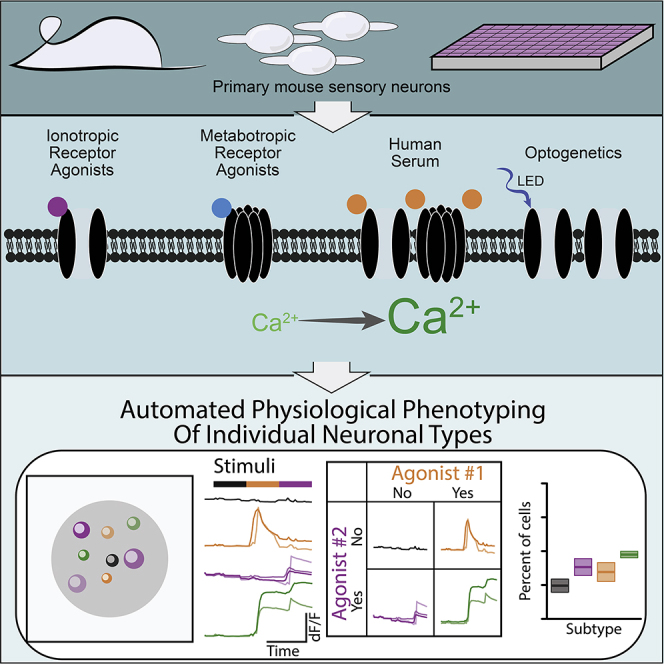
Summary
High-throughput physiological assays lose single-cell resolution, precluding subtype-specific analyses of activation mechanism and drug effects. We demonstrate APPOINT (automated physiological phenotyping of individual neuronal types), a physiological assay platform combining calcium imaging, robotic liquid handling, and automated analysis to generate physiological activation profiles of single neurons at large scale. Using unbiased techniques, we quantify responses to sequential stimuli, enabling subgroup identification by physiology and probing of distinct mechanisms of neuronal activation within subgroups. Using APPOINT, we quantify primary sensory neuron activation by metabotropic receptor agonists and identify potential contributors to pain signaling. We expand the role of neuroimmune interactions by showing that human serum directly activates sensory neurons, elucidating a new potential pain mechanism. Finally, we apply APPOINT to develop a high-throughput, all-optical approach for quantification of activation threshold and pharmacologically validate contributions of ion channel families to optical activation.
Keywords: high-throughput calcium imaging, machine learning, dorsal root ganglia, nociceptor physiology, all-optical physiology, optogenetics
Graphical abstract
Highlights
-
•
APPOINT enables high-throughput calcium imaging of individual neurons
-
•
Human serum directly activates calcium flux in primary sensory neurons
-
•
Optogenetic activation recruits native ion channels that modulate optical rheobase
Motivation
Physiological assays are typically small scale, such as patch clamp and traditional calcium imaging, whereas larger-scale techniques lose cellular resolution and thus have limited value in heterogeneous neuronal populations. Applications that require analysis of large cell numbers, including screens or experiments that address neuronal diversity, require larger-scale physiological characterization at single-cell resolution. In addition, small-scale tools are limited by experimental and analytical biases, and the desired platform would reduce these biases. We combine high-content longitudinal calcium imaging with liquid handling and an unbiased machine-learning-based analysis pipeline to generate a tool for larger-scale granular physiological investigation.
DuBreuil et al. demonstrate an integrated platform for high-throughput analysis of calcium responses elicited in individual neurons by sequential stimuli. The automated pipeline leverages machine learning classifiers and will facilitate large-scale studies, including phenotyping of neuronal subtypes, evaluating drug effects within subtypes, and quantifying drug-induced changes in optogenetic rheobase.
Introduction
Modern molecular and genetic tools reveal neuronal subtypes in stunning granularity (Hrvatin et al., 2018; Lin et al., 2015; Lubeck and Cai, 2012), but physiological techniques lag in their ability to provide a combination of large scale and high resolution. Rig-based patch clamp and calcium imaging remain the most common approaches for physiological analyses because of the wealth of pharmacological and kinetic information generated (Wainger et al., 2015), but cannot capture neuronal diversity or evaluate large compound libraries. Plate reader-based calcium imaging provides high throughput, but only field-wide metrics, and can neither distinguish signal from noise nor quantify responses among individual cells (Brenneis et al., 2014; Stacey et al., 2018). High-throughput techniques for direct measurement of action potential firing, such as multi-electrode array (Moakley et al., 2019; Wainger et al., 2014, 2015) or automated patch-clamp recording (Dunlop et al., 2008), lose single-cell resolution, require large numbers of cells, or are incompatible with critical primary cell types, such as neurons. A technique that quantifies neuronal activity at large scale while retaining single-cell resolution would yield substantial benefit for both hypothesis testing and drug screening.
Such a tool might be particularly valuable for identifying new targets and treatments for chronic pain, as the lack of adequate treatments underlies the more than 50 million adults in the United States suffering from chronic pain and fuels the ongoing opioid epidemic (Dahlhamer et al., 2018; Okie, 2010). Genetic and physiologic evidence (Cao et al., 2016; Cox et al., 2006) supports the therapeutic potential of blocking activation of nociceptors, first-order sensory neurons that initiate acute and chronic pathological pain (Basbaum et al., 2009; Hucho and Levine, 2007). Nociception can be interrogated in vitro using primary rodent, human postmortem (Valtcheva et al., 2016), and human stem cell-based models (Schwartzentruber et al., 2018; Wainger et al., 2015), but two considerations demonstrate the need for physiological techniques with improved throughput and single-cell resolution. First, independent subgroups of sensory neurons mediate and distinguish diverse sensory percepts, including pain, pleasant touch, and itch (Huang et al., 2018; Li et al., 2016; Seal et al., 2009; Usoskin et al., 2014; Zheng et al., 2019). Second, individual receptors and ion channels transduce signals in a context-dependent manner based on the cellular and molecular environment of distinct neuronal subtypes (Hill et al., 2018; Rush et al., 2006). These considerations build a strong case for a phenotypic approach to target and drug identification by using sensory neurons with sufficient scale to assess effects in both common and rare nociceptor types.
Here, we use a high-throughput platform for automated physiological phenotyping of individual neuronal types (APPOINT) to quantify the activation of primary and human induced pluripotent stem cell (iPSC)-derived neurons in response to automated application of chemical and optogenetic stimuli. APPOINT utilizes unbiased hierarchical clustering and random forest machine learning approaches to distinguish positive and negative responses to each of several sequential stimuli, and then uses the response patterns for each individual cell to assemble its physiological phenotype. We apply APPOINT to quantify the extent and breadth of activation of sensory neuron subtypes by a panel of ionotropic and metabotropic receptor agonists, demonstrate a new potential pain mechanism by quantifying sensory neuron activation by human blood serum, and develop an approach for measuring optogenetic activation threshold with pharmacological validation of contributions from physiologically relevant ion channels. APPOINT is a flexible platform enabling diverse high-throughput physiological assays for neuronal excitation and holds promise for identification and testing of novel activation mechanisms and development of non-opioid analgesics.
Results
Setup of high-throughput calcium imaging assay
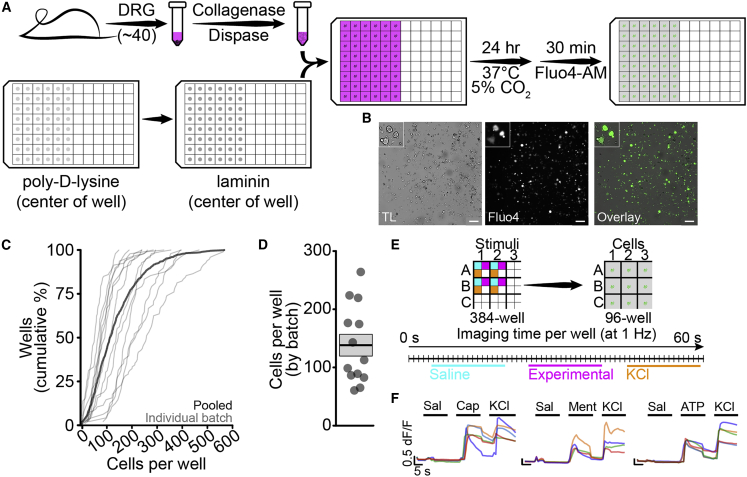
We first validated primary sensory neuron isolation and plating protocols to ensure reproducibility within and among neuronal batches. We isolated primary mouse dorsal root ganglia (DRG) and plated dissociated neurons into 96-well plates (Figure 1A). Cells were imaged in a single central field for each well (Figure 1B) and neuronal cell bodies were counted automatically using a custom analysis script (Figure S1), reducing bias from site or cell selection. Across a sample of 859 wells in 13 independent batches, we analyzed an average of 137.2 ± 3.5 cells per well, or 13,000 cells per 96-well batch (Figures 1C and 1D). Within batches, the coefficient of variation was low and few wells had fewer than 20 cells (Figure S2). We did not observe a significant difference between edge and center wells in neuronal density (main effect of edge by univariate ANOVA, n = 316 edge wells per 543 center wells, p = 0.484); therefore, we continued to use all wells per plate. Traditional rig-based experiments generally use fewer than 100 cells per group (Bataille et al., 2020; Baykara et al., 2019; Tonello et al., 2017); thus, using the lowest average cell count per well, we would only require two wells per group to surpass current typical sample sizes.
Figure 1.
Sensory neuron isolation, plating, and automated stimulation
(A) DRG isolation and plating in 96-well format.
(B) Dissociated DRG neurons visualized by transmitted light (TL) (gray, left) and Fluo4 (green, middle) with overlay (right). Scale bars, 200 μm; inset shows detail.
(C) Cumulative distribution of cells per well for 13 plates individually (thin lines) and pooled (thick line).
(D) Average number of cells per well by plate (points), with mean ± SEM.
(E) Overview of liquid handling and imaging protocol for sequential stimuli.
(F) Traces from individual cells showing consecutive activation by either capsaicin (200 nM, left), menthol (300 μM, middle), or meATP (250 μM, right) and KCl (100 mM). Scale bars, 5 s, 0.5 dF/F.
Consecutive application of chemical stimuli in rig-based microperfusion systems has allowed for cell-type-specific analysis of neuronal function (Caterina et al., 2000; Chen et al., 1995; Yin et al., 2018). We developed a stimulation protocol to apply multiple stimuli to cells in each imaging well, with the goal of facilitating physiological phenotyping of functional cell types. Three chemical stimuli were applied from 384-well plates to each well of cells in 96-well plates (Figure 1E). This configuration allows up to eight chemical stimuli using two stimulus plates, potentially enabling high-throughput constellation pharmacology approaches for cell type identification and drug discovery (Teichert et al., 2015). Each cell was exposed to negative and positive control stimuli—saline and high K+—as well as capsaicin, menthol, or αβ-methyleneadenosine 5′-triphosphate (meATP), which activate TrpV1, TrpM8, and P2X3 receptors, respectively (Figure 1F). Within individual cells, we observed large amplitude responses to both K+ and all three ionotropic receptor agonists.
Unbiased quantification of high-throughput calcium imaging using machine learning techniques
Current methods for collecting and analyzing calcium imaging data using rig-based platforms require experimenter site and cell selection as well as subjective amplitude thresholds for distinguishing responsive and non-responsive cells (Barabas et al., 2012; Than et al., 2013). These choices increase bias, reduce reproducibility, and decrease throughput. Using our approach, we have already removed empirical site and cell selection via automated imaging and we next developed an unbiased pipeline for quantification of neuronal activation.
