SUMMARY
To investigate circuit mechanisms underlying locomotor behavior, we used serial-section electron microscopy (EM) to acquire a synapse-resolution dataset containing the ventral nerve cord (VNC) of an adult female Drosophila melanogaster. To generate this dataset, we developed GridTape, a technology that combines automated serial-section collection with automated high-throughput transmission EM. Using this dataset, we studied neuronal networks that control leg and wing movements by reconstructing all 507 motor neurons that control the limbs. We show that a specific class of leg sensory neurons synapses directly onto motor neurons with the largest-caliber axons on both sides of the body, representing a unique pathway for fast limb control. We provide open access to the dataset and reconstructions registered to a standard atlas to permit matching of cells between EM and light microscopy data. We also provide GridTape instrumentation designs and software to make large-scale EM more accessible and affordable to the scientific community.
Keywords: serial-section electron microscopy, connectomics, limb control, sensory feedback, campaniform sensilla, motor neurons
Graphical Abstract
ETOC
GridTape, an automated transmission electron microscopy pipeline, is used to reconstruct all motor neurons controlling the limb at synapse resolution from an adult Drosophila ventral nerve cord to reveal pathways in motor control.
INTRODUCTION
To navigate a complex world, an animal’s nervous system must stimulate precise patterns of muscle contractions to produce coordinated body movements. Humans have more than 100,000 motor neurons (MNs) that innervate more than 100 million muscle fibers (Kanning et al., 2010). Limb MNs reside in the spinal cord, where neuronal networks integrate signals from the brain with sensory feedback from the body to coordinate limb movements. A century of studies in mammals has revealed many principles of spinal cord organization and development (Kiehn, 2011). However, we still lack a comprehensive understanding of the neuronal circuits that control motor output, largely because we do not know their wiring and connectivity.
The fruit fly, Drosophila melanogaster, is a particularly appealing system for studying mechanisms of motor control. Insects have a ventral nerve cord (VNC) that is homologous to the vertebrate spinal cord (Niven et al., 2008), but they lack vertebrae, making the VNC more experimentally accessible. Moreover, many insects have neurons that are uniquely identifiable across individuals, making insects well-established models for understanding the physiology of motor circuits (Burrows, 1996; Buschges et al., 2008). More specifically, Drosophila is a genetically accessible model system with complex and well-characterized behaviors including walking, flight, escape responses, grooming, and courtship. Recent technical advances have enabled in vivo electrophysiological recordings and calcium imaging of genetically identified VNC neurons in behaving adult Drosophila, providing insight into motor-related activity patterns during behavior (Chen et al., 2018; Mamiya et al., 2018; Tuthill and Wilson, 2016b). Furthermore, the small size of the Drosophila nervous system makes it suitable for comprehensive connectivity mapping using electron microscopy (EM). EM-based connectivity mapping was previously undertaken for a larval Drosophila nervous system (Ohyama et al., 2015; Schneider-Mizell et al., 2016) and an adult brain (Scheffer et al., 2020; Takemura et al., 2013; Tobin et al., 2017; Zheng et al., 2018), but not yet for an adult VNC. A VNC ‘connectome’—a map of its neurons and their synaptic connections—would enhance our understanding of how VNC circuits control muscles of the legs (Soler et al., 2004), neck (Strausfeld et al., 1987), wings (O'Sullivan et al., 2018), and halteres (Dickerson et al., 2019) to give rise to behavior.
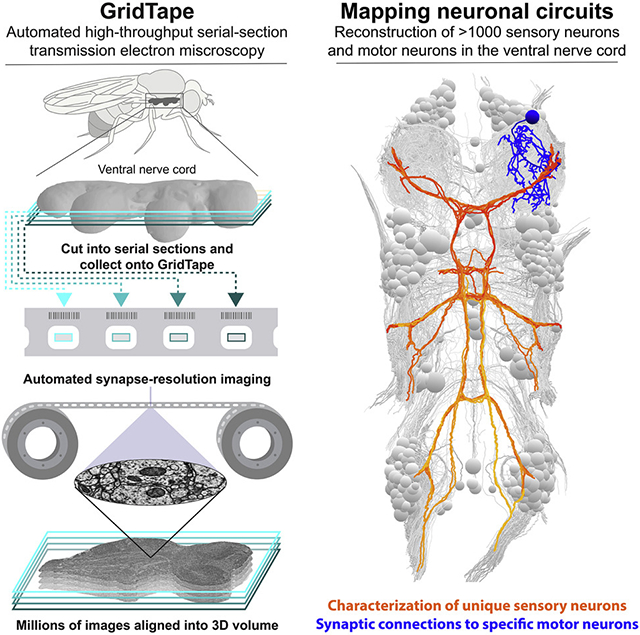
EM is the gold standard for mapping structural connectivity within neuronal circuits (Sjostrand, 1958; White et al., 1986). However, even seemingly small tissue volumes (1 mm3) acquired at synaptic resolution (e.g., 4×4×40 nm3 per voxel) produce massive datasets (>1500 teravoxels) that require automated methods for reliable acquisition in a reasonable amount of time. Recent developments in scanning EM (SEM) methods have enabled connectomic analyses of multiple circuits (Briggman et al., 2011; Hildebrand et al., 2017; Kasthuri et al., 2015; Kornfeld et al., 2017; Morgan et al., 2016; Schmidt et al., 2017; Tapia et al., 2012; Wanner et al., 2016). Compared to SEM, transmission EM (TEM) allows for higher spatial resolution (Nakane et al., 2020; Yip et al., 2020), an order of magnitude greater signal-to-noise (Xu et al., 2017; Zheng et al., 2018), and straightforward parallelization (Bock et al., 2011; Lee et al., 2016; Tobin et al., 2017; Zheng et al., 2018). Although there have been recent developments in motorized TEM section collection (Lee et al., 2018) and automated high-throughput TEM imaging (Zheng et al., 2018), we lack an end-to-end platform for automated large-scale TEM section collection and imaging. To address this, we designed a tape-based data acquisition pipeline that combines automated sectioning with a TEM-compatible collection substrate and an automated, reel-to-reel imaging stage. This technology, called GridTape, accelerates section collection and enables fully automated TEM imaging for a fraction of the cost of alternative systems.
Here, we used GridTape to produce a synapse-resolution EM dataset of an adult female Drosophila melanogaster VNC. We then reconstructed over 1,000 sensory and motor neurons in a neuronal network that controls limb movements to investigate wiring principles such as the organization of peripheral nerves and the uniqueness and bilateral symmetry of leg MNs. We registered the EM dataset to a light microscopy (LM)–based atlas, allowing us to find genetically identified neurons in the EM dataset based on their morphology. Through EM reconstruction, we found a class of leg proprioceptive neuron, the bilaterally projecting campaniform sensillum (bCS) neurons, that provide direct synaptic input onto the MNs with the largest-caliber axons in multiple legs. We identified the functionally characterized "fast" tibia flexor MN as a major synaptic target of bCS neurons based on registration of EM and LM morphology. Finally, we generated intersectional driver lines to genetically label bCS neurons, revealing their location in the leg, confirming their morphology in the VNC, and providing tools for future studies. We provide the EM dataset, neuron reconstructions, and designs for GridTape instrumentation as freely available resources.
RESULTS
GridTape: an accessible TEM platform for connectomics
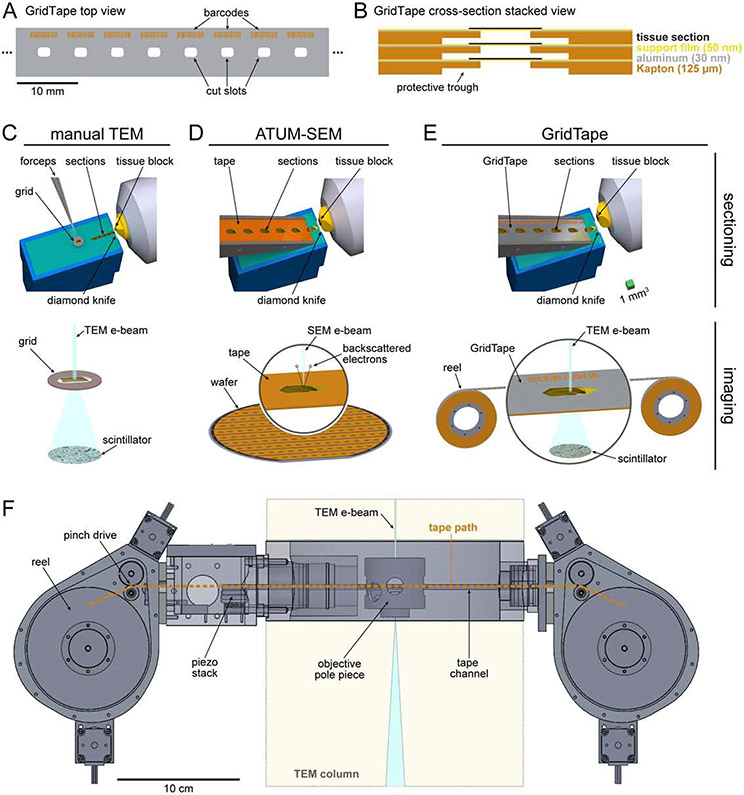
We developed GridTape, a TEM-compatible tape substrate (Fig. 1A-B) that combines advantages of section collection from the automated tape-collecting ultra-microtome SEM (ATUM-SEM) approach (Hayworth et al., 2014) with the advantages of TEM imaging (Fig. 1C-E). To produce GridTape, regularly spaced 2×1.5 mm2 holes resembling slots in conventional TEM grids are laser-milled through aluminum-coated polyimide tape (Fig. 1A). A 4 mm-wide trough is also milled from the uncoated surface so that the tape can be safely layered upon itself (Fig. 1B). The milled tape is coated with a 50 nm-thick film (LUXFilm®) that spans the slots to provide support for section collection. We collected sections onto GridTape using an ATUM modified for compatibility with GridTape (Fig. S1A-C). The tape is positioned near the knife edge so that sections consistently adhere to the moving tape as they are being cut. By monitoring the ultramicrotome cutting speed and adjusting the tape speed, the movement of GridTape slots is locked in-phase with cutting. This closed-loop sectioning approach permits automated collection of >4,000 sections per day, >10 times faster than manual section collection (Bock et al., 2011; Lee et al., 2016; Tobin et al., 2017; Zheng et al., 2018), with reliable positioning of sections over film-coated slots.
Figure 1. A high-throughput serial-section transmission electron microscopy (TEM) pipeline built around GridTape.
(A) Regularly spaced holes and barcodes are laser-milled into a length of tape to produce GridTape, a substrate for collection of serial sections.
(B) Schematic of stacked GridTape layers in cross-section. Tape thickness is exaggerated for clarity.
(C-E) Schematics of sectioning (top) and imaging (bottom) for different serial-section EM approaches. Bottom schematics do not share the same scale.
(C) Manual serial-section collection and TEM imaging. Samples are serially sectioned and manually picked up onto coated slot grids (3 mm outer diameter).
(D) Automated tape-collecting ultra-microtome (ATUM) serial-section collection and SEM imaging. Sections are collected automatically onto tape (8 mm wide). Tape is then cut into strips and adhered to a wafer (bottom) for imaging. Bottom inset: zoomed-in view of a section on tape.
(E) GridTape serial-section collection and TEM imaging. Samples are sectioned using a GridTape-compatible ATUM. Sections adhere to GridTape (8 mm wide) immediately after being cut and are targeted to land over film-coated holes in the tape. GridTape-collected sections are imaged using a reel-to-reel system. Bottom inset: zoomed-in view of a section on GridTape.
(F) Schematic of the GridTape reel-to-reel stage. Reels of GridTape are inserted into the custom stage, which positions sections under the electron beam. Portions of the TEM microscope column in beige. Electron beam in light blue (not to scale).
Scale bars, 10 mm (A), 10 cm (F). Scale box, 1 mm (C-E, top).
Collecting sections onto thin films enables widefield TEM imaging. To automate the imaging process, we engineered a stage that attaches to standard TEM microscopes and houses GridTape reels in vacuum (Fig. 1F). Tape housings were added on opposite sides of the microscope column to allow motors to feed the tape between the two sides and position sections under the electron beam. To image large areas at synaptic resolution, the microscope automatically montages each section using piezoelectric nanopositioners. The tape is then translated to position the next section for montaging, enabling continuous unattended operation. Using a 2×2 camera array (Bock et al., 2011), we achieve effective imaging rates of >40 Mpixels per second (Table S1). This microscope, termed TEMCA-GT (Transmission Electron Microscope with a Camera Array and GridTape), provides high-throughput imaging at a relatively low cost of ~US$300,000 per microscope (Table S1-S2).
GridTape enables rapid acquisition of a VNC EM dataset
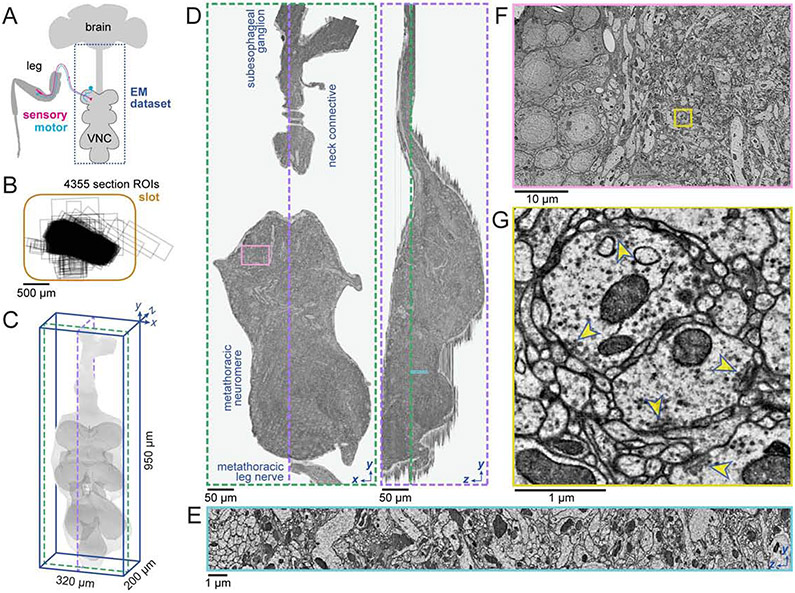
To map circuits underlying motor control, we acquired a dataset encompassing an adult female Drosophila VNC that consists of 86.3 trillion voxels and spans 21 million μm3 (Fig. 2). The dataset includes the VNC, neck connective, and a portion of the brain’s subesophageal ganglion (Fig. 2A, C-G). The imaged volume was captured from 4355 serial coronal sections, each cut around 45 nm thick and collected onto GridTape continuously over 27 hr (22.1 sec per section). Of these sections, 98% were positioned within 0.37 mm of the average section position, with only six sections having 20% or more of the VNC off the imageable slot area (Figs. 2B, S1D; STAR Methods). An additional three sections were lost before imaging due to support film damage. No off-slot or lost sections were consecutive. Imaging at 4.3×4.3 nm2 per pixel resolution required 60 continuous days on one TEMCA-GT at a rate of 42.73±3.04 Mpixels per sec (mean±SD across sections), an order of magnitude faster than most volumetric EM approaches (Table S1). This amounted to 20.6 million images, or 172.6 TB of raw data.
Figure 2. An adult Drosophila ventral nerve cord (VNC) dataset.
(A) Schematic of the adult Drosophila central nervous system and leg. The synapse-resolution EM dataset presented here contains the VNC and its connection to the brain (dashed outline).
(B) The VNC was cut into 4355 thin sections and collected onto GridTape. Each black rectangle indicates the imaged region for a single section relative to the slot (orange outline). Two sections collected off-slot are not shown.
(C) Volumetric rendering of the VNC dataset. Light grey, outline of all imaged tissue. Dark grey, outline of VNC neuropil.
(D) A single coronal section (left, section 1228) and sagittal reslice through the aligned image volume (right). Green and purple dashed lines in (C) and (D) indicate the slice locations. The imaged region spans from the subesophageal ganglion in the ventral brain, across the neck connective to the metathoracic neuromere and the metathoracic leg nerve.
(E) Zoomed-in sagittal reslice of the region (cyan box) in (D).
(F) Zoom-in of the region (pink box) in (D).
(G) Zoom-in of the region (yellow box) in (F) showing synapses. Yellow arrowheads indicate presynaptic specializations known as T-bars.
Scale bars, 500 μm (B), 50 μm (D), 10 μm (F), 1 μm (E, G).
We adapted a software pipeline (STAR Methods) to align the images into a three-dimensional (3D) volume (Video S1). While alignment of serial-section TEM data has historically been a challenge due to irregularities in the sectioning process, GridTape section collection provided consistent placement of serial sections, with few partial, lost, or damaged sections (STAR Methods). This facilitated the high-quality alignment of serial sections (Fig. 2E) required for efficient and accurate connectomic reconstruction.
Motor and sensory neurons occupy distinct domains within peripheral nerves
After alignment, we searched for peripheral nerves carrying axon bundles to and from the VNC. We found all previously described nerves that innervate the legs, wings, halteres, and neck (Court et al., 2020; Power, 1948). For individual neurons passing through each nerve, we reconstructed skeletonized models of their projections within the VNC. Reconstructed neurons fell into two major morphological categories corresponding to MNs and sensory neurons (Baek and Mann, 2009; Brierley et al., 2012; Mamiya et al., 2018; Tsubouchi et al., 2017). MNs had cell bodies located in the VNC, projected to a dorsal layer of the VNC, and did not contain synaptic vesicles or presynaptic specializations within the neuropil (Fig. 3A, Video S2). Sensory neurons did not have cell bodies in the VNC, arborized more ventrally, and made synaptic outputs within the neuropil (Fig. 3B, Video S2). An additional 20 neurons did not fall into one of these two main categories (Fig. S2). Consistent with previous reports, we also counted 3738 axons travelling between the brain and VNC via the neck connective (Coggshall et al., 1973).
