Abstract
The C-X-C motif (CXC) chemokines are a family of chemotactic molecules that have been identified as potential prognostic markers and prospective therapeutic targets for many kinds of cancer types. Increasing evidence shows that CXC chemokines are associated with the progression of colorectal cancer (CRC); however, the correlations of CXC chemokines with prognostic and immune infiltrates in CRC remain to be clarified. In this study, we analyzed the mRNA expression level, prognostic data and immune infiltrates of CXC chemokines in CRC patients from the Gene Expression Profiling Interactive Analysis, Oncomine, cBioPortal and databases using GeneMANIA, STRING, DAVID 6.8, and TIMER. Our results showed that CXCL1/2/3/4/5/8/9/10/11/13/14/16 were significantly overexpressed in CRC tissues. Furthermore, expression of CXCL1/2/3/9/10/11 was associated with tumor stage in CRC. A significant association was also identified between the co-expression of CXCL16 with EGFR, KRAS and NRAS. In addition, the survival analysis suggested that high CXCL2/3/8/9/10/11/14 expression is correlated with clinical outcomes of CRC patients. Moreover, a significant association was observed between the CXCL8/9/10/11 expression and immune infiltration in colonic and rectal adenocarcinoma. Overall, CXC chemokines are not only implicated as prognostic biomarkers for CRC patients, but may also influence the immune status of CRC tissues.
Keywords: comprehensive analysis, CXC chemokines, colorectal cancer, immune infiltration
INTRODUCTION
Colorectal cancer (CRC), including colon cancer and rectal cancer, is the second most common malignancy and the third cause of cancer-related deaths worldwide [1]. In 2020, approximately 147,950 new cases of CRC were diagnosed, including an estimated 53,200 CRC-related deaths in the United States [2]. Despite significant progress and considerable efforts aimed at the development of multidisciplinary therapies, the quality of life and survival of patients with severe CRC remain unsatisfactory [3]. Furthermore, the detailed molecular mechanisms in CRC pathogenesis are still not fully understood [4]. Therefore, it is necessary to identify prognostic markers and characterize the underlying immune infiltration in CRC to efficiently improve prognosis and individualized treatments.
The C-X-C motif (CXC) chemokines are a group of approximately 50 chemotactic molecules of low-molecular-weight that are known to function as the basic regulators of the directional migration of leukocytes [5]. Chemokines are chemoattractants secreted by leukocytes, tumor cells and other cell types, such as epithelial cells, fibroblasts, and immune cells [6]. These small proteins play a crucial role in many biological processes associated with cancers, such as tumor growth, progression, metastasis, and angiogenesis, and influence therapeutic effect, and patient outcomes [4, 6]. According to the relative spacing of conserved cysteine residues in the N-terminal region of the peptide chain, chemokines have been classified into four broad groups, C, CC, CXC, and CX3C chemokines, in which the X represents any amino acid [4]. As a vital member of the chemokine family of proteins, CXC chemokines have been identified as potential prognostic markers and prospective therapeutic targets in many kinds of cancers, including breast cancer and kidney cancer [3, 7, 8].
To date, sixteen CXC chemokines have been detected in mammalian cells and named to reflect the order of their discovery (CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17) [6]. Multiple studies have indicated that CXC chemokines play vital roles in the development and progression of CRC [4, 9]. For instance, CXCL1 has been reported to be associated with CRC progression and metastasis by inducing glycolysis [10]. Additionally, recent studies have indicated that CXCL8 promotes angiogenesis and cell migration in CRC [11]. Furthermore, Lin et al. reported that CXCL14 participates in the progression of CRC and is encoded by a candidate cancer suppressor gene [12]. However, the role of CXC chemokine members in CRC tumor progression and development is highly complex. Further studies are required to investigate the role of the intricate CXC chemokine network in tumor development to provide new insights for therapeutic applications in patients with CRC. In this study, we conducted a comprehensive bioinformatics analysis of the expression and mutational activation of CXC chemokine members and their link with potential prognosis and immune infiltrates in CRC patients.
MATERIALS AND METHODS
Ethics statement
The collection of colorectal tissues was approved by the Ethics Committee of the Taizhou People’s Hospital, Suzhou, China (project no. KY 202005401), and was conducted according to the relevant guidelines. The datasets were obtained from the relevant published literature.
Oncomine analysis
The transcription levels of CXC chemokines in different cancers were downloaded from a cancer microarray database (Oncomine, https://www.oncomine.org). The differences in expression levels of CXC chemokines at the mRNA level in CRC and corresponding normal tissues were evaluated by Student’s t-test. The cutoffs for the identification of differentially expressed genes were set as P < 0.0001 and a fold change in expression ≥2.
Gene expression profiling interactive analysis (GEPIA) dataset
GEPIA, which is an interactive web server, was used to assess the RNA sequencing expression, co-expression, and survival data of 8, 587 normal samples and 9, 736 tumors from the Genotype Tissue Expression projects and The Cancer Genome Atlas (TCGA) according to the standard processing pipeline (http://gepia.cancer-pku.cn). GEPIA was used to compare the mRNA expression levels of CXC chemokines between CRC and normal samples. The correlation between CXC chemokines expression and patient prognosis in colon adenocarcinoma (COAD), rectal adenocarcinoma (READ), and colon and rectal adenocarcinoma (COAD + READ) were analyzed in terms of disease-free survival (DFS), overall survival (OS), and the tumor stage. Moreover, the relationships between CXC chemokine expression and EGFR, KRAS and NRAS of COAD, READ, and COAD + READ were evaluated.
Immunohistochemistry (IHC) analysis
All samples were obtained from CRC patients who underwent initial surgical resection in Taizhou People’s Hospital. CRC samples and adjacent noncancerous colorectal tissues were collected for IHC analysis using the following primary detection antibodies: anti-CXCL2 (Bioss, Cat#bs200208R, China), anti-CXCL3 (Bioss, Cat#bs2547R), anti-CXCL8 (PTG, Cat#27095-1-AP, China), anti-CXCL9 (PTG, Cat#22355-1-AP, China), anti-CXCL10 (PTG, Cat#10937-1-AP, China), anti-CXCL11 (PTG, Cat#10707-1-AP, China), and anti-CXCL14 (PTG, Cat#10468-1-AP, China). Immunohistochemistry analysis was used following the manufacturer’s protocol.
cBioPortal for cancer genomics analysis
The CRC datasets (TCGA, Firehose legacy) involving data from 640 cases with pathology reports were selected for further analysis of CXC chemokines using cBioPortal (https://www.cbioportal.org/). The genomic profiles included protein expression z-scores (RPPA), mutations, mRNA expression z-scores (RNA Seq V2 RSEM), and putative copy-number alterations from genomic identification of significant targets in cancer (GISTIC). Moreover, co-expression was assessed according to cBioPortal and using the R package “ggcorrplot”.
GeneMANIA and STRING
GeneMANIA, which is a web-based dataset, was used to analyze the internal associations of gene sets (https://genemania.org/). STRING is a database of predicted protein-protein interactions, including direct (physical) and indirect (functional) associations (https://string-db.org/). Therefore, the interactions of CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 were analyzed at the gene and protein expression levels by GeneMANIA and STRING.
Database for annotation, visualization and integrated discovery (DAVID) analysis
As a bioinformatics resource, DAVID (version 6.8) is an open database focusing on gene functional annotations and enrichment pathway analysis (https://david.ncifcrf.gov/home.jsp). In our study, DAVID was used to annotate the cellular component (CC), gene ontology (GO) terms for biological process, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and molecular function categories (MF) of CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17.
KEGG analysis
The KEGG (https://www.kegg.jp/) is a database resource for understanding the high-level functions of biological systems. The signaling pathway enrichments of CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 were identified and adjusted according to KEGG and Cytoscape.