We used a large dataset of 58,635 neurons from 10 batches of mouse primary sensory neurons, with each neuron stimulated by three stimuli, including saline, one nociceptor-specific ionotropic receptor ligand (capsaicin, menthol, or meATP), and high K+. We measured the peak amplitude and maximum rate of rise for each response independently, pooled together all responses to saline or high K+, ignoring responses to capsaicin, menthol, and meATP, and used agglomerative hierarchical clustering to form three clusters of responses. We examined traces sorted into each cluster and found that, in every batch tested, a single cluster contained primarily negative responses and both other clusters contained positive responses (Figure 2A). We considered all responses sorted into the cluster with the smallest amplitude and shallowest slope as negative, and responses in both other clusters as positive. Positive responses generally had larger amplitudes than negative responses, as expected (Figure 2B), although some response amplitudes yield both positive and negative classifications. Manual checks of positive and negative responses near 0.5 dF/F (Figure 2C) revealed striking differences, with the negative responses visually indicative of stimulus artifacts from liquid injection. Notably, use of a fixed threshold, as is currently the standard approach for such analyses (Blanchard et al., 2015; Wainger et al., 2015), which would have classified these artifacts as positive responses, and thus would have resulted in increased false-positive responses.
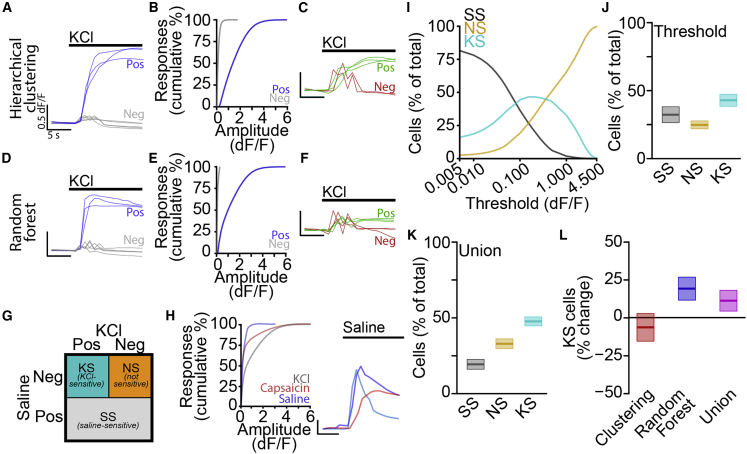
Figure 2.
Machine learning-enabled classification of automated calcium imaging responses
(A) Traces of KCl-dependent responses classified as positive (blue) and negative (gray) by hierarchical clustering. Scale bars, 5 s, 0.5 dF/F.
(B) Cumulative distributions of positive (blue) and negative (gray) response amplitudes.
(C) Traces of small amplitude KCl-dependent responses classified as negative (red) and positive (green) by hierarchical clustering. Scale bars, 5 s, 0.5 dF/F.
(D) Traces of KCl-dependent responses classified as positive (blue) and negative (gray) by random forest model. Scale bars, 5 s, 0.5 dF/F.
(E) Cumulative distributions of positive (blue) and negative (gray) response amplitudes.
(F) Traces of small amplitude KCl-dependent responses classified as negative (red) and positive (green) by random forest model. Scale bars, 5 s, 0.5 dF/F.
(G) Cell classification scheme.
(H) Cumulative distributions of response amplitudes (left) for saline (blue), capsaicin (red), and KCl (gray) with traces (right, colors indicate individual cells). Scale bars, 5 s, 0.5 dF/F.
(I) Percentage of total cells classified as saline sensitive (SS) (gray), not sensitive (NS) (orange), and KCl-sensitive (KS) (cyan).
(J) Percentage of cells classified as SS (gray), NS (orange), and KS (cyan) by using standard threshold of 0.15 dF/F as mean ± SEM.
(K) Percentage of cells classified as SS (gray), NS (orange), and KS (cyan) by using the union of hierarchical clustering and random forest classifiers mean ± SEM.
(L) Percent change from threshold-based analysis in percentage of KS cells by using hierarchical clustering (red) or random forest (blue) classifiers or the union (magenta) as mean ± SEM.
We repeated this analysis, including responses to capsaicin, menthol, and meATP along with saline and high K+ (Figure S3). Positive responses to each of the three ionotropic receptor activators qualitatively matched responses to high K+ and were easily distinguished from negative responses. Thus, hierarchical clustering permits unbiased classification of calcium responses and decreases false-positive calls by integrating amplitude and rate of change metrics.
Hierarchical clustering is an unsupervised machine learning approach, and we thought that a supervised approach might complement clustering-based classification. We trained random forest models, one for each neuronal batch, to identify positive and negative responses. Using measurements of peak amplitude and maximum rate of change, as well as raw baseline and peak intensities, we trained each model with less than 5% of responses from each batch and then classified the remaining 95% of responses. We examined responses classified as positive and negative and qualitatively verified accuracy for high K+ and ionotropic receptor responses (Figures 2D and S3). There was less overlap between cumulative amplitude distributions for positive and negative responses (Figure 2E) compared with the hierarchical clustering method, and large amplitude stimulus artifacts were again properly classified as negative (Figure 2F). To assess the robustness of our response models, we tested whether classifiers trained on individual batches could correctly identify responses when tested on each other batch. For every pair of batches, considering the within-batch classification as ground truth for each response, cross-batch model accuracy was significantly above chance (87.6% ± 0.6% correct, Table S1), indicating robust classification across batches.
These approaches use multiple features of each calcium response and rely on distinct methodologies for classifying responses; however, they were remarkably consistent. Across all 117,270 individual stimulus responses, the two approaches agreed on 88.4% of responses, with a sensitivity of 82.3% and specificity of 96.6%. For the remainder of the study, we considered all responses classified as positive by both approaches as true-positive responses.
Application of unbiased response classifications for phenotyping individual cells
To compare effects of response classification approaches on overall cell type classification, we sorted a sample of 58,635 neurons from 10 batches of mouse primary sensory neurons into 1 of 3 groups based on their responses to K+ and saline (Figure 2G). Saline-responsive cells, regardless of K+ sensitivity, were grouped together (saline sensitive [SS]) and the remaining cells were classified as either not responsive (NS) or K+ sensitive (KS). Positive responses to saline were relatively rare compared with capsaicin and high K+, but large amplitude responses were clearly distinguishable from stimulus artifacts or noise (Figure 2H). Mechanosensitive channels likely contribute to these saline-dependent responses, but the multitude of potential mechanisms complicates investigation of their physiological relevance (Beaulieu-Laroche et al., 2020; Coste et al., 2012; Murthy et al., 2018). To compare our method with traditional threshold-based approaches, we identified positive responses using several thresholds and quantified the relative proportion of cells in each category (Figure 2I). With low thresholds, most cells were classified as SS, whereas, at higher thresholds, most cells were NS. Between these extremes, KS cells comprised the largest group.
Using a typical amplitude threshold of 0.15 dF/F, we observed that roughly equal proportions of cells were sorted into each category (SS 32.3% ± 5.8% per batch, NS 24.8% ± 2.9% per batch, KS 42.9% ± 4.5% per batch; Figure 2J). The hierarchical clustering approach generated a similar pattern (SS 17.1% ± 3.1% per batch, NS 43.0% ± 3.3% per batch, KS 39.8% ± 3.2% per batch; Figure S4A), but, using the random forest approach or the union of the two approaches, we observed a clear increase in cells classified as KS relative to SS or NS (random forest: SS 21.3% ± 3.3% per batch, NS 27.6% ± 2.4% per batch, KS 51.1% ± 3.3% per batch; Figure S4B; union: SS 23.2% ± 3.7% per batch, NS 26.8% ± 2.4% per batch, KS 50.0% ± 3.5% per batch; Figure 2K). Furthermore, when directly comparing the three unbiased approaches to the conventional threshold approach, we observed that the random forest and union approaches categorized more cells as KS (random forest: 19.0% ± 7.7% increase; union: 16.5% ± 8.1% increase, n = 10 batches; Figure 2L) and fewer cells as SS (random forest: 34.0% ± 10.1% decrease; union: 28.0% ± 11.3% decrease, n = 10 batches; Figure S4C), with no change in NS cells (random forest: 11.4% ± 9.6% increase; union: 8.0% ± 9.8% increase, n = 10 batches, Figure S4D). These results demonstrate that unbiased hierarchical clustering and random forest machine learning classifiers enable high-throughput classification of neurons based on physiological responses to consecutive stimuli.
Neuronal subtype determination by physiological activation of cell-type-specific ionotropic receptors
We next applied to APPOINT to quantify neuronal activation in response to increasing doses of K+ or ionotropic receptor agonists. Increasing concentrations of K+ elicited progressively larger-amplitude calcium responses (Figure 3A) and recruited larger populations of neurons (Figure 3B). The percentage of neurons that was activated by K+ reached a plateau of 57.8% ± 5.5% at around 35 mM K+, with a half-maximal effective concentration (EC50) of 16.4 ± 3.6 mM K+. We also applied capsaicin, menthol, and meATP at five doses each and observed robust activation (Figures 3C and 3D). Both response amplitude distributions and neuron recruitment showed strong dose sensitivity, with increasing doses yielding larger responses and increased numbers of responding neurons (number of responsive cells assessed by univariate ANOVA, n = 6 batches, capsaicin: p = 1.3 × 10−7; menthol: p = 2.2 × 10−10; meATP: p = 0.016).
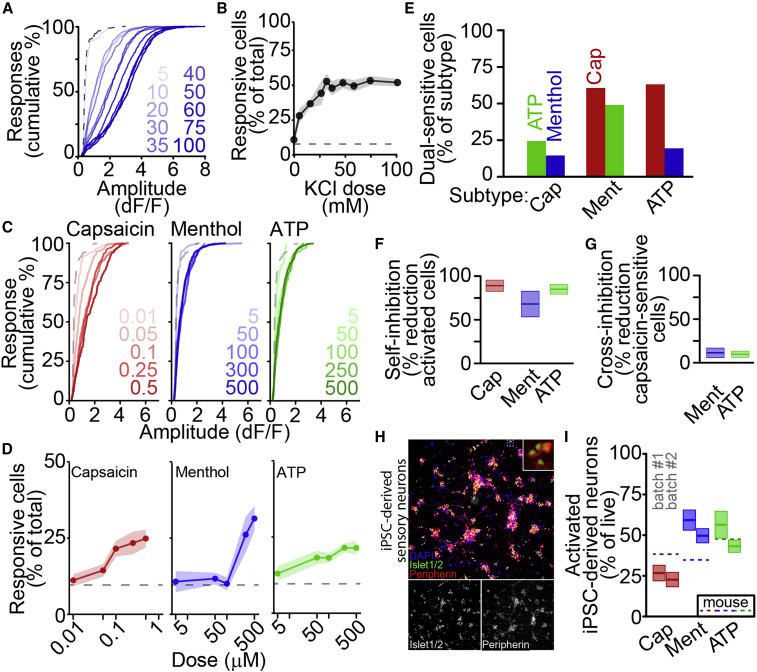
Figure 3.
Quantification of sensory neuron subtypes by ionotropic receptor activation
(A) Cumulative distributions of peak amplitudes after stimulation by increasing doses of KCl (mM, blue) or saline (dashed).
(B) Percentage of neurons activated by increasing doses of KCl, as mean ± SEM.
(C) Cumulative distributions of positive response amplitudes after stimulation by increasing doses (in μM) of capsaicin (red), menthol (blue), and meATP (green), with saline (dashed).