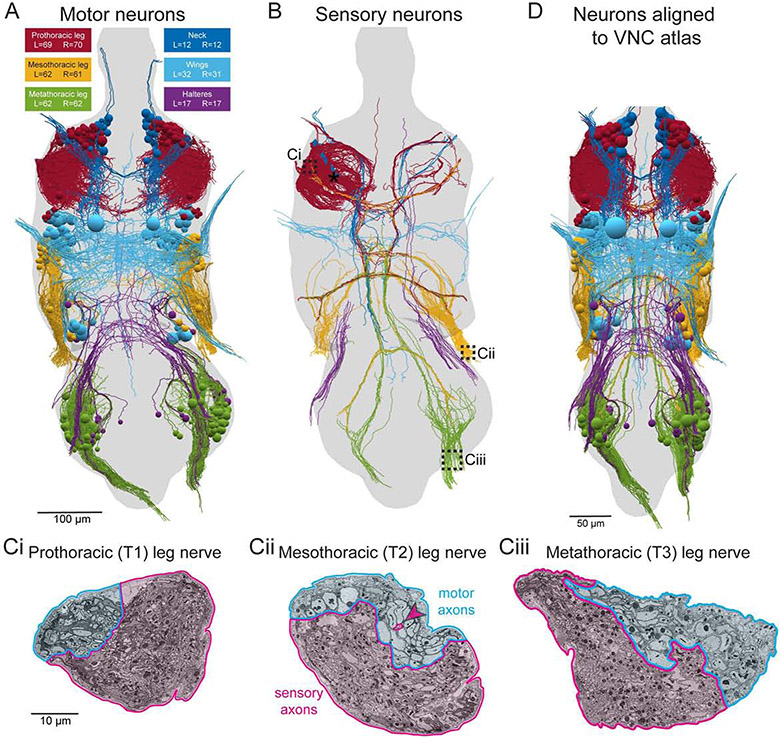
Figure 3. Reconstruction of motor and sensory neurons reveals precise functional domains in nerves.
(A) All 507 motor neurons (MNs) in the VNC’s thoracic segments were reconstructed from the EM dataset. Each MN projects its axon out one peripheral nerve, leaving the EM dataset, to innervate muscles. Spheres represent cell bodies. Unless otherwise noted, all renderings are viewed from the dorsal side of the VNC.
(B) 655 reconstructed sensory axons. Reconstruction included some neurons from all limbs but focused primarily on the left T1 neuromere (asterisk). Same color code as (A).
(C) Sections through the prothoracic (T1), mesothoracic (T2), and metathoracic (T3) leg nerves, which contain most of the sensory and motor axons connecting the VNC to the front, middle, and hind legs, respectively. Section locations indicated by dashed boxes in (B). The leg nerves have distinct domains containing the axons of MNs (cyan) and sensory neurons (magenta). The only intermingling between motor and sensory axons is a group of three sensory axons within the motor domain of the T2 leg nerve (magenta arrowhead).
(D) Reconstructions transformed into a standard atlas coordinate space (Fig. S3). Renderings of EM reconstructions in subsequent figures were transformed into the atlas space.
Scale bars, 100 μm (A-B), 10 μm (C) 50 μm (D).
We focused on reconstructing neurons projecting through the VNC’s peripheral nerves. We found that motor and sensory axons segregated into distinct spatial domains within leg nerves (Fig. 3C), consistent with findings in larger insects (Zill et al., 1980). Sensory axons outnumbered MN axons by an order of magnitude in most nerves. For example, we found 863 sensory and 42 MN axons in the left prothoracic leg nerve (ProLN). By reconstructing neurons in the motor domain of each nerve, we identified and partially reconstructed a total of 507 MNs in the VNC’s thoracic segments (Fig. 3A, Video S2). Together with 13 octopaminergic unpaired median (UM) neurons (Duch et al., 1999) and two “multinerve” neurons (Fig. S2), these reconstructions encompass the complete set of neurons that this VNC used to control the muscles of the legs, wings, halteres, and neck (Fig. 3A). Additionally, of the >6500 sensory neurons we counted in the sensory domains of nerves, we first partially reconstructed 655 (Fig. 3B, Video S2). Of the ~900 sensory neurons entering the left T1 neuromere, we reconstructed the main branches of 392 (Fig. 3B, asterisk), focusing primarily on proprioceptive sensory neurons.
Registering the EM dataset to a standard atlas
We registered the EM dataset to a standard atlas to place the EM reconstructions into a reference coordinate system (Figs. 3D, S3). The VNC atlas is a map of synapse density based on light microscopy (LM) imaging of fluorescently labeled presynaptic sites (Bogovic et al., 2019). To register the EM dataset to the atlas, we first estimated synapse density across the EM dataset using an artificial neural network (Buhmann et al., 2019) trained to identify synapses based on their ultrastructural features (Fig. S3; STAR Methods). The synapse predictions were as accurate (precision: 71.4%, recall: 72.8%) as those in a whole-brain TEM dataset (Buhmann et al., 2019). We used the synapse density map derived from these predictions to align the EM dataset to the atlas (Figs. 3D, S3G, Video S3). Registering to the atlas compensated for asymmetries introduced by specimen preparation and facilitated quantitative comparisons between EM and LM reconstructions for neuron identification (Figs. 4, 7)
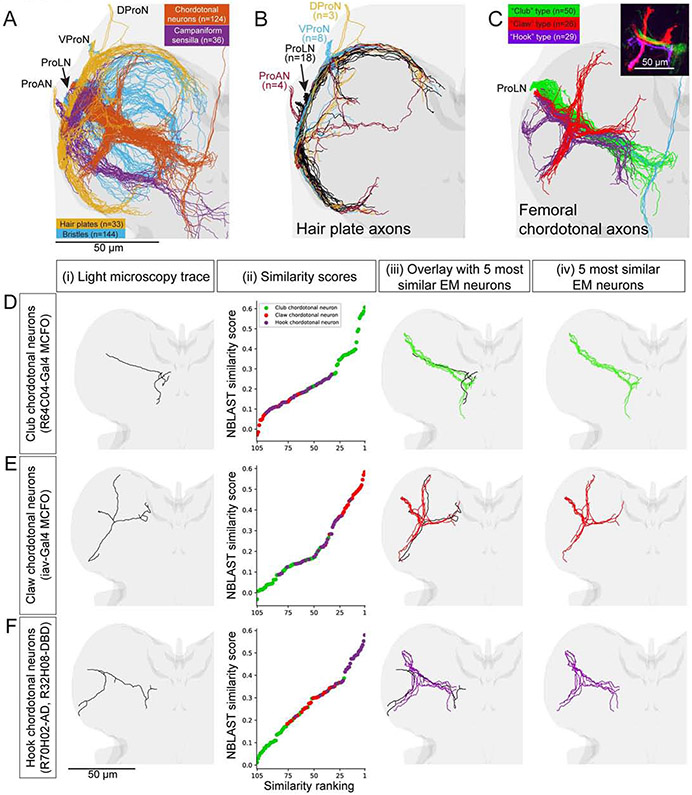
Figure 4. Identification of sensory neuron subtypes.
(A) Reconstruction of the main branches of sensory axons for the front left leg. The four main functional subtypes of sensory neurons (different colors) are identifiable from their projection patterns. Light grey, VNC. Darker grey, neuropil. ProAN, prothoracic accessory nerve; ProLN, prothoracic leg nerve; VProN, ventral prothoracic nerve; DProN, dorsal prothoracic nerve.
(B) Organization of hair plate neuron projections. Hair plate axons enter the T1 neuromere through four different nerves (different colors) and branch to encircle the neuromere.
(C) Femoral chordotonal organ (FeCO) neuron subtypes. Inset: Different subtypes, characterized previously with light microscopy (LM), encode different aspects of leg kinematics (adapted from Mamiya et al., 2018).
(D-E) Comparison of EM reconstructions with LM reconstructions from genetic driver lines that specifically label FeCO neurons (Mamiya et al. 2018, Chen et al. in preparation). (i) Rendering of LM reconstruction. (ii) Ranked distribution of NBLAST similarity scores (worst to best, left to right) color coded by FeCO neuron subtype (as in C). (iii) Overlay of the LM reconstruction and the five most similar EM reconstructions. (iv) The five most similar EM reconstructions alone.
(D) A club FeCO neuron (MCFO from R64C04-Gal4).
(E) A claw FeCO neuron (MCFO from iav-Gal4).
(F) A hook FeCO neuron (R70H02-AD, R32H08-DBD).
Scale bars, 50 μm (A-F).
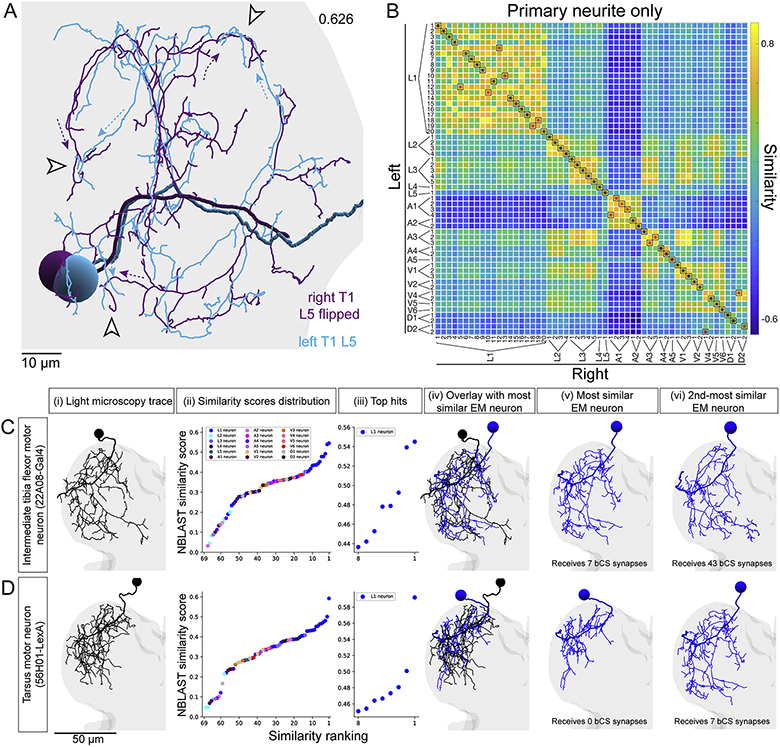
Figure 7. A fast tibia flexor motor neuron is a major synaptic target of bCS neurons.
(A-C) MNs reconstructed from LM matched to the most similar neurons reconstructed from EM. (i) Rendering of LM reconstruction. (ii) Ranked distribution of NBLAST similarity scores (worst to best, left to right) color coded by MN bundle (key, Aii top). (iii) Zoom-in on the 8 highest similarity scores. (iv) Overlay of the LM reconstruction and the most similar EM reconstruction. (v) The most similar EM reconstruction. (vi) The second-most similar EM reconstruction.
(A) A fast tibia flexor MN (81A07-Gal4). The two most similar EM reconstructions both receive strong synaptic input from the two left and two right T1 bCS neurons.
(B) A slow tibia flexor MN (35C09-Gal4). The two most similar EM reconstructions receive minimal synaptic input from T1 bCS neurons.
(C) A MN innervating the tibia long tendon muscle, which controls movements of the tarsus (21G01-LexA). The two most similar EM reconstructions receive no synaptic input from T1 bCS neurons.
Scale bars, 50 μm (A-C).
Identifying sensory neuron classes
Reconstructed leg sensory axons typically had a projection pattern corresponding to one of the four most numerous classes of sensory neurons (Tsubouchi et al., 2017; Tuthill and Wilson, 2016b): hair plate (Merritt and Murphey, 1992), chordotonal (Mamiya et al., 2018), campaniform sensillum (CS) (Merritt and Murphey, 1992), and bristle (Murphey et al., 1989) neurons (Fig. 4A, Video S4; STAR Methods for classification criteria). We reconstructed the main branches of every proprioceptive axon originating from the left front leg and arborizing in the left T1 neuromere (Fig. 4A-C). We found numbers of hair plate (n=33), chordotonal (n=124), and CS (n=36) neurons consistent with previous reports (Kuan et al., 2020; Mamiya et al., 2018; Merritt and Murphey, 1992). We also reconstructed 144 of ~600 putative bristle neuron axons. An additional 55 of 392 left T1 sensory axon reconstructions did not clearly fall into one of these classes.
The chordotonal axons could be further divided into subtypes matching the “club” (n=50), “claw” (n=26), and “hook” (n=29) morphologies (Fig. 4C) known to encode leg vibration, position, and velocity, respectively (Mamiya et al., 2018). Five additional chordotonal axons ascended directly to the brain (Tsubouchi et al., 2017). Another 14 of 124 left T1 chordotonal axons did not fall clearly into one of these subtypes.
We tested our ability to identify neuron classes quantitatively by comparing EM reconstructions with LM reconstructions (Figs. 4C-F, S4). We used LM data from Gal4 and split-Gal4 fly lines that label known subtypes of sensory neurons, as well as data from MultiColor FlpOut (MCFO) experiments (Mamiya et al., 2018; Meissner et al., 2020). By performing NBLAST similarity searches (Costa et al., 2016) between the registered EM and LM reconstructions (STAR Methods), we confirmed our identifications of the major leg sensory neuron classes (Fig. S4) and chordotonal subtypes (Fig. 4D-F).
Bilaterally projecting leg sensory neurons co-activate motor neurons innervating different legs
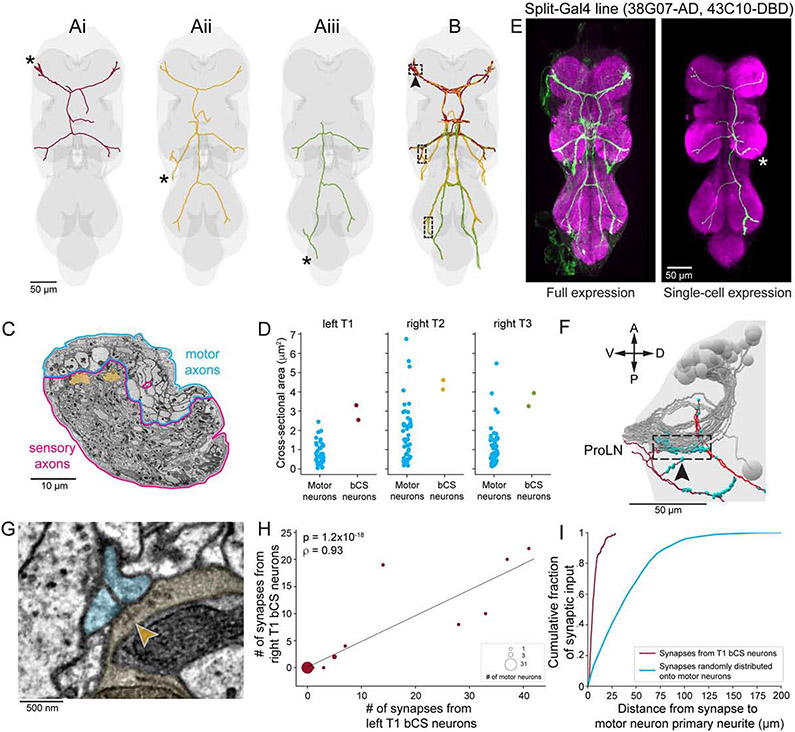
Campaniform sensilla are proprioceptive mechanoreceptors that encode load on a fly’s leg by detecting mechanical strain in the exoskeleton (Pringle, 1938; Tuthill and Azim, 2018; Zill and Moran, 1981). We identified CS axons in the EM volume by their similarity to three morphological types described in larger fly species (Merritt and Murphey, 1992). The first type projects only to the neuromere corresponding to its leg of origin. The second projects to ipsilateral neuromeres corresponding to other legs on the same side of the body. The third—which we call bilateral CS (bCS) neurons—projects to multiple ipsilateral and contralateral neuromeres. bCS neurons had multiple striking features. They were the only type of leg sensory neuron to project across the midline (Fig. 5A-B). Their axons had the largest caliber of any leg sensory neuron (Fig. 5C), exceeding the caliber of most MN axons (Fig. 5C-D). Their average cross-sectional area was 3.63±0.74 μm2 (mean±SD deviation, n=6 axons in 3 nerves), larger than 94–100% (Fig. 5D) of MN axons in each respective nerve. By reconstructing the sensory neurons from each leg with the largest caliber axons and examining their projections, we found 12 bCS neurons in total in the VNC, with two originating in each of the six legs (Fig. 5A-B). All bCS neurons from the front legs projected to the front and middle leg neuromeres (Fig. 5Ai), those from the middle leg projected to all neuromeres (Fig. 5Aii), and those from the hind legs projected to the hind and middle leg neuromeres (Fig. 5Aiii).
Figure 5. Bilateral campaniform sensillum (bCS) neurons from both sides of the body directly connect to MNs near their spike-initiation zones.
(A) Single bCS axons from the front (i), middle (ii), and hind (iii) left legs. Asterisks denote where each axon enters the VNC.
(B) Two neurons with the morphologies shown in (A) originate from each of the six legs. Dashed boxes indicate a ~25 μm-long tract where bCS axons originating from one leg travel alongside bCS axons originating from other legs.