Tumor IMmune Estimation Resource (TIMER)
TIMER2.0 is a comprehensive resource for the systematic analysis of immune infiltrates across different cancer types from TCGA (http://timer.cistrome.org/) [13, 14]. We identified the association between CXCL2/3/8/9/10/11/14 expression in COAD and READ and the abundances of six immune infiltrates, including CD4+ T cells, CD8+ T cells, B cells, macrophages, dendritic cells (DCs), and neutrophils. TIMER2.0 generates a heatmap table of the Spearman’s correlations between the expression of CXCL2/3/8/9/10/11/14 and the abundance of immune cell type as well as its subtypes across COAD and READ. Moreover, correlations between genetic markers of tumor-infiltrating immune cells and CXCL2/3/8/9/10/11/14 expression were evaluated using correlation modules (Gene_Corr). According to TIMER2.0, the positive correlation is defined as P < 0.05, rho > 0; the negative correlation is defined as P < 0.05, rho < 0; not significant is defined as P ≥ 0.05.
RESULTS
Transcription levels of CXC chemokines in patients with CRC
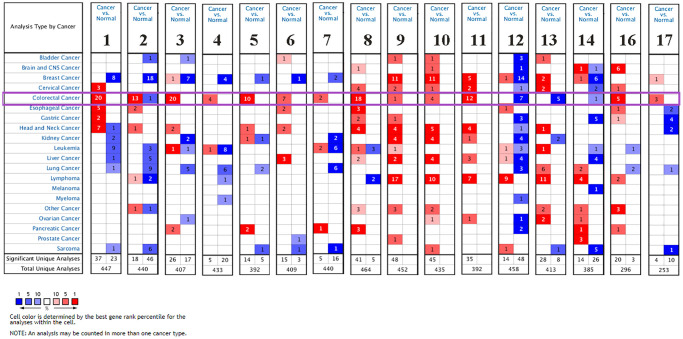
The transcription levels of CXC chemokine family members (excluding CXCL15) were assessed in human cancers. As indicated as Table 1 and Figure 1, we used Oncomine to compare the mRNA expression levels of CXC chemokines in CRC with those in normal samples. This analysis showed that the transcription levels of CXCL1/2/3/4/5/6/7/8/9/10/11/16/17 were significantly upregulated in CRC patients. However, the mRNA expression levels of CXCL12/13/14 were significantly downregulated in CRC patients.
Table 1. The significant changes of CXCL expression in transcription level between different types of CRC and normal colorectal tissues (Oncomine database).
| Type of colorectal cancer | Normal | Fold change | P Value | t Test | Dataset | |
| CXCL1 | Colon adenocarcinoma | Colon | 12.328 | 7.62E-9 | 7.379 | Notterman colon |
| Colon carcinoma | Colon | 5.695 | 2.71E-5 | 7.410 | Zou colon | |
| Colorectal adenoma epithelia | Colorectal | 2.641 | 4.71E-8 | 6.147 | Gaspar colon | |
| Colon carcinoma epithelia | Colon | 13.052 | 8.47E-12 | 21.652 | Skrzypczak colorectal 2 | |
| Colon mucinous adenocarcinoma | Colon | 9.298 | 2.14E-13 | 10.386 | TCGA colorectal | |
| CXCL2 | Colon adenoma | Colon | 12.924 | 7.11E-10 | 15.037 | Skrzypczak colorectal 2 |
| Colorectal carcinoma | Colorectal | 2.821 | 2.82E-7 | 6.75 | Grauden colon | |
| Rectal adenocarcinoma | Rectal | 7.368 | 2.82E-33 | 16.274 | Gaedcke colorectal | |
| Colorectal carcinoma | Colorectal | 10.114 | 9.83E-12 | 9.634 | Skrzypczak colorectal | |
| Rectal adenocarcinoma | Rectal | 8.625 | 1.62E-5 | 7.632 | Kaiser colon | |
| CXCL3 | Colorectal adenoma epithelia | Colorectal | 2.117 | 5.34E-8 | 6.255 | Gaspar colon |
| Colon carcinoma epithelia | Colon | 9.918 | 1.89E-9 | 13.837 | Skrzypczak colorectal 2 | |
| Rectal adenocarcinoma | Rectal | 10.56 | 3.02E-6 | 8.107 | Kaiser colon | |
| Colon adenocarcinoma | Colon | 4.193 | 4.28E-16 | 9.795 | Ki colon | |
| Colorectal carcinoma | Colorectal | 11.017 | 5.55E-12 | 10.192 | Skrzypczak colorectal | |
| CXCL4 | Colon carcinoma | Colon | 5.228 | 5.84E-10 | 15.615 | Skrzypczak colorectal 2 |
| Colorectal adenocarcinoma | Colorectal | 2.704 | 9.38E-11 | 7.697 | Skrzypczak colorectal | |
| Colon adenoma | Colon | 5.012 | 1.96E-11 | 8.964 | Sabates-Bellver colon | |
| CXCL5 | Rectosigmoid adenocarcinoma | rectosigmoid | 11.693 | 8.34E-14 | 16.245 | TCGA colorectal |
| Colorectal carcinoma | Colorectal | 11.504 | 2.50E-11 | 8.148 | Skrzypczak colorectal | |
| Colon adenocarcinoma | Colon | 3.682 | 1.19E-10 | 8.196 | Kaiser colon | |
| Rectal adenocarcinoma | Rectal | 12.096 | 8.89E-23 | 12.435 | Gaedcke colorectal | |
| Colon adenoma | Colon | 4.184 | 5.17E-5 | 4.637 | Sabates-Bellver colon | |
| CXCL6 | Colorectal carcinoma | Colorectal | 4.156 | 3.39E-10 | 7.401 | Skrzypczak colorectal |
| Colon adenocarcinoma | Colon | 2.428 | 7.22E-9 | 6.53 | Ki colon | |
| Colon carcinoma epithelia | Colon | 6.937 | 5.37E-7 | 16.403 | Skrzypczak colorectal 2 | |
| Rectal adenocarcinoma | Rectal | 3.095 | 5.73E-17 | 10.672 | Gaedcke colorectal | |
| Colon mucinous adenocarcinoma | Colon | 5.107 | 4.92E-8 | 7.029 | TCGA colorectal | |
| CXCL7 | Colorectal carcinoma | Colorectal | 14.815 | 4.41E-10 | 7.328 | Hong colorectal |
| Colorectal carcinoma | Colorectal | 6.016 | 1.49E-6 | 5.489 | Skrzypczak colorectal | |
| CXCL8 | Colon mucinous adenocarcinoma | Colon | 17.688 | 6.96E-10 | 13.138 | Kaiser colon |
| Colon adenocarcinoma | Colon | 3.617 | 2.66E-5 | 4.518 | Alon colon | |
| Colon adenocarcinoma | Colon | 4.661 | 2.99E-5 | 4.584 | Notterman colon | |
| Colon adenoma | Colon | 6.531 | 4.39E-9 | 14.898 | Skrzypczak colorectal 2 | |
| Rectosigmoid adenocarcinoma | rectosigmoid | 18.411 | 2.83E-9 | 8.982 | TCGA colorectal | |
| CXCL9 | Colon carcinoma epithelia | Colon | 7.004 | 2.28E-7 | 9.52 | Skrzypczak colorectal 2 |
| CXCL10 | Colorectal carcinoma | Colorectal | 6.673 | 3.40E-10 | 8.062 | Skrzypczak colorectal |
| Colon adenocarcinoma | Colon | 2.574 | 2.13E-8 | 6.061 | Ki colon | |
| Colon carcinoma epithelia | Colon | 17.521 | 2.44E-7 | 9.735 | Skrzypczak colorectal 2 | |
| Rectal adenocarcinoma | Rectal | 3.816 | 1.08E-17 | 10.009 | Gaedcke colorectal | |
| Colon adenocarcinoma | Colon | 2.694 | 0.012 | 2.39 | Notterman colon | |
| CXCL11 | Colon carcinoma epithelia | Colon | 29.164 | 9.03E-13 | 30.893 | Skrzypczak colorectal 2 |