(D) Percentage of cells activated by increasing doses of capsaicin (red), menthol (blue), and meATP (green) as mean ± SEM, with saline (dashed).
(E) Percentage of nociceptor subtypes functionally co-expressing other subtype-specific receptors.
(F) Self-inhibition of capsaicin (red, 100 nM), menthol (blue, 300 μM), or meATP (green, 250 μM) activation by prior stimulus as mean ± SEM.
(G) Cross-inhibition of capsaicin (100 nM) responses by prior stimulation with menthol (blue, 300 μM) or meATP (green, 250 μM) as mean ± SEM.
(H) iPSC-derived sensory neurons at 5 weeks in culture stained with DAPI (blue), anti-Islet1/2 (green), and anti-Peripherin (red). Overlay inset shows detail.
(I) Percentage of live iPSC-derived neurons activated by ionotropic receptor agonists at 9 weeks in culture as mean ± SEM for two differentiations. Dashed lines show average activation of primary mouse sensory neurons.
These agonists identify broad subpopulations of sensory neuron types, but there is disagreement regarding the extent of their functional co-expression (Li et al., 2016; Usoskin et al., 2014). Therefore, we quantified functional co-expression of pairwise combinations of receptors. We applied each pair of agonists, with equal wells for each pair and each unique stimulus order and observed co-expression of each receptor pair (Figure 3E). Capsaicin-sensitive cells were rarely activated by meATP (24.4%) or menthol (14.5%), whereas menthol-activated cells were frequently activated by capsaicin (60.7%) or meATP (49.1%). Although we observed positive responses to each stimulus, whether applied first or second, we also quantified inhibition resulting from consecutive stimulation (Figures 3F–3G). As expected, activation by each stimulus was strongly inhibited by prior application of the same stimulus (capsaicin: 89.0% ± 6.4%; menthol: 68.0% ± 14.7%; meATP: 85.0% ± 5.7%; Figure 3F); however, activation by capsaicin was very weakly inhibited by prior application of either menthol (10.1.0% ± 6.4%) or meATP (9.2% ± 4.1%). Thus, although there is the potential for cross-stimulus inhibition, as with conventional rig-based calcium imaging platforms, APPOINT permits robust quantification of interference.
Although mouse primary sensory neurons provide a useful and easy source of neurons for analysis, higher-throughput applications of APPOINT will require expandable cell types, such as human iPSC-derived neurons. We quantified subtypes of iPSC-derived sensory neurons using physiological responses to cell-type-specific activators. Sensory neurons were differentiated from a human iPSC line according to an established protocol (Schwartzentruber et al., 2018) and expression of the sensory neuron markers Islet and Peripherin was verified at 5 weeks in culture (Figure 3H). Across two independent batches, positive responses to each ionotropic receptor agonist were observed after 9 weeks in culture, with similar proportions of each nociceptor subtype across batches (Figure 3I, for batch no. 1;batch no. 2, n = 8 wells;12 wells per stimulus, capsaicin: 26.7% ± 4.8%;22.7% ± 4.4% of live cells, p = 0.9996; menthol: 59.3% ± 6.2%;49.6% ± 4.4% of live cells, p = 0.927; meATP: 56.4% ± 8.3%/43.2% ± 3.8% of live cells, p = 0.714; significance assessed as batch × stimulus interaction by univariate ANOVA followed by Tukey HSD). Comparing receptor expression in iPSC-derived and primary mouse neuronal cultures, iPSC-derived sensory neuron cultures generated notably fewer capsaicin-sensitive and more menthol-sensitive cells, a common trend in iPSC-derived sensory neurons (Blanchard et al., 2015; Chambers et al., 2012; Nickolls et al., 2020).
Systematic identification of metabotropic receptors mediating calcium flux and pain
Metabotropic receptors contribute to inflammatory and neuropathic pain by modulating sensory neuron excitability (Eskander et al., 2015; Gibbs et al., 2007; Hendrich et al., 2012; Melemedjian et al., 2010; Mendieta et al., 2016). Most studies focus on nociceptor sensitization by inflammatory mediators, but some mediators, such as bradykinin and prostaglandin E2 (PGE2), directly activate calcium flux and cause pain in humans and rodents (Hong and Abbott, 1994; Mørk et al., 2003; Oh et al., 2001). The broad ability of inflammatory mediators to activate sensory neurons, and thereby potentially inflammatory pain, has not been addressed systematically; thus, we applied APPOINT to quantify nociceptor activation by a pool of metabotropic receptor agonists.
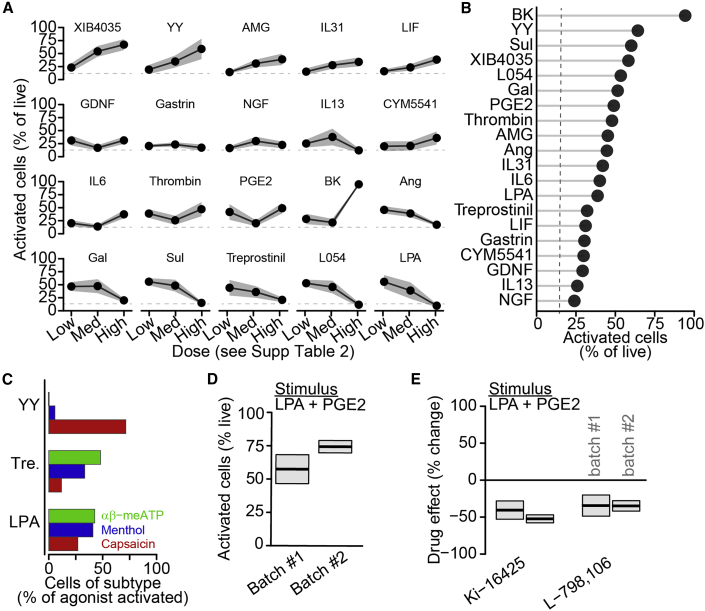
We performed an unbiased analysis of transcriptomic datasets to identify metabotropic receptors potentially involved in pain signaling based on high expression levels in DRG (GEO: GSE63576, Li et al., 2016), enriched expression in DRG (GEO: GSE10246, Lattin et al., 2008), and nociceptors (GEO: GSE55114, Chiu et al., 2014), and changes in expression after nerve injury (GEO: GSE89224, Cobos et al., 2018). We selected receptors with available and validated activators and obtained a pool of 18 receptor agonist pairs (Table S2). Identified receptors activate downstream signaling pathways via phospholipase C, protein kinase A, and Jak-Stat signaling, some of which might directly mobilize intracellular calcium, whereas others might act more indirectly. We tested three doses for each compound (Table S2), including bradykinin and PGE2 as positive control stimuli (Figure 4A) (Hong and Abbott, 1994; Linhart et al., 2003; Mørk et al., 2003). Bradykinin was the most effective agonist (Figure 4B), but several other agonists, including neuropeptide Y (YY), sulprostone, and L-054,264, activated at least as many sensory neurons as PGE2 and some elicited strong activation near the target receptor EC50. Calcium flux in these neurons is likely due to activation of intracellular signaling leading to intracellular calcium release, which might be related to neuronal sensitization (Cesare et al., 1999; Coderre, 1992). Every agonist tested activated more neurons than saline; however, several candidates with prominent roles in pain, including nerve growth factor (Denk et al., 2017) and interleukin-6 (Zhou et al., 2016), activated surprisingly small subpopulations.
Figure 4.
Metabotropic receptor activation elicits calcium flux in primary mouse sensory neurons
(A) Percentage of live mouse primary sensory neurons activated by metabotropic receptor agonists (Table S2) as mean ± SEM with saline activation (dashed).
(B) Rank ordering of metabotropic receptor agonists at single dose activating highest percentage of neurons.
(C) Activation of nociceptor subtypes by selected metabotropic receptor agonists.
(D) Percentage of live primary sensory neurons activated by a cocktail of metabotropic receptor agonists, LPA (200 μM) and PGE2 (1 mM) as mean ± SEM of two batches.
(E) Percent change in cocktail-activated cells by receptor-specific antagonists, Ki-16425 (LPA3, 10 μM) and L-798,106 (EP3, 10 μM), relative to vehicle as mean ± SEM.
We then examined the specific types of sensory neurons that were activated by a subset of these metabotropic receptor agonists: YY, treprostinil, and lysophosphatidic acid (LPA). YY selectively activated capsaicin-sensitive nociceptors, whereas both treprostinil and LPA activated all three subtypes (Figure 4C).
To assess blockers of metabotropic receptor activation, we combined LPA and PGE2 into a single stimulus cocktail and quantified inhibition by blockers of each target receptor, LPA-3 and EP3. Across two batches of primary neurons, we observed consistent neuronal activation by the LPA + PGE2 cocktail (Figure 4D, for batch no. 1;batch no. 2, n = 16 wells;14 wells, mean ± SEM: 57.5% ± 10.8%;74.5% ± 4.6% of live cells, main effect of batch by univariate ANOVA, p = 0.182) and pretreatment with either Ki-16425, an inhibitor of LPA-1 and LPA-3 receptors (Velasco et al., 2017), or L-798,106, an inhibitor of EP3 receptors (Shridas et al., 2014), decreased activation relative to vehicle (Figure 4E, for batch no. 1;batch no. 2, n = 16 wells per batch per drug, Ki-16425: 40.7% ± 12.6%;52.7% ± 5.5% decrease, one-sample t test, p = 0.005/8.9 × 10−8; L-798,106: 34.6% ± 14.5%;35.0% ± 7.1% decrease, one-sample t test, p = 0.030;0.0002). Within each drug, we did not observe a significant difference in inhibition across batches (Ki-16425: n = 16 wells per batch, p = 0.853; L-798,106: n = 16 wells per batch, p = 0.999; drug × batch interaction by univariate ANOVA with Tukey HSD). We conclude that many metabotropic receptors directly elicit calcium flux in subgroups of sensory neurons, demonstrating the power of this approach to identify activators and inhibitors of metabotropic receptor-mediated calcium flux.
