technical control to exclude any effects of the electroporation procedure used for silencing. The tissues were electroporated using the P3 Primary Cell 4D-Nucleofector™ X Kit L and the Nucleofactor™ 2b device (Lonza, Switzerland) following the manufacturer’s kit protocol. First trimester villous explants were cultured overnight in Dulbecco’s modified Eagle’s medium/Ham’s F-12 nutrient mixture (DMEM/F-12; 1:1; Life Technologies; Grand Island, NY) containing 10% fetal bovine serum (FBS; Life Technologies) and 1% Gibco™ antibiotic-antimycotic at 8% O2. On day 2, the explants were placed in the cuvette (2 explants/cuvette) along with 100uL of electroporation solution (10uL of silencing probe mix + 90uL of electroporation buffer). The program U017 was used for electroporation after which the explants were taken out and cultured in fresh media for 48 hours. After the culture period, one set of explants were processed for immunohistochemistry by fixing in 4% paraformaldehyde and another set was frozen in 700uL of Qiazol to be used for RNA expression studies. The media was collected and frozen to be used later for sFLT1 analysis using ELISA.
Statistical Analysis
All statistical analysis was performed with GraphPad Prism 7.0 software. Raw mRNA and protein expressions were normalized to respective housekeeping genes or protein. ELISA data was normalized based on semi dry-tissue weight. Relative expression/secretion values from untreated tissues (Figure 1) were analyzed by student’s t-test after determination if samples are normally distributed and an F-test was applied to determine variances between groups which was then used in the parameters for the t-test. Raw mRNA and protein expression values from tissues treated with either DMSO, Rosiglitazone and T0070907, or GCM1-siRNA and scramble siRNA (Figures 2–4) were normalized to respective housekeeping genes or protein. Relative expression or secretion values for each tissue sets were subsequently normalized to respective DMSO (vehicle control, set equal to 1) or scramble siRNA control (set equal to 1) represented by a dotted line on the graphs. Groups were then analyzed by student’s t-test, after determination if samples are normally distributed and an F-test was applied to determine variances between groups which was then used in the parameters for the t-test. p<0.05 is considered significant and is indicated with (*) on each graph. Data is reported as Mean±S.E.M [51]. All sample numbers are reported as per group, for example, n=6 designates 6 samples per treatment/group.
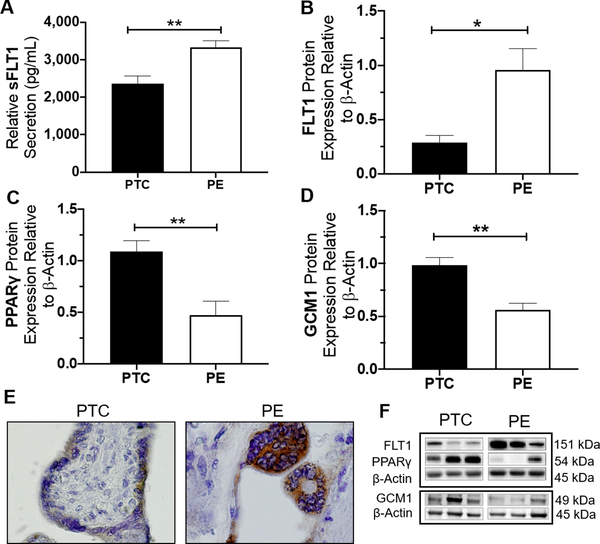
Figure 1. Placentas from women with sPE exhibit higher sFLT1 secretion and FLT1 expression accompanied with lower expressions of PPARγ and GCM1.
sFLT1 secretion was significantly higher secreted by sPE compared to gestational-age matched control (PTC) (2361.3±198pg vs. 3326.8±178.8pg, p=0.0067, n=5) (A). This finding corresponds with higher expression of FLT1 protein in sPE compared to control (p=0.0167, n=6) (B). Immunostaining for total-FLT1 (FLT1 and sFLT1 variants) shows light staining in the syncytiotrophoblast of PTC tissues compared to a more intense signal in sPE placenta (E). Immunoblotting assessment revealed sPE placentas exhibit lower protein expression of PPARγ (p=0.0042, n=6) and GCM1 (p=0.000, n=14) compared to PTC (C, D). Representative western blots are shown in (F). (Relative mRNA and protein expression were determine by normalization to housekeeping genes or protein, followed by a student’s t-test to determine significant differences between groups, * p<0.05, ** p<0.01, Rosi=Rosiglitazone, T007=T0070907, bar plots and data reported are presented mean ± SEM, PE = preeclampsia, PTC = preterm healthy control, SN = Syncytial Knots, scale bar = 50 μm, bar plots are presented as mean ± SEM.).