(C) Right mesothoracic (T2) leg nerve. bCS axons (yellow) have large-caliber axons compared to other leg sensory and motor neurons.
(D) Cross-sectional areas of bCS axons and MN primary neurites for three different legs.
(E) A split-Gal4 line labeling bCS neurons. Full expression pattern (left) and a single bCS axon labeled using MultiColor FlpOut (Nern et al., 2015).
(F) Lateral view of the location indicated by the arrowhead in (B). A, anterior; P, posterior; V, ventral; D, dorsal. In the boxed region, bCS axons originating from left T1 (dark red), right T1 (light red), and left and right T2 (not shown) converge, traveling directly alongside primary neurites of ProLN MNs (grey; same neurons as Fig. S6C). bCS output synapses denoted by cyan spheres.
(G) Synapse from a right T2 bCS axon (yellow) onto two left T1 MNs (cyan). Arrowhead indicates presynaptic T-bar structure. All 12 bCS neurons synapse onto MNs in each neuromere to which they project.
(H-I) Analysis of all synaptic connections made by left and right T1 bCS axons along the ~25 μm stretch indicated in (F).
(H) Connections from left T1 versus right T1 bCS axons onto left ProLN MNs. The two left bCS axons and two right bCS axons largely target the same MNs.
(I) Distribution of distances from each bCS synapse to each postsynaptic MN’s primary neurite (red, n=264 postsynaptic sites) compared to synapses randomly distributed across MN dendrites (cyan).
Scale bars, 50 μm (A-B, E-F), 10 μm (C), 500 nm (G).
Guided by the EM reconstructions, we generated two independent split-Gal4 driver lines that genetically target the bCS neurons. Imaging fluorescent reporters driven by these lines confirmed that our EM reconstructions included all major branches in the VNC (Fig. 5E) and further revealed that bCS neurons innervate campaniform sensilla in a proximal leg segment, the trochanter (Fig. S5A).
Studies in other insects have shown that trochanter CS detect increased load on the body and activate muscle synergies to increase grip on surfaces (Zill et al., 2015). Notably, bCS axons had branches and made synapses directly alongside many leg MN primary neurites—the main branch between the cell body and the axon (Fig. 5F)—where MN action potentials are likely initiated (Gwilliam and Burrows, 1980). Therefore, we hypothesized that bCS neurons provide direct input to MNs to increase substrate grip using multiple legs. To test this, we first asked whether bCS axons synapse directly onto leg MNs. Indeed, all 12 bCS axons made synapses onto MNs in all the neuromeres to which they projected (Fig. 5G). To determine how frequently bCS synapses targeted MNs, we reconstructed all synapses from left and right T1 bCS neurons along the ~25 μm-long branch in left T1 where their axons travel alongside MN primary neurites (Fig. 5F). In this region, left T1 bCS axons made 96 synapses and right T1 bCS axons made 49, of which 98.6% (143 of 145) had at least one MN as a postsynaptic partner. There were 3.01±1.23 (mean±SD) postsynaptic partners per synapse, totaling 437 postsynaptic sites. Of these, 64.8% belonged to MNs, 21.1% belonged to central neurons (whose arborizations remained within the central nervous system), and 14.2% could not be classified (STAR Methods). Compared to MNs, central neurons received far fewer synapses from bCS neurons (Fig. S5B). Most postsynaptic central neurons (Fig. S5C, asterisk) appeared to be members of lineage 19A (Harris et al., 2015), but others exhibited a variety of dendritic and axonal arborization patterns (Fig. S5C-D).
Next, we next analyzed the connectivity with MNs, the predominant target of the reconstructed bCS synapses. We first found that the two bCS axons from the same leg synapsed onto the same MN subpopulation. Specifically, the number of synapses a given MN received from each left T1 bCS neuron was highly correlated (Fig. S5E, Spearman’s ρ=0.95, p=2.5×10−22, n=42 neurons). Inputs from the two right T1 bCS neurons were also correlated (Fig. S5F, Spearman’s ρ=0.93, p=2.6×10−19, n=42 neurons). Notably, left and right T1 bCS axons synapsed onto the same MNs. The five MNs receiving the most synapses from left T1 bCS neurons were also the top five targets of the right T1 bCS neurons (Fig. 5H, Spearman’s ρ=0.93, p=1.2×10−18, n=42 neurons). As predicted from their proximity, bCS neurons synapsed directly onto the MN primary neurite or onto short (<10 μm) second-order branches (Fig. 5I), making bCS synapses well-positioned to stimulate spikes in MNs.
Uniqueness of leg motor neurons
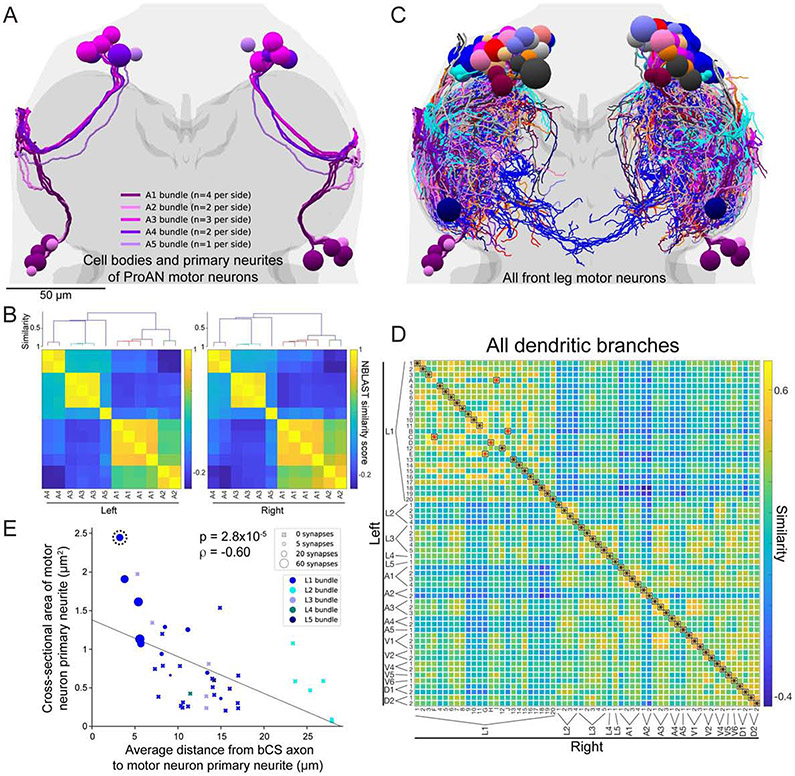
Our next goal was to identify which MNs were postsynaptic targets of bCS neurons. Leg MNs originate from ~15 developmental lineages (Baek and Mann, 2009; Brierley et al., 2012), and neurons from the same lineage have their primary neurites bundled together (Shepherd et al., 2016). Consistent with this, the EM-reconstructed front leg MN primary neurites appeared spatially clustered, forming 18 distinct bundles within the left and right T1 neuromeres (Figs. 6A, S6A-B). The spatial organization of these 18 bundles also appeared to be mirror symmetric, matching one-to-one between left and right sides (Figs. 6A, S6A,C-E). The largest right-side bundle contained 30 primary neurites, one more than the 29 found in the largest left-side bundle, but the other 17 bundles contained identical numbers on both sides. We used a clustering analysis to quantitatively demonstrate the existence and symmetry of these bundles (Fig. 6B, S6C-E). Additionally, their spatial clustering was maintained in peripheral nerves, where members of each bundle remained adjacent with no intermixing (Fig. S6F). Consistent with the hypothesis that these bundles correspond to lineages, the largest bundles we identified (L1 in Fig. S6C) closely matched previous reports of the largest MN lineage Lin15B in gross morphology and number of neurons per hemisphere (Brierley et al., 2012). Notably, all 11 MNs receiving synaptic input from bCS neurons were members of this largest bundle, with no bCS connections to other bundles. The remaining 17 clusters may correspond to the other 14 MN lineages (Baek and Mann, 2009), but determining their correspondence requires future work.
Figure 6. MN bundles, symmetry, uniqueness, and bCS connectivity.
(A) Reconstruction of cell bodies and primary neurites of the 24 ProAN MNs (12 per side). Primary neurites travel through the neuromere in five distinct and highly symmetric bundles (numbered A1 through A5, colored in shades of purple). See also Fig. S6.
(B) Quantitative analysis of bundles of MN primary neurites. ProAN MNs on each side of the VNC were clustered by the similarity in primary neurite positions (STAR Methods). Top, dendrogram from hierarchical clustering. Members of each bundle cluster together. Bottom, matrix of NBLAST similarity scores.
(C) Branching patterns of all 139 MNs arborizing in the T1 neuromeres were reconstructed and transformed into the atlas coordinate system (Fig. S3).
(D) Identification of left–right homologous pairs of front leg MNs. Of the 69 left and 70 right T1 MNs, expert annotators identified 61 symmetric left–right pairs. A global pairwise assignment of NBLAST similarity scores agreed on 92% (56 of 61) of identified pairs. Black asterisks, agreements. Red asterisks, disagreements.
(E) Relationship between four anatomical properties of leg MNs: proximity to bCS axons (x-axis), primary neurite cross-sectional area (y-axis), primary neurite bundle (marker color), and number of synapses received from bCS neurons (marker type and size). MNs closer to bCS axons have larger-caliber primary neurites. Only MNs in the L1 bundle received any synapses from bCS neurons. Within the L1 bundle, those receiving the most synapses have large-caliber primary neurites and are closest to bCS axons. Dashed circle indicates the MN whose morphology is most similar to a functionally characterized fast flexor MN (Fig. 7A).
Scale bars, 50 μm (A, C).
We next asked whether MNs that receive bCS input had uniquely identifiable morphologies, which would permit their correspondence with LM reconstructions. Most MNs have unique and stereotyped innervations of muscle fibers in the leg (Baek and Mann, 2009; Brierley et al., 2012), but their dendritic arborizations in the VNC are more complex. MNs innervating the same leg segment have similar dendritic arborizations (Baek and Mann, 2009; Brierley et al., 2012), so the number of leg MNs with uniquely identifiable dendritic morphologies is unclear. To investigate this, we first extended our reconstructions of all 139 MNs for both front legs to include their largest dendritic branches (Fig. 6C). We then tested whether MNs were individually identifiable by searching for left–right pairs of MNs with unique, symmetric dendritic arbors. Of 69 possible left–right pairings, we manually identified 61 putative homologous pairs with distinct, matching branching patterns (Video S5). To quantitatively confirm our assignments, we computed NBLAST similarity scores between left and right front leg MNs after transforming each into the atlas coordinate space and reflecting right-side neurons across the midplane. From the similarity scores, we used an algorithm to generate a globally optimal, one-to-one assignment of left–right pairs (STAR Methods). These assignments matched 92% (56 of 61) of the manual pairings, demonstrating that most leg MNs have uniquely identifiable and symmetric dendritic morphologies (Fig. 6D). Of 11 MNs receiving bCS synapses, eight had uniquely identifiable morphologies by these criteria, including all five of those receiving >20 bCS synapses.
While dendritic arbors of left–right pairs were largely mirror symmetric, there was some variability in their branches. We often observed higher-order branches following different paths to reach the same terminal zones (Fig. S7A), matching observations in larval Drosophila (Schneider-Mizell et al., 2016). In contrast, primary neurites were sufficiently symmetric both within and across bundles that most homologous pairs could be identified based on their primary neurites alone (Fig. S7B). In summary, most MNs have uniquely identifiable dendritic arbors and their primary neurites are positioned precisely within bundles, but finer dendritic branches are often variable.
Fast flexor MNs are major postsynaptic targets of bCS neurons
To determine the rules governing how bCS neurons distribute synapses onto MNs, we measured two additional characteristics of each ProLN MN primary neurite: cross-sectional area and average distance from the bCS axon segments indicated in Figure 5F. These variables were correlated, such that MNs with the largest-caliber primary neurites were more posterior (closer to the bCS axons) than those with smaller caliber primary neurites (Fig. 6E, Spearman’s ρ=−0.60, p=2.8×10−5, n=42 neurons). Both of these characteristics were correlated with the number of synaptic inputs from bCS neurons (bCS inputs vs primary neurite area: Spearman’s ρ=0.52, p=3.7×10−4; bCS inputs vs distance: Spearman’s ρ=−0.52, p=4.0×10−4, both n=42 neurons). The five most highly connected MNs had large-caliber and posteriorly positioned primary neurites (Fig. 6E). Additionally, bCS synapses targeted 11 of 29 MNs in the largest bundle, but none of the 13 MNs in the other four bundles of ProLN MNs despite some having large, posterior primary neurites (Fig. 6E). Taken together, primary neurite bundle identity, positioning along the anterior–posterior axis, and caliber are all strong predictors for whether a MN receives synaptic input from bCS axons.
Because bCS neurons synapse onto MNs with large-caliber primary neurites, we hypothesized that bCS neurons target “fast” MNs that control large ballistic movements, but not “slow” MNs that control small postural movements (Azevedo et al., 2020). To investigate this, we genetically targeted one fast and one slow MN controlling the tibia flexor muscles of the front leg for whole-cell recording, filled them with dye via the patch pipette, and imaged them using LM. We then reconstructed the neurons, registered the LM reconstructions to the atlas (Fig. S3), and calculated NBLAST similarity scores with the 69 EM-reconstructed left front leg MNs to search for morphologies resembling the fast or slow tibia flexor MNs (Fig. 7A-B). For the fast MN, the highest-scoring EM reconstruction had a highly similar dendritic structure, the largest-caliber primary neurite of any searched EM reconstructions (dashed circle in Fig. 6E), and was one of the five major synaptic targets of T1 bCS neurons (Fig. 7A, Video S6). For the slow MN, the highest scoring EM reconstruction had a highly similar structure, a small-caliber primary neurite, and received no synapses from bCS neurons (Fig. 7B, Video S6).
We repeated the matching process for one functionally characterized “intermediate” tibia flexor MN (Azevedo et al., 2020) and two MNs controlling movements of a different leg segment, the tarsus (Figs. 7C, S7C-D). Neither tarsus MN matched EM reconstructions receiving strong input from bCS neurons (Figs. 7C, S7D). Whether the intermediate MN received bCS input was less conclusive (Fig. S7C). Overall, in four of five cases, we successfully matched MN morphologies between EM and LM to link connectivity with functional identity. In this way, we identified a fast tibia flexor MN as a major synaptic target of bCS neurons.
DISCUSSION
Large-scale neuronal wiring diagrams at synapse resolution will be a crucial element of future progress in neuroscience. Here, we present GridTape, a technology for accelerating large-scale EM data acquisition. We demonstrated the power of this approach by acquiring a dataset encompassing an adult female Drosophila VNC. We then used this dataset to identify a monosynaptic circuit that directly links a specialized proprioceptive cell type, the bCS neurons, with specific motor neurons. Our results highlight how EM datasets can be used to characterize cell types and guide development of cell type-specific driver lines. The public release of this dataset provides a resource for studying the circuit connectivity underlying motor control and demonstrates the rapid advances that can be powered by the GridTape approach.
An accessible TEM pipeline for connectomics
Data acquisition remains a rate-limiting step in generating EM connectomics datasets. Manual sectioning for TEM is slow, imprecise, and unreliable. Meanwhile, SEM approaches that circumvent the need for manual sectioning have slow imaging speeds or require massive parallelization of expensive electron optics to acquire comparable datasets (Table S1). GridTape builds on previous efforts toward TEM parallelization and automation (Bock et al., 2011; Peltier et al., 2005; Zheng et al., 2018), but overcomes the need for manual sectioning, allowing faster and more consistent section collection and imaging. Because imaging is nondestructive, GridTape is compatible with enhancement by post-section labeling and allows for re-imaging. By eliminating the need to separately handle thousands of fragile sections, GridTape reduces data loss and artifact frequency. This results in better alignment of sections into a coherent, high signal-to-noise image volume, leading to efficient and accurate reconstructions.
GridTape is also less expensive than high-throughput SEM platforms. For the current price of one commercial multi-beam SEM system (Eberle et al., 2015), ten TEMCA-GTs can be built, and samples collected on GridTape can be distributed across microscopes for simultaneous imaging. The fixed microscope hardware costs are accompanied by consumable costs associated with support film coating (~USD$4 per slot, or ~$18,000 for this study), but we expect this cost to decrease due to technological improvements and economies of scale.
In the future, GridTape acquisition rates will increase as cameras and imaging sensors improve. Because TEM imaging is a widefield technique, imaging throughput can be increased by using larger camera arrays and brighter electron sources. Moreover, sections larger than current slot dimensions could be accommodated with wider tape and larger slots, although custom microscopes may be necessary for very large samples and slot size will depend on material properties of the support film (STAR Methods).
A synapse-resolution adult Drosophila VNC dataset
The EM dataset presented here provides a public resource for understanding how the Drosophila nervous system generates behavior. We chose to image an adult Drosophila VNC because it is an ideal test case for generating and validating a connectomic dataset. The circuit is genetically and electrophysiologically accessible and neurons are identifiable across individuals. The VNC is compact, containing about a third of the neurons in the adult central nervous system, but contains neuronal networks for executing complex motor behaviors. Because the brain controls behavior via descending projections to the VNC (Namiki et al., 2018), it is critical to be able to study neuronal circuits in both the brain and the VNC at synaptic resolution. Notably, this VNC dataset complements the recent release of an EM dataset comprising the complete adult female Drosophila brain (Zheng et al., 2018).