Human serum directly activates calcium flux in primary sensory neurons
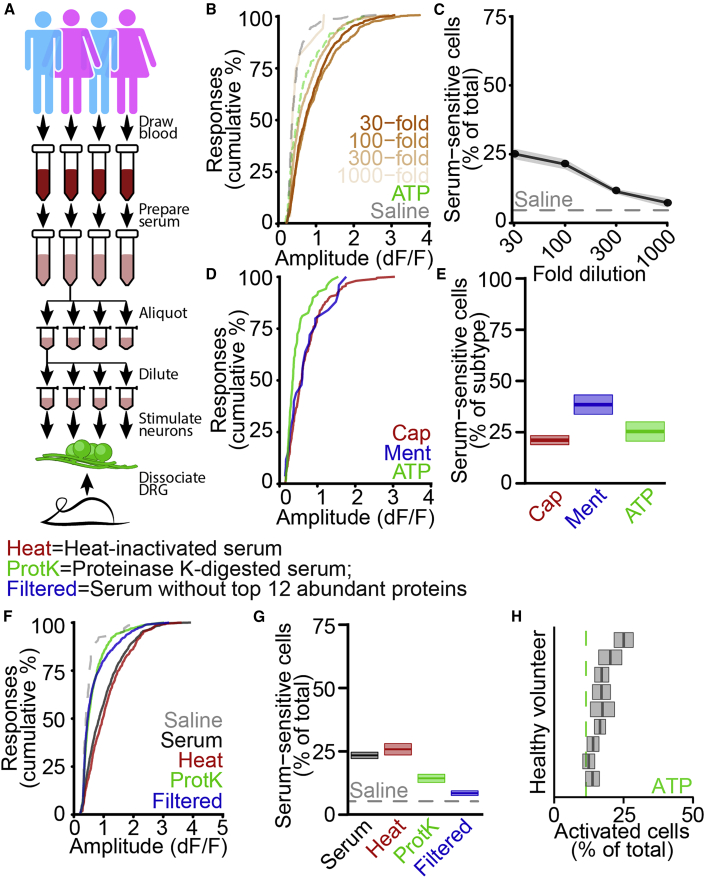
Growing evidence suggests a dynamic interaction between vascular permeability and pain via inflammatory mediators released locally by invading immune cells (Chiu et al., 2012; Pinho-Ribeiro et al., 2017); however, we hypothesized that blood itself might directly elicit calcium flux in sensory neurons. To test this, we obtained serum samples from nine healthy volunteer participants (Figure 5A) and used APPOINT to quantify the effects of serum application on primary mouse sensory neurons. Serum elicited large amplitude calcium responses that qualitatively resembled responses to ionotropic and metabotropic receptor agonists (Figure S5A). Initially, we quantified responses in all neurons, regardless of sensitivity to saline and observed strong dose-response relationships for both response amplitude (Figure S5B) and the percent of cells activated by serum (Figure S5C). Because of the substantial activation even at 1,000-fold dilution, we then discarded from analysis all cells that responded to saline, which might respond non-specifically to liquid handling. Regardless, we observed robust dose-dependence for both response amplitudes (Figure 5B) and percent of cells activated (Figure 5C). Responses to serum were of similar amplitude (Figure 5D) in sensory neurons sensitive to capsaicin, menthol, and meATP, and the percentage of serum-sensitive cells was similar across all three subtypes (Figure 5E, capsaicin: 20.6% ± 2.2%, n = 31 wells, 3 batches; menthol: 37.9% ± 4.7%, n = 18 wells, 3 batches; meATP: 24.8% ± 4.7%, n = 18 wells, 2 batches). We then sought to identify the active components of serum that contribute to neuronal activation by (1) heat inactivation of the complement system (Soltis et al., 1979), (2) digestion of proteins by using proteinase K, and (3) removal of the 12 most abundant serum proteins by spin column filtration (Figures 5F–5G). Heat inactivation reduced neither the amplitude (Figure 5F) nor the percentage of cells activated (Figure 5G, original: 23.4% ± 1.3%, n = 78 wells, 3 batches; heat inactivated: 25.8% ± 2.2%, n = 26 wells, 1 batch), indicating no contribution of complement components. By contrast, both proteinase K treatment and removal of abundant serum proteins decreased the amplitude of serum-mediated calcium responses and the percentage of activated neurons (proteinase K: 14.3 + 1.2%, n = 52 wells, 2 batches; filtered: 8.3% ± 0.9%, n = 82 wells, 2 batches). Finally, we quantified activation by serum samples from each volunteer individually (Figure 5H). Serum-mediated activation was observed by using all nine individual samples, indicating that the ability to activate sensory neurons is a general feature of human blood serum and might suggest a potential new pain mechanism.
Figure 5.
Activation of primary mouse nociceptors by human blood serum
(A) Protocol for stimulating primary mouse sensory neurons with human serum.
(B) Cumulative distributions of positive response amplitudes after stimulation by serum dilutions (as fold dilution, orange), along with meATP (dashed green) and saline (dashed gray), excluding cells sensitive to initial saline.
(C) Dose-response curve of sensory neuron activation by serum as mean ± SEM along with saline (dashed), excluding cells sensitive to initial saline.
(D) Cumulative distributions of amplitudes of serum-dependent responses in capsaicin-sensitive (red), menthol-sensitive (blue), or meATP-sensitive (green) neurons.
(E) Percentage of capsaicin- (red), menthol- (blue), and meATP-sensitive (green) sensory neurons activated by serum (300-fold dilution) as mean ± SEM.
(F) Cumulative distributions of amplitudes of serum-dependent responses after heat inactivation (Heat, red), proteinase K digestion (ProtK, green), or spin column removal of top 12 abundant proteins (Filtered, blue), relative to unmodified serum (black) and saline (gray).
(G) Percentage of sensory neurons activated by unmodified serum (black), heat-inactivated serum (Heat, red), proteinase K-digested serum (ProtK, green), or serum after abundant protein removal (Filtered, blue), with saline activation, as mean ± SEM.
(H) Percentage of live primary mouse sensory neurons activated by serum samples from nine healthy volunteers (300-fold dilution) as mean ± SEM, along with meATP (250 μM, dashed green).
A high-throughput platform for quantification and pharmacological modulation of activation threshold
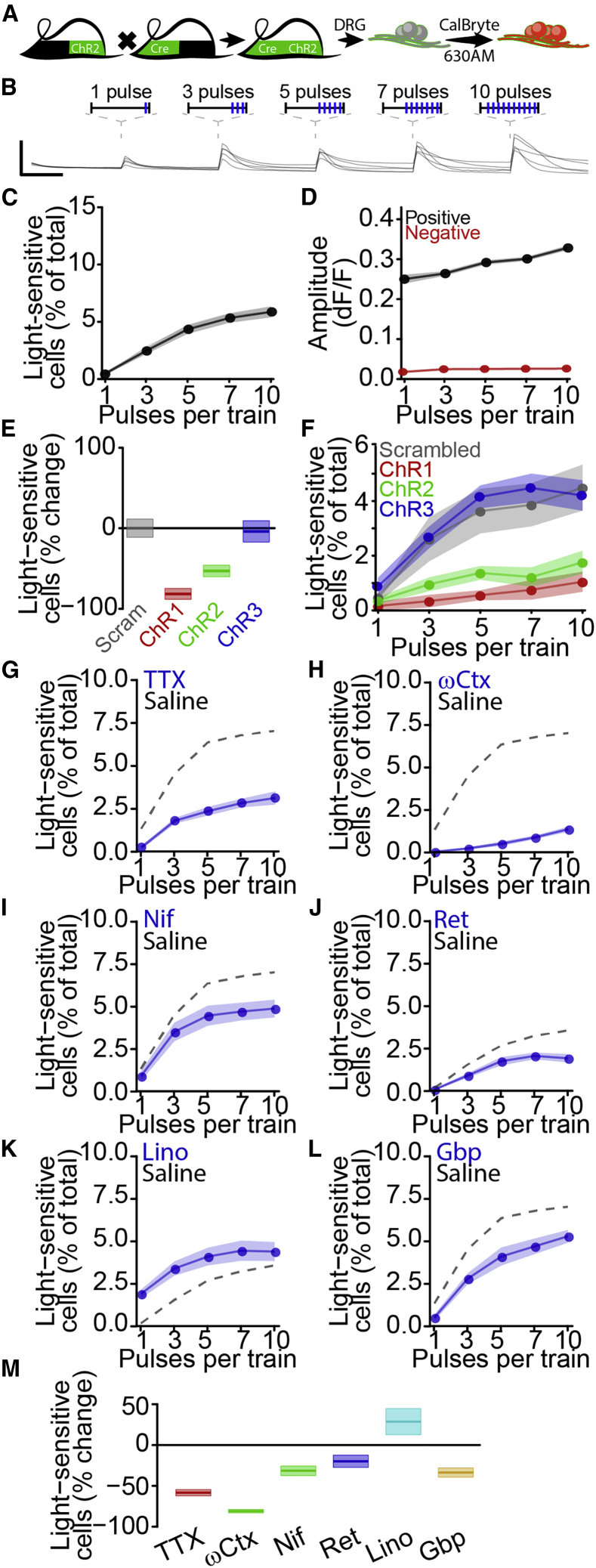
Based on successful application of our unbiased classification approach for identifying calcium responses to ionotropic (Figure 3) and metabotropic (Figure 4) receptor agonists, as well as serum (Figure 5), we hypothesized that a similar approach might be suitable for classification of optogenetic responses. To test this, we developed an all-optical approach to quantify activation threshold, which might provide a more robust and general measure of neuronal excitability independent of specific receptors. We expressed ChR2-EYFP in nociceptors by using transgenic mouse strains (Figure 6A) (Cavanaugh et al., 2011a, 2011b; Madisen et al., 2012) and monitored intracellular calcium by using a red fluorescent calcium-sensitive dye, CalBryte-630AM (Yang et al., 2019).To activate neurons, we applied five trains of 1 ms light pulses, increasing the number of pulses with each successive stimulus train (Figure 6B). We observed positive responses to at least one of the five stimulus trains in 6.4% ± 0.5% of all neurons (n = 28 wells across 6 independent batches) with an average amplitude of 0.301 ± 0.003 dF/F (n = 1,671 responses). The percentage of light-sensitive cells increased with the number of stimuli per train (Figure 6C), as did the amplitude of positive responses (Figure 6D). To confirm the dependence of these responses on ChR2-EYFP expression, we applied three different antisense oligonucleotides (ASOs) (Table 1) (10 μM) targeting the ChR2-EYFP transgene and a scrambled ASO control to dissociated sensory neurons. Seven day treatment reduced the percentage of light-sensitive cells relative to control ASO treatment for two ChR2-EYFP targeting ASOs (ChR1: 81.6% ± 6.7% decrease, n = 10 wells, 2 batches; ChR2: 53.1% ± 7.1% decrease, n = 10 wells, 2 batches), with no effect of the third (ChR3: 4.0% ± 13.2% decrease, n = 10 wells, 2 batches; Figures 6E and 6F). None of the ASO treatments affected the amplitude of light-activated calcium responses (Figure S6).
Figure 6.
Development of high-throughput activation threshold analysis
(A) All-optical sensory neuron activation.
(B) Traces of light-dependent responses in individual sensory neurons. Scale bars (lines), 5 s, 0.5 dF/F.
(C) Percentage of cells responding to each optical train as mean ± SEM.
(D) Average response amplitude for positive (black) and negative (red) responses to each optical train as mean ± SEM.
(E) Percent change in light-sensitive sensory neurons after 7-day treatment by ChR2-targeting ASOs (red, green, blue, 10 μM) or scrambled ASO (gray, 10 μM) as mean ± SEM.
(F) Percentage of cells responding to each optical train after 7-day treatment by ChR2-targeting ASOs (red, green, blue) or scrambled ASO (gray) as mean ± SEM.
(G–L) Inhibition of optogenetic activation by tetrodotoxin (G) (TTX, 300 nM), ω-conotoxin MVIIA (H) (ωCtx, 3 μM), nifedipine (I) (Nif, 10 μM), retigabine (J) (Ret, 10 μM), linopirdine (K), (Lino, 50 μM), or gabapentin (L) (Gbp, 300 μM) with saline (dashed) as mean ± SEM.
(M) Percent change from saline in optogenetic activation by any stimulus train as mean ± SEM.
Table 1.