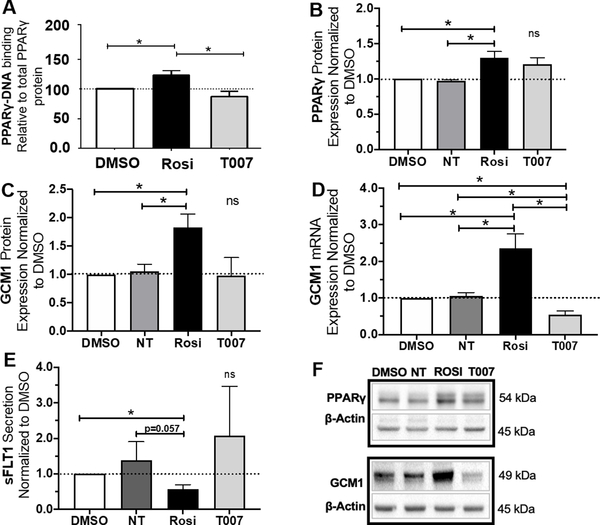
Figure 2. Rosiglitazone increases PPARγ activity and GCM1 mRNA expression while reducing sFLT1 secretion in the first trimester placenta.
Treatment with Rosiglitazone caused a significant increase in PPARγ activity (p<0.05, n=3) (A). T0070907 did not cause a significant change in PPARγ activity (A). Rosiglitazone significantly upregulated PPARγ (p=0.0446, n=4) and GCM1 (p=0.0402, n=4) protein expression (B, C, F). T0070907 did not cause a significant change in PPARγ or GCM1 protein expression (B, C, F). Rosiglitazone significantly upregulated GCM1 mRNA expression (p=0.0433, n=4) (D). T0070907 significantly reduced GCM1 mRNA expression (p=0.02, n=4) (D). Rosiglitazone also caused a significant reduction in sFLT1 secretion (p=0.025, n=3) (E). Antagonizing PPARγ by T0070907 did not cause a statistically significant change in sFLT1 secretion (E). (Relative mRNA and protein expression were determine by normalization to housekeeping genes or protein. Relative expression values for individual tissue sets were normalized to DMSO (vehicle control, dotted line, set equal to 1) and subsequent statistical analysis was performed by student’s t-test to determine significant differences between groups,* p<0.05, ns=p>0.05, NT=not treated, Rosi=Rosiglitazone, T007=T0070907, bar plots and data reported are reported as mean ± SEM).
Figure 4. Rosiglitazone increases expression of PPARγ and GCM1 while simultaneously decreasing sFLT1 in sPE placenta.
PPARγ protein expression significantly increased in sPE placenta after Rosiglitazone treatment (p=0.0051, n=4) (A/B). GCM1 mRNA significantly increased in the sPE placenta by Rosiglitazone treatment (p=0.0162, n=9). GCM1 mRNA expression significantly reduced in the sPE placenta after treatment with T0070907 (p=0.001, n=9) (C). sFLT1 mRNA was significantly reduced in the sPE placenta after Rosiglitazone treatment. No significant change in sFLT1 mRNA expression was observed after exposure to T0070907 (B).
(Relative mRNA/protein expressions were determined by normalization to respective housekeeping genes/protein. Relative expression values for individual tissue sets were normalized to DMSO (vehicle control, dotted line, set equal to 1) and subsequent statistical analysis was performed by student’s t-test to determine significant differences between groups, *p<0.05, **p<0.01, ns=p>0.05, NT= not treated, Rosi=Rosiglitazone, T007=T0070907, bar plots are reported as mean ± SEM).
Results
Cultured sPE placentas show increased protein expression of FLT1, increased secretion of sFLT1 and reduced protein expression of PPARγ and GCM1 compared to PTC controls.
sFLT1 secretion into the placenta culture media was measured by ELISA after 48 hours of culture for sPE and gestational age-matched preterm control (PTC) placentas. We observed significantly higher secretion of sFLT1 from sPE placentas compared to PTC (3327±198 pg/mL vs. 2361±198 pg/mL, p=0.0067, n=5, Fig. 1A). Placental protein expression of FLT1, PPARγ, and GCM1 were measured by western blotting. FLT1 protein expression was significantly upregulated in sPE placentas comparison to PTC (0.96±0.2 vs. 0.28±0.06 relative expression values, p=0.0167, n=6, Fig. 1B) which mirrored representative immunohistochemical staining patterns showing enhanced localized expression of total-FLT1 (which includes all FLT1 and sFLT1 variants) in the syncytiotrophoblast layer of the sPE placenta (Figure 1E). sPE placentas showed a significant reduction of PPARγ protein expression (0.476±0.13 vs. 1.09±0.1, p=0.0042, n=6, Fig. 1C, F) and a significant reduction of GCM1 protein expression (0.56±0.06 vs. 0.99±0.07, p=0.0001, n=14, Fig. 1D, F) compared to PTC.