We validate the VNC dataset by automatically mapping its synapses with high accuracy, successfully registering the predicted synapse density map to a standard atlas (Fig. S3) and finding a high degree of similarity between EM and LM reconstructed neurons (Figs. 4, S4, 5, 7, S7). We demonstrate a pipeline for identifying cells of interest in the dataset by comparing EM reconstructions to LM data. Finally, as a foundation for future work, we make publicly available >1000 neuron reconstructions and their connectivity. While these reconstructions were generated manually, advances in automated segmentation approaches are dramatically accelerating analysis of serial-section TEM data (Dorkenwald et al., 2020; Li et al., 2019).
Direct sensory feedback to motor neurons
Flexible motor control relies heavily on feedback from proprioceptors, a class of sensory neurons that measure body position, velocity, and load. In both vertebrates and invertebrates, proprioceptive feedback is processed by the central nervous system to tune motor output (Tuthill and Azim, 2018). In insects, morphologically distinct subclasses of chordotonal neurons encode different features of leg movement such as position, velocity, and vibration (Mamiya et al., 2018). Campaniform sensilla encode load signals similar to mammalian Golgi tendon organs (Pringle, 1938; Tuthill and Azim, 2018; Zill and Moran, 1981). Although we know the main proprioceptor types and the signals they encode, we are now poised to understand how motor circuits integrate proprioceptive inputs to control the body by mapping the complete wiring diagram of an adult Drosophila VNC.
EM datasets also enable the discovery of cell types and synaptic connections that may be overlooked by other methods. For instance, our targeted reconstruction of sensory afferents revealed that the leg sensory neurons with the largest-caliber axons are the bCS neurons, which make direct synapses onto large-caliber leg MNs (Figs. 5, 6E). This connection is monosynaptic and bCS inputs are specifically located near the putative MN spike initiation zone (Fig. 5F, I), suggesting that speed and reliability are essential for the function of these connections.
The unique bilateral and intersegmental projections of bCS neurons suggests that they directly influence multiple limbs on both sides of the body (Fig. 5A-B). This leads to several hypotheses about their function. Prior work suggested that campaniform sensilla encode information about step timing that could drive the transition between stance and swing phases of walking (Dallmann et al., 2017; Ridgel et al., 1999). However, we observe that bCS neurons synapse onto the same MNs on both sides of the body (Fig 5H), suggesting they drive symmetric movements of left and right legs. This makes it unlikely that bCS neurons contribute to walking, which involves antiphase movement of contralateral legs (DeAngelis et al., 2019). Instead, bCS neurons may underlie a fast reflex where multiple legs flex in response to bCS activation. CS can signal either increases or decreases in load, depending on the sensillum’s placement and orientation on the leg (Zill and Moran, 1981; Zill et al., 1981). Therefore, bCS neuron activation could forcefully stabilize posture in response to additional weight (e.g., to prevent the body from being crushed) or to grip a surface in response to a loss of load (e.g., to prevent being blown away by a gust of wind). The genetic tools we created to target bCS neurons (Figs. 5E, S5A) will enable future analyses of their function.
Monosynaptic sensory-to-motor neuron connectivity is infrequent in larval Drosophila (Zarin et al., 2019), but has been observed in other adult insects (Burrows, 1996). Direct sensory feedback may be key in adults for precise control of their segmented limbs. The absence of such connections in larvae may indicate that controlling a limbless body relies less on sensory feedback and more on feedforward processing (Fushiki et al., 2016). As adult flies move much faster than larvae, another possibility is that fast monosynaptic sensory feedback is crucial for fast-moving animals. Indeed, research on escape responses has demonstrated that high-velocity movements are often controlled by the fastest neuronal pathways (Eaton et al., 1977; Trimarchi and Schneiderman, 1995).
Diversity and stereotypy within complete leg motor neuron populations
MNs have diverse but stereotyped functions, reflecting the array of muscles and muscle fibers they innervate. Some MNs have unique and reproducible transcription factor signatures that underlie their physiological properties and axonal morphology (Enriquez et al., 2015; Venkatasubramanian et al., 2019). These unique transcription factor patterns specify morphologies that are fairly stereotyped across animals (Baek and Mann, 2009; Brierley et al., 2012). Our results extend these findings by quantitatively demonstrating that most dendritic arborizations of leg MNs are sufficiently stereotyped to be individually identifiable by structure alone. Because we reconstructed the complete population of MNs controlling the two front legs, we were able to show that mirror symmetry in primary neurite number and position is a systematic principle of MN populations (Fig. 6). In contrast, sensory neurons have more redundant copies and variable copy numbers (Takemura et al., 2015; Tobin et al., 2017).
Adult Drosophila as a model system for studying circuit mechanisms of motor control
Previously, comprehensive neuronal connectivity maps were acquired for the nerve cords of other organisms including C. elegans (White et al., 1986), leeches (Stent et al., 1978), lampreys (Buchanan and Grillner, 1987; Grillner, 2003), and Drosophila larvae (Cardona et al., 2010; Fushiki et al., 2016; Ohyama et al., 2015; Zwart et al., 2016). These maps enabled a more complete understanding of how the nervous system controls locomotor rhythms underlying swimming and crawling. Less is known about the connectivity underlying motor control in limbed animals. The EM dataset we present here as a public resource will enable complete connectivity mapping for the circuits that control the legs and wings of an adult Drosophila. Combined with recent advances in recording activity from genetically identified VNC neurons during behavior (Azevedo et al., 2020; Chen et al., 2018; Mamiya et al., 2018), adult Drosophila is emerging as a powerful system for studying motor control. With these tools, we expect that a deeper understanding of the circuit basis for complex motor control is within reach.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wei-Chung Allen Lee (wei-chung_lee@hms.harvard.edu).
Materials availability
All unique reagents generated in this study are available from the Lead Contact.
Data and code availability
The EM dataset and reconstructions are freely available. EM image data and neuron tracings are publicly viewable at Virtual Fly Brain (https://fanc.catmaid.virtualflybrain.org/). Neuron reconstructions can be downloaded from (https://github.com/htem/GridTape_VNC_paper/tree/master/neuron_reconstructions). EM image data can be viewed and downloaded from BossDB (https://bossdb.org/project/phelps_hildebrand_graham2020). EM image data can be downloaded in CATMAID-ready JPEG tiles from a public Google Cloud bucket (gs://vnc1_r066/alignmentV3/jpgs_for_catmaid) using Google Cloud Console, the API Link, or gsutil (https://cloud.google.com/storage/docs/access-public-data). Reel-to-reel instrumentation designs and software are available at (https://github.com/htem/GridTapeStage). Code for analysis and figures is available at (https://github.com/htem/GridTape_VNC_paper/tree/master/figures_and_analysis/). Additional code is available at (https://github.com/htem/GridTape_VNC_paper, https://github.com/htem, https://www.lee.hms.harvard.edu/resources), or upon reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster were raised on a standard cornmeal and molasses medium and kept at 25°C in a 12:12 hour light:dark cycle. The specimen used for the EM dataset was an adult female aged 1-2 days post-eclosion, genotype y,w/w[1118]; +; P{VT025718-Gal4}attP2/P{pBI-UASC–3×MYC–sbAPEX2–dlg-S97}18.
METHOD DETAILS
Specimen preparation
All procedures involving animals were conducted in accordance with the ethical guidelines of the NIH and approved by the IACUC at Harvard Medical School. The Standing Committee on the Use of Animals in Research and Training of Harvard University approved all animal experiments.
We fixed and stained the central nervous system of one adult female Drosophila melanogaster (aged 1–2 days post-eclosion, genotype y,w/w[1118]; +; P{VT025718-Gal4}attP2/P{pBI-UASC–3×MYC–sbAPEX2–dlg-S97}18). Following fixation (2% paraformaldehyde/2.5% glutaraldehyde) and dissection (Tobin et al., 2017), the specimen was reacted with diaminobenzadine (DAB) and H2O2 as described previously (Zhang et al., 2019), but an EM-dense label was not observed in this sample. The dissected central nervous system was then post-fixed and stained with 1% osmium tetroxide/1.5% potassium ferrocyanide, followed by 1% thiocarbohydrazide, a subsequent incubation in 2% osmium tetroxide, then 1% uranyl acetate, followed by lead aspartate (Walton, 1979), then dehydrated with a graded ethanol series. The specimen was then embedded in epoxy resin (TAAB 812 Epon, Canemco), positioned in a cutout of mouse cortex (Hildebrand et al., 2017) processed for EM using the same protocol without the DAB reaction. Sections cut from this specimen were not post-section stained.
The mouse thalamus specimen shown in supplementary data (Fig. S1F) was prepared as previously described (Deerinck et al., 2010; Hua et al., 2015) and post-section stained with stabilized lead citrate (Ultrastain II, Leica).
For matching cells in the EM dataset with genetically identified cell types, transgenic Drosophila lines, husbandry, and LM imaging are described in (Azevedo et al., 2020; Mamiya et al., 2018; Meissner et al., 2020). We generated LM data using female flies 1 to 5 days post-eclosion. Genotypes for the flies used in Figs. 4, 5, 7, S4, and S7 were:
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{y[+t7.7] w[+mC]=GMR81A07-GAL4}attP2/+
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{y[+t7.7] w[+mC]=GMR35C09-GAL4}attP2/+.
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{y[+t7.7] w[+mC]=GMR22A08-GAL4}attP2/+
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{y[+t7.7] w[+mC]=GMR74F07-GAL4}attP2/+.
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{y[+t7.7] w[+mC]=GMR22E04-GAL4}attP2/+
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/ P{y[+t7.7] w[+mC]=R38G07-p65.AD}attP40; P{y[+t7.7] w[+mC]=R43C10-GAL4.DBD}attP2/+.
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/ P{y[+t7.7] w[+mC]=R60B12-p65.AD}attP40; P{y[+t7.7] w[+mC]=R70C02-GAL4.DBD}attP2/+.
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{w[+mC]=iav-GAL4.K}3
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{y[+t7.7] w[+mC]=R64C04-GAL4}attP2/+.
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/P{y[+t7.7] w[+mC]=R70H02-p65.AD}attP40; P{y[+t7.7] w[+mC]=R32H08-GAL4.DBD}attP2/+ (from Chen et al., in preparation)
w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40/+; P{y[+t7.7] w[+mC]=GMR21D12-GAL4}attP2/+.
w[1118] P{y[+t7.7] w[+mC]=13xLexAop2-IVS-myr::smGdP-FLAG}su(Hw)attP8 ; P{y[+t7.7] w[+mC]=GMR21G01-lexA}attP40/+
w[1118] P{y[+t7.7] w[+mC]=13xLexAop2-IVS-myr::smGdP-FLAG}su(Hw)attP8; P{y[+t7.7] w[+mC]=GMR56H01-lexA}attP40/+
w[1118] P{y[+t7.7] w[+mC]=hs-FLPG5.PEST}attP3 ; +/ P{y[+t7.7] w[+mC]=R38G07-p65.AD}attP40; PBac{y[+mDint2] w[+mC]=10xUAS(FRT.stop) myr::smGdP-HA}VK00005 P{y[+t7.7] w[+mC]=10xUAS(FRT.stop) myr::smGdP-V5-THS-10xUAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1/ P{y[+t7.7] w[+mC]=R43C10-GAL4.DBD}attP2
Substrate production
GridTape was produced from 125 μm-thick aluminum-coated Kapton® film (Dunmore) slit into 8 mm-wide reels of 35 m length (Metlon Corporation). This stock tape was modified using a custom laser-milling system consisting of a reel-to-reel tape positioning machine and commercial 1 W ultraviolet laser marking system (Samurai, DPSS Lasers). Control software triggered laser milling of a 30 mm length of tape, used custom computer vision to check the result of the cutting, advanced the tape 30 mm and finally adjusted the position of the tape to align the next 30 mm of tape to cut. This system enabled the autonomous production of >30 m lengths of cut tape containing over 5000 slots. Following laser milling, the cut tape was cleaned by wiping it with isopropyl alcohol-soaked lint-free wipes. Finally, the cut tape was coated with a 50 nm-thick TEM support film (LUXFilm®, Luxel Corporation).
We used a slot geometry based on conventional TEM slot grids with rounded rectangular holes. GridTape holes are 1.5 × 2 mm2 compared to 1 × 2 mm2 for conventional TEM slot grids. Customized slot geometries are possible and we have used slots as large as 2 × 3 mm2. Larger slot geometries could be used, but will depend on material properties of the thin film and may require modification of TEM column hardware.
Sample block trimming
In preparation for sectioning, embedded tissue blocks were trimmed (Trim 90, Diatome) into an oblong hexagonal shape (Fig. 1E) with 3.5–4 mm height, 1–2 mm width, a greater than 90° degree bottom tip angle and less than 90° top tip angle.
Serial sectioning
An ultramicrotome (UC7, Leica) and diamond knife (4 mm, 35° Ultra or Ultra-Maxi, Diatome) were used to cut ~45 nm-thick serial sections from prepared samples. These sections were collected using a modified automated tape-collecting ultramicrotome (ATUM; (Hayworth et al., 2014)). All tape guides and rollers on the ATUM were modified by adding a 4 mm-wide channel to prevent contact with the TEM support film spanning GridTape slots. Additionally, an optical interrupter (GP1A57HRJ00F, Sharp Electronics) was affixed to the ATUM to detect the passage of GridTape slots (Fig. S1B). A hall-effect sensor (A1301EUA-T, Allegro MicroSystems) and magnet were attached to the microtome swing arm to detect the cutting of sections (Fig. S1A). Custom software monitored the period and relative phase-offset of these two sensors during section collection. By setting the microtome to a fixed cutting speed and varying the ATUM tape speed, effective phase-locking at a fixed offset was achieved (Fig. S1A-C).
For sectioning of the Drosophila specimen to reach stable conditions, an initial stretch of 45 sections was collected while adjustments were made to the tape speed and fixed offset to optimize section placement. Of these 45 sections, 21 were off-slot and thus not imageable with TEM. The 24 that were on-slot contained small portions of the abdominal ganglion and were imaged and included in the dataset. Of the 4355 serial sections subsequently collected, the ventral nerve cord (VNC) was completely off-slot in two sections and partially off-slot in four sections (20%, 30%, 70%, and 90% off-slot). Due to support film breakage, three sections were completely lost before imaging, and four were partially lost (10%, 10%, 20%, and 40%). One additional section was partially lost (10%) because it cut very thinly and a portion was distorted. No further sections had substantial data loss.
Note that sections collected onto GridTape but off-slot can still be acquired using the traditional ATUM-SEM approach (Fig. S1F). Because of the high reliability of the section placement (Fig. 2B and S1D), SEM imaging was not required for the VNC dataset.
Measuring section placement consistency
Section placement was measured by first capturing overview photographs (Flea3 FL3-U3-13E4C-C, PointGrey) of each slot. Collimated low-angle illumination (MWWHL4, Thorlabs) enhanced the visibility of sections on GridTape. Using these images, the location of the slot was found using the Fiji plugin “Template Matching and Slice Alignment” (https://sites.google.com/site/qingzongtseng/template-matching-ij-plugin), selecting the slot as the template. Any failures to automatically find the slot (<1% occurrence) were corrected manually in Fiji (Schindelin et al., 2012). Subsequently, the location of the tissue section was found using the same plugin, selecting a prominent feature of the tissue section as the template. The VNC tissue’s shape and appearance changed significantly across the 4355 section series, so template matching was performed on batches of ~500–1000 sections, with a separate feature chosen for template matching in each batch. This approach enabled automatic identification of the tissue’s placement for ~98% of sections. The remaining ~2% of sections that were not correctly identified were located manually. The sections needing this manual correction mainly fell into two categories: sections that were cut very thin, causing the tissue to have reduced visibility, or sections with the template feature placed near the slot edge.
TEM imaging
To perform TEM imaging of sections collected onto GridTape, a custom in-vacuum, reel-to-reel stage was constructed (Fig. 1F) and attached to a TEMCA system (Bock et al., 2011) consisting of a TEM (JEOL 1200 EX) with a 2×2 array of sCMOS cameras (Zyla 4.2, Andor). The stage allows a 7500-slot, 45 m-long reel of GridTape to be loaded into the microscope for imaging under vacuum. After loading and pump-down, a set of pinch drives (one on each side of the TEM column) allows linear movement of GridTape to exchange and position sections under the electron beam in preparation for imaging. After positioning, both pinch drives dispense a small amount of GridTape towards the center of the column, introducing slack on both sides of the sample held under the beam. This allows an XY stack of piezo nanopositioners (SLC-1720, SmarAct) to make the many small movements necessary to montage large areas. Individual camera captures were 2048×2048 pixel 16-bit images. At 4.3 nm lateral resolution, the 2×2 camera array’s field of view for a single location was just over 16 μm square. By capturing many images at slightly overlapping regions (typically 20–30%) for a single section, square millimeter-sized regions of interest could be imaged. Imaging regions for each section were selected using the overview photographs using a custom graphical user interface in MATLAB (MathWorks). Magnification at the microscope was 2500×, accelerating potential was 120 kV, and beam current was ~90 μA through a tungsten filament. The VNC dataset was acquired at a net sustained imaging rate of 42.73±3.04 Mpixels per second (mean±SD across sections), equivalent to a “burst” imaging rate of ~160 Mpixels per second for a single microscope.