| Colorectal carcinoma | Colorectal | 10.483 | 9.20E-14 | 9.523 | Skrzypczak colorectal | |
| Colorectal carcinoma | Colorectal | 2.112 | 2.00E-5 | 4.957 | Graudens colon | |
| Rectal adenocarcinoma | Rectal | 12.5 | 3.15E-7 | 8.481 | Sabates-Bellver colon | |
| Colorectal carcinoma | Colorectal | 5.68 | 1.21E-8 | 7.927 | Hong colorectal | |
| CXCL12 | Colon carcinoma epithelia | Colon | 1.248 | 0.007 | 3.131 | Skrzypczak colorectal 2 |
| CXCL14 | Rectosigmoid adenocarcinoma | rectosigmoid | 2.585 | 0.021 | 3.666 | TCGA colorectal |
| CXCL16 | Colorectal carcinoma | Colorectal | 2.636 | 2.08E-18 | 13.425 | Hong colorectal |
| Colon adenoma | Colon | 2.382 | 1.42E-13 | 10.197 | Sabates-Bellver colon | |
| Colon carcinoma epithelia | Colon | 3.244 | 1.64E-5 | 6.191 | Skrzypczak colorectal 2 | |
| Colon mucinous adenocarcinoma | Colon | 2.443 | 1.04E-5 | 6.809 | Kaiser colon | |
| Colorectal adenocarcinoma | Colorectal | 1.822 | 6.21E-11 | 8.45 | Skrzypczak colorectal | |
| CXCL17 | Rectosigmoid adenocarcinoma | rectosigmoid | 2.664 | 1.25E-5 | 7.31 | TCGA colorectal |
| Colorectal carcinoma | Colorectal | 1.301 | 7.56E-5 | 4.067 | Skrzypczak colorectal | |
| Rectosigmoid adenocarcinoma | rectosigmoid | 1.237 | 0.013 | 2.552 | Kaiser colon | |
| Rectal adenocarcinoma | Rectal | 1.35 | 2.72E-5 | 4.316 | Gaedcke colorectal | |
| Colon carcinoma epithelia | Colon | 1.248 | 0.032 | 2.242 | Skrzypczak colorectal 2 |
Figure 1.
The mRNA expression levels of CXC chemokines in different types of cancers (Oncomine). The figure presents the numbers of datasets with statistically significant mRNA over-expression (red) or downregulated expression (blue) of CXC chemokines.
The transcription levels of CXCL1 in colorectal carcinoma were higher than those in colorectal tissues in all 10 datasets. In the Notterman colon dataset, the mRNA expression levels of CXCL1 in colon adenocarcinoma were increased (fold change = 12.328, P = 7.62E-9). In the Skrzypczak colorectal 2 dataset, CXCL2 was overexpressed at the mRNA level in colon adenoma (fold change = 12.924, P = 7.11E-10), colon epithelial carcinoma (fold change = 7.451, P = 2.29E-10), colon epithelial adenoma (fold change = 10.404, P = 3.08E-7), and colon carcinoma (fold change = 8.527, P = 6.09E-9). In the Gaspar colon dataset, CXCL3 was overexpressed in colon epithelial adenoma (fold change = 2.117, P = 5.34E-8) compared with that in colon tissues. In the Skrzypczak colorectal 2 dataset, CXCL4 was overexpressed in colon carcinoma (fold change = 5.228, P = 5.84E-10) and colon epithelial adenoma (fold change = 3.165, P = 8.43E-7). The mRNA levels of CXCL5 in rectosigmoid adenocarcinoma (fold change = 11.693, P = 8.34E-14), colon adenocarcinoma (fold change = 2.739, P = 3.60E-22), colon mucinous adenocarcinoma (fold change = 23.08, P = 9.97E-11), and rectal adenocarcinoma (fold change = 2.538, P = 3.20E-13) were significantly higher in TCGA dataset. In addition, analysis of the Skrzypczak colorectal dataset indicated the mRNA expression levels of CXCL6 were significantly unregulated in patients with colorectal carcinoma (fold change = 4.156, P = 3.39E-10).
A similar trend was found in the Hong and Skrzypczak colorectal datasets, with mRNA expression levels of CXCL7 found to be significantly higher in CRC (fold change = 14.815, P = 4.41E-10 and fold change = 6.016, P = 1.49E-6, respectively). The results from nine datasets indicated that the mRNA expression levels of CXCL8 were significantly upregulated in patients with colorectal carcinoma. In the Skrzypczak colorectal 2 dataset, CXCL9 was overexpressed in colon epithelial carcinoma (fold change = 7.004, P = 2.28E-7) compared with that in non-cancerous colon tissues. Furthermore, in the Skrzypczak colorectal 2 dataset, the mRNA expression levels of CXCL10 were upregulated in colon carcinoma (fold change = 6.673, P = 3.40E-10). In the Skrzypczak colorectal 2 dataset, the mRNA levels of CXCL11 were increased in colon epithelial carcinoma (fold change = 29.164, P = 9.03E-13), colon carcinoma (fold change = 32.137, P = 3.09E-9), colon adenoma (fold change = 4.306, P = 6.60E-7), and colon epithelial adenoma (fold change = 5.543, P = 2.83E-5). In the Hong colorectal dataset, CXCL16 was upregulated in colon carcinoma (fold change = 2.636, P = 2.08E-18). In addition, the results from TCGA colorectal dataset indicated that the mRNA expression levels of CXCL17 were significantly higher in rectosigmoid adenocarcinoma (fold change = 2.664, P = 1.25E-5), rectal mucinous adenocarcinoma (fold change = 3.492, P = 1.60E-5), and colon mucinous adenocarcinoma (fold change = 4.178, P = 2.10E-7).
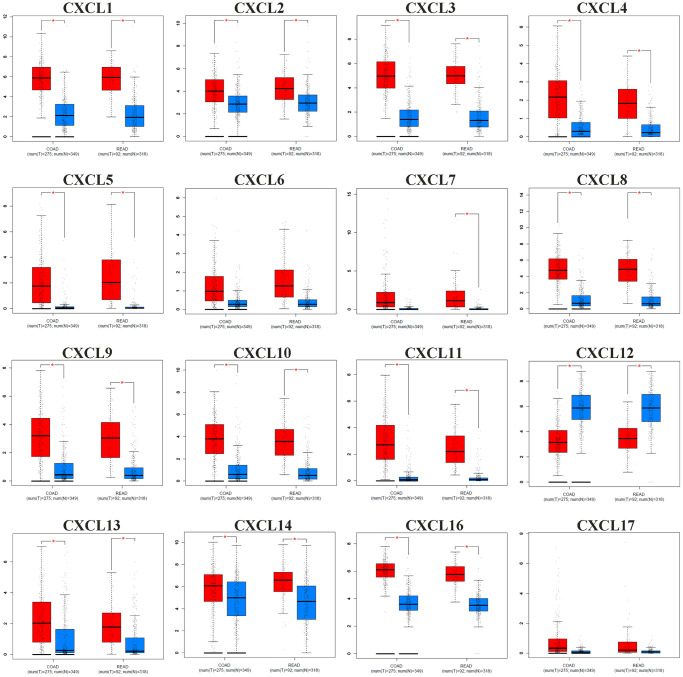
Expression levels and co-expression analysis of CXC chemokines in patients with CRC
The comparative analysis of the CXC mRNA expression in normal vs. COAD and READ tissues was conducted according to the information from the GEPIA database. As shown as Figure 2, the mRNA expression levels of CXCL1/2/3/4/5/8/9/10/11/13/14/16 were significantly upregulated in COAD and READ tissues vs. normal tissue, while CXCL12 expression was downregulated. In addition, the results shown in Supplementary Table 1 revealed significant associations in COAD + READ between tumor stage and expression of CXCL1 (P = 0.0345), CXCL2 (P = 0.0156), CXCL3 (P = 0.0344), CXCL9 (P = 0.0122), CXCL10 (P = 0.0108), and CXCL11 (P = 0.0227) expression.