Activation of PPARγ by Rosiglitazone induces GCM1 expression and lowers placental sFLT1 secretion in first trimester placental explants.
First trimester villous placental explants were used as a model to understand how sFLT1 changes in response to modulating of the PPARγ–GCM1 axis. These tissues were cultured for 18 hours with PPARγ agonist, Rosiglitazone (10mM), or antagonist, T0070907 (1mM). Successful activation of PPARγ by Rosiglitazone was confirmed by a 28±5% increase in PPARγ global DNA-binding activity in the Rosiglitazone treated explants (128±5 vs. 100, p<0.05, n=3, Fig. 2A) as measured by an ELISA-based transcription factor binding assay. There was no significance difference in PPARγ DNA-binding activity in the T0070907 treated explants (Fig. 2A). PPARγ and GCM1 protein expressions in first trimester tissues were measured by western blotting. Rosiglitazone significantly increased PPARγ protein expression (1.3±0.09 vs 1, p=0.0446, n=4, Fig. 2B, F) and GCM1 protein expression (1.83±0.2 vs. 1, p=0.0402, n=4, Fig. 2C, F). Rosiglitazone significantly increased GCM1 mRNA expression (2.35±0.4 vs. 1, p=0.04, n=4) while T0070907 significantly decreased GCM1 mRNA expression (0.55±0.1 vs. 1, p=0.02, n=4) (Fig. 2D). Rosiglitazone significantly decreased sFLT1 secretion into the culture media (0.57±0.07 vs. 1, p=0.025, n=3, Fig. 2E). Treatment with T0070907 did not result in a significant change in sFLT1 secretion from the first trimester explants (Fig. 2E).
Silencing of GCM1 upregulates FLT1 and sFLT1 in human first trimester explants.
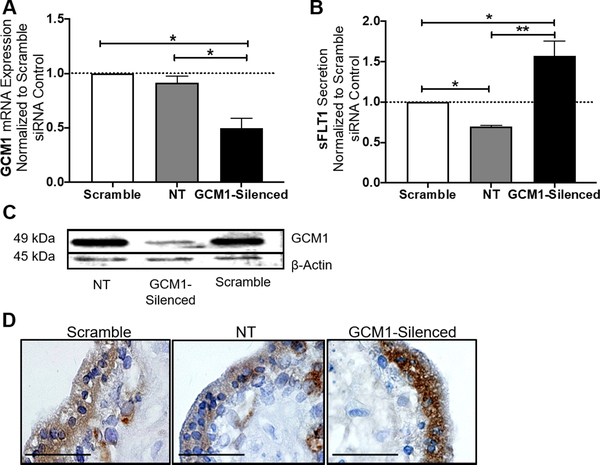
We have shown that activating PPARγ in the placenta leads to a significant induction of GCM1 mRNA and protein expression and a significant reduction of sFLT1 secretion from the first trimester placenta. Since it is already established that PPARγ acts as an upstream transcriptional regulator for GCM1 [32], we aimed to determine if GCM1 has a role in this potential pathway for regulation of sFLT1 secretion. We further used first trimester villous explants as a model for siRNA-mediated repression of GCM1. GCM1 siRNA caused a significant reduction of GCM1 mRNA expression (0.49±0.09, p=0.031, n=3, Fig. 3A) and reduced GCM1 protein as compared to tissues transfected with the scramble siRNA (Fig. 3C). The GCM1-silenced explants secreted significantly more sFLT1 (1.59±0.13 vs. 1, p=0.0389, n=3, Fig. 3B). A representative immunohistochemistry staining shows higher expression of total-FLT1 protein (FLT1 and sFLT1 variants) in the syncytiotrophoblast of the GCM1-silenced explants compared to scramble siRNA and no-treatment controls (Fig. 3D).
Figure 3. GCM1 reduction increases total-FLT1 expression and sFLT1 secretion in first trimester explants.
In our first trimester explant model, siRNA-mediated silencing of GCM1 caused a significant reduction of GCM1 mRNA expression (p=0.031, n=3) (A). Similarly, western blot shows GCM1 protein expression appeared to be decreased in the GCM1-silenced tissues in comparison to the scramble siRNA and no-treatment controls (C). sFLT1 secretion was significantly increased in the GCM1-silenced explants (p=0.0389, n=3) (B). Similarly, placental expression of total-FLT1 protein (FLT1 and sFLT1 variants) was induced in the syncytiotrophoblast of the first trimester explants, after GCM1 knockdown (D). (Relative mRNA expression was normalized to housekeeping genes. sFLT1 secretion (pg/mL) data were normalized based on tissue weight. Relative expression/secretion values for individual tissue sets were normalized to the scramble siRNA control (dotted line, set equal to 1) and subsequent statistical analysis was performed by student’s t-test was performed to determine significant differences between groups, NT=not treated, * p<0.05, ns= p>0.05, bar plots are reported as mean ± SEM).