Section stitching and serial-section alignment
Image alignment for the VNC dataset was performed with a custom software pipeline that deployed AlignTK's image alignment functions (https://mmbios.pitt.edu/aligntk-home) in parallel on a computing cluster (Bock et al., 2011; Lee et al., 2016; Tobin et al., 2017). After acquisition, camera images for each section were virtually stitched together into seamless montages. Subsequently, section-to-section alignment was performed on 8× downscaled versions of these section montages. To align the 4355 stitched sections into a three-dimensional volume, an initial volume comprised of every 25th section was first generated. The only features consistently recognizable across gaps of 25 sections were neuronal nuclei, so this initial volume positioned every 25th section in a location that ensured a given nucleus would stay at the same (x,y) location across the ~150 sections in which each nucleus was visible. This positioning of every 25th section was used as a global constraint on the full dataset’s alignment (using the absolute_maps option in AlignTK’s align function).
Due to the small number of sections with artifacts or missing data, elastic alignment (AlignTK’s register function) between neighboring sections was sufficient for generating a high-quality global alignment, except for 27 sections where alignment to secondary neighbors was necessary. EM artifacts that can generate alignment errors include knife marks, cracks, folds, debris, and missing regions where the section or film was physically damaged. These can introduce alignment errors where regions close to artifacts on adjacent sections get locally misaligned or warped. However, regularization included in the elastic alignment ensured that defects were typically isolated and dissipated within 1–2 sections from an artifact. Because misalignments were local and sparse, reconstruction across the vast majority of the dataset was not impeded by errors in alignment. Additionally, no sections were mis-ordered during section collection or imaging, eliminating the need for a section order correction step. Relative to manually collected series, the consistency of GridTape section collection simplified the alignment process substantially and enabled the final volume to have high quality alignment (Video S1).
Neuron reconstruction
We reconstructed neurons in the EM dataset as described previously (Lee et al., 2016; Tobin et al., 2017). We used CATMAID (Saalfeld et al., 2009) to manually place a series of marker points down the middle of each neurite to generate skeletonized models of neuronal arbors. We annotated neurons passing through each peripheral nerve and reconstructed those neurons into the VNC. Neurons that had a cell body in the VNC and received synaptic inputs in the neuropil were considered motor neurons (MNs). Neurons that made synaptic outputs in the neuropil and did not have cell bodies in the VNC were considered sensory neurons. The few exceptions to these categorizations are shown in Figure S2. Neurons with projections and cell bodies in the VNC but that did not pass through a peripheral nerve were considered central neurons.
In peripheral nerves, axons of MNs were clustered together (Figs. 3C and S6F). After finding a single motor axon in a given nerve, we reconstructed its neighbors, continuing to reconstruct further neighbors until all MNs in the nerve were reconstructed. We confirmed that sensory neurons near the motor domain were in fact sensory neurons by reconstructing them into the VNC, and we additionally reconstructed large-caliber axons in the sensory domain that we suspected could be MNs despite their position. No MN axons were found in the sensory domain of any peripheral nerve. We found one case where three sensory neurons had axons located in the motor domain of the right mesothoracic leg nerve (Fig. 3Cii). With this reconstruction approach, we identified all 507 MNs in all thoracic nerves. We did not reconstruct from the abdominal nerves, which do not contain limb MNs.
Completion state of motor neuron reconstructions:
All 507 MNs were first reconstructed from their peripheral nerve through their primary neurites and to their cell bodies. For front leg MNs, dendritic branches were reconstructed until multiple expert annotators were able to independently identify left–right homologous pairs based on their symmetrical morphology (Fig. 6). The amount of dendritic reconstruction required for unambiguous pair identification varied across the population (Video S5). For front leg MNs where left–right pair identification required minimal or no dendritic reconstruction, we nevertheless reconstructed the largest dendritic branches. Additionally, we completely reconstructed the microtubule-containing backbone for the L5-bundle MNs (Fig. S7A). While performing reconstructions and identifying homologous pairs, annotators were blind to the left–right pair predictions generated algorithmically through analysis of NBLAST similarity scores (Figs. 6D, S7B). MNs controlling the middle and hind legs were reconstructed only until their cell body was located. Some dendritic branches were reconstructed during this process, but their reconstruction was not otherwise continued. For MNs controlling the neck, wings, and halteres, the largest-caliber dendritic branches were reconstructed to confirm that each neuron arborized in the neck, wing, or haltere neuropils (Court et al., 2020; Namiki et al., 2018).
Completion state of sensory neuron reconstructions:
All 655 sensory neurons shown in Figure 3B were reconstructed from a peripheral nerve into the VNC until a synaptic output was identified within the neuron, at which point its identity as a sensory neuron was considered confirmed. For the 392 sensory neuron reconstructions in the left T1 neuromere (Fig. 4A), we reconstructed their axonal projections until each neuron could be unambiguously identified as one of the four major subtypes of sensory neurons (see “Neuron subtype classification”), or until it the axonal morphology deviated from one of these four subtypes. We reconstructed the 12 bCS neurons to near completion, only excluding very fine processes (Fig. 5).
Transsynaptic reconstruction:
We identified synapses using a combination of ultrastructural criteria, specifically the existence of a presynaptic T-bar, presynaptic vesicles, and postsynaptic densities. For each of the 12 bCS neurons, we annotated output synapses and reconstructed the postsynaptic twigs back to their parent neuron until we confirmed that each bCS neuron synapsed onto at least one MN in each neuromere. Then, we annotated all output synapses in the two left and two right T1 bCS neurons within the branch of their axons indicated in Figure 5F. Multiple independent annotators reviewed these synapse annotations to ensure accuracy and completeness. We reconstructed all postsynaptic twigs at each of those synapses back to their parent neuron. 62 out of 437 postsynaptic twigs (14.2%) were orphaned, meaning their connection to a neuronal backbone could not be found. The other 375 (85.8%) were successfully connected either to a MN reconstruction or a neuronal backbone that made output synapses, which identified it as a central neuron. We never observed a sensory neuron postsynaptic to a bCS synapse. The postsynaptic MNs included 11 ProLN MNs in the L1 bundle and one VProN MN. Analysis in Figure 5I was restricted to the nine ProLN MNs receiving five or more synapses from bCS neurons. Analysis in Figures 5H, 6E, and S5E-F included all ProLN MNs.
Completion state of central neuron reconstructions:
For each of the 17 central neurons receiving two or more synaptic inputs from bCS neurons, we reconstructed its morphology until we located its cell body (Fig. S5C). For each of the five central neurons receiving five or more synaptic inputs from bCS neurons, we determined where it arborized in the VNC by reconstructing all large- and medium-caliber branches, but fine processes remain unreconstructed.
Together, these procedures follow a previously described and validated protocol for reconstructing neurons in serial-section TEM datasets (Schneider-Mizell et al., 2016).
Neuron subtype classification
We identified sensory neuron subtypes by their stereotyped projection patterns in the VNC, which corresponded well with previous observations of these neurons using light microscopy (Baek and Mann, 2009; Brierley et al., 2012; Harris et al., 2015; Mamiya et al., 2018; Merritt and Murphey, 1992; Tsubouchi et al., 2017). Bristle neuron axons traveled along either the anterior, posterior, or ventral edge of the neuromere without significant branching. Hair plate neuron axons trifurcated upon entering the VNC and projected along the anterior, posterior, and lateral edges of the neuromere. Chordotonal neuron axons projected through the middle of the neuromere toward the midline. Campaniform sensillum axons projected down the oblique tract, located posterior to the chordotonal neuron axons.
The cluster of central neurons postsynaptic to T1 bCS neurons (Fig. S5C, asterisk) was identified as lineage 19A based on comparing cell body location and general arborization pattern with LM data (Harris et al., 2015).
Automated synapse prediction and atlas alignment
To transform EM reconstructions into the atlas space, we computationally generated a “neuropil stain” (Heinrich et al., 2018) by automatically detecting postsynaptic specializations in the EM volume that would be apposed to presynaptic specializations labeled by immunostaining (Kittel et al., 2006). Specifically, we trained and deployed a convolutional neural network to automatically identify synaptic locations across the entire EM dataset (Buhmann et al., 2019). To produce ground truth data to use for training, we densely annotated pre- and post-synaptic sites in 9 cubes of image data, each 3 × 3 × 3 μm3 or 768 × 768 × 75 voxels in size. We selected an additional 11 cubes with no synapses. The ground truth annotations were turned into training data by creating a mask of pixel locations within 10 nm of each postsynaptic annotation. We used mean-squared loss to train the network to predict these mask values, with the network’s input being four-fold downsampled EM image data (effective voxel size 17.2 × 17.2 × 45 nm3). We augmented the training data with random x,y transpositions, x,y flips, continuous rotations around the z-axis, and section-wise elastic deformations and intensity changes. We used a 3D U-Net (Falk et al., 2019), comprised of four resolution levels with downsample factors in x, y, z of (2, 2, 1), (2, 2, 1), and (2, 2, 3). The topmost level contained eight feature maps and the number of feature maps in subsequent levels increased by a factor of five. Convolutional passes were comprised of two convolutions with kernel sizes of (3, 3, 3) followed by a rectified linear unit (ReLU) activation function. A final convolution with kernel size (1, 1, 1) produced the map of predicted postsynaptic sites. The network was trained to 700,000 iterations using 75% of the data from each of the 20 ground-truth cubes, with the remaining 25% held out for performance evaluation. We saved the network weights every 5,000 iterations between 600,000 and 700,000 iterations, and found that the best performance on the held-out data was achieved using the weights at 610,000 iterations (precision: 71.4%, recall: 72.8%). We deployed the network using those weights to predict postsynaptic locations throughout the entire VNC EM dataset.
The density of the predicted synapses matched the spatial extents of the VNC neuropil. There was a low density of synapses predicted in regions of the dataset containing cell bodies and fasciculated neuronal tracts, reflecting the high precision of the predictions (Fig. S3E-F). We subsequently downsampled and Gaussian blurred (σ = 900 nm) the predicted synapse locations to produce a synapse density map at the approximate resolution of LM data (Fig. S3G). We used this synapse density map to register the EM dataset to the JRC 2018 VNC Female atlas (Bogovic et al., 2019) using elastix (http://elastix.isi.uu.nl/) (Video S3). After registering the synapse density map to the atlas, reconstructed neurons were transformed and imported into a CATMAID project using custom code (see DATA AND CODE AVAILABILITY). Confocal microscopy data was also transformed into the same VNC atlas coordinate system using elastix (Figs. 4, S4, 5, 7 and S7). The elastix-based atlas registration pipeline is made available (see DATA AND CODE AVAILABILITY).
Our initial goal in predicting synapses across the VNC EM dataset was to use the synapse predictions to align the EM dataset with a standard light-level atlas to bridge EM and LM data. Combined with future automated neuron segmentation (Li et al., 2019), these synapse predictions can be used for automated connectome reconstruction (Scheffer et al., 2020).
Light microscopy-based cell matching
To match identified neurons between LM and EM, we reconstructed neurons from confocal microscopy data, either intracellularly filled with dyes (Fig. 7A-B, S7C) or expressing fluorescent proteins (Figs. 4D-F, 7C, S4, and S7D). Intracellular labeling, immunohistochemistry, confocal microscopy, and tracing of genetically identified and physiologically characterized MNs was performed as previously described (Azevedo et al., 2020). Briefly, targeted neurons were labeled during whole-cell patch pipette recordings with 13 mM neurobiotin in the internal solution. After whole-cell recordings, the dissected VNC was lightly fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min. The tissue was then washed in PBST (PBS + Triton, 0.2% w/w), incubated in blocking solution (PBST + 5% normal goat serum) for 20 min, and then incubated for 24 hr in blocking solution containing a primary antibody for neuropil counterstain (1:50 mouse anti-Bruchpilot, Developmental Studies Hybridoma Bank, nc82). After a subsequent PBST wash, the tissue was incubated in blocking solution containing secondary antibodies for 24 hr (streptavidin AlexaFluor conjugate, Invitrogen; 1:250 goat anti-mouse AlexaFluor conjugate, Invitrogen). Other genetically identified neurons expressing fluorescent proteins were processed similarly, but without whole-cell intracellular labeling. Following staining, the tissue was mounted in Vectashield (Vector Labs). Legs (Fig. S5A) were fixed in for 20 minutes in 4% formaldehyde in PBS, rinsed in PBST 3x, and mounted in Vectashield (Vector Labs) between two coverslips with spacers. Confocal stacks were acquired using a Zeiss 510 confocal microscope. Cell morphologies were traced in Fiji (Schindelin et al., 2012), using the Simple Neurite Tracer plugin (Longair et al., 2011). Neuron traces were registered and transformed into the VNC atlas space using an elastix-based atlas registration pipeline (see DATA AND CODE AVAILABILITY) and imported into CATMAID.
QUANTIFICATION AND STATISTICAL ANALYSIS
Clustering and symmetry analysis
For the primary neurite clustering analysis (Figs. 6B and S6C-E), EM reconstructions were first transformed into the VNC atlas space using the registration described above. Then, neurons were pruned to exclude any parts of the reconstruction falling outside the VNC neuropil. This retained the neurites in the neuropil, but excluded cell bodies, which are known to have variable positions across individuals even for identified neurons and are therefore not reliable indicators of neuron identity (Baek and Mann, 2009). Neurons were further pruned to only include their primary neurite (Fig. S6B). NBLAST similarity scores (Costa et al., 2016) were calculated between each pair of pruned neurons in both forward and reverse directions (i.e. neuron A to neuron B and neuron B to neuron A) and normalized such that the similarity score of each neuron with itself is equal to 1. The forward and reverse scores were then averaged to generate a final similarity score for each pair of neurons. Hierarchical clustering with single linkage was performed on similarity scores for MNs of each peripheral nerve using the SciPy Python package. The clustering dendrograms and neuron reconstructions were visually inspected, and a cut height on each dendrogram was chosen that separated MN bundles traveling along distinct trajectories.
For symmetry analysis, neurons were transformed into the VNC atlas space and pruned to exclude any parts of the reconstruction falling outside the VNC neuropil. Dendritic branches emerging from the primary neurite were included (Fig. 6D) or pruned (Fig. S7B). Neurons on the right side of the dataset were reflected across the midplane of the atlas to enable comparison with neurons on the left side. NBLAST similarity scores were calculated between each left-side MN and each reflected right-side MN. Scores ranged from −0.43 (most dissimilar pair) to 0.69 (most similar pair). Based on these scores, we used the Munkres algorithm (Munkres, 1957) in MATLAB (MathWorks) to compute a globally optimal pairwise assignment that maximized the sum of similarity scores for assigned pairs of MNs on the left and right sides of the VNC. We provide code for performing this workflow (see DATA AND CODE AVAILABILITY).
Measurement of cross-sectional areas of bCS axons and motor neuron primary neurites
We selected three sections distributed across the region of the dataset where the left ProLN traveled directly perpendicular to the sectioning plane. In each of the three sections, the polygon selection tool in Fiji was used to manually measure the area of each bCS axon and each MN primary neurite. Measured areas were averaged across the three sections to produce final values (Fig. 5D “left T1” and Fig. 6E). This procedure was repeated for the bCS neurons and MNs in the right mesothoracic leg nerve (Fig. 5D “right T2”) and right metathoracic leg nerve (Fig. 5D “right T3”).
Analysis of synaptic connectivity for bCS neurons
Analysis was performed in Python using pymaid (https://pymaid.readthedocs.io/en/latest/index.html) for pulling reconstructions from CATMAID, SciPy for linear regression, and matplotlib for visualization. For measuring distances between synapses and the primary neurites of their postsynaptic MNs (Fig. 5I), geodesic or “along-the-arbor” distance was calculated. To determine the distribution of distances between possible synaptic locations and the primary neurite and putative spike initiation (Gwilliam and Burrows, 1980), we computed the distances from all positions on the MN arbor to the primary neurite (Fig. 5I), assuming that all locations on the MN were equally likely to receive synaptic input. In reality, synapses are preferentially positioned on the distal branches of neurons (Schneider-Mizell et al., 2016), so the random distributions presented here likely underestimate the distances from the primary neurite at which synaptic inputs are found. This implies that the bias in which bCS synapses target regions close to the primary neurite relative to randomly positioned input is likely to be even stronger than suggested by our analysis.
To measure the proximity of ProLN MNs to bCS neurons (Fig. 6E), we first computed the minimum Euclidean distance between each MN’s primary neurite and each of the 145 bCS synapses reconstructed within the segment indicated in Figure 5F. For each MN, we averaged these measurements across all 145 synapses to produce a final measurement of the average distance between a primary neurite and the reconstructed synaptic sites within bCS axons.
In Figures 5H, 6F, and S5E-F, the line of best fit was calculated using SciPy’s stats.linregress function. Spearman’s ρ and p-value were calculated using SciPy’s stats.spearmanr function. The 42 left ProLN MNs were included in each regression.
Supplementary Material
Video S1 (Related to Figure 2). Fly-through of the aligned VNC EM dataset.
Zoomed-out (left) and zoomed-in (right, white boxed region from the left) views of aligned image data. Section number in the upper left corner.
Video S2 (Related to Figure 3). VNC motor and sensory neuron reconstructions.
Skeleton reconstructions of motor neurons (left) and sensory neurons (middle) from the VNC EM dataset (gray surface rendering). Sensory and motor neuron reconstructions registered to a standard VNC atlas (right, see also Fig. S3). Spheres represent neuronal cell bodies. These neurons connect the VNC to the front (red), middle (yellow), and hind (green) legs as well as the neck (blue), wings (cyan), and halteres (purple). Anterior is up and posterior is down.
Video S3 (Related to Figures 3-4, 6-7). Alignment of the EM dataset to a standard VNC atlas.
An artificial neural network was used to predict the location of synapses in the VNC EM dataset (Fig. S3). Those predictions were blurred and downsampled to produce a map of synapse densities across the VNC dataset (left). This enabled the EM dataset to be aligned (middle) to a standard VNC atlas (right).