ASO sequences
| ASO | Sequence |
|---|---|
| ChR1 | mU∗mU∗mG∗mU∗mA∗C∗T∗C∗C∗A∗G∗C∗T∗T∗G∗mU∗mG∗mC∗mC∗mC |
| ChR2 | mC∗mC∗mA∗mU∗mG∗G∗G∗T∗C∗T∗G∗C∗T∗T∗G∗mU∗mG∗mU∗mC∗mU |
| ChR3 | mC∗mU∗mC∗mA∗mG∗G∗T∗A∗G∗T∗G∗G∗T∗T∗G∗mU∗mC∗mG∗mG∗mG |
| Scrambled | mA∗mG∗mC∗mC∗mC∗C∗A∗C∗A∗T∗C∗T∗T∗T∗G∗mC∗mU∗mA∗mC∗mC |
Channelrhodopsin-mediated neuronal activation has been used to probe neural circuits, but studies examining contributions of physiologically relevant ion channels to activation have been limited (Zhang et al., 2016). Sensitivity of ChR2 responses to ion channel blockers and activators would support the feasibility of leveraging optically evoked calcium responses to evaluate mechanisms of neuronal activation and identify blocking drugs (Figures 6G–6M). The voltage-gated sodium channel inhibitor tetrodotoxin (TTX) (300 nM) reduced calcium responses to optogenetic stimulation, indicating recruitment of TTX-sensitive sodium channels by optogenetic stimulation (TTX: 57.3% ± 3.9% decrease, n = 8 wells/drug) (Blair and Bean, 2002). Voltage-gated calcium channel blockers ω-conotoxin MVIIA and nifedipine both reduced optogenetic activation (ωCtx: 81.0% ± 2.1% decrease; Nif: 31.6% ± 5.9% decrease, n = 8 wells per drug), consistent with contributions of N- and L-type calcium channels to sensory neuron activation (Andrade et al., 2010; Barzan et al., 2016). We found that the KV7 channel activator retigabine and the KV7 channel inhibitor linopirdine exerted opposing effects on optogenetic activation, with linopirdine facilitating responses to brief optical trains and retigabine suppressing responses to strong stimuli (Ret: 20.0% ± 7.5% decrease; Lino: 28.6% ± 16.0% increase, n = 8 wells per drug) (Brown and Passmore, 2009). These data implicate physiologically relevant ion channels in calcium responses to optical stimuli and suggest the potential of this approach for identifying novel analgesics. To directly assess this application, we tested the well-established chronic pain treatment gabapentin (Sills, 2006) and observed robust inhibition of optogenetic activation (33.8% ± 5.6% decrease, n = 8 wells per drug), indicating the potential for identification and validation of analgesic compounds by using this approach.
Discussion
We developed an experimental platform, APPOINT, that combines high-content imaging, robotic liquid handling, and an unbiased analysis pipeline to interrogate the physiology of individual neurons and leverage their physiological profiles to resolve neuronal subtypes. We focused on applications related to nociceptors, on account of the acute need for novel mechanistic insight into pain pathophysiology and development of non-opioid treatment (Dahlhamer et al., 2018; Okie, 2010). We identified new components of sensory neuroimmune signaling, including the direct activation of nociceptors by inflammatory mediators as well as human serum, the latter providing a potential novel pain mechanism. Using APPOINT, we developed an approach for high-throughput quantification of optical activation threshold, expanding all-optical physiology options at large scale, and quantified effects of individual ion channels on ChR2-dependent calcium flux in neurons.
Contributions of potential pain targets to pain perception depend on molecular context and cell type; thus, traditional target-based screens might be of limited value for analgesic development (Albisetti et al., 2019; Hill et al., 2018; Huang et al., 2018; Rush et al., 2006; Seal et al., 2009). Instead, phenotypic screens, which focus on biologically relevant phenotypes rather than specific proteins isolated from their biological context, have received increasing enthusiasm (Wagner and Schreiber, 2016; Moffat et al., 2017); however, the high-throughput tools required to perform physiology-based phenotypic screens remain limited (Bradley and Strock, 2019; Sidders et al., 2018). Few physiology-based phenotypic screens of sensory neurons utilizing calcium imaging have been executed (Brenneis et al., 2014; Stacey et al., 2018) and these do not provide single-cell resolution to evaluate heterogeneous populations (Hill et al., 2018). APPOINT, by comparison, enables diverse screening applications while maintaining readouts on cellular subtypes.
In contrast to typical high-throughput physiology approaches, traditional rig-based physiological techniques provide detailed phenotypes of individual cells (Wainger et al., 2015) but are restricted by the small numbers of neurons and compounds that can be studied. As well as throughput, rig-based imaging approaches generally harbor bias from the manual choice of imaging field, post-hoc selection of neurons for analysis, and identification of responding cells by using only an empirically selected amplitude threshold. APPOINT automates each of these, reducing bias. We demonstrate APPOINT as an integrated method for both in vitro calcium imaging and data analysis; however, implementation of the unbiased analysis pipeline alone by using traditional rig-based acquisition might have advantages for both increasing throughput and reducing bias.
Direct activation of nociceptors offers a clear explanation for an increasingly large number of neuroimmune processes (Chiu et al., 2013), and metabotropic activation of nociceptors causes pain (Liu et al., 2010; Mørk et al., 2003). Combining APPOINT with unbiased in silico identification and ranking of candidate metabotropic receptors revealed metabotropic receptor agonists that elicited calcium flux selectively in physiologically defined subtypes of sensory neurons. We relied on well-validated physiological markers of nociceptor subtypes, which are insufficient to capture the full diversity of primary sensory neurons. Nonetheless, genetic-labeling strategies (Cavanaugh et al., 2011a; Stirling et al., 2005; Wang and Zylka, 2009; Zheng et al., 2019), subtype-specific GPCRs (Dong et al., 2001), or even intersectional subtype classification (Teichert et al, 2014, 2015), would enable interrogation of responses within increasingly restricted sensory neuron subtypes.
Abnormalities of vascular beds contribute to a large number of painful conditions, ranging from common bruises to hematomas, subarachnoid hemorrhage, and migraine. Although mechanisms for enhancing pain signaling have been demonstrated in specific diseases (Dodick, 2018), the fundamental initiation of pain signaling—how first-order nociceptor neurons become activated—remains unclear. Extravascular leakage of blood components is a unifying feature of these painful conditions and we observed that serum elicited large amplitude calcium responses in a large population of sensory neurons. Osmolarity and pH changes from serum dilution could not have accounted for such strong activation (Viana et al., 2001; Wemmie et al., 2013) and the decrease in serum-mediated activation by proteinase K treatment or depletion of abundant serum proteins supports specific activation by a proteinaceous component in serum. Regulation of vascular permeability and blood leakage, which occurs through an array of neuroimmune interactions (Pinho-Ribeiro et al., 2017), might be a direct mechanism of nociceptor activation. Although serum injection into healthy volunteers was reported to be painless, factors released from platelets can elicit robust action potential firing in rodent sensory neuron axons, as well as cause pain in healthy individuals (Ringkamp et al., 1994; Schmelz et al., 1997). Furthermore, alterations in serum composition in the context of pathological pain, as well as sensitization of sensory neurons, might increase the pain induced by serum injection in patients. Elucidating the specific ligands and targets responsible for the observed activation will require further work. Nonetheless, should nociceptor activation by serum differ between healthy volunteers and patients with neuropathic or inflammatory pain, serum-dependent activation could be used as a physiological biomarker for pathological pain.
Increased neuronal excitability is a pathognomonic feature of pathological pain states (Hucho and Levine, 2007; Ma et al., 2006) as well as other neurological diseases (Devinsky et al., 2013; Geevasinga et al., 2016). A previous elegant study using HEK cells equipped with heterologously expressed voltage-gated sodium channels and leak potassium channels showed how optogenetic activation can reveal properties of sodium channel modulation (Zhang et al., 2016). Here, we show that graded optogenetic stimulation preserves sensitivity to endogenous sodium, calcium, and potassium channel modulators. Although we focus on a calcium readout here, we anticipate more closely approximating traditional rheobase measurements by incorporating recent fluorescent voltage indicators that have substantially improved sensitivities (Abdelfattah et al., 2016). The broad availability of transgenic mouse lines enabling targeted expression of optogenetic actuator proteins in discrete functional classes of sensory neurons (Zheng et al., 2019) will allow for granular analysis of optical activation. Finally, future studies can now examine nociceptor-dependent processes involved in pathological pain, such as peripheral sensitization, as well as phenotypic screens to reduce nociceptor activation.
Limitations of the study
Although the analytical pipeline from APPOINT is applicable to any calcium-based acquisition technique, the full platform was developed by using a commercial high-content imager and associated liquid handling and software. Adaptations to other image segmentation and perfusion systems could require modification; changes, such as continuous liquid administration, might provide a somewhat altered profile of neuronal subtypes, potentially improving the resolution of neurons that responded to vehicle in our system.
Sensory neuron subtypes, particularly nociceptor subgroups, can be defined physiologically to a first approximation using cell-type-specific agonists (Caterina et al., 1997, Caterina et al., 2000; Chen et al., 1995; Yin et al., 2018), but these are unable to capture broad sensory neuron diversity (Li et al., 2016; Usoskin et al., 2014; Zheng et al., 2019). Although we chose sensory neurons on the basis of the utility of specific ion channels and ionotropic receptors, these tools for subtype dissection are not available for most neuronal subtypes. Thus, we anticipate that APPOINT could be used in characterizing large numbers of genetically labeled neurons and identifying physiologically defined subgroups.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Fluo4-AM | ThermoFisher | Cat# F14201 |
| CalBryte-630AM | AAT Bioquest | Cat# 20720 |
| Critical commercial assays | ||
| Top 12 Abundant Protein Depletion Spin Columns | Thermo Fisher | Cat# 85165 |
| Deposited data | ||
| Mouse Gene Atlas | NCBI GEO | GEO: GSE10246 |
| Transcriptome analysis of somatosensory neuron types | NCBI GEO | GEO: GSE55114 |
| Single-cell RNA-seq of somatosensory neuron types | NCBI GEO | GEO: GSE63576 |
| Expression profiling after neuropathic nerve injury | NCBI GEO | GEO: GSE89224 |
| Experimental models: organisms/strains | ||
| Mouse: C57Bl6/J | Jackson Labs | IMSR Cat# JAX:000664; RRID:IMSR_JAX:000664 |
| Mouse: Trpv1-Cre: B6.129-Trpv1tm1(cre)Bbm/J | Jackson Labs | IMSR Cat# JAX:017769; RRID:IMSR_JAX:017769 |
| Mouse: LSL-ChR2-EYFP: B6.Cg-Gt(ROSA)26Sortm32(CAG-COP4∗H134R/EYFP)Hze/J | Jackson Labs | IMSR Cat# JAX:024109; RRID:IMSR_JAX:024109 |
| Oligonucleotides | ||
| See Table 1 for ASO sequences | Integrated DNA Technologies | |
| Software and algorithms | ||
| MetaXpress | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metaxpress |
| R | The R Project for Statistical Computing | https://www.r-project.org/ |
| APPOINT analysis pipeline | This paper | https://github.com/waingerlab/APPOINT_analysis_script |
| Other | ||
| ImageXPress Micro Confocal | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/high-content-imaging/imagexpress-micro-confocal |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brian Wainger (brian.wainger@mgh.harvard.edu)
Materials availability
This study did not generate new unique reagents.
Data and code availability
The complete analysis pipeline for APPOINT (including MetaXPress cell segmentation script and R response classification script) are available at GitHub (https://github.com/waingerlab/APPOINT_analysis_script).