Rosiglitazone restores PPARγ and GCM1 expression and downregulates sFLT1 in the severe preeclamptic placenta.
To test if the PPARγ-GCM1 axis can be modulated in the sPE placenta, we pre-treated sPE placentas with Rosiglitazone (10μM) or T0070809 (1μM) for 24 hours. Rosiglitazone significantly increased PPARγ protein expression in sPE (1.34±0.04 vs. 1, p=0.051, n=4) and treatment with T0070907 did not have a significant effect on PPARγ protein expression. Rosiglitazone restored GCM1 by increasing mRNA expression the sPE placenta (1.28±0.09 vs. 1, p=0.0162, n=9, Fig 4A). Treatment with T0070907 caused a significant reduction of GCM1 mRNA expression in the sPE placenta (0.49±0.05 vs. 1, p=0.001, n=9, Fig. 4A). Rosiglitazone significantly decreased sFLT1 mRNA expression in the sPE placenta (0.655±0 vs. 1, p=0.0058, n=7, Fig 4B). Treatment with T0070907 did not cause a significant change in sFLT1 mRNA expression in the sPE placenta (Fig. 4B).
Discussion
In this study, we provide novel molecular evidence to demonstrate that FLT1 and its secreted splice variant, sFLT1, are regulated by a PPARγ–GCM1 axis in the trophoblast layer covering human placental villi. Under physiologic conditions, this axis mediates orderly syncytiotrophoblast formation via asymmetric divisions of progenitor villous cytotrophoblasts [52], which in turn promotes PlGF synthesis and release into maternal blood, and in tandem, represses sFLT1 [18, 24, 25, 29]. Conversely, under sub-optimal PPARγ–GCM1 signaling, the placental villi show both structural and molecular defects characterized by defective syncytial fusion, repressed PlGF and aberrant release of sFLT1 into maternal blood [28, 40–43]. sFLT1 plays a major role in PE pathology as it promotes wide-spread endothelial dysfunction which largely contributes to the multi-organ dysfunction in the mother [7, 53, 54]. Highly secreted sFLT1 and its anti-angiogenic properties are well characterized in sPE placentas, where syncytial knot formation is one of the hallmarks of the maternal-vascular malformation disease [55, 56]. While FLT1 is known to be increased in tissues during vascular reoxygenation in response to hypoxic/ischemic insult [57] which similarly occurs in the PE placenta [58], the regulatory mechanism(s) of FLT1 and sFLT1 in the placenta remains unclear.
We observed higher sFLT1 secretion and higher protein expression of FLT1 in the PE placenta. Additionally, our immunohistochemistry staining identified that total-FLT1 protein (FLT1 and sFLT1 variants) exclusively localized to the syncytiotrophoblast in both control and sPE placenta, and appears to have increased localization in the syncytial knots found in sPE tissue. Our data validates previous findings by Taché et al., who showed a correlation between high syncytiotrophoblast sFLT1 levels and PE disease severity [55]. In addition, Jebbink et al. showed syncytiotrophoblast specific localization of sFLT1 mRNA transcripts and higher sFLT1 mRNA in the PE placenta [22]. Together, these data support the notion that the abnormal syncytiotrophoblast layer in PE may be the major source of high secretion of sFLT1.
Our data shows an inverse correlation between high FLT1 expression and sFLT1 secretion and low PPARγ and GCM1 expression in the PE placenta. This finding aligns with previous reports in the literature [25, 28, 29]. The relationship between PPARγ and sFLT1/FLT1 in the placenta was previously shown in a study by McCarthy et al., where a reduced uteroplacental perfusion pressure (RUPP) model was established to mimic PE in pregnant rats [59]. These RUPP animals showed significantly elevated levels of sFLT1 [59]. When treated with Rosiglitazone, these animals showed decreased blood pressure and decreased levels of sFLT1 [59]. In a separate study, McCarthy et al. treated pregnant rats with the PPARγ antagonist, T0070907, which caused these animals to develop PE-like symptoms such as elevated blood pressure and proteinuria accompanied by decreased VEGF and increased plasma levels of sFLT1 [44].