Video S4. (Related to Figure 4) Reconstructed sensory and motor neuron subtypes.
Skeleton reconstructions and subtype classification of sensory and motor neurons in the left T1 neuromere. Sensory neurons are color coded by type as in Figure 4A: chordotonal (orange), campaniform sensilla (purple), hair plates (yellow), bristles (light blue). Primary neurites and cell bodies of the motor neurons controlling the front left leg are colored by bundle as in Figure S6A. Anterior is up and posterior is down.
Video S5 (Related to Figure 6). Gallery of left–right homologous motor neuron pairs.
Rendering of the 56 left–right pairs of T1 motor neurons whose pairings were agreed upon by manual and algorithmic assignments (Fig. 6D). Neurons are color coded by primary neurite bundle (see Figs. 6A and S6C-E for color code). Outline of the VNC in light grey. Outline of the VNC’s neuropil in darker grey. Spheres represent motor neuron cell bodies.
Video S6 (Related to Figures 5, 7). Bilateral campaniform sensillum (bCS) neurons synapse onto a fast flexor motor neuron.
Left and right T1 bCS axons (dark red and light red, respectively) make output synapses (cyan spheres) directly onto the EM reconstruction most similar to a fast tibia flexor motor neuron (blue, 81A07-Gal4). bCS axons do not synapse onto the EM reconstruction most similar to a slow tibia flexor motor neuron (yellow, 35C09-Gal4). Notice the close proximity of the fast motor neuron’s primary neurite to the bCS axons.
Figure S1 (Related to Figure 1). The GridTape imaging platform.
(A) Photograph of the magnet and hall effect sensor (dashed circle) attached to the microtome cutting arm to measure cutting frequency.
(B) Photograph of the digital opto-interrupter (dashed circle) used to detect the slot frequency in the tape.
(C) The analog signal from the hall effect sensor (left) and digital signal from the opto-interrupter (right) are used to perform closed-loop, phase-locked collection of sections onto GridTape slots.
(D) Cumulative distribution of section placement deviation (Euclidean distance from the average section position) across the VNC dataset.
(E) Photograph of a reel of GridTape containing serial sections spanning an adult female Drosophila VNC.
(F) Regions of sections not collected over slots can be imaged with SEM as in the ATUM-SEM approach. SEM images of a stretch of GridTape carrying mouse thalamus sections (left), a single section over a slot (middle), and tissue collected over the edge of the slot (right). This approach was not required for the VNC dataset due to the low number of off-slot sections.
Scale bars, 20 mm (A-B), 10 s (C), 10 mm (E), 1 mm (F, left), 500 μm (F, middle), 1 μm (F, right).
Figure S2 (Related to Figure 3). Efferent neurons with unusual features.
(A) The peripherally synapsing interneuron (PSI). (i) Dorsal view. (ii) Anterior view. (iii) Cross-section through the posterior dorsal mesothoracic nerve at the location indicated by the dashed line in (ii), showing the PSI (red) fasciculated with four motor neurons (asterisks). (iv) Higher resolution view of the region indicated in (iii). The PSI makes synaptic outputs onto neighboring motor neurons (white arrowheads) and receives reciprocal synapses back from those motor neurons (yellow arrowheads), as described previously (King and Wyman, 1980). Presynaptic specializations were not observed within any other efferent or motor neurons.
(B) Unpaired median (UM) neurons (Duch et al., 1999) with cell bodies organized into 3 clusters. (i) Dorsal view. (ii) Lateral view. Colors indicate target organ (see color key in Fig. 3A).
(C) The “multinerve” neurons. (i) Dorsal view. (ii) Anterior view. Like UM neurons but unlike motor neurons, the axon of each multinerve neuron branches to exit the VNC through multiple peripheral nerves, in this case the ipsilateral dorsal prothoracic nerve (DProN) and ipsilateral prothoracic accessory nerve (ProAN). Their dendritic branches are positioned on the dorsal-most surface of the VNC and include a contralateral projection. Like motor neurons, multinerve neurons do not make synaptic outputs within the VNC. Multinerve neurons are unique to T1, as similar neurons were not found in T2 or T3. The full dendritic backbone of the dark blue neuron (asterisk) was completely reconstructed. The other three neurons were partially reconstructed.
Scale bars, 50 μm (Ai-ii, B-C), 1 μm (Aiii-iv).
Figure S3 (Related to Figures 3-4, 6-7). Automated synapse prediction and registration to a standard VNC atlas.
(A-F) Automatic identification of postsynaptic sites using an artificial neural network.
(A) All presynaptic sites (small red dots) and postsynaptic sites (large orange dots) were manually annotated in 9 ground-truth cubes of image data spread throughout the dataset. These annotations were used to train an artificial neural network to identify postsynaptic locations (STAR Methods).
(B) EM data from (A) overlaid with postsynaptic site predictions (purple) produced by the trained network.
(C) EM data from a region of the dataset not used for training. Same data as Fig. 2G.
(D) EM data from (C) overlaid by postsynaptic site predictions (purple).
(E) A wider field of EM data. Yellow square corresponds to region in (C).
(F) EM data from (E) overlaid with postsynaptic site predictions (purple). Note that the region with a high density of postsynaptic site predictions is synaptic neuropil, whereas low density regions contain either cell bodies or axon bundles.
(G) Atlas registration strategy for comparing morphologies of neurons imaged using light microscopy (LM) with neurons reconstructed in the EM dataset. First, the trained network predicted the location of postsynaptic sites throughout the EM dataset. Next, the predicted synapse locations were 3D Gaussian blurred (σ = 900 nm) and downsampled to produce a synapse density map across the entire EM dataset. The blurred and downsampled EM synapse-density predictions were elastically transformed to register them to a LM-based standard VNC atlas based on synapse density (also see Video S3; Bogovic et al. 2019). This elastic transform was then applied to EM reconstructions to register them to the atlas. Fluorescently labeled neurons were registered to the same atlas using a neuropil counterstain. Having EM and LM reconstructions both in the same reference coordinate system enabled quantitative comparisons using NBLAST (Costa et al. 2016) (see Figs. 4, 7, S4, S7).
Scale bars, 1 μm (A-D), 10 μm (E-F), 50 μm (G).
Figure S4 (Related to Figure 4). LM to EM sensory neuron identification.
(A-G) Sensory neurons reconstructed from LM data matched to the most similar neurons reconstructed in the EM dataset. (i) Rendering of a genetically identified sensory neuron reconstructed from LM. Dorsal view of the left T1 neuromere (A-F) or the T1 and T2 neuromeres (G). (ii) Ranked distribution of NBLAST similarity scores (worst to best, left to right) between the LM neuron and the 337 left T1 leg sensory neurons reconstructed and classified in the EM dataset (Fig. 4A), color coded by sensory neuron type. (iii) Overlay of the LM-reconstructed neuron and the five highest scoring EM-reconstructed neurons. (iv) The five highest scoring EM reconstructions. Each LM sensory neuron’s identity was validated with LM imaging of the sensory neuron’s location in the leg.
(A) A club femoral chordotonal neuron (MCFO from R64C04-Gal4).
(B) A claw femoral chordotonal neuron (MCFO from iav-Gal4).
(C) A hook femoral chordotonal neuron (R70H02-AD, R32H08-DBD).
(D) A hook femoral chordotonal neuron from a different fly line (MCFO from R21D12-Gal4) than (C).
(E) A campaniform sensillum neuron (MCFO from 22E04-Gal4).
(F) A campaniform sensillum neuron from a different fly line (MCFO from R74F07-Gal4) than (E).
(G) A bilateral campaniform sensillum (bCS) neuron (MCFO from R43C10-Gal4). Only the top 2 highest scoring EM neurons are rendered. See Figure 5 for characterization of this cell type.
Scale bars, 50 μm (A-G).
Figure S5 (Related to Figure 5) bCS neuron anatomy and connectivity.
(A) Split-Gal4 labeling of campaniform sensilla in the leg. (i) Fly leg schematic from the anterior view. Red boxed region represented in (ii-iv). (ii) Surface rendering of the trochanter showing campaniform sensilla (circular domes) of cluster TrCS5 and cluster TrCS8 that were unlabeled (white) or labeled (green) by two split-Gal4 lines (iii-iv). Rendering was generated using leg reconstructions reported in (Kuan et al. 2020). (iii-iv) Fluorescence imaging of sensory neurons labeled in the trochanter by lines 38G07-AD, 43C10-DBD (iii) and 60B12-AD, 70C02-DBD (iv).
(B) Types of neurons postsynaptic to bCS neurons, ordered by number of synapses received.
(C-D) Reconstruction of central neurons receiving 2 or more synaptic inputs (C) or 5 or more synaptic inputs (D) from the reconstructed branch of bCS axons (Fig. 5F). Asterisk denotes cluster of putative Lin19A cell bodies.
(E) The two bCS axons originating from the left T1 leg (arbitrarily designated 1 and 2) target a subset of the left ProLN motor neurons. 31 out of 42 motor neurons receive no input from the reconstructed bCS axons. For any given motor neuron, the number of synaptic inputs from the two bCS axons are significantly correlated.
(F) Similar to (E), but for the two right bCS axons onto left ProLN motor neurons.
Scale bars, 10 μm (A), 50 μm (C-D).
Figure S6 (Related to Figure 6). Motor neuron bundles and nerve organization.
(A) EM reconstructions of the cell bodies and primary neurites of the front leg motor neurons. Primary neurites travel through the T1 neuromere in distinct bundles before leaving the VNC through one of four different peripheral nerves.
(B) After being transformed into the VNC atlas coordinate system, reconstructions were pruned to exclude regions outside of the neuropil. The remaining portions included only the primary neurite and were used for calculation of all NBLAST similarity scores in Figures 6B, 6E and in (C-E) below.
(C-E) (i) Rendering of primary neurites and cell bodies, color coded by bundle. (ii) Hierarchical clustering dendrograms and matrices of similarity scores, as in Fig. 6A-B but for motor neurons of different nerves.
(C) Prothoracic leg nerve (ProLN) motor neurons.
(D) Ventral prothoracic nerve (VProN) motor neurons.
(E) Dorsal prothoracic nerve (DProN) motor neurons.
(F) Positions of the subtypes of sensory and motor neuron axons within the ProLN (Fig. 3Ci). Axons of each bundle from (C) are spatially clustered within the motor domain. Axons of sensory neurons also cluster by subtype within the nerve. Chordotonal axons, arising mostly from a single sensory organ in the femur (Mamiya et al. 2018), are more clustered than other types, which arise from sensory organs distributed across different leg segments. L1-L5: L1-L5 bundle motor neurons; UM: Unpaired median neurons; br: bristle, hp: hair plate, cho: chordotonal, cs: campaniform sensillum; unc: unclassified; NR: not reconstructed.
Scale bars, 50 μm (A-E), 10 μm (F).
Figure S7 (Related to Figures 6 and 7). Motor neuron similarity, variance, and identification.
(A) Rendering of the left and right L5-bundle motor neurons, with the right-side motor neuron reconstruction reflected across the midline. These motor neurons had cell bodies in a unique location (L5 bundle in Fig. S6Ci) that allowed these two neurons to be unambiguously identified as a left–right homologous pair. NBLAST similarity score in the upper right corner. Note that while these neurons project to similar locations within the neuromere, their branches sometimes have different trajectories. Empty arrowheads point to example branch terminations with similar positions, with arrows depicting their different paths. Similarity scores between other left–right homologous pairs typically ranged between 0.5 and 0.7. Slight variability in dendritic morphology even between homologous pairs, such as the variability illustrated here, may underlie why left–right homologous neuron pairs do not have a similarity score closer to 1.
(B) A global pairwise assignment algorithm based on NBLAST similarity scores of primary neurites alone agreed with 73% (41 of 56) of front leg homologous motor neuron pairs. Black asterisks indicate agreements, red asterisks disagreements.
(C-D) Additional motor neuron reconstructions from LM matched to most similar neuron reconstructions in the EM dataset. Same layout as Figure 7.
(C) An intermediate tibia flexor motor neuron (22A08-Gal4). The highest scoring EM reconstruction receives some synaptic input from bCS neurons. The second highest scoring EM reconstruction has a similar NBLAST score but receives strong synaptic input from bCS neurons.
(D) A motor neuron innervating the tibia long tendon muscle, which controls movements of the tarsus (56H01-LexA). Different fly line and neuron than Figure 7C.
Scale bars, 10 μm (A-B), 50 μm (C-D).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Adult female Drosophila ventral nerve cord electron microscopy dataset | This paper | https://bossdb.org/project/phelps_hildebrand_graham2020 |
| Adult female Drosophila ventral nerve cord electron microscopy dataset with neuron reconstructions | This paper | https://fanc.catmaid.virtualflybrain.org |
| Experimental Models: Organisms/Strains | ||
| Drosophila genotype y,w/w[1118]; +; P{VT025718-Gal4}attP2/P{pBI-UASC–3×MYC–sbAPEX2–dlg-S97}18 | This paper | N/A |
| Drosophila genotype w[1118]; P{JFRC7-20XUAS-IVS-mCD8::GFP} attp40 | Bloomington | RRID: BDSC_32194 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=GMR81A07-GAL4}attP2 | Bloomington | RRID: BDSC_40100 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=GMR35C09-GAL4}attP2 | Bloomington | RRID: BDSC_49901 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=GMR22A08-GAL4}attP2 | Bloomington | RRID: BDSC_47902 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=GMR74F07-GAL4}attP2 | Bloomington | RRID: BDSC_39864 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=GMR22E04-GAL4}attP2 | Bloomington | RRID: BDSC_49873 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=R43C10-GAL4.DBD}attP2 | Bloomington | RRID: BDSC_69610 |
| Drosophila genotype w*; P{w[+mC]=iav-GAL4.K}3 | Bloomington | RRID: BDSC_52273 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=R64C04-GAL4}attP2 | Bloomington | RRID: BDSC_70035 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=R70H02-p65.AD}attP40 | Bloomington | RRID: BDSC_70794 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=R32H08-GAL4.DBD}attP2 | Bloomington | RRID: BDSC_69119 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=GMR21D12-GAL4}attP2 | Bloomington | RRID: BDSC_48946 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=13xLexAop2-IVS-myr::smGdP-FLAG}su(Hw)attP8 | Bloomington | RRID:BDSC_62116 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=GMR21G01-lexA}attP40 | Bloomington | RRID: BDSC_61521 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=R38G07-p65.AD}attP40 | Bloomington | RRID: BDSC_70666 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=R70C02-GAL4.DBD}attP2 | Bloomington | RRID: BDSC_69783 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=R60B12-p65.AD}attP40 | Bloomington | RRID: BDSC_75889 |
| Drosophila genotype w[1118]; P{y[+t7.7] w[+mC]=hs-FLPG5.PEST}attP3; +; PBac{y[+mDint2] w[+mC]=10xUAS(FRT.stop)myr::smGdP-HA}VK00005 P{y[+t7.7] w[+mC]=10xUAS(FRT.stop)myr::smGdP-V5-THS-10xUAS(FRT.stop)myr::smGdP-FLAG}su(Hw)attP1 | Bloomington | RRID: BDSC_64085 |
| Antibodies | ||
| Mouse anti-Bruchpilot (nc82) | Developmental Studies Hybridoma Bank | RRID: AB_2314866 |
| Software and Algorithms | ||
| TEMCA-GT control software | This paper | https://github.com/htem/GridTapeStage |
| Fiji | (Schindelin et al., 2012) | RRID: SCR_002285 |
| Template Matching and Slice Alignment (Fiji plugin) | (Tseng et al., 2011) | https://sites.google.com/site/qingzongtseng/template-matching-ij-plugin |
| Simple Neurite Tracer (Fiji plugin) | (Longair et al., 2011) | https://github.com/fiji/SNT |
| MATLAB | MathWorks | RRID: SCR_001622 |
| AlignTK | (Bock et al., 2011) | https://mmbios.pitt.edu/aligntk-home |
| CATMAID | (Saalfeld et al., 2009; Schneider-Mizell et al., 2016) | RRID:SCR_006278 |
| Synaptic location prediction network | (Buhmann et al., 2019) | https://github.com/funkelab/synful |
| elastix | (Klein et al., 2010) | RRID:SCR_009619 |
| elastix-based atlas registration pipeline | This paper | https://github.com/htem/GridTape_VNC_paper/tree/master/template_registration_pipeline/run_elastix |
| NBLAST | (Costa et al., 2016) | RRID:SCR_015884 |
| Munkres (Hugarian) algorithm for linear assignment | (Munkres, 1957) | https://www.mathworks.com/matlabcentral/fileexchange/20652-hungarian-algorithm-for-linear-assignment-problems-v2-3 |
| pymaid | Open-source | https://pymaid.readthedocs.io/; https://github.com/schlegelp/PyMaid |
| SciPy | Open-source | RRID:SCR_008058 |
| Other | ||
| GridTape | This paper | https://luxel.com/gridtape/ |
| TEMCA-GT hardware designs | This paper | https://github.com/htem/GridTapeStage |
| Automated tape-collecting ultramicrotome (ATUM) | (Hayworth et al., 2014) | N/A |
| TEMCA-GT cameras | Andor | Zyla 4.2 sCMOS |
| Vacuum extension | (Bock et al., 2011) | Custom part |
| TEMCA-GT scintillator, 6 mg/cm2 P43 on 5 mm Mylar | Grant Scientific | Custom part |
| Drosophila female ventral nerve cord template (JRC 2018 VNC Female) | (Bogovic et al., 2019) | https://www.janelia.org/open-science/jrc-2018-brain-templates |
Highlights.
An automated tape-based transmission electron microscopy pipeline for connectomics.