Experimental model and subject details
Mice
All animal protocols were approved by the MGH IACUC. Male and female mice (C57Bl6/J) from 2 to 8 weeks old were used for all experiments. For optogenetic activation experiments, Trpv1-Cre (Jax 017769) male mice were crossed with LSL-ChR2-EYFP (Jax 024109) female mice and first-generation Trpv1/ChR2-EYFP pups were used for preparation of sensory neurons. Mice were euthanized by CO2 asphyxiation, followed by decapitation. DRG (C1-L6, left and right) were quickly dissected into ice-cold DMEM/F-12 (Thermo Fisher 11320082) and dissociated using a solution of collagenase A (Sigma-Aldrich 10103578001, 2 mg/mL) and dispase (Thermo Fisher 17105041, 2 mg/mL) diluted in Hank’s Balanced Salt Solution (Thermo Fisher 14185052) for 60–90 min at 37°C, followed by mechanical trituration using a flame-polished Pasteur pipette. Dissociated cells were filtered using a 70 μm cell strainer (Thermo Fisher 22363548) and BSA gradient (Sigma-Aldrich A9576, 10% in PBS, centrifuge 12 min, 200 rcf). Cells were cultured in neurobasal media (Thermo Fisher 21103049) supplemented with B27 (Thermo Fisher 17504044), GlutaMax (Thermo Fisher 35050061), Pen/Strep (Thermo Fisher 15070063) and AraC (R&D Systems 4520) overnight at 37°C in 96-well plates (Ibidi 89626) treated with poly-d-lysine(Sigma-Aldrich A-003-E, 1–2 hr at 37°C, 2 μL/well) followed by laminin (Thermo Fisher 23017015, 1–2 hr at 37°C, 2 μL/well). All primary sensory neurons were stained and imaged at 1 day in vitro.
Induced pluripotent stem cells
Induced pluripotent stem cells (iPSCs) of a single line, 446, from a healthy individual (Lee et al., 2009) were obtained under MGH IRB approval and differentiated into sensory neurons according to an established protocol (Schwartzentruber et al., 2018). Frozen stocks of a single passage were thawed and expanded in Matrigel (Corning 354277) coated 6-well plates (Corning 353046) for 3 days using mTESR1 media (Stem Cell 85850) before initiating differentiation. On day 1 to day 3 of differentiation, cells received hESC media [DMEM/F12 media with Knock-out Serum Replacement (Thermo Fisher 10828-028), non-essential amino acids (Corning 25-025-CI), GlutaMax, and β-mercaptoethanol (Thermo Fisher 31350010)] supplemented with LDN-193189 (Stemgent 04-0074-02) and SB431542 (DNSK International DNSK-KI-12). Beginning on day 4, hESC was supplemented with the combination of SU5402 (DNSK International DNSK-KI-11), CHIR 99021(Tocris 4423), and DAPT (DNSK International DNSK-EI-01), in addition to LDN and SB. For days 5-10, NB media [Neurobasal with N2 (Thermo Fisher 17502048), B27, GlutaMax, and β-mercaptoethanol] was increasingly added to to hESC media every 2 days, such that media for days 5 and 6 constituted 75% hESC/25% NB, for days 7 and 8 constituted 50% hESC/50% NB, and for days 9 and 10 25% hESC/75% NB. Supplementation with SU, CHIR, and DAPT was maintained during this transition, but LDN and SB were removed after day 7. On day 11, immature neurons were dissociated using accutase (Worthington LK003178) and replated at a density of 3∗106 cells/well in Matrigel-coated 6-well plates containing NB media supplemented with BDNF (Thermo Fisher PHC7074), GDNF (Thermo Fisher PHC7044), NGF (R&D 256-GF), NT-3 (Thermo Fisher PHC7036), and ascorbic acid (Sigma-Aldrich A4403). Media was changed twice weekly thereafter until 32 or 63 days in vitro for staining and physiology, respectively.
Method details
Calcium imaging
Primary mouse and human iPSC-derived sensory neurons were stained with the calcium indicator Fluo4-AM for monitoring intracellular calcium concentration. For both cell types, cells were washed once with physiological saline (in mM: 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 D-glucose, 10 HEPES and pH 7.3-7.4 with NaOH) followed by incubation with Fluo4-AM (Thermo Fisher F14201, 3 μg/mL in 0.3% DMSO) in culture media in the dark at room temperature for 30 min. After 30 min, dye-containing media was removed and replaced with 100 μL of saline and immediately transferred to the imaging chamber. For optogenetics experiments, loading with the calcium indicator CalBryte-630AM (AAT Bioquest 20720, 3 μg/mL in 0.3% DMSO) followed the same protocol.
All calcium imaging experiments were conducted using an ImageXPress micro confocal high content imaging system (Molecular Devices) with automated robotic liquid handling. Cells were maintained at 37°C and with 5% CO2/O2 during imaging and were imaged (60 s/well) using widefield mode with 10x magnification at a frequency of 1 Hz in the center of each well. Asynchronous liquid dispensing was used for all stimulus applications, allowing for continuous cell visualization during stimulation. All stimuli were a total volume of 20 μL and delivered into the bath solution at a rate of 5 μL/s. Stimulus concentrations are reported as the initial concentration in the stimulus plate, not the final concentration after mixing with the cell bath solution. Doses for all stimuli are indicated in the main text and all stimuli and treatments were diluted in physiological saline, with DMSO (maximum 0.1%) or ethanol (maximum 0.1%) when necessary. Saline stimulation used the same physiological saline, with 0.1% DMSO or 0.1% ethanol when appropriate. DMSO and ethanol did not elicit any direct activation compared with saline alone.
Optogenetic stimuli consisted of five trains of increasing numbers of stimuli (1,3,5,7, and 10 pulses per train) of 1 ms each, delivered at 50 Hz with 10 s between successive trains. CalBryte-630 intensity was measured at 5 Hz.
Quantification of neuronal activation
All custom analysis scripts will be made available upon request. Initial identification and quantification of Fluo4 intensity was performed using a custom journal in MetaXPress (Molecular Devices) analysis software. Briefly, Flou4-positive cells in each well were identified using a minimum projection of the timelapse image stack based on size and Fluo4 intensity. Each cell automatically generated an individual ROI, which was transferred back to the original timelapse stack and the mean intensity of each ROI was calculated for every image. Raw data by cell for each well were saved as a csv file for further analysis. Non-neuronal cells remaining from initial dissociation and not eliminated by AraC treatment were reliably excluded during cell segmentation based on their substantially smaller cell body size (Figure S2).
Analysis of intensity and quantification of neuronal activation was performed in R (v3.5.0). Briefly, raw intensity values for each cell were normalized to the baseline intensity of the initial 5 images. Peak response amplitude was calculated as the median of 3 intensity values centered on the peak intensity within 15 s after stimulus onset, with independent baselines of the 5 s preceding stimulus onset for each stimulus. Maximum rate of rise of Fluo4 was calculated as the peak of the derivative of intensity values within 15 s after stimulus onset.
For hierarchical clustering analysis, pooled responses were filtered to center clustering around typical thresholds (peak amplitudes between 0.075 and 0.225 dF/F; maximum rate of rise between 0.5 and 1.5 dF/F/s). Hierarchical clustering used the euclidean distance between observations and Ward D2 agglomeration algorithm. Random forest modeling was accomplished using caret (v6.0) and randomForest (v4.6) R packages using default settings and selecting the simplest model within one standard error of the optimal model.
Immunohistochemistry
iPSC-derived sensory neurons were dissociated at 32 days in vitro using papain for 25-35 min and replated into 96-well plates (Ibidi 89626). They were maintained in NB supplemented with BDNF, GDNF, NGF, NT3 and ascorbic acid for 3 days, followed by fixation with 4% paraformaldehyde (Thermo Fisher 50-980-495). Cells were blocked for 4 hrs at 4°C, incubated with primary antibodies for Isl1/2 (mouse, DSHB 39.4D5, 1:500) and Peripherin (rabbit, Millipore AB1530, 1:200) overnight at 4°C, followed with donkey anti-mouse (Thermo Fisher a21202) and donkey anti-rabbit (Thermo Fisher A10042) secondary antibodies at 23°C for 4 h. Cells were imaged using ImageXPress micro confocal high content imaging system (Molecular Devices).
ASO design
ChR1, ChR2, ChR3 and scrambled ASOs (Integrated DNA Technologies) were constructed using a gapmer design, phosophorothioate bonds (indicated by ∗ in sequence), and 2’-O-methyl modified RNA bases (indicated by preceding “m”) flanking unmodified DNA bases (indicated by underline). ASO sequences are listed in Table 1 and all ASOs were applied at 10 μM for 7 days to sensory neurons on day 0 of dissociation. Culture media was supplemented with NGF to promote neuronal health during extended culture.
In silico analysis of transcriptomic datasets
Read count data for GSE10246, GSE55114, GSE63576, and GSE89224 were downloaded from GEO and loaded into R (v3.5.0). The data were filtered to include only genes present in all four datasets and fold change and adjusted p values from each dataset were pooled. Mean expression counts from GSE63576 were used to reflect relative expression strength in mouse DRG. Genes were then filtered to identify relevant genes with expression regulated by nerve injury (adjusted p value spared nerve injury v sham<0.001), or preferential expression in DRG relative to other tissues (adjusted p value DRG v Other<0.005 and expression > 10), or preferential expression in nociceptors relative to proprioceptors (adjusted p value Nociceptors v Proprioceptors<0.002 and expression >3). Metabotropic receptors were identified using all genes associated with gene ontology terms (0030594, 0005030, 0038187, 0001653, 0004888). Each remaining gene was then ranked using the weighted average of fold-change enrichment and raw expression (weights: SNI enrichment=0.3, Nociceptor enrichment=0.3, DRG enrichment=0.2, raw expression=0.2).
Serum-dependent activation
Blood samples were collected from healthy individuals (5 male and 4 female, 25 to 85 years old, mean age=47 years) under MGH IRB approval. Serum was prepared and frozen in aliquots at -80°C. For activation, serum was thawed and split into four portions for: 1) unmodified serum stimulation; 2) heat inactivation at 56°C for 1 h; 2) proteinase K digestion (New England Biolabs P8107S) at 37°C for 18 h; 3) depletion of top 12 abundant serum proteins (Thermo Fisher 85165) according to manufacturer instructions. The 12 most abundant serum proteins include: albumin, IgG, α1-acid glycoprotein, α1-antitrypsin, α2-macroglobulin, ApoAI, ApoAII, fibrinogen, haptoglobin, IgA, IgM, transferrin.
Quantification and statistical analysis
Mean and standard error of the mean are reported throughout, unless specified. Details of specific statistical analyses are included in the main text. For differences between cumulative distributions, we used the two-sample Kolmogorov-Smirnov test of the hypothesis that both individual distributions are drawn from the same underlying distribution. Significant changes in activation are calculated as percent change from baseline or vehicle condition by batch and significance assessed as one-sample t-test of the hypothesis that the net change across batches is zero. Batch effects are assessed using univariate ANOVA, followed by Tukey HSD correction for multiple comparisons when appropriate.
Acknowledgments
Funding from NIH New Innovator Award (1DP2-NS106664-01), the New York Stem Cell Foundation, and MGH Departments of Neurology and Anesthesia, Critical Care & Pain Medicine (to B.J.W.). B.J.W. is a New York Stem Cell Foundation – Robertson Investigator. We thank Marco Loggia for providing samples, and Anne-Louise Oaklander and members of the Albers and Lagier-Tourenne labs for helpful discussions.
Author contributions
D.M.D., B.M.C., K.Z., P.F., X.L., and Y.S. performed the experiments. D.M.D. and B.J.W designed the experiments, analyzed and interpreted the data, and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published: April 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100004.