From this finding and our observations that PPARγ and GCM1 are inversely correlated with FLT1 expression, we hypothesized that a molecular connection exists between PPARγ, GCM1, and FLT1/sFLT1 in the human placenta, via repression of syncytin-mediated syncytial fusion. We used our first trimester placental explant model to test if this pathway can be modulated in the placenta. We used the PPARγ agonist, Rosiglitazone, and PPARγ antagonist, T0070907, to modulate this pathway. T0070907 antagonizes PPARγ by selectively binding to the PPARγ ligand binding pocket, preventing its activation by another ligand [60]. It is suggested that repressive ligands such as T0070907 do not have an effect on PPARγ protein expression levels [60], which could explain our finding that PPARγ protein expression was not significantly different in first trimester placenta or in sePE placentas when treated with T0070907. Rosiglitazone acts as a chemical ligand for PPARγ by increasing its DNA-binding activity to influence gene expression [34] and is also shown to increase PPARγ transcription [61]. We confirmed that Rosiglitazone caused an increased in PPARγ activity in the first trimester explants and this coincided with a significant increase in PPARγ protein expression in the first trimester explants and in the sePE placenta. We observed that Rosiglitazone significantly upregulated GCM1 mRNA and significantly reduced sFLT1 secretion into placental media culture in our first trimester explant model. These results suggest that the PPARγ-GCM1 axis has a role in regulating the angiogenic environment of the placenta via sFLT1.
We further questioned whether the changes observed in sFLT1 secretion were a direct result from PPARγ activation or if GCM1 may serve as an intermediate in this pathway to modulate FLT1 expression, since PPARγ transcriptionally regulates GCM1 through two PPARγ response elements in the promoter region of GCM1 [32]. We show that siRNA mediated repression of GCM1 in our first trimester villous explants caused a significant upregulation of sFLT1 secretion, as well, caused an increase in FLT1 protein expression in the syncytiotrophoblast, observed through immunohistochemistry. Our findings align with previous studies that showed a heterozygous knockdown of Gcm1 in the mouse led to increased secretion of sFLT1 [29]. These results suggest that GCM1 may be involved in FLT1 regulation and a decrease of GCM1 expression may contribute to the anti-angiogenic state during PE through upregulation of sFLT1. We lastly show that this pathway is not unique to the first trimester placenta and can be modulated in the diseased severely preeclamptic placenta. We found that activation of PPARγ in sPE placenta increased GCM1 and reduced sFLT1 mRNA expression.
In the current study, we show for the first time that activation of PPARγ can modulate the angiogenic environment of the human placenta by altering expression of GCM1, FLT1 and sFLT1. More studies are needed for better understanding of how this pathway impacts placental function and physiology during the second and third trimesters of pregnancy, when the high anti-angiogenic environment in PE becomes largely problematic. We acknowledge the limitations of our study, such as small sample size which necessitates follow up studies with larger cohorts. We were unable to observe an effect based on mode of delivery however this should be considered as a confounding factor in future studies. To our knowledge, we report novel findings of PPARγ, GCM1 and sFLT1 in the placenta, although we have not clearly demonstrated that a direct or indirect interaction between these molecules exist. We performed in silico prediction of DNA-binding sites for GCM1 in the promoter region FLT1 using data from HOmo sapiens COmprehensive MOdel COllection (HOCOMOCO) v9 [62]. No putative binding site for GCM1 was found in the proximity of the FLT1 active promoter however there are two putative GCM1 binding sites approximately ~50kb from the FLT1 promoter (Supplemental Figure S3). Further studies should focus on potential long-range effects of GCM1 at these regions to regulate FLT1 promoter elements, co-factors or splicing machinery to directly influence gene expression or protein secretion of FLT1 and its splice variants. Moreover, future studies should consider how modulating this pathway could affect overall angiogenic balance, such as through increasing secretion of PIGF and VEGF from the placenta. Detailed molecular studies investigating these targets potentially using both in vivo and in vitro models with knockdown or knockout of PPARγ and GCM1 are needed to fully understand this potential mechanism and their effects on overall placental function.
Perspectives
Abnormal trophoblast differentiation and turnover poses a major threat to placental function in preeclampsia (PE) and is driven by dysregulation of the PPARγ-GCM1 syncytialization axis. We identify that high FLT1 expression localizes to the damaged syncytial membrane in the preeclamptic placenta. Silencing of GCM1 in the human placenta significantly upregulates sFLT1 secretion and causes intense FLT1 localization to the GCM1-deficient syncytiotrophoblast, further suggesting that dysregulation of villous trophoblast turnover is responsible for the high FLT1 and pathologic levels sFLT1 observed in PE. Our data position PPARγ proximal to GCM1 to regulate its expression, as we show that induction of PPARγ restores GCM1 while providing a physiologic brake on sFLT1 production, and further affords the PPARγ-GCM1 axis a therapeutic target status. Our study highlights the importance of understanding the molecular control of villous trophoblast turnover as a key for improving placental and maternal health in PE. Future directions should investigate the detailed workings of how GCM1 acts to repress FLT1 and sFLT1 in the placenta.
Supplementary Material
Novelty and Significance.
What Is New?