Figure 2.
The mRNA expression pattern of CXC chemokines (CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17) from GEPIA between CRC tissues (red) and normal tissues (blue). The P-value was set at 0.05, * indicate that the results are statistically significant.
We also analyzed potential correlations of CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 co-expression with EGFR, KRAS and NRAS according to the mRNA expression levels using the GEPIA database for CRC. The results shown in Table 2 indicated that EGFR expression was significantly correlated with the expression of CXCL3, CXCL12 and CXCL16 in COAD + READ. In addition, a significant association was identified between the expression of KRAS and CXCL16 in COAD + READ. Furthermore, NRAS expression was significantly correlated with the expression of CXCL14, CXCL16 and CXCL17 in COAD + READ.
Table 2. The co-expression of different expressed CXC chemokines with EGFR, KRAS and NRAS of CRC patients (GEPIA).
| P-value (EGFR) | P-value (KRAS) | P-value (NRAS) | |||||||
| COAD + READ | COAD | READ | COAD + READ | COAD | READ | COAD + READ | COAD | READ | |
| CXCL1 | 0.52 | 0.55 | 0.88 | 0.3 | 0.87 | 0.025 | 0.54 | 0.98 | 0.063 |
| CXCL2 | 0.099 | 0.11 | 0.78 | 0.48 | 0.45 | 0.062 | 0.41 | 0.57 | 0.49 |
| CXCL3 | 0.011 | 0.11 | 0.96 | 0.48 | 0.48 | 0.012 | 0.97 | 0.6 | 0.15 |
| CXCL4 | 0.24 | 0.21 | 0.65 | 0.62 | 0.59 | 0.7 | 0.5 | 0.68 | 0.83 |
| CXCL5 | 0.42 | 0.91 | 0.041 | 0.94 | 0.74 | 0.89 | 0.81 | 0.84 | 0.78 |
| CXCL6 | 0.19 | 0.31 | 0.28 | 0.3 | 0.29 | 0.82 | 0.061 | 0.037 | 0.99 |
| CXCL7 | 0.35 | 0.4 | 0.73 | 0.49 | 0.48 | 0.64 | 0.21 | 0.26 | 0.54 |
| CXCL8 | 0.79 | 0.85 | 0.39 | 0.081 | 0.46 | 0.21 | 0.15 | 0.12 | 0.65 |
| CXCL9 | 0.47 | 0.81 | 0.032 | 0.79 | 0.64 | 0.82 | 0.68 | 0.83 | 0.27 |
| CXCL10 | 0.87 | 0.73 | 0.25 | 0.78 | 0.7 | 0.76 | 0.72 | 0.85 | 0.27 |
| CXCL11 | 0.65 | 0.52 | 0.26 | 1 | 0.79 | 0.96 | 0.82 | 0.79 | 0.53 |
| CXCL12 | 0.0045 | 0.019 | 0.068 | 0.4 | 0.21 | 0.98 | 0.13 | 0.067 | 0.78 |
| CXCL13 | 0.6 | 0.35 | 0.046 | 0.74 | 0.83 | 0.42 | 0.098 | 0.088 | 0.79 |
| CXCL14 | 0.28 | 0.29 | 0.79 | 0.51 | 0.2 | 0.92 | 0.0028 | 0.002 | 0.62 |
| CXCL16 | 1.10E-10 | 4.70E-07 | 1.50E-07 | 0.0086 | 0.014 | 0.055 | 0.0028 | 0.027 | 0.0047 |
| CXCL17 | 0.52 | 0.94 | 0.2 | 0.96 | 0.64 | 0.77 | 0.0047 | 0.78 | 0.077 |
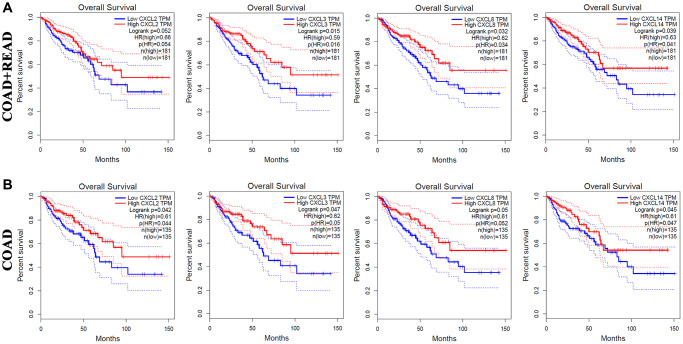
Association of CXC chemokine expression with the prognosis of CRC
Using GEPIA, we also analyzed the correlation between CXC chemokine expression and the OS and DFS of patients with CRC (Supplementary Table 2). The OS curves shown in Figure 3A demonstrated that high mRNA expression levels of CXCL2 (P = 0.052), CXCL3 (P = 0.015), CXCL8 (P = 0.032), and CXCL14 (P = 0.039) were correlated with longer OS in 181 CRC patients (COAD + READ). As illustrated as Figure 3B, increased mRNA expression levels of CXCL2 (P = 0.042), CXCL3 (P = 0.047), CXCL8 (P = 0.05), and CXCL14 (P = 0.045) were significantly correlated with longer OS in 145 COAD patients. These results indicated that CXCL2/3/8/14 are independent risk factor for OS among CRC patients.
Figure 3.
The prognostic value of different expressed CXC chemokines in CRC patients in OS (GEPIA). The OS curve of CXCL2/3/8/14 in (A) COAD + READ and (B) COAD.
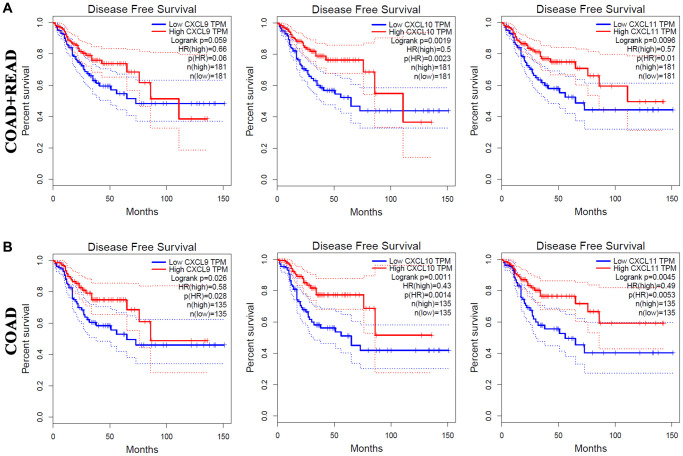
The correlations of CXC chemokine expression with the DFS of CRC patients were also investigated (Figure 4.) As shown in in Figure 4A, increased mRNA expression levels of CXCL9 (P = 0.059), CXCL10 (P = 0.0019), and CXCL11 (P = 0.0096) were associated with longer DFS in COAD + READ patients. Moreover, high mRNA levels of CXCL9 (P = 0.026), CXCL10 (P = 0.0011), and CXCL11 (P = 0.0045) were significantly associated with longer DFS in COAD patients (Figure 4B).
Figure 4.
