ABSTRACT
Serine incorporator 5 (SERINC5) reduces the infectivity of progeny HIV-1 virions by incorporating into the outer host-derived viral membrane during egress. To counter SERINC5, the HIV-1 accessory protein Nef triggers SERINC5 internalization by engaging the adaptor protein 2 (AP-2) complex using the [D/E]xxxL[L/I]167 Nef dileucine motif. Nef also engages AP-2 via its dileucine motif to downregulate the CD4 receptor. Although these two Nef functions are related, the mechanisms governing SERINC5 downregulation are incompletely understood. Here, we demonstrate that two primary Nef isolates, referred to as 2410 and 2391 Nef, acquired from acutely HIV-1 infected women from Zimbabwe, both downregulate CD4 from the cell surface. However, only 2410 Nef retains the ability to downregulate cell surface SERINC5. Using a series of Nef chimeras, we mapped the region of 2391 Nef responsible for the functional uncoupling of these two antagonistic pathways to the dileucine motif. Modifications of the first and second x positions of the 2410 Nef dileucine motif to asparagine and aspartic acid residues, respectively (ND164), impaired cell surface SERINC5 downregulation, which resulted in reduced infectious virus yield in the presence of SERINC5. The ND164 mutation additionally partially impaired, but did not completely abrogate, Nef-mediated cell surface CD4 downregulation. Furthermore, the patient infected with HIV-1 encoding 2391 Nef had stable CD4+ T cell counts, whereas infection with HIV-1 encoding 2410 Nef resulted in CD4+ T cell decline and disease progression.
IMPORTANCE A contributing factor to HIV-1 persistence is evasion of the host immune response. HIV-1 uses the Nef accessory protein to evade the antiviral roles of the adaptive and intrinsic innate immune responses. Nef targets SERINC5, a restriction factor which potently impairs HIV-1 infection by triggering SERINC5 removal from the cell surface. The molecular determinants underlying this Nef function remain incompletely understood. Recent studies have found a correlation between the extent of Nef-mediated SERINC5 downregulation and the rate of disease progression. Furthermore, single-residue polymorphisms outside the known Nef functional motifs can modulate SERINC5 downregulation. The identification of a naturally occurring Nef polymorphism impairing SERINC5 downregulation in this study supports a link between Nef downregulation of SERINC5 and the rate of plasma CD4+ T cell decline. Moreover, the observed functional impairments of this polymorphism could provide clues to further elucidate unknown aspects of the SERINC5 antagonistic pathway via Nef.
KEYWORDS: HIV, Nef, SERINC5, CD4, membrane trafficking, infectivity
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) accessory protein Nef is a virulence factor that is critical for accelerating disease progression from acute HIV-1 infection to AIDS (1, 2). Nef mechanistically enhances virion infectivity by countering the antiviral functions of the serine incorporator 5 (SERINC5) restriction factor (3, 4).
SERINC5 belongs to the SERINC protein family, a well-conserved protein family in eukaryotes with uncharacterized physiological cellular functions (5, 6). SERINC5 exerts a potent antiviral effect by incorporating into the outer host-derived lipid viral membrane of egressing progeny virions (3, 4). When incorporated into virions, SERINC5 impedes HIV-1 cellular entry at the membrane fusion step (3, 4, 7), as demonstrated by the minimal reverse transcription activity or de novo HIV-1 viral protein synthesis upon target cell infection (3). The presence of SERINC5 can interfere with membrane fusion by increasing the energy barrier required for enlarging the fusion pore necessary for successful infection (7). Nef counters SERINC5 primarily by triggering its removal from the cell surface through linking the restriction factor to the endolysosomal