Strong FLT1 production occurs in dysregulated areas of syncytiotrophoblast, accompanied by low expression of PPARγ and GCM1 which regulate villous trophoblast syncytialization in the preeclamptic placenta.
Silencing of GCM1 in the first trimester placenta leads to an induction of sFLT1.
We are the first to show that induction of PPARγ increases GCM1 and reduces sFLT1 expression in the placenta.
What Is Relevant?
Restoring the PPARγ-GCM1 syncytialization axis provides a physiological ‘break’ on sFLT1 production from severely preeclamptic placenta.
Summary.
Aberrant villous trophoblast turnover contributes to pathologic levels of sFLT1 and this can be halted by restoration of the PPARγ-GCM1 syncytialization axis in the preeclamptic placenta.
Acknowledgments
We thank the Spectrum Health Accelerator of Research Excellence (SHARE) biorepository (Spectrum Health, Grand Rapids, MI) for providing de-identified biospecimens for this study.
Sources of Funding
This research was supported by the National Institutes of Health under award number HL128628 and the March of Dimes Foundation awarded to S.D. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number T32HD087166, MSU AgBio Research and Michigan State University, awarded to B.A. Support for this research was contributed by grant number CIHR MOP 64302 awarded to Dr. John Kingdom. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures
None.
References
- 1.Lindheimer MD, CHESLEY’S HYPERTENSIVE DISORDERS IN PREGNANCY THIRD EDITION Preface. Chesley’s Hypertensive Disorders in Pregnancy, 3rd Edition, 2009: p. Ix–X. [Google Scholar]
- 2.Knofler M, et al. , Human placenta and trophoblast development: key molecular mechanisms and model systems. Cellular and Molecular Life Sciences, 2019. 76(18): p. 3479–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armistead B, et al. , Placental Regulation of Energy Homeostasis During Human Pregnancy. Endocrinology, 2020. 161(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zur RL, et al. , The Placental Basis of Fetal Growth Restriction. Obstet Gynecol Clin North Am, 2020. 47(1): p. 81–98. [DOI] [PubMed] [Google Scholar]
- 5.Phipps EA, et al. , Pre-eclampsia: pathogenesis, novel diagnostics and therapies. Nat Rev Nephrol, 2019. 15(5): p. 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho AEP, et al. , T2* Placental Magnetic Resonance Imaging in Preterm Preeclampsia: An Observational Cohort Study. Hypertension, 2020. 75(6): p. 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rana S, et al. , Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ Res, 2019. 124(7): p. 1094–1112. [DOI] [PubMed] [Google Scholar]
- 8.Armistead B, et al. , The Role of NFkappaB in Healthy and Preeclamptic Placenta: Trophoblasts in the Spotlight. Int J Mol Sci, 2020. 21(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers JE, et al. , Angiogenic factors combined with clinical risk factors to predict preterm pre-eclampsia in nulliparous women: a predictive test accuracy study. BJOG, 2013. 120(10): p. 1215–23. [DOI] [PubMed] [Google Scholar]
- 10.Levine RJ, et al. , Circulating angiogenic factors and the risk of preeclampsia. N Engl J Med, 2004. 350(7): p. 672–83. [DOI] [PubMed] [Google Scholar]
- 11.Doherty A, et al. , Altered hemodynamics and hyperuricemia accompany an elevated sFlt-1/PlGF ratio before the onset of early severe preeclampsia. J Obstet Gynaecol Can, 2014. 36(8): p. 692–700. [DOI] [PubMed] [Google Scholar]
- 12.McLaughlin K, et al. , PlGF (Placental Growth Factor) Testing in Clinical Practice: Evidence From a Canadian Tertiary Maternity Referral Center. Hypertension, 2021: p. HYPERTENSIONAHA12117047. [DOI] [PubMed] [Google Scholar]
- 13.Zeisler H, et al. , Soluble fms-Like Tyrosine Kinase-1-to-Placental Growth Factor Ratio and Time to Delivery in Women With Suspected Preeclampsia. Obstet Gynecol, 2016. 128(2): p. 261–269. [DOI] [PubMed] [Google Scholar]
- 14.Zeisler H, et al. , Predictive Value of the sFlt-1:PlGF Ratio in Women with Suspected Preeclampsia. N Engl J Med, 2016. 374(1): p. 13–22. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin K, et al. , Placental Growth Factor Testing In Clinical Practice: Evidence From A Canadian Tertiary Maternity Referral Centre 2021: Hypertension. [DOI] [PubMed] [Google Scholar]
- 16.Ganss R, Maternal Metabolism and Vascular Adaptation in Pregnancy: The PPAR Link. Trends Endocrinol Metab, 2017. 28(1): p. 73–84. [DOI] [PubMed] [Google Scholar]