The prognostic value of different expressed CXC chemokines in CRC patients in DFS (GEPIA). The DFS curve of CXCL9/10/11 in (A) COAD + READ and (B) COAD.
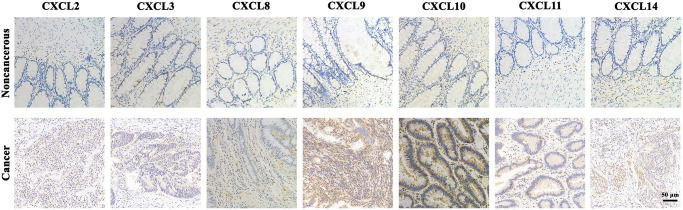
IHC analysis
As shown in Figure 5, IHC analysis of CXCL2/3/8/9/10/11/14 proteins revealed overexpression of CXCL2/3/8/9/10/11/14 proteins in CRC tissues compared with the adjacent noncancerous tissues.
Figure 5.
The expression protein of CXCL2/3/8/9/10/11/14 in CRC tissues and noncancerous tissues (IHC).
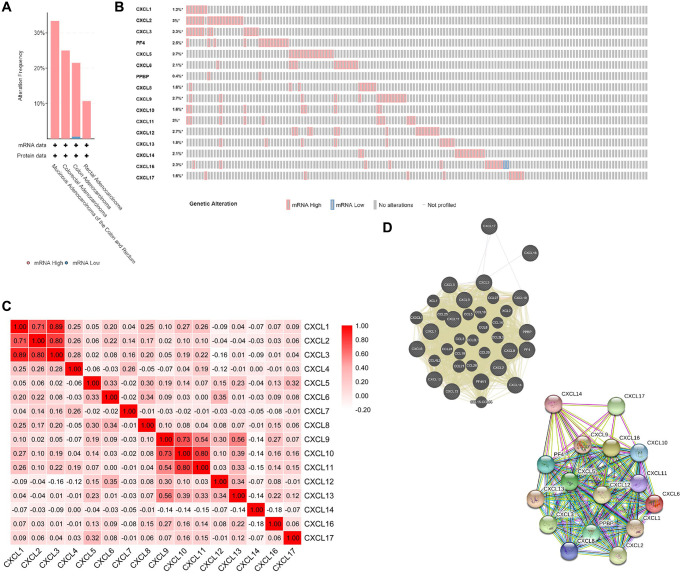
Alteration, co-expression and interaction analyses of CXC chemokines at the gene and protein levels in patients with CRC
We investigated the alterations and correlations of CXC chemokine expression at the gene and protein levels using the cBioPortal database for CRC (TCGA, Firehose legacy). CXC chemokine expression was were altered in 131 of 640 (21%) of CRC samples. As shown as in Figure 6A and 6B, we observed alterations in the expression of CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 in 1.2%, 3%, 2.3%, 2.5%, 2.7%, 2.1%, 0.4%, 1.6%, 2.7%, 1.6%, 2%, 2.7%, 1.8%, 2.1%, 2.3%, and 1.6% of the queried CRC samples, respectively. We also analyzed the co-expression patterns among the chemokines (CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17) at the mRNA level using the cBioPortal database for CRC. The results shown in Figure 6C indicated a low to moderate correlation in the expression patterns among CXCL4/5/6/7/8/11/13/14/15/16/17; a moderate to high correlation in the expression of CXCL9 with CXCL11, and CXCL9 with CXCL12; and a high correlation in the expression of CXCL1 with CXCL2, CXCL1 with CXCL3, CXCL2 with CXCL3, CXCL9 with CXCL10, and CXCL10 with CXCL11.
Figure 6.
CXC chemokines gene expression and mutation analysis in CRC (cBioPortal). (A) Summary of alterations in CXC chemokines (cBioPortal). (B) OncoPrint visual summary of alteration on a query of CXC chemokines (cBioPortal). (C) Correction between different CXC chemokines in CRC (cBioPortal). (D) Gene-gene interaction network (GeneMANIA) and protein-protein interaction network (STRING) among CXC chemokines.
Using GeneMANIA, we identified the relationships among CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 at the gene expression level. As shown as in Figure 6D, several relationships were identified among CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17, including co-localization, co-expression, and genetic interactions. In addition, we analyzed the interactions among CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 at the protein expression level using STRING. In total, 111 edges and 16 nodes were identified in resulting the network.
Cellular functions and pathways of CXC chemokines in patients with CRC
As shown in Supplementary Figure 1, the functions of CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 were predicted by analyzing the GO terms and KEGG pathways using DAVID. The GO enrichment terms were classified into three aspects of function: CC (3 items), MF (5 items), BP (10 items), and KEGG pathway (9 items). The CC analysis demonstrated a significant enrichment of genes related to the extracellular space, external side of plasma membrane, and extracellular region (Supplementary Figure 1A). The MF analysis revealed enrichment in chemokine activity, CXCR3 chemokine receptor binding, heparin binding, CXCR chemokine receptor binding, and receptor binding (Supplementary Figure 1B). In addition, BP was associated with cell-cell signaling, chemotaxis, regulation of cell proliferation, positive regulation of leukocyte chemotaxis, immune response, chemokine-mediated signaling pathway, inflammatory response, response to lipopolysaccharide, cell chemotaxis, and G-protein coupled receptor signaling pathway (Supplementary Figure 1C). As shown as Supplementary Figure 1D, we also found significant enrichment in KEGG pathways, including chemokine signaling pathway (Supplementary Figure 2A), cytokine-cytokine receptor interaction (Supplementary Figure 2B), legionellosis, Salmonella infection, toll-like receptor signaling pathway, NOD-like receptor signaling pathway, rheumatoid arthritis, pertussis, and TNF signaling pathway.
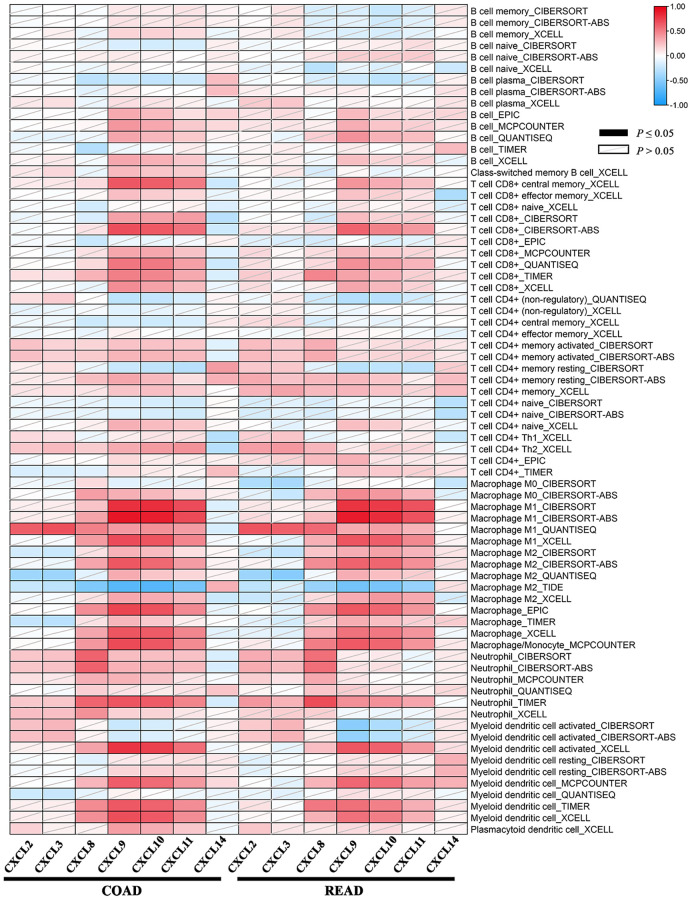
Correlation analysis between CXC chemokine expression and immune infiltration levels in CRC
Tumor-infiltrating lymphocytes are used to analyze the sentinel lymph node status and survival in cancers. According to TIMER2.0, we observed the correlation between the CXCL2/3/8/9/10/11/14 expression and the abundance of immune cell type as well as its subtypes estimated by all six algorithms across COAD and READ. As shown in Figure 7, the heatmap table represented the purity-adjusted partial spearman’s rho values as the degree of correlation. Moreover, a significant association was observed between the CXCL8/9/10/11 expression and immune infiltration in COAD and READ.