Supplemental information
References
- Abdelfattah A.S., Farhi S.L., Zhao Y., Brinks D., Zou P., Ruangkittisakul A., Platisa J., Pieribone V.A., Ballanyi K., Cohen A.E., Campbell R.E. A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J. Neurosci. 2016;36:2458–2472. doi: 10.1523/JNEUROSCI.3484-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albisetti G.W., Pagani M., Platonova E., Hösli L., Johannssen H.C., Fritschy J.-M., Wildner H., Zeilhofer H.U. Dorsal horn gastrin-releasing peptide expressing neurons transmit spinal itch but not pain signals. J. Neurosci. 2019;39:2238–2250. doi: 10.1523/JNEUROSCI.2559-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade A., Denome S., Jiang Y.-Q., Marangoudakis S., Lipscombe D. Opioid inhibition of N-type Ca2+ channels and spinal analgesia couple to alternative splicing. Nat. Neurosci. 2010;13:1249–1256. doi: 10.1038/nn.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabas M.E., Kossyreva E.A., Stucky C.L. TRPA1 is functionally expressed primarily by IB4-binding, non-peptidergic mouse and rat sensory neurons. PLoS One. 2012;7:e47988. doi: 10.1371/journal.pone.0047988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzan R., Pfeiffer F., Kukley M. N- and L-type voltage-gated calcium channels mediate fast calcium transients in axonal shafts of mouse peripheral nerve. Front. Cell Neurosci. 2016;10 doi: 10.3389/fncel.2016.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum A.I., Bautista D.M., Scherrer G., Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille A., Leschiera R., L’Hérondelle K., Pennec J.-P., Le Goux N., Mignen O., Sakka M., Plée-Gautier E., Brun C., Oddos T., et al. In vitro differentiation of human skin-derived cells into functional sensory neurons-like. Cells. 2020;9 doi: 10.3390/cells9041000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baykara M., Ozcan M., Bilgen M., Kelestimur H. Effects of gadolinium and gadolinium chelates on intracellular calcium signaling in sensory neurons. Neurosci. Lett. 2019;707:134295. doi: 10.1016/j.neulet.2019.134295. [DOI] [PubMed] [Google Scholar]
- Beaulieu-Laroche L., Christin M., Donoghue A., Agosti F., Yousefpour N., Petitjean H., Davidova A., Stanton C., Khan U., Dietz C., et al. TACAN is an ion channel involved in sensing mechanical pain. Cell. 2020;180:956–967.e17. doi: 10.1016/j.cell.2020.01.033. [DOI] [PubMed] [Google Scholar]
- Blair N.T., Bean B.P. Roles of tetrodotoxin (TTX)-sensitive Na+ current, TTX-resistant Na+ current, and Ca2+ current in the action potentials of nociceptive sensory neurons. J. Neurosci. 2002;22:10277–10290. doi: 10.1523/JNEUROSCI.22-23-10277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J.W., Eade K.T., Szűcs A., Lo Sardo V., Tsunemoto R.K., Williams D., Sanna P.P., Baldwin K.K. Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 2015;18:25–35. doi: 10.1038/nn.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J.A., Strock C.J. Screening for neurotoxicity with microelectrode array. Curr. Protoc. Toxicol. 2019;79:e67. doi: 10.1002/cptx.67. [DOI] [PubMed] [Google Scholar]
- Brenneis C., Kistner K., Puopolo M., Jo S., Roberson D., Sisignano M., Segal D., Cobos E.J., Wainger B.J., Labocha S., et al. Bupivacaine-induced cellular entry of QX-314 and its contribution to differential nerve block. Br. J. Pharmacol. 2014;171:438–451. doi: 10.1111/bph.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.A., Passmore G.M. Neural KCNQ (Kv7) channels. Br. J. Pharmacol. 2009;156:1185–1195. doi: 10.1111/j.1476-5381.2009.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., McDonnell A., Nitzsche A., Alexandrou A., Saintot P.-P., Loucif A.J.C., Brown A.R., Young G., Mis M., Randall A., et al. Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons and patients with inherited erythromelalgia. Sci. Transl. Med. 2016;8:335ra56. doi: 10.1126/scitranslmed.aad7653. [DOI] [PubMed] [Google Scholar]
- Caterina M.J., Leffler A., Malmberg A.B., Martin W.J., Trafton J., Petersen-Zeitz K.R., Koltzenburg M., Basbaum A.I., Julius D. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- Cavanaugh D.J., Chesler A.T., Braz J.M., Shah N.M., Julius D., Basbaum A.I. Restriction of transient receptor potential vanilloid-1 to the peptidergic subset of primary afferent neurons follows its developmental downregulation in nonpeptidergic neurons. J. Neurosci. 2011;31:10119–10127. doi: 10.1523/JNEUROSCI.1299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh D.J., Chesler A.T., Jackson A.C., Sigal Y.M., Yamanaka H., Grant R., O’Donnell D., Nicoll R.A., Shah N.M., Julius D., Basbaum A.I. Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J. Neurosci. 2011;31:5067–5077. doi: 10.1523/JNEUROSCI.6451-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare P., Dekker L.V., Sardini A., Parker P.J., McNaughton P.A. Specific involvement of PKC-epsilon in sensitization of the neuronal response to painful heat. Neuron. 1999;23:617–624. doi: 10.1016/s0896-6273(00)80813-2. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Qi Y., Mica Y., Lee G., Zhang X.-J., Niu L., Bilsland J., Cao L., Stevens E., Whiting P., et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C., Akopian A.N., Sivilotti L., Colquhoun D., Burnstock G., Wood J.N. A P2X purinoceptor expressed by a subset of sensory neurons. Nature. 1995;377:428–431. doi: 10.1038/377428a0. [DOI] [PubMed] [Google Scholar]
- Chiu I.M., Barrett L.B., Williams E.K., Strochlic D.E., Lee S., Weyer A.D., Lou S., Bryman G.S., Roberson D.P., Ghasemlou N., et al. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. ife. 2014;3 doi: 10.7554/eLife.04660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu I.M., Heesters B.A., Ghasemlou N., Von Hehn C.A., Zhao F., Tran J., Wainger B., Strominger A., Muralidharan S., Horswill A.R., et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature. 2013;501:52–57. doi: 10.1038/nature12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu I.M., von Hehn C.A., Woolf C.J. Neurogenic inflammation and the peripheral nervous system in host defense and immunopathology. Nat. Neurosci. 2012;15:1063–1067. doi: 10.1038/nn.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos E.J., Nickerson C.A., Gao F., Chandran V., Bravo-Caparrós I., González-Cano R., Riva P., Andrews N.A., Latremoliere A., Seehus C.R., et al. Mechanistic differences in neuropathic pain modalities revealed by correlating behavior with global expression profiling. Cell Rep. 2018;22:1301–1312. doi: 10.1016/j.celrep.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coderre T.J. Contribution of protein kinase C to central sensitization and persistent pain following tissue injury. Neurosci. Lett. 1992;140:181–184. doi: 10.1016/0304-3940(92)90097-q. [DOI] [PubMed] [Google Scholar]
- Coste B., Xiao B., Santos J.S., Syeda R., Grandl J., Spencer K.S., Kim S.E., Schmidt M., Mathur J., Dubin A.E., et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483:176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.J., Reimann F., Nicholas A.K., Thornton G., Roberts E., Springell K., Karbani G., Jafri H., Mannan J., Raashid Y., et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlhamer J., Lucas J., Zelaya C., Nahin R., Mackey S., DeBar L., Kerns R., Von Korff M., Porter L., Helmick C. Prevalence of chronic pain and high-Impact chronic pain among adults—United States, 2016. MMWR Morb. Mortal. Wkly. Rep. 2018;67:1001–1006. doi: 10.15585/mmwr.mm6736a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk F., Bennett D.L., McMahon S.B. Nerve growth factor and pain mechanisms. Annu. Rev. Neurosci. 2017;40:307–325. doi: 10.1146/annurev-neuro-072116-031121. [DOI] [PubMed] [Google Scholar]
- Devinsky O., Vezzani A., Najjar S., De Lanerolle N.C., Rogawski M.A. Glia and epilepsy: excitability and inflammation. Trends Neurosci. 2013;36:174–184. doi: 10.1016/j.tins.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Dodick D.W. Migraine. Lancet. 2018;391:1315–1330. doi: 10.1016/S0140-6736(18)30478-1. [DOI] [PubMed] [Google Scholar]
- Dong X., Han S., Zylka M.J., Simon M.I., Anderson D.J. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–632. doi: 10.1016/S0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- Dunlop J., Bowlby M., Peri R., Vasilyev D., Arias R. High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology. Nat. Rev. Drug Discov. 2008;7:358–368. doi: 10.1038/nrd2552. [DOI] [PubMed] [Google Scholar]
- Eskander M.A., Ruparel S., Green D.P., Chen P.B., Por E.D., Jeske N.A., Gao X., Flores E.R., Hargreaves K.M. Persistent nociception triggered by nerve growth factor (NGF) is mediated by TRPV1 and oxidative mechanisms. J. Neurosci. 2015;35:8593–8603. doi: 10.1523/JNEUROSCI.3993-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geevasinga N., Menon P., Özdinler P.H., Kiernan M.C., Vucic S. Pathophysiological and diagnostic implications of cortical dysfunction in ALS. Nat. Rev. Neurol. 2016;12:651–661. doi: 10.1038/nrneurol.2016.140. [DOI] [PubMed] [Google Scholar]
- Gibbs J.L., Diogenes A., Hargreaves K.M. Neuropeptide Y modulates effects of bradykinin and prostaglandin E2 on trigeminal nociceptors via activation of the Y1 and Y2 receptors. Br. J. Pharmacol. 2007;150:72–79. doi: 10.1038/sj.bjp.0706967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich J., Alvarez P., Chen X., Levine J.D. GDNF induces mechanical hyperalgesia in muscle by reducing I(BK) in isolectin B4-positive nociceptors. Neuroscience. 2012;219:204–213. doi: 10.1016/j.neuroscience.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R.Z., Morita T., Brem R.B., Bautista D.M. S1PR3 mediates itch and pain via distinct TRP channel-dependent pathways. J. Neurosci. 2018;38:7833–7843. doi: 10.1523/JNEUROSCI.1266-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Abbott F.V. Behavioural effects of intraplantar injection of inflammatory mediators in the rat. Neuroscience. 1994;63:827–836. doi: 10.1016/0306-4522(94)90527-4. [DOI] [PubMed] [Google Scholar]
- Hrvatin S., Hochbaum D.R., Nagy M.A., Cicconet M., Robertson K., Cheadle L., Zilionis R., Ratner A., Borges-Monroy R., Klein A.M., et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 2018;21:120–129. doi: 10.1038/s41593-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-C., Yang W., Guo C., Jiang H., Li F., Xiao M., Davidson S., Yu G., Duan B., Huang T., et al. Anatomical and functional dichotomy of ocular itch and pain. Nat. Med. 2018;24:1268–1276. doi: 10.1038/s41591-018-0083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucho T., Levine J.D. Signaling pathways in sensitization: toward a nociceptor cell biology. Neuron. 2007;55:365–376. doi: 10.1016/j.neuron.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Lattin J.E., Schroder K., Su A.I., Walker J.R., Zhang J., Wiltshire T., Saijo K., Glass C.K., Hume D.A., Kellie S., Sweet M.J. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A., et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-L., Li K.-C., Wu D., Chen Y., Luo H., Zhao J.-R., Wang S.-S., Sun M.-M., Lu Y.-J., Zhong Y.-Q., et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26:83–102. doi: 10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-R., Fallahi-Sichani M., Sorger P.K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 2015;6:8390. doi: 10.1038/ncomms9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart O., Obreja O., Kress M. The inflammatory mediators serotonin, prostaglandin E2 and bradykinin evoke calcium influx in rat sensory neurons. Neuroscience. 2003;118:69–74. doi: 10.1016/s0306-4522(02)00960-0. [DOI] [PubMed] [Google Scholar]