- 17.Fan X, et al. , Endometrial VEGF induces placental sFLT1 and leads to pregnancy complications. J Clin Invest, 2014. 124(11): p. 4941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang M, et al. , Glial cell missing 1 regulates placental growth factor (PGF) gene transcription in human trophoblast. Biology of reproduction, 2008. 78(5): p. 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drewlo S, et al. , Heparin promotes soluble VEGF receptor expression in human placental villi to impair endothelial VEGF signaling. J Thromb Haemost, 2011. 9(12): p. 2486–97. [DOI] [PubMed] [Google Scholar]
- 20.Kendall RL and Thomas KA, Inhibition of vascular endothelial cell growth factor activity by an endogenously encoded soluble receptor. Proceedings of the National Academy of Sciences, 1993. 90(22): p. 10705–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendall RL, Wang G, and Thomas KA, Identification of a Natural Soluble Form of the Vascular Endothelial Growth Factor Receptor, FLT-1, and Its Heterodimerization with KDR. Biochemical and Biophysical Research Communications, 1996. 226(2): p. 324–328. [DOI] [PubMed] [Google Scholar]
- 22.Jebbink J, et al. , Expression of placental FLT1 transcript variants relates to both gestational hypertensive disease and fetal growth. Hypertension, 2011. 58(1): p. 70–6. [DOI] [PubMed] [Google Scholar]
- 23.Thomas CP, et al. , A recently evolved novel trophoblast-enriched secreted form of fms-like tyrosine kinase-1 variant is up-regulated in hypoxia and preeclampsia. J Clin Endocrinol Metab, 2009. 94(7): p. 2524–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baczyk D, et al. , Glial cell missing-1 transcription factor is required for the differentiation of the human trophoblast. Cell Death and Differentiation, 2009. 16(5): p. 719–727. [DOI] [PubMed] [Google Scholar]
- 25.Baczyk D, et al. , Complex patterns of GCM1 mRNA and protein in villous and extravillous trophoblast cells of the human placenta. Placenta, 2004. 25(6): p. 553–559. [DOI] [PubMed] [Google Scholar]
- 26.Anson-Cartwright L, et al. , The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nature Genetics, 2000. 25: p. 311. [DOI] [PubMed] [Google Scholar]
- 27.Parast MM, et al. , PPARgamma regulates trophoblast proliferation and promotes labyrinthine trilineage differentiation. PLoS One, 2009. 4(11): p. e8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CP, et al. , Decreased Placental GCM1 (Glial Cells Missing) Gene Expression in Pre-eclampsia. Placenta, 2004. 25(5): p. 413–421. [DOI] [PubMed] [Google Scholar]
- 29.Bainbridge SA, et al. , Effects of Reduced Gcm1 Expression on Trophoblast Morphology, Fetoplacental Vascularity, and Pregnancy Outcomes in Mice. Hypertension, 2012. 59(3): p. 732–739. [DOI] [PubMed] [Google Scholar]
- 30.Mohammadi H, et al. , HIV antiretroviral exposure in pregnancy induces detrimental placenta vascular changes that are rescued by progesterone supplementation. Sci Rep, 2018. 8(1): p. 6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruebner M, et al. , Regulation of the human endogenous retroviral Syncytin-1 and cell-cell fusion by the nuclear hormone receptors PPARgamma/RXRalpha in placentogenesis. J Cell Biochem, 2012. 113(7): p. 2383–96. [DOI] [PubMed] [Google Scholar]
- 32.Levytska K, et al. , PPAR-gamma Regulates Trophoblast Differentiation in the BeWo Cell Model. Ppar Research, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelrahman M, Sivarajah A, and Thiemermann C, Beneficial effects of PPAR-gamma ligands in ischemia-reperfusion injury, inflammation and shock. Cardiovascular Research, 2005. 65(4): p. 772–781. [DOI] [PubMed] [Google Scholar]
- 34.Kadam L, Kohan-Ghadr HR, and Drewlo S, The balancing act - PPAR-gamma’s roles at the maternal-fetal interface. Syst Biol Reprod Med, 2015. 61(2): p. 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, et al. , ANGPTL4 mediates the protective role of PPAR. activators in the pathogenesis of preeclampsia. Cell Death & Disease, 2017. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barak Y, et al. , PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell, 1999. 4(4): p. 585–95. [DOI] [PubMed] [Google Scholar]
- 37.Tache V, et al. , Hypoxia and trophoblast differentiation: a key role for PPARgamma. Stem Cells Dev, 2013. 22(21): p. 2815–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane SL, et al. , Pharmacological activation of peroxisome proliferator-activated receptor gamma (PPAR-gamma) protects against hypoxia-associated fetal growth restriction. FASEB J, 2019. 33(8): p. 8999–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Julian CG, et al. , Inhibition of peroxisome proliferator-activated receptor gamma: a potential link between chronic maternal hypoxia and impaired fetal growth. FASEB J, 2014. 28(3): p. 1268–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodie VA, et al. , Human Placental Peroxisome Proliferator-Activated Receptor δ and γ Expression in Healthy Pregnancy and in Preeclampsia and Intrauterine Growth Restriction. Journal of the Society for Gynecologic Investigation, 2005. 12(5): p. 320–329. [DOI] [PubMed] [Google Scholar]