An adult Drosophila ventral nerve cord at synapse resolution made publicly available.
>1000 motor neuron and sensory neuron reconstructions registered to a standard atlas.
A unique class of load-sensing neurons synapse onto specific leg motor neurons.
ACKNOWLEDGEMENTS
We thank A. DeCoursey, T. Pedersen, R. Xu, and A. Yeager for reconstructions; the HMS EM Core for technical support; S. Gerhard, G. Hood, X. Chen, and R. Zheng for software support; A. Bleckert, D. Brittain, R. Torres, N. da Costa, and R. C. Reid for their feedback and support; K. Hayworth for his vision, inspiration, and advice; J. Lichtman, R. Schalek, and K. Hayworth for an ATUM device and SEM support; O. Mazor and P. Gorelik for engineering support; R. Court, J. Matelsky, W. Gray-Roncal, and V. Jain for data hosting; L. Cheadle and M. Greenberg for tissue samples; R. Fetter for histology advice; J. Bogovic for atlas alignment advice; B. Mark for synapse annotation; N. Perrimon, L. Ventakasubramanian, R. Mann, S. Rayshubskiy, and R. Wilson for Drosophila lines; C. Chen and P. Gurung for LM data; H. Somhegyi for dissection assistance; M. Pecot for fly husbandry resources; T. Ayers, R. Smith, and Luxel Corporation for coating tape; and R. Wilson, D. Ginty, E. Raviola, and the Lee laboratory for comments on the manuscript. This work was supported by NIH grants (R21NS085320, RF1MH114047, RF1MH117808), the Bertarelli Program in Translational Neuroscience and Neuroengineering, Edward R. and Anne G. Lefler Center, Stanley and Theodora Feldberg Fund, a Genise Goldenson Award, and the Intelligence Advanced Research Projects Activity (IARPA) of the Department of Interior/Interior Business Center (DoI/IBC) through contract number D16PC00004 to W-C.A.L.; and by NIH grants (T32MH20017, T32HL007901) for D.G.C.H. Portions of this research were conducted on the Orchestra High Performance Compute Cluster at Harvard Medical School partially provided through NIH NCRR grant 1S10RR028832-01. The views and conclusions contained herein are those of the authors and should not be interpreted as representing the official policies or endorsements, either expressed or implied, of the funding sources including NIH, IARPA, DoI/IBC, or the U.S. Government.
Footnotes
DECLARATION OF INTERESTS
The authors declare that Harvard University filed patent applications for GridTape (WO2017184621A1) and the GridTape stage (WO2018089578A1) on behalf of B.J.G., D.G.C.H., and W-C.A.L. and negotiated licensing agreements with interested partners.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Azevedo AW, Dickinson ES, Gurung P, Venkatasubramanian L, Mann RS, and Tuthill JC (2020). A size principle for recruitment of Drosophila leg motor neurons. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, and Mann RS (2009). Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. J Neurosci 29, 6904–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock DD, Lee WCA, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, and Reid RC (2011). Network anatomy and in vivo physiology of visual cortical neurons. Nature 471, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogovic JA, Otsuna H, Heinrich L, Ito M, Jeter J, Meissner G, Nern A, Colonell J, Malkesman O, Ito K, et al. (2019). An unbiased template of the Drosophila brain and ventral nerve cord. bioRxiv, 376384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley DJ, Rathore K, VijayRaghavan K, and Williams DW (2012). Developmental origins and architecture of Drosophila leg motoneurons. J Comp Neurol 520, 1629–1649. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Helmstaedter M, and Denk W (2011). Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188. [DOI] [PubMed] [Google Scholar]
- Buchanan JT, and Grillner S (1987). Newly identified 'glutamate interneurons' and their role in locomotion in the lamprey spinal cord. Science 236, 312–314. [DOI] [PubMed] [Google Scholar]
- Buhmann J, Sheridan A, Gerhard S, Krause R, Nguyen T, Heinrich L, Schlegel P, Lee W-CA, Wilson R, Saalfeld S, et al. (2019). Automatic Detection of Synaptic Partners in a Whole-Brain Drosophila EM Dataset. bioRxiv, 2019.2012.2012.874172. [Google Scholar]
- Burrows M (1996). The neurobiology of an insect brain (Oxford University Press on Demand; ). [Google Scholar]
- Buschges A, Akay T, Gabriel JP, and Schmidt J (2008). Organizing network action for locomotion: insights from studying insect walking. Brain Res Rev 57, 162–171. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Preibisch S, Schmid B, Cheng A, Pulokas J, Tomancak P, and Hartenstein V (2010). An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Hermans L, Viswanathan MC, Fortun D, Aymanns F, Unser M, Cammarato A, Dickinson MH, and Ramdya P (2018). Imaging neural activity in the ventral nerve cord of behaving adult Drosophila. Nat Commun 9, 4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggshall J, Boschek C, and Buchner S (1973). Preliminary Investigations on a Pair of Giant Fibers in the Central Nervous System of Dipteran Flies. Zeitschrift für Naturforschung C 28, 783–784b. [Google Scholar]
- Costa M, Manton JD, Ostrovsky AD, Prohaska S, and Jefferis GS (2016). NBLAST: Rapid, Sensitive Comparison of Neuronal Structure and Construction of Neuron Family Databases. Neuron 91, 293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court R, Namiki S, Armstrong JD, Borner J, Card G, Costa M, Dickinson M, Duch C, Korff W, Mann R, et al. (2020). A Systematic Nomenclature for the Drosophila Ventral Nerve Cord. Neuron 107, 1071–1079 e1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann CJ, Hoinville T, Durr V, and Schmitz J (2017). A load-based mechanism for inter-leg coordination in insects. Proc Biol Sci 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis BD, Zavatone-Veth JA, and Clark DA (2019). The manifold structure of limb coordination in walking Drosophila. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deerinck TJ, Bushong EA, Thor A, and Ellisman MH (2010). NCMIR methods for 3D EM: a new protocol for preparation of biological specimens for serial block face scanning electron microscopy. Microscopy, 6–8. [Google Scholar]
- Dickerson BH, de Souza AM, Huda A, and Dickinson MH (2019). Flies Regulate Wing Motion via Active Control of a Dual-Function Gyroscope. Curr Biol 29, 3517–3524 e3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorkenwald S, McKellar C, Macrina T, Kemnitz N, Lee K, Lu R, Wu J, Popovych S, Mitchell E, Nehoran B, et al. (2020). FlyWire: Online community for whole-brain connectomics. bioRxiv, 2020.2008.2030.274225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duch C, Mentel T, and Pfluger HJ (1999). Distribution and activation of different types of octopaminergic DUM neurons in the locust. J Comp Neurol 403, 119–134. [DOI] [PubMed] [Google Scholar]
- Eaton RC, Bombardieri RA, and Meyer DL (1977). The Mauthner-initiated startle response in teleost fish. J Exp Biol 66, 65–81. [DOI] [PubMed] [Google Scholar]
- Eberle AL, Mikula S, Schalek R, Lichtman J, Tate MLK, and Zeidler D (2015). High-resolution, high-throughput imaging with a multibeam scanning electron microscope. J Microsc 259, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez J, Venkatasubramanian L, Baek M, Peterson M, Aghayeva U, and Mann RS (2015). Specification of individual adult motor neuron morphologies by combinatorial transcription factor codes. Neuron 86, 955–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk T, Mai D, Bensch R, Cicek O, Abdulkadir A, Marrakchi Y, Bohm A, Deubner J, Jackel Z, Seiwald K, et al. (2019). U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods 16, 67–70. [DOI] [PubMed] [Google Scholar]
- Fushiki A, Zwart MF, Kohsaka H, Fetter RD, Cardona A, and Nose A (2016). A circuit mechanism for the propagation of waves of muscle contraction in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner S (2003). The motor infrastructure: from ion channels to neuronal networks. Nat Rev Neurosci 4, 573–586. [DOI] [PubMed] [Google Scholar]
- Gwilliam G, and Burrows M (1980). Electrical characteristics of the membrane of an identified insect motor neurone. J Exp Biol 86, 49–61. [Google Scholar]
- Harris RM, Pfeiffer BD, Rubin GM, and Truman JW (2015). Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayworth KJ, Morgan JL, Schalek R, Berger DR, Hildebrand DG, and Lichtman JW (2014). Imaging ATUM ultrathin section libraries with WaferMapper: a multi-scale approach to EM reconstruction of neural circuits. Front Neural Circuits 8, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayworth KJ, Xu CS, Lu Z, Knott GW, Fetter RD, Tapia JC, Lichtman JW, and Hess HF (2015). Ultrastructurally smooth thick partitioning and volume stitching for large-scale connectomics. Nat Methods 12, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich L, Funke J, Pape C, Nunez-Iglesias J, and Saalfeld S (2018). Synaptic Cleft Segmentation in Non-isotropic Volume Electron Microscopy of the Complete Drosophila Brain (Cham: Springer International Publishing; ). [Google Scholar]
- Hildebrand DGC, Cicconet M, Torres RM, Choi W, Quan TM, Moon J, Wetzel AW, Scott Champion A, Graham BJ, Randlett O, et al. (2017). Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Laserstein P, and Helmstaedter M (2015). Large-volume en-bloc staining for electron microscopy-based connectomics. Nat Commun 6, 7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanning KC, Kaplan A, and Henderson CE (2010). Motor neuron diversity in development and disease. Annu Rev Neurosci 33, 409–440. [DOI] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vazquez-Reina A, Kaynig V, Jones TR, et al. (2015). Saturated Reconstruction of a Volume of Neocortex. Cell 162, 648–661. [DOI] [PubMed] [Google Scholar]
- Kiehn O (2011). Development and functional organization of spinal locomotor circuits. Curr Opin Neurobiol 21, 100–109. [DOI] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al. (2006). Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054. [DOI] [PubMed] [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA, and Pluim JP (2010). elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29, 196–205. [DOI] [PubMed] [Google Scholar]
- Knott G, Marchman H, Wall D, and Lich B (2008). Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci 28, 2959–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld J, Benezra SE, Narayanan RT, Svara F, Egger R, Oberlaender M, Denk W, and Long MA (2017). EM connectomics reveals axonal target variation in a sequence-generating network. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan AT, Phelps JS, Thomas LA, Nguyen TM, Han J, Chen C-L, Azevedo AW, Tuthill JC, Funke J, Cloetens P, et al. (2020). Dense neuronal reconstruction through X-ray holographic nano-tomography. Nat Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Kumar A, Balwani AH, Brittain D, Kinn S, Tovey CA, Dyer EL, da Costa NM, Reid RC, Forest CR, et al. (2018). Large-scale neuroanatomy using LASSO: Loop-based Automated Serial Sectioning Operation. PLoS One 13, e0206172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WCA, Bonin V, Reed M, Graham BJ, Hood G, Glattfelder K, and Reid RC (2016). Anatomy and function of an excitatory network in the visual cortex. Nature 532, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PH, Lindsey LF, Januszewski M, Zheng Z, Bates AS, Taisz I, Tyka M, Nichols M, Li F, Perlman E, et al. (2019). Automated Reconstruction of a Serial-Section EM Drosophila Brain with Flood-Filling Networks and Local Realignment. bioRxiv, 605634. [Google Scholar]
- Longair MH, Baker DA, and Armstrong JD (2011). Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454. [DOI] [PubMed] [Google Scholar]
- Mamiya A, Gurung P, and Tuthill JC (2018). Neural Coding of Leg Proprioception in Drosophila. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner GW, Dorman Z, Nern A, Forster K, Gibney T, Jeter J, Johnson L, He Y, Lee K, Melton B, et al. (2020). An image resource of subdivided Drosophila GAL4-driver expression patterns for neuron-level searches. bioRxiv, 2020.2005.2029.080473. [Google Scholar]
- Merritt DJ, and Murphey RK (1992). Projections of leg proprioceptors within the CNS of the fly Phormia in relation to the generalized insect ganglion. J Comp Neurol 322, 16–34. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Berger DR, Wetzel AW, and Lichtman JW (2016). The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus. Cell 165, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkres J (1957). Algorithms for the assignment and transportation problems. Journal of the society for industrial and applied mathematics 5, 32–38. [Google Scholar]
- Murphey RK, Possidente D, Pollack G, and Merritt DJ (1989). Modality-specific axonal projections in the CNS of the flies Phormia and Drosophila. J Comp Neurol 290, 185–200. [DOI] [PubMed] [Google Scholar]
- Nakane T, Kotecha A, Sente A, McMullan G, Masiulis S, Brown P, Grigoras IT, Malinauskaite L, Malinauskas T, Miehling J, et al. (2020). Single-particle cryo-EM at atomic resolution. Nature 587, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki S, Dickinson MH, Wong AM, Korff W, and Card GM (2018). The functional organization of descending sensory-motor pathways in Drosophila. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. PNAS 112, E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niven JE, Graham CM, and Burrows M (2008). Diversity and evolution of the insect ventral nerve cord. Annu Rev Entomol 53, 253–271. [DOI] [PubMed] [Google Scholar]
- O’Sullivan A, Lindsay T, Prudnikova A, Erdi B, Dickinson M, and von Philipsborn AC (2018). Multifunctional Wing Motor Control of Song and Flight. Curr Biol 28, 2705–2717 e2704. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Schneider-Mizell CM, Fetter RD, Aleman JV, Franconville R, Rivera-Alba M, Mensh BD, Branson KM, Simpson JH, Truman JW, et al. (2015). A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639. [DOI] [PubMed] [Google Scholar]
- Peltier S, Bouwer J, Jin L, Khodjasaryan K, Geist S, Xuong N, and Ellisman M (2005). Design of a new 8k x 8k lens coupled detector for wide-field, high-resolution transmission electron microscopy. Microsc Microanal, 610–611. [Google Scholar]
- Power ME (1948). The thoracico-abdominal nervous system of an adult insect, Drosophila melanogaster. J Comp Neurol 88, 347–409. [DOI] [PubMed] [Google Scholar]
- Pringle J (1938). Proprioception in insects: II. The action of the campaniform sensilla on the legs. J Exp Biol 15, 114–131. [Google Scholar]
- Ridgel AL, Frazier SF, Dicaprio RA, and Zill SN (1999). Active signaling of leg loading and unloading in the cockroach. J Neurophysiol 81, 1432–1437. [DOI] [PubMed] [Google Scholar]
- Saalfeld S, Cardona A, Hartenstein V, and Tomancak P (2009). CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25, 1984–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, et al. (2020). A connectome and analysis of the adult Drosophila central brain. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Gour A, Straehle J, Boergens KM, Brecht M, and Helmstaedter M (2017). Axonal synapse sorting in medial entorhinal cortex. Nature 549, 469–475. [DOI] [PubMed] [Google Scholar]
- Schneider-Mizell CM, Gerhard S, Longair M, Kazimiers T, Li F, Zwart MF, Champion A, Midgley FM, Fetter RD, Saalfeld S, et al. (2016). Quantitative neuroanatomy for connectomics in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd D, Harris R, Williams DW, and Truman JW (2016). Postembryonic lineages of the Drosophila ventral nervous system: Neuroglian expression reveals the adult hemilineage associated fiber tracts in the adult thoracic neuromeres. J Com Neurol 524, 2677–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrand FS (1958). Ultrastructure of retinal rod synapses of the guinea pig eye as revealed by three-dimensional reconstructions from serial sections. J Ultrastruct Res 2, 122–170. [DOI] [PubMed] [Google Scholar]
- Soler C, Daczewska M, Da Ponte JP, Dastugue B, and Jagla K (2004). Coordinated development of muscles and tendons of the Drosophila leg. Development 131, 6041–6051. [DOI] [PubMed] [Google Scholar]
- Stent GS, Kristan WB Jr., Friesen WO, Ort CA, Poon M, and Calabrese RL (1978). Neuronal generation of the leech swimming movement. Science 200, 1348–1357. [DOI] [PubMed] [Google Scholar]
- Strausfeld NJ, Seyan H, and Milde J (1987). The neck motor system of the fly Calliphora erythrocephala-I. Muscles and motor neurons. J Comp Physiol A 160, 205–224. [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. (2013). A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Xu CS, Lu Z, Rivlin PK, Parag T, Olbris DJ, Plaza S, Zhao T, Katz WT, Umayam L, et al. (2015). Synaptic circuits and their variations within different columns in the visual system of Drosophila. PNAS 112, 13711–13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia JC, Wylie JD, Kasthuri N, Hayworth KJ, Schalek R, Berger DR, Guatimosim C, Seung HS, and Lichtman JW (2012). Pervasive synaptic branch removal in the mammalian neuromuscular system at birth. Neuron 74, 816–829. [DOI] [PubMed] [Google Scholar]
- Tobin WF, Wilson RI, and Lee WCA (2017). Wiring variations that enable and constrain neural computation in a sensory microcircuit. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JR, and Schneiderman AM (1995). Flight initiations in Drosophila melanogaster are mediated by several distinct motor patterns. J Comp Physiol A 176, 355–364. [DOI] [PubMed] [Google Scholar]
- Tseng Q, Wang I, Duchemin-Pelletier E, Azioune A, Carpi N, Gao J, Filhol O, Piel M, Thery M, and Balland M (2011). A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab Chip 11, 2231–2240. [DOI] [PubMed] [Google Scholar]
- Tsubouchi A, Yano T, Yokoyama TK, Murtin C, Otsuna H, and Ito K (2017). Topological and modality-specific representation of somatosensory information in the fly brain. Science 358, 615–623. [DOI] [PubMed] [Google Scholar]
- Tuthill JC, and Azim E (2018). Proprioception. Curr Biol 28, R194–R203. [DOI] [PubMed] [Google Scholar]
- Tuthill JC, and Wilson RI (2016a). Mechanosensation and adaptive motor control in insects. Current Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, and Wilson RI (2016b). Parallel transformation of tactile signals in central circuits of Drosophila. Cell 164, 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubramanian L, Guo Z, Xu S, Tan L, Xiao Q, Nagarkar-Jaiswal S, and Mann RS (2019). Stereotyped terminal axon branching of leg motor neurons mediated by IgSF proteins DIP-alpha and Dpr10. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J (1979). Lead aspartate, an en bloc contrast stain particularly useful for ultrastructural enzymology. J Histochem Cytochem 27, 1337–1342. [DOI] [PubMed] [Google Scholar]
- Wanner AA, Genoud C, Masudi T, Siksou L, and Friedrich RW (2016). Dense EM-based reconstruction of the interglomerular projectome in the zebrafish olfactory bulb. Nat Neurosci 19, 816–825. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, and Brenner S (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans Royal Soc London B 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Xu CS, Hayworth KJ, Lu Z, Grob P, Hassan AM, Garcia-Cerdan JG, Niyogi KK, Nogales E, Weinberg RJ, and Hess HF (2017). Enhanced FIB-SEM systems for large-volume 3D imaging. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip KM, Fischer N, Paknia E, Chari A, and Stark H (2020). Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161. [DOI] [PubMed] [Google Scholar]
- Zarin AA, Mark B, Cardona A, Litwin-Kumar A, and Doe CQ (2019). A multilayer circuit architecture for the generation of distinct locomotor behaviors in Drosophila. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Lee WCA, Paul DL, and Ginty DD (2019). Multiplexed peroxidase-based electron microscopy labeling enables simultaneous visualization of multiple cell types. Nat Neurosci 22, 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, Torrens O, Price J, Fisher CB, Sharifi N, et al. (2018). A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell 174, 730–743 e722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill SN, Chaudhry S, Buschges A, and Schmitz J (2015). Force feedback reinforces muscle synergies in insect legs. Arthropod Struct Dev 44, 541–553. [DOI] [PubMed] [Google Scholar]
- Zill SN, and Moran DT (1981). The exoskeleton and insect proprioception. I. Responses of tibial campaniform sensilla to external and muscle-generated forces in the American cockroach, Periplaneta americana. J Exp Biol 91, 1–24. [Google Scholar]
- Zill SN, Moran DT, and Varela FG (1981). The exoskeleton and insect proprioception: II. Reflex effects of tibial campaniform sensilla in the American cockroach, Periplaneta americana. J Exp Biol 94, 43–55. [Google Scholar]
- Zill SN, Underwood MA, Rowley JC 3rd, and Moran DT (1980). A somatotopic organization of groups of afferents in insect peripheral nerves. Brain Res 198, 253–269. [DOI] [PubMed] [Google Scholar]
- Zwart MF, Pulver SR, Truman JW, Fushiki A, Fetter RD, Cardona A, and Landgraf M (2016). Selective Inhibition Mediates the Sequential Recruitment of Motor Pools. Neuron 91, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1 (Related to Figure 2). Fly-through of the aligned VNC EM dataset.