- Liu B., Linley J.E., Du X., Zhang X., Ooi L., Zhang H., Gamper N. The acute nociceptive signals induced by bradykinin in rat sensory neurons are mediated by inhibition of M-type K+ channels and activation of Ca2+-activated Cl– channels. J. Clin. Invest. 2010;120:1240–1252. doi: 10.1172/JCI41084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E., Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Greenquist K.W., LaMotte R.H. Inflammatory mediators enhance the excitability of chronically compressed dorsal root ganglion neurons. J. Neurophysiol. 2006;95:2098–2107. doi: 10.1152/jn.00748.2005. [DOI] [PubMed] [Google Scholar]
- Madisen L., Mao T., Koch H., Zhuo J., Berenyi A., Fujisawa S., Hsu Y.-W.A., Garcia A.J., Gu X., Zanella S., et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian O.K., Asiedu M.N., Tillu D.V., Peebles K.A., Yan J., Ertz N., Dussor G.O., Price T.J. IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J. Neurosci. 2010;30:15113–15123. doi: 10.1523/JNEUROSCI.3947-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendieta D., De la Cruz-Aguilera D.L., Barrera-Villalpando M.I., Becerril-Villanueva E., Arreola R., Hernández-Ferreira E., Pérez-Tapia S.M., Pérez-Sánchez G., Garcés-Alvarez M.E., Aguirre-Cruz L., et al. IL-8 and IL-6 primarily mediate the inflammatory response in fibromyalgia patients. J. Neuroimmunol. 2016;290:22–25. doi: 10.1016/j.jneuroim.2015.11.011. [DOI] [PubMed] [Google Scholar]
- Moakley D., Koh J., Pereira J.D., DuBreuil D.M., Devlin A.-C., Berezovski E., Zhu K., Wainger B.J. Pharmacological profiling of purified human stem cell-derived and primary mouse motor neurons. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- Mørk H., Ashina M., Bendtsen L., Olesen J., Jensen R. Experimental muscle pain and tenderness following infusion of endogenous substances in humans. Eur. J. Pain. 2003;7:145–153. doi: 10.1016/S1090-3801(02)00096-4. [DOI] [PubMed] [Google Scholar]
- Murthy S.E., Loud M.C., Daou I., Marshall K.L., Schwaller F., Kühnemund J., Francisco A.G., Keenan W.T., Dubin A.E., Lewin G.R., Patapoutian A. The mechanosensitive ion channel Piezo2 mediates sensitivity to mechanical pain in mice. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickolls A.R., Lee M.M., Espinoza D.F., Szczot M., Lam R.M., Wang Q., Beers J., Zou J., Nguyen M.Q., Solinski H.J., et al. Transcriptional programming of human mechanosensory neuron subtypes from pluripotent stem cells. Cell Rep. 2020;30:932–946.e7. doi: 10.1016/j.celrep.2019.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.B., Tran P.B., Gillard S.E., Hurley R.W., Hammond D.L., Miller R.J. Chemokines and glycoprotein120 produce pain hypersensitivity by directly exciting primary nociceptive neurons. J. Neurosci. 2001;21:5027–5035. doi: 10.1523/JNEUROSCI.21-14-05027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okie S. A flood of opioids, a rising tide of deaths. N. Engl. J. Med. 2010;363:1981–1985. doi: 10.1056/NEJMp1011512. [DOI] [PubMed] [Google Scholar]
- Pinho-Ribeiro F.A., Verri W.A., Chiu I.M. Nociceptor sensory neuron-immune interactions in pain and inflammation. Trends Immunol. 2017;38:5–19. doi: 10.1016/j.it.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringkamp M., Schmelz M., Kress M., Allwang M., Ogilvie A., Reeh P.W. Activated human platelets in plasma excite nociceptors in rat skin, in vitro. Neurosci. Lett. 1994;170:103–106. doi: 10.1016/0304-3940(94)90249-6. [DOI] [PubMed] [Google Scholar]
- Rush A.M., Dib-Hajj S.D., Liu S., Cummins T.R., Black J.A., Waxman S.G. A single sodium channel mutation produces hyper- or hypoexcitability in different types of neurons. Proc. Natl. Acad. Sci. U S A. 2006;103:8245–8250. doi: 10.1073/pnas.0602813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz M., Osiander G., Blunk J., Ringkamp M., Reeh P.W., Handwerker H.O. Intracutaneous injections of platelets cause acute pain and protracted hyperalgesia. Neurosci. Lett. 1997;226:171–174. doi: 10.1016/S0304-3940(97)00269-3. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J., Foskolou S., Kilpinen H., Rodrigues J., Alasoo K., Knights A.J., Patel M., Goncalves A., Ferreira R., Benn C.L., Wilbrey A., Bictash M., Impey E., Cao L., Lainez S., Loucif A.J., Whiting P.J., HIPSCI Consortium. Gutteridge A., Gaffney D.J. Molecular and functional variation in iPSC-derived sensory neurons. Nat. Genet. 2018;50:54–61. doi: 10.1038/s41588-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal R.P., Wang X., Guan Y., Raja S.N., Woodbury C.J., Basbaum A.I., Edwards R.H. Injury-induced mechanical hypersensitivity requires C-low threshold mechanoreceptors. Nature. 2009;462:651–655. doi: 10.1038/nature08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shridas P., Zahoor L., Forrest K.J., Layne J.D., Webb N.R. Group X secretory phospholipase A2 regulates insulin secretion through a cyclooxygenase-2-dependent mechanism. J. Biol. Chem. 2014;289:27410–27417. doi: 10.1074/jbc.M114.591735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidders B., Karlsson A., Kitching L., Torella R., Karila P., Phelan A. Network-based drug discovery: coupling network pharmacology with phenotypic screening for neuronal excitability. J. Mol. Biol. 2018;430:3005–3015. doi: 10.1016/j.jmb.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Sills G. The mechanisms of action of gabapentin and pregabalin. Curr. Opin. Pharmacol. 2006;6:108–113. doi: 10.1016/j.coph.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Soltis R.D., Hasz D., Morris M.J., Wilson I.D. The effect of heat inactivation of serum on aggregation of immunoglobulins. Immunology. 1979;36:37–45. [PMC free article] [PubMed] [Google Scholar]
- Stacey P., Wassermann A.M., Kammonen L., Impey E., Wilbrey A., Cawkill D. Plate-based phenotypic screening for pain using human iPSC-derived sensory neurons. SLAS Discov. 2018;23:585–596. doi: 10.1177/2472555218764678. [DOI] [PubMed] [Google Scholar]
- Stirling L.C., Forlani G., Baker M.D., Wood J.N., Matthews E.A., Dickenson A.H., Nassar M.A. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain. 2005;113:27–36. doi: 10.1016/j.pain.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Teichert R.W., Memon T., Aman J.W., Olivera B.M. Using constellation pharmacology to define comprehensively a somatosensory neuronal subclass. Proc. Natl. Acad. Sci. U S A. 2014;111:2319–2324. doi: 10.1073/pnas.1324019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert R.W., Schmidt E.W., Olivera B.M. Constellation pharmacology: a new paradigm for drug discovery. Annu. Rev. Pharmacol. Toxicol. 2015;55:573–589. doi: 10.1146/annurev-pharmtox-010814-124551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than J.Y.-X.L., Li L., Hasan R., Zhang X. Excitation and modulation of TRPA1, TRPV1, and TRPM8 channel-expressing sensory neurons by the pruritogen chloroquine. J. Biol. Chem. 2013;288:12818–12827. doi: 10.1074/jbc.M113.450072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonello R., Fusi C., Materazzi S., Marone I.M., De Logu F., Benemei S., Gonçalves M.C., Coppi E., Castro-Junior C.J., Gomez M.V., et al. The peptide Phα1β, from spider venom, acts as a TRPA1 channel antagonist with antinociceptive effects in mice. Br. J. Pharmacol. 2017;174:57–69. doi: 10.1111/bph.13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D., Furlan A., Islam S., Abdo H., Lönnerberg P., Lou D., Hjerling-Leffler J., Haeggström J., Kharchenko O., Kharchenko P.V., et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2014;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Valtcheva M.V., Copits B.A., Davidson S., Sheahan T.D., Pullen M.Y., McCall J.G., Dikranian K., Gereau R.W. Surgical extraction of human dorsal root ganglia from organ donors and preparation of primary sensory neuron cultures. Nat. Protoc. 2016;11:1877–1888. doi: 10.1038/nprot.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco M., O’Sullivan C., Sheridan G.K. Lysophosphatidic acid receptors (LPARs): potential targets for the treatment of neuropathic pain. Neuropharmacology. 2017;113:608–617. doi: 10.1016/j.neuropharm.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Viana F., de la Peña E., Pecson B., Schmidt R.F., Belmonte C. Swelling-activated calcium signalling in cultured mouse primary sensory neurons. Eur. J. Neurosci. 2001;13:722–734. doi: 10.1046/j.0953-816x.2000.01441.x. [DOI] [PubMed] [Google Scholar]
- Wagner B.K., Schreiber S.L. The power of sophisticated phenotypic screening and modern mechanism-of-action methods. Cell Chem. Biol. 2016;23:3–9. doi: 10.1016/j.chembiol.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger B.J., Buttermore E.D., Oliveira J.T., Mellin C., Lee S., Saber W.A., Wang A.J., Ichida J.K., Chiu I.M., Barrett L., et al. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat. Neurosci. 2015;18:17–24. doi: 10.1038/nn.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger B.J., Kiskinis E., Mellin C., Wiskow O., Han S.S.W., Sandoe J., Perez N.P., Williams L.A., Lee S., Boulting G., et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zylka M.J. Mrgprd-expressing polymodal nociceptive neurons innervate most known classes of substantia gelatinosa neurons. J. Neurosci. 2009;29:13202–13209. doi: 10.1523/JNEUROSCI.3248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie J.A., Taugher R.J., Kreple C.J. Acid-sensing ion channels in pain and disease. Nat. Rev. Neurosci. 2013;14:461–471. doi: 10.1038/nrn3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang X., Li L., Yang G., Ouyang X., Xiang J., Zhang T., Min X. LASS2 inhibits proliferation and induces apoptosis in HepG2 cells by affecting mitochondrial dynamics, the cell cycle and the nuclear factor-κB pathways. Oncol. Rep. 2019 doi: 10.3892/or.2019.7058. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wu M., Zubcevic L., Borschel W.F., Lander G.C., Lee S.-Y. Structure of the cold- and menthol-sensing ion channel TRPM8. Science. 2018;359:237–241. doi: 10.1126/science.aan4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Reichert E., Cohen A.E. Optical electrophysiology for probing function and pharmacology of voltage-gated ion channels. eLife. 2016;5:e15202. doi: 10.7554/eLife.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Liu P., Bai L., Trimmer J.S., Bean B.P., Ginty D.D. Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron. 2019;103:598–616.e7. doi: 10.1016/j.neuron.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.-Q., Liu Z., Liu Z.-H., Chen S.-P., Li M., Shahveranov A., Ye D.-W., Tian Y.-K. Interleukin-6: an emerging regulator of pathological pain. J. Neuroinflammation. 2016;13:141. doi: 10.1186/s12974-016-0607-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete analysis pipeline for APPOINT (including MetaXPress cell segmentation script and R response classification script) are available at GitHub (https://github.com/waingerlab/APPOINT_analysis_script).