- 41.Waite LL, Louie RE, and Taylor RN, Circulating activators of peroxisome proliferator-activated receptors are reduced in preeclamptic pregnancy. J Clin Endocrinol Metab, 2005. 90(2): p. 620–6. [DOI] [PubMed] [Google Scholar]
- 42.Holdsworth-Carson SJ, et al. , Peroxisome proliferator-activated receptors are altered in pathologies of the human placenta: Gestational diabetes mellitus, intrauterine growth restriction and preeclampsia. Placenta, 2010. 31(3): p. 222–229. [DOI] [PubMed] [Google Scholar]
- 43.Permadi W, et al. , Differences in expression of Peroxisome Proliferator-activated Receptor-gamma in early-onset preeclampsia and late-onset preeclampsia. BMC Res Notes, 2020. 13(1): p. 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy FP, et al. , Evidence Implicating Peroxisome Proliferator-Activated Receptor-gamma in the Pathogenesis of Preeclampsia. Hypertension, 2011. 58(5): p. 882–U447. [DOI] [PubMed] [Google Scholar]
- 45.ACOG Practice Bulletin No. 202: Gestational Hypertension and Preeclampsia. Obstet Gynecol, 2019. 133(1): p. e1–e25. [DOI] [PubMed] [Google Scholar]
- 46.Baczyk D, et al. , Bi-potential behaviour of cytotrophoblasts in first trimester chorionic villi. Placenta, 2006. 27(4–5): p. 367–74. [DOI] [PubMed] [Google Scholar]
- 47.Chang CW, Wakeland AK, and Parast MM, Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J Endocrinol, 2018. 236(1): p. R43–R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker J, et al. , Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol, 2013. 20(4): p. 525–31. [DOI] [PubMed] [Google Scholar]
- 49.Drewlo S, Levytska K, and Kingdom J, Revisiting the housekeeping genes of human placental development and insufficiency syndromes. Placenta, 2012. 33(11): p. 952–954. [DOI] [PubMed] [Google Scholar]
- 50.Leach RE, et al. , High throughput, cell type-specific analysis of key proteins in human endometrial biopsies of women from fertile and infertile couples. Human Reproduction, 2012. 27(3): p. 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altman DG and Bland JM, Standard deviations and standard errors. BMJ, 2005. 331(7521): p. 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knofler M and Pollheimer J, Human placental trophoblast invasion and differentiation: a particular focus on Wnt signaling. Front Genet, 2013. 4: p. 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maynard SE, et al. , Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest, 2003. 111(5): p. 649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahul S, et al. , Circulating Antiangiogenic Factors and Myocardial Dysfunction in Hypertensive Disorders of Pregnancy. Hypertension, 2016. 67(6): p. 1273–1280. [DOI] [PubMed] [Google Scholar]
- 55.Tache V, et al. , Placental expression of vascular endothelial growth factor receptor-1/soluble vascular endothelial growth factor receptor-1 correlates with severity of clinical preeclampsia and villous hypermaturity. Hum Pathol, 2011. 42(9): p. 1283–8. [DOI] [PubMed] [Google Scholar]
- 56.Rajakumar A, et al. , Transcriptionally active syncytial aggregates in the maternal circulation may contribute to circulating soluble fms-like tyrosine kinase 1 in preeclampsia. Hypertension, 2012. 59(2): p. 256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashar-Patel A, et al. , FLT1 and transcriptome-wide polyadenylation site (PAS) analysis in preeclampsia. Sci Rep, 2017. 7(1): p. 12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aouache R, et al. , Oxidative Stress in Preeclampsia and Placental Diseases. Int J Mol Sci, 2018. 19(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarthy FP, et al. , Peroxisome Proliferator-Activated Receptor-gamma as a Potential Therapeutic Target in the Treatment of Preeclampsia. Hypertension, 2011. 58(2): p. 280–U319. [DOI] [PubMed] [Google Scholar]
- 60.Brust R, et al. , A structural mechanism for directing corepressor-selective inverse agonism of PPARgamma. Nat Commun, 2018. 9(1): p. 4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee G, et al. , T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities. J Biol Chem, 2002. 277(22): p. 19649–57. [DOI] [PubMed] [Google Scholar]
- 62.Kulakovskiy IV, et al. , HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res, 2016. 44(D1): p. D116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.