Figure 7.
The association between the CXC chemokines expression and immune infiltration level of multiple immune cells types estimated by all six algorithms in ad heatmap table across COAD and READ. The red indicates a statistically significant positive association (P ≤ 0.05, rho > 0), and the blue indicates a statistically significant negative association (P ≤ 0.05, rho < 0). White denotes a non-significant result (P > 0.05).
Correlation analysis between CXC chemokine expression and immune markers
To further investigate the association between the expression of CXCL2/3/8/9/10/11/14 and the diverse immune cell infiltration in CRC patients, we analyzed the relationship between CXC chemokines and sets of markers of various immune cells based on TIMER2.0, including tumor-associated macrophages (TAMs), neutrophils, DCs, and T helper 1 (Th1), Th2 and regulatory T (Treg) cells (Figure 8). Specifically, the CXCL 8/9/10/11 expression in COAD and READ was correlated with high levels of infiltration by TAMs, DCs, neutrophils, Th1, Th2 and Tregs. In addition, the results shown in Figure 8 confirmed the role of CXCL2/3/8/9/10/11/14 in the colorectal adenocarcinoma microenvironment.
Figure 8.
The association between the CXC chemokines expression and immune markers in ad heatmap table across COAD and READ. The red indicates a statistically significant positive association (P ≤ 0.05, rho > 0), and the blue indicates a statistically significant negative association (P ≤ 0.05, rho < 0). White denotes a non-significant result (P > 0.05).
DISCUSSION
CXC chemokines have been reported to function as inflammatory mediators and drivers of leukocytes. Although the crucial role of CXC chemokines in tumor cell proliferation, tumorigenesis, tumor metastasis, and prognosis of several cancers has been partially elucidated, the biological functions, and prognostic value of these molecules in CRC remain to be fully characterized [3]. We used an integrative bioinformatics approach to investigate the mRNA expression, prognostic values (OS and DFS), biological functions, and correlations with immune cells of different CXC chemokines in CRC.
In the Oncomine database, we found that the expression levels of CXCL1/2/3/4/5/6/7/8/9/10/11/16/17 were significantly upregulated while that of CXCL12/13/14 were downregulated in CRC patients. These results were majorly consistent with our findings from the GEPIA database, which indicated that the CXCL1/2/3/4/5/8/9/10/11/13/14/16 expression was significantly upregulated in COAD and READ tissues relative to that in the normal tissue, while the CXCL12 expression was downregulated. Our results were also consistent with those reported previously. For example, Zhuo et al. reported that the higher CXCL1 expression correlated with cancer progression and metastasis in CRC patients [15]. Mechanistically, the knockdown of CXCL1 could reduce the levels of some glycolytic enzymes (HK2, GLUT1, and LDHA). In addition, Doll et al. reported that the expression of CXCL2, CXCL3, and CXCL8 in the colon cancer tissues was significantly higher than that in the normal tissues [16]. Zhao et al. also demonstrated that CXCL5 was upregulated in the tumor tissues, which was associated with poor prognosis and advanced tumor stage in CRC patients [17]. Moreover, Zhang et al. discovered that CXCL5 was highly expressed in CRC, and that its overexpression could reverse the suppressive effect of miR-363-3p on CRC progression [18]. Overall, CXCL1/2/3/4/5/8/9/10/11/13/14/16 are highly expressed in CRC patients, thereby serving as an oncogene in CRC in addition to their association with prognosis.
Several past studies have reported a correlation between CXC chemokines expression and the prognosis of CXC patients. For example, Xiong et al. noted that the high expression of CXCL3 was closely associated with poor OS in CRC patients from the TCGA dataset [19]. Survival analysis conducted on GEO cohorts and Guangxi Medical University revealed that the high CXCL3 expression levels were associated with a shorter OS in colon cancer patients [20]. However, Doll et al. recorded no significant correlation between the CXCL3 expression levels and CRC survival [16]. Wu et al. also reported that the higher of CXCL9 mRNA and protein expression was significantly associated with OS of CRC patients [21]. Zeng et al. reported that the elevated CXCL14 expression in the CRC tissues from stage III/IV patients was associated with worse OS, thus representing a potential therapeutic target in CRC patients after curative resection [22]. However, other research groups have indicated that high levels of CXCL14 are correlated with a good prognosis [12]. Matsushita et al. reported that the high levels of preoperative serum levels of soluble CXCL16 in CRC patients were associated with a poor prognosis [23]. Due to the different sources of CRC patients, different studies recorded varied expression levels of CXC chemokines within the CRC tissues. In the present study, we analyzed the correlation between the CXC chemokine expression and the OS and DFS of patients with CRC using GEPIA. The results demonstrated that the high mRNA expression levels of CXCL2/3/8/14 were correlated with longer OS in CRC patients. In addition, the increased mRNA expression levels of CXCL9/10/11 were associated with longer DFS in CRC patients. Moreover, the expression of CXCL1/2/3/9/10/11 was associated with the tumor stage in CRC. A significant association was also identified for the co-expression of CXCL16 with EGFR, KRAS and NRAS. These results together suggest that CXC chemokines act as potential biomarkers for predicting CRC prognosis. Nevertheless, these results should be investigated in future studies using in vitro and in vivo experimental setting for validation.
The mechanisms underlying the role of CXC chemokines on CRC remain complex. For instance, Chen et al. found that metastasis suppressor 1 (MTSS1) may play an important suppressive role in CRC metastasis and that the underlying mechanisms may involve the downregulation of the CXCR4/CXCL12 signaling axis [24]. Zhang et al. discovered that CXCL5 was highly expressed in CRC, and that circCTNNA1 could act as a ceRNA for miR-363-3p to facilitate the progression of CRC through the promotion of the CXCL5 expression [18]. Zhao et al. found that the overexpression of CXCL5 could induce the epithelial-mesenchymal transition (EMT) to enhance the invasion and migration of CRC cells by the activation of the AKT/GSK3β/β-catenin and the ERK/Elk-1/Snail pathways in a CXCR2-dependent manner [17]. Xia et al. indicated that cisatracurium could suppress the viability, metastasis, and tumor growth of CRC by regulating the CXCR4/let-7a-5p axis through the inhibition of the TGF-β/SMAD2/3 signaling pathway [25]. GO and KEGG enrichment analyses were conducted for the CXC chemokines to evaluate their biological functions of them. In line with previous results, we found that the most-enriched GO terms were the chemokine activity, CXCR3 chemokine receptor binding, CXCR chemokine receptor binding, chemotaxis, regulation of cell proliferation, positive regulation of leukocyte chemotaxis, immune response, chemokine-mediated signaling pathway, inflammatory response, cell chemotaxis, and G-protein coupled receptor signaling pathway. The most commonly enriched KEGG pathways were the chemokine signaling pathway, cytokine-cytokine receptor interaction, toll-like receptor signaling pathway, NOD-like receptor signaling pathway, rheumatoid arthritis, pertussis, and the TNF signaling pathway. Therefore, further investigations are warranted to comprehend the biological functions of CXC chemokine.