Zoomed-out (left) and zoomed-in (right, white boxed region from the left) views of aligned image data. Section number in the upper left corner.
Video S2 (Related to Figure 3). VNC motor and sensory neuron reconstructions.
Skeleton reconstructions of motor neurons (left) and sensory neurons (middle) from the VNC EM dataset (gray surface rendering). Sensory and motor neuron reconstructions registered to a standard VNC atlas (right, see also Fig. S3). Spheres represent neuronal cell bodies. These neurons connect the VNC to the front (red), middle (yellow), and hind (green) legs as well as the neck (blue), wings (cyan), and halteres (purple). Anterior is up and posterior is down.
Video S3 (Related to Figures 3-4, 6-7). Alignment of the EM dataset to a standard VNC atlas.
An artificial neural network was used to predict the location of synapses in the VNC EM dataset (Fig. S3). Those predictions were blurred and downsampled to produce a map of synapse densities across the VNC dataset (left). This enabled the EM dataset to be aligned (middle) to a standard VNC atlas (right).
Video S4. (Related to Figure 4) Reconstructed sensory and motor neuron subtypes.
Skeleton reconstructions and subtype classification of sensory and motor neurons in the left T1 neuromere. Sensory neurons are color coded by type as in Figure 4A: chordotonal (orange), campaniform sensilla (purple), hair plates (yellow), bristles (light blue). Primary neurites and cell bodies of the motor neurons controlling the front left leg are colored by bundle as in Figure S6A. Anterior is up and posterior is down.
Video S5 (Related to Figure 6). Gallery of left–right homologous motor neuron pairs.
Rendering of the 56 left–right pairs of T1 motor neurons whose pairings were agreed upon by manual and algorithmic assignments (Fig. 6D). Neurons are color coded by primary neurite bundle (see Figs. 6A and S6C-E for color code). Outline of the VNC in light grey. Outline of the VNC’s neuropil in darker grey. Spheres represent motor neuron cell bodies.
Video S6 (Related to Figures 5, 7). Bilateral campaniform sensillum (bCS) neurons synapse onto a fast flexor motor neuron.
Left and right T1 bCS axons (dark red and light red, respectively) make output synapses (cyan spheres) directly onto the EM reconstruction most similar to a fast tibia flexor motor neuron (blue, 81A07-Gal4). bCS axons do not synapse onto the EM reconstruction most similar to a slow tibia flexor motor neuron (yellow, 35C09-Gal4). Notice the close proximity of the fast motor neuron’s primary neurite to the bCS axons.
Figure S1 (Related to Figure 1). The GridTape imaging platform.
(A) Photograph of the magnet and hall effect sensor (dashed circle) attached to the microtome cutting arm to measure cutting frequency.
(B) Photograph of the digital opto-interrupter (dashed circle) used to detect the slot frequency in the tape.
(C) The analog signal from the hall effect sensor (left) and digital signal from the opto-interrupter (right) are used to perform closed-loop, phase-locked collection of sections onto GridTape slots.
(D) Cumulative distribution of section placement deviation (Euclidean distance from the average section position) across the VNC dataset.
(E) Photograph of a reel of GridTape containing serial sections spanning an adult female Drosophila VNC.
(F) Regions of sections not collected over slots can be imaged with SEM as in the ATUM-SEM approach. SEM images of a stretch of GridTape carrying mouse thalamus sections (left), a single section over a slot (middle), and tissue collected over the edge of the slot (right). This approach was not required for the VNC dataset due to the low number of off-slot sections.
Scale bars, 20 mm (A-B), 10 s (C), 10 mm (E), 1 mm (F, left), 500 μm (F, middle), 1 μm (F, right).
Figure S2 (Related to Figure 3). Efferent neurons with unusual features.
(A) The peripherally synapsing interneuron (PSI). (i) Dorsal view. (ii) Anterior view. (iii) Cross-section through the posterior dorsal mesothoracic nerve at the location indicated by the dashed line in (ii), showing the PSI (red) fasciculated with four motor neurons (asterisks). (iv) Higher resolution view of the region indicated in (iii). The PSI makes synaptic outputs onto neighboring motor neurons (white arrowheads) and receives reciprocal synapses back from those motor neurons (yellow arrowheads), as described previously (King and Wyman, 1980). Presynaptic specializations were not observed within any other efferent or motor neurons.
(B) Unpaired median (UM) neurons (Duch et al., 1999) with cell bodies organized into 3 clusters. (i) Dorsal view. (ii) Lateral view. Colors indicate target organ (see color key in Fig. 3A).
(C) The “multinerve” neurons. (i) Dorsal view. (ii) Anterior view. Like UM neurons but unlike motor neurons, the axon of each multinerve neuron branches to exit the VNC through multiple peripheral nerves, in this case the ipsilateral dorsal prothoracic nerve (DProN) and ipsilateral prothoracic accessory nerve (ProAN). Their dendritic branches are positioned on the dorsal-most surface of the VNC and include a contralateral projection. Like motor neurons, multinerve neurons do not make synaptic outputs within the VNC. Multinerve neurons are unique to T1, as similar neurons were not found in T2 or T3. The full dendritic backbone of the dark blue neuron (asterisk) was completely reconstructed. The other three neurons were partially reconstructed.
Scale bars, 50 μm (Ai-ii, B-C), 1 μm (Aiii-iv).
Figure S3 (Related to Figures 3-4, 6-7). Automated synapse prediction and registration to a standard VNC atlas.
(A-F) Automatic identification of postsynaptic sites using an artificial neural network.
(A) All presynaptic sites (small red dots) and postsynaptic sites (large orange dots) were manually annotated in 9 ground-truth cubes of image data spread throughout the dataset. These annotations were used to train an artificial neural network to identify postsynaptic locations (STAR Methods).
(B) EM data from (A) overlaid with postsynaptic site predictions (purple) produced by the trained network.
(C) EM data from a region of the dataset not used for training. Same data as Fig. 2G.
(D) EM data from (C) overlaid by postsynaptic site predictions (purple).
(E) A wider field of EM data. Yellow square corresponds to region in (C).
(F) EM data from (E) overlaid with postsynaptic site predictions (purple). Note that the region with a high density of postsynaptic site predictions is synaptic neuropil, whereas low density regions contain either cell bodies or axon bundles.
(G) Atlas registration strategy for comparing morphologies of neurons imaged using light microscopy (LM) with neurons reconstructed in the EM dataset. First, the trained network predicted the location of postsynaptic sites throughout the EM dataset. Next, the predicted synapse locations were 3D Gaussian blurred (σ = 900 nm) and downsampled to produce a synapse density map across the entire EM dataset. The blurred and downsampled EM synapse-density predictions were elastically transformed to register them to a LM-based standard VNC atlas based on synapse density (also see Video S3; Bogovic et al. 2019). This elastic transform was then applied to EM reconstructions to register them to the atlas. Fluorescently labeled neurons were registered to the same atlas using a neuropil counterstain. Having EM and LM reconstructions both in the same reference coordinate system enabled quantitative comparisons using NBLAST (Costa et al. 2016) (see Figs. 4, 7, S4, S7).
Scale bars, 1 μm (A-D), 10 μm (E-F), 50 μm (G).
Figure S4 (Related to Figure 4). LM to EM sensory neuron identification.
(A-G) Sensory neurons reconstructed from LM data matched to the most similar neurons reconstructed in the EM dataset. (i) Rendering of a genetically identified sensory neuron reconstructed from LM. Dorsal view of the left T1 neuromere (A-F) or the T1 and T2 neuromeres (G). (ii) Ranked distribution of NBLAST similarity scores (worst to best, left to right) between the LM neuron and the 337 left T1 leg sensory neurons reconstructed and classified in the EM dataset (Fig. 4A), color coded by sensory neuron type. (iii) Overlay of the LM-reconstructed neuron and the five highest scoring EM-reconstructed neurons. (iv) The five highest scoring EM reconstructions. Each LM sensory neuron’s identity was validated with LM imaging of the sensory neuron’s location in the leg.
(A) A club femoral chordotonal neuron (MCFO from R64C04-Gal4).
(B) A claw femoral chordotonal neuron (MCFO from iav-Gal4).
(C) A hook femoral chordotonal neuron (R70H02-AD, R32H08-DBD).
(D) A hook femoral chordotonal neuron from a different fly line (MCFO from R21D12-Gal4) than (C).
(E) A campaniform sensillum neuron (MCFO from 22E04-Gal4).
(F) A campaniform sensillum neuron from a different fly line (MCFO from R74F07-Gal4) than (E).
(G) A bilateral campaniform sensillum (bCS) neuron (MCFO from R43C10-Gal4). Only the top 2 highest scoring EM neurons are rendered. See Figure 5 for characterization of this cell type.
Scale bars, 50 μm (A-G).
Figure S5 (Related to Figure 5) bCS neuron anatomy and connectivity.
(A) Split-Gal4 labeling of campaniform sensilla in the leg. (i) Fly leg schematic from the anterior view. Red boxed region represented in (ii-iv). (ii) Surface rendering of the trochanter showing campaniform sensilla (circular domes) of cluster TrCS5 and cluster TrCS8 that were unlabeled (white) or labeled (green) by two split-Gal4 lines (iii-iv). Rendering was generated using leg reconstructions reported in (Kuan et al. 2020). (iii-iv) Fluorescence imaging of sensory neurons labeled in the trochanter by lines 38G07-AD, 43C10-DBD (iii) and 60B12-AD, 70C02-DBD (iv).
(B) Types of neurons postsynaptic to bCS neurons, ordered by number of synapses received.
(C-D) Reconstruction of central neurons receiving 2 or more synaptic inputs (C) or 5 or more synaptic inputs (D) from the reconstructed branch of bCS axons (Fig. 5F). Asterisk denotes cluster of putative Lin19A cell bodies.
(E) The two bCS axons originating from the left T1 leg (arbitrarily designated 1 and 2) target a subset of the left ProLN motor neurons. 31 out of 42 motor neurons receive no input from the reconstructed bCS axons. For any given motor neuron, the number of synaptic inputs from the two bCS axons are significantly correlated.
(F) Similar to (E), but for the two right bCS axons onto left ProLN motor neurons.
Scale bars, 10 μm (A), 50 μm (C-D).
Figure S6 (Related to Figure 6). Motor neuron bundles and nerve organization.
(A) EM reconstructions of the cell bodies and primary neurites of the front leg motor neurons. Primary neurites travel through the T1 neuromere in distinct bundles before leaving the VNC through one of four different peripheral nerves.
(B) After being transformed into the VNC atlas coordinate system, reconstructions were pruned to exclude regions outside of the neuropil. The remaining portions included only the primary neurite and were used for calculation of all NBLAST similarity scores in Figures 6B, 6E and in (C-E) below.
(C-E) (i) Rendering of primary neurites and cell bodies, color coded by bundle. (ii) Hierarchical clustering dendrograms and matrices of similarity scores, as in Fig. 6A-B but for motor neurons of different nerves.
(C) Prothoracic leg nerve (ProLN) motor neurons.
(D) Ventral prothoracic nerve (VProN) motor neurons.
(E) Dorsal prothoracic nerve (DProN) motor neurons.
(F) Positions of the subtypes of sensory and motor neuron axons within the ProLN (Fig. 3Ci). Axons of each bundle from (C) are spatially clustered within the motor domain. Axons of sensory neurons also cluster by subtype within the nerve. Chordotonal axons, arising mostly from a single sensory organ in the femur (Mamiya et al. 2018), are more clustered than other types, which arise from sensory organs distributed across different leg segments. L1-L5: L1-L5 bundle motor neurons; UM: Unpaired median neurons; br: bristle, hp: hair plate, cho: chordotonal, cs: campaniform sensillum; unc: unclassified; NR: not reconstructed.
Scale bars, 50 μm (A-E), 10 μm (F).
Figure S7 (Related to Figures 6 and 7). Motor neuron similarity, variance, and identification.
(A) Rendering of the left and right L5-bundle motor neurons, with the right-side motor neuron reconstruction reflected across the midline. These motor neurons had cell bodies in a unique location (L5 bundle in Fig. S6Ci) that allowed these two neurons to be unambiguously identified as a left–right homologous pair. NBLAST similarity score in the upper right corner. Note that while these neurons project to similar locations within the neuromere, their branches sometimes have different trajectories. Empty arrowheads point to example branch terminations with similar positions, with arrows depicting their different paths. Similarity scores between other left–right homologous pairs typically ranged between 0.5 and 0.7. Slight variability in dendritic morphology even between homologous pairs, such as the variability illustrated here, may underlie why left–right homologous neuron pairs do not have a similarity score closer to 1.
(B) A global pairwise assignment algorithm based on NBLAST similarity scores of primary neurites alone agreed with 73% (41 of 56) of front leg homologous motor neuron pairs. Black asterisks indicate agreements, red asterisks disagreements.
(C-D) Additional motor neuron reconstructions from LM matched to most similar neuron reconstructions in the EM dataset. Same layout as Figure 7.
(C) An intermediate tibia flexor motor neuron (22A08-Gal4). The highest scoring EM reconstruction receives some synaptic input from bCS neurons. The second highest scoring EM reconstruction has a similar NBLAST score but receives strong synaptic input from bCS neurons.
(D) A motor neuron innervating the tibia long tendon muscle, which controls movements of the tarsus (56H01-LexA). Different fly line and neuron than Figure 7C.
Scale bars, 10 μm (A-B), 50 μm (C-D).
Data Availability Statement
The EM dataset and reconstructions are freely available. EM image data and neuron tracings are publicly viewable at Virtual Fly Brain (https://fanc.catmaid.virtualflybrain.org/). Neuron reconstructions can be downloaded from (https://github.com/htem/GridTape_VNC_paper/tree/master/neuron_reconstructions). EM image data can be viewed and downloaded from BossDB (https://bossdb.org/project/phelps_hildebrand_graham2020). EM image data can be downloaded in CATMAID-ready JPEG tiles from a public Google Cloud bucket (gs://vnc1_r066/alignmentV3/jpgs_for_catmaid) using Google Cloud Console, the API Link, or gsutil (https://cloud.google.com/storage/docs/access-public-data). Reel-to-reel instrumentation designs and software are available at (https://github.com/htem/GridTapeStage). Code for analysis and figures is available at (https://github.com/htem/GridTape_VNC_paper/tree/master/figures_and_analysis/). Additional code is available at (https://github.com/htem/GridTape_VNC_paper, https://github.com/htem, https://www.lee.hms.harvard.edu/resources), or upon reasonable request.