Moreover, we identified a significant association between the CXCL2/3/8/9/10/11/14 expression and immune infiltration in CRC using TIMER2.0. CXCL8 plays an important role in inflammatory responses, leukocyte chemotaxis, and infectious diseases [11, 26, 27]. Li et al. reported that targeting the CXCL8-CXCR1/CXCR2 axis can act as a key factor in mediating the antitumor effects on CRC by impeding DC activation or recruitment [28]. In line with the results of some previous studies, we demonstrated that the CXCL8 expression was upregulated. Moreover, the CXCL8 expression level in COAD and READ was significantly and positively associated with infiltration by neutrophils, macrophage, and DCs. CXCL9/10/11 exerted their biological effects related to EMT via CXCR3, which was upregulated in the invasive front of the CRC tumor tissues [29]. CXCL9/10/11 and CXCR3 play pivotal roles in the activation, differentiation, and effector T cell function. CXCR3 is important for CD8 effectors, CD4 Th1 cells, memory cells, and for the functioning of natural killer T cells and natural killer in the CRC tissues. Our study also revealed that CXCL9/10/11 was overexpressed, and positively associated with PFS of CRC patients. More importantly, infiltration by immune cells, including CD8+ T cells, neutrophils, macrophages, and DCs, was found to be associated with the CXCL9/10/11 expression. Thus, these results together indicate that CXCLs are closely related to the immune infiltration of CRC, thereby providing new concepts for immunotherapy.
CXC chemokines not only serve as prognostic biomarkers and participate in the regulation of the tumor microenvironment, but also represent the potential therapeutic targets for CRC patients. It has been reported that the activation of the CXCR1/CXCR2 or CXCR4/CXCR7 pathways is associated with tumor aggressiveness and poor prognosis. Therefore, specific inhibition of these receptors can be considered as a possible therapeutic strategy [30]. For example, Wang et al. found that prostaglandin E2 (PGE2) induced of CXCL1 expression, thus implicating CXCL1 inhibitors as potential anti-angiogenic agents in the treatment of CRC [31]. Liu et al. discovered that the LPS-induced CXCR4 expression and EMT activation via the NF-κB signaling pathway promoted CRC progression [32]. Liao et al. indicated that the anti-PD-1 resistance of KRAS-expressing tumors could be overcome to some extent by inhibiting the CXCR2 expression [33]. Ning et al. revealed that CXCL8 regulated OXA-resistance of CRC cell lines by binding to CXCR2 and activating the AKT/MAPK/NF-κB signaling pathway [11]. Jamieson et al. reported that CXCR2 antagonists exert therapeutic effects in a mouse model of colon cancer [34]. Murakami et al. found that targeting CXCR3 and CXCR4 inhibited CRC metastasis to the liver and lung [35]. Moreover, Rupertus et al. indicated that CXCL11 and CXCL12 may play important roles in tumor angiogenesis, and blocking their expression inhibited tumor angiogenesis and growth [36]. In addition, the CXCL4/9/10/11 expression levels were closely associated with chemotherapy and radiation therapy in CRC patients [37–39]. However, the role of other CXC chemokines, such as CXCL16/17, remains unclear. Overall, although the studies on CXC chemokines as therapeutic targets in CRC are presently preliminary, with ensuing researches, their roles in the treatment of CRC are expected to become increasingly clear, thereby improving CRC patient survival by targeting CXC chemokines.
CONCLUSION
In this study, we conducted a systematic analysis of the expression, prognostic value, and signaling pathways of CXC chemokines, to provide a comprehensive understanding of the molecular biological properties of CRC. The mRNA expression levels of CXCL1/2/3/4/5/8/9/10/11/13/14/16 were significantly elevated in CRC tissues and we observed a significant correlation between the expression of CXCL1/2/3/9/10/11 and the tumor stage in CRC. Moreover, we also identified a significant association between the co-expression of CXCL16 with EGFR, KRAS and NRAS. Our findings suggest that the high CXCL2/3/8/9/10/11/14 expression is correlated with the clinical outcomes CRC patients. Interestingly, a significant correlation was identified between CXCL8/9/10/11 expression and tumor infiltration by immune cell types. Overall, CXC chemokines are not only valuable prognostic markers for CRC patients, but may also influence immune status of CRC tissues.
Supplementary Materials
AUTHOR CONTRIBUTIONS: Qing Guo, Junxing Huang, and Xinchen Sun conceived this study; Feng Sheng, Yirong Xu, and Liu Jiao extracted the data; Ling Gao and Ju Yang interpreted the data; Xi Yang and Yuanfeng Wei analyzed the data and wrote the manuscript. All authors reviewed the manuscript.
CONFLICTS OF INTEREST: The authors declare no conflicts of interest related to this study.
FUNDING: This research was supported by Innovative Team of Jiangsu Province (CXTDA2017042), Taizhou science and technology support plan (Social Development) project (TS201903), China Postdoctoral Science Foundation (2019M663505), and Postdoctoral Interdisciplinary Innovation Foundation, Sichuan University (No. 0040204153243).
This corresponding author has a verified history of publications using a personal email address for correspondence
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69:7–34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020; 70:145–64. 10.3322/caac.21601 [DOI] [PubMed] [Google Scholar]
- 3.Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008; 267:226–44. 10.1016/j.canlet.2008.04.050 [DOI] [PubMed] [Google Scholar]
- 4.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004; 4:540–50. 10.1038/nrc1388 [DOI] [PubMed] [Google Scholar]
- 5.Atretkhany KN, Drutskaya MS, Nedospasov SA, Grivennikov SI, Kuprash DV. Chemokines, cytokines and exosomes help tumors to shape inflammatory microenvironment. Pharmacol Ther. 2016; 168:98–112. 10.1016/j.pharmthera.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 6.Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res. 2014; 2:1125–31. 10.1158/2326-6066.CIR-14-0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen E, Qin X, Peng K, Xu X, Li W, Cheng X, Tang C, Cui Y, Wang Z, Liu T. Identification of Potential Therapeutic Targets Among CXC Chemokines in Breast Tumor Microenvironment Using Integrative Bioinformatics Analysis. Cell Physiol Biochem. 2018; 45:1731–46. 10.1159/000487782 [DOI] [PubMed] [Google Scholar]
- 8.Zeng Q, Sun S, Li Y, Li X, Li Z, Liang H. Identification of Therapeutic Targets and Prognostic Biomarkers Among CXC Chemokines in the Renal Cell Carcinoma Microenvironment. Front Oncol. 2020; 9:1555. 10.3389/fonc.2019.01555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell A, Hasanali SL, Morera DS, Baskar R, Wang X, Khan R, Talukder A, Li CS, Manoharan M, Jordan AR, Wang J, Bollag RJ, Singh N, et al. A chemokine/chemokine receptor signature potentially predicts clinical outcome in colorectal cancer patients. Cancer Biomark. 2019; 26:291–301. 10.3233/CBM-190210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv J, Li L. Hub Genes and Key Pathway Identification in Colorectal Cancer Based on Bioinformatic Analysis. Biomed Res Int. 2019; 2019:1545680. 10.1155/2019/1545680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ning Y, Manegold PC, Hong YK, Zhang W, Pohl A, Lurje G, Winder T, Yang D, LaBonte MJ, Wilson PM, Ladner RD, Lenz HJ. Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. Int J Cancer. 2011; 128:2038–49. 10.1002/ijc.25562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin K, Zou R, Lin F, Zheng S, Shen X, Xue X. Expression and effect of CXCL14 in colorectal carcinoma. Mol Med Rep. 2014; 10:1561–68. 10.3892/mmr.2014.2343 [DOI] [PubMed] [Google Scholar]
- 13.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016; 17:174. 10.1186/s13059-016-1028-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020; 48:W509–14. 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuo C, Wu X, Li J, Hu D, Jian J, Chen C, Zheng X, Yang C. Chemokine (C-X-C motif) ligand 1 is associated with tumor progression and poor prognosis in patients with colorectal cancer. Biosci Rep. 2018; 38:BSR20180580. 10.1042/BSR20180580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doll D, Keller L, Maak M, Boulesteix AL, Siewert JR, Holzmann B, Janssen KP. Differential expression of the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and their impact on metastatic disease and survival. Int J Colorectal Dis. 2010; 25:573–81. 10.1007/s00384-010-0901-1 [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Ou B, Han D, Wang P, Zong Y, Zhu C, Liu D, Zheng M, Sun J, Feng H, Lu A. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/Snail and AKT/GSK3β/β-catenin pathways. Mol Cancer. 2017; 16:70. 10.1186/s12943-017-0629-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Zheng S, Liao N, Huang H, Chen W, Wu Z, Wu D. CircCTNNA1 acts as a ceRNA for miR-363-3p to facilitate the progression of colorectal cancer by promoting CXCL5 expression. J Biol Res (Thessalon). 2021; 28:7. 10.1186/s40709-021-00135-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong Y, You W, Wang R, Peng L, Fu Z. Prediction and Validation of Hub Genes Associated with Colorectal Cancer by Integrating PPI Network and Gene Expression Data. Biomed Res Int. 2017; 2017:2421459. 10.1155/2017/2421459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan GT, Gong YZ, Liao XW, Wang S, Huang W, Wang XK, Zhu GZ, Liao C, Gao F. Diagnostic and prognostic values of C-X-C motif chemokine ligand 3 in patients with colon cancer. Oncol Rep. 2019; 42:1996–2008. 10.3892/or.2019.7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Z, Huang X, Han X, Li Z, Zhu Q, Yan J, Yu S, Jin Z, Wang Z, Zheng Q, Wang Y. The chemokine CXCL9 expression is associated with better prognosis for colorectal carcinoma patients. Biomed Pharmacother. 2016; 78:8–13. 10.1016/j.biopha.2015.12.021 [DOI] [PubMed] [Google Scholar]
- 22.Zeng J, Yang X, Cheng L, Liu R, Lei Y, Dong D, Li F, Lau QC, Deng L, Nice EC, Xie K, Huang C. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J Transl Med. 2013; 11:6. 10.1186/1479-5876-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushita K, Toiyama Y, Tanaka K, Saigusa S, Hiro J, Uchida K, Inoue Y, Kusunoki M. Soluble CXCL16 in preoperative serum is a novel prognostic marker and predicts recurrence of liver metastases in colorectal cancer patients. Ann Surg Oncol. 2012. (Suppl 3); 19:S518–27. 10.1245/s10434-011-1993-8 [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Chen Q, Wu Y, Zhu M, Hu J, Zhuang Z. MTSS1 inhibits colorectal cancer metastasis by regulating the CXCR4/CXCL12 signaling axis. Int J Mol Med. 2021; 47:65. 10.3892/ijmm.2021.4898 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Xia YZ, Shan GF, Yang H, Zha J, Wang L, Chen JM, Zhang XS. Cisatracurium regulates the CXCR4/let-7a-5p axis to inhibit colorectal cancer progression by suppressing TGF-β/SMAD2/3 signalling. Chem Biol Interact. 2021; 339:109424. 10.1016/j.cbi.2021.109424 [DOI] [PubMed] [Google Scholar]
- 26.Li J, Liu Q, Huang X, Cai Y, Song L, Xie Q, Liu F, Chen X, Xu P, Zeng F, Chu Y, Zeng F. Transcriptional Profiling Reveals the Regulatory Role of CXCL8 in Promoting Colorectal Cancer. Front Genet. 2020; 10:1360. 10.3389/fgene.2019.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen T, Yang Z, Cheng X, Xiao Y, Yu K, Cai X, Xia C, Li Y. CXCL8 induces epithelial-mesenchymal transition in colon cancer cells via the PI3K/Akt/NF-κB signaling pathway. Oncol Rep. 2017; 37:2095–100. 10.3892/or.2017.5453 [DOI] [PubMed] [Google Scholar]
- 28.Li E, Yang X, Du Y, Wang G, Chan DW, Wu D, Xu P, Ni P, Xu D, Hu Y. CXCL8 Associated Dendritic Cell Activation Marker Expression and Recruitment as Indicators of Favorable Outcomes in Colorectal Cancer. Front Immunol. 2021; 12:667177. 10.3389/fimmu.2021.667177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abron JD, Singh NP, Murphy AE, Mishra MK, Price RL, Nagarkatti M, Nagarkatti PS, Singh UP. Differential role of CXCR3 in inflammation and colorectal cancer. Oncotarget. 2018; 9:17928–36. 10.18632/oncotarget.24730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabrero-de Las Heras S, Martínez-Balibrea E. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J Gastroenterol. 2018; 24:4738–49. 10.3748/wjg.v24.i42.4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Wang H, Brown J, Daikoku T, Ning W, Shi Q, Richmond A, Strieter R, Dey SK, DuBois RN. CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. J Exp Med. 2006; 203:941–51. 10.1084/jem.20052124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu WT, Jing YY, Yan F, Han ZP, Lai FB, Zeng JX, Yu GF, Fan QM, Li R, Zhao QD, Wu MC, Wei LX. LPS-induced CXCR4-dependent migratory properties and a mesenchymal-like phenotype of colorectal cancer cells. Cell Adh Migr. 2017; 11:13–23. 10.1080/19336918.2015.1134404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, Li J, Wang G, Lan Z, Li J, Tang M, Jiang S, Ma X, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell. 2019; 35:559–72.e7. 10.1016/j.ccell.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamieson T, Clarke M, Steele CW, Samuel MS, Neumann J, Jung A, Huels D, Olson MF, Das S, Nibbs RJ, Sansom OJ. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J Clin Invest. 2012; 122:3127–44. 10.1172/JCI61067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami T, Kawada K, Iwamoto M, Akagami M, Hida K, Nakanishi Y, Kanda K, Kawada M, Seno H, Taketo MM, Sakai Y. The role of CXCR3 and CXCR4 in colorectal cancer metastasis. Int J Cancer. 2013; 132:276–87. 10.1002/ijc.27670 [DOI] [PubMed] [Google Scholar]
- 36.Rupertus K, Sinistra J, Scheuer C, Nickels RM, Schilling MK, Menger MD, Kollmar O. Interaction of the chemokines I-TAC (CXCL11) and SDF-1 (CXCL12) in the regulation of tumor angiogenesis of colorectal cancer. Clin Exp Metastasis. 2014; 31:447–59. 10.1007/s10585-014-9639-4 [DOI] [PubMed] [Google Scholar]
- 37.Agostini M, Janssen KP, Kim IJ, D'Angelo E, Pizzini S, Zangrando A, Zanon C, Pastrello C, Maretto I, Digito M, Bedin C, Jurisica I, Rizzolio F, et al. An integrative approach for the identification of prognostic and predictive biomarkers in rectal cancer. Oncotarget. 2015; 6:32561–74. 10.18632/oncotarget.4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Wang Z, Liu F, Zhu J, Yang L, Cai G, Zhang Z, Huang W, Cai S, Xu Y. CXCL10 mRNA expression predicts response to neoadjuvant chemoradiotherapy in rectal cancer patients. Tumour Biol. 2014; 35:9683–91. 10.1007/s13277-014-2234-0 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Gao J, Wang X, Deng S, Ye H, Guan W, Wu M, Zhu S, Yu Y, Han W. CXCL4 mediates tumor regrowth after chemotherapy by suppression of antitumor immunity. Cancer Biol Ther. 2015; 16:1775–83. 10.1080/15384047.2015.1095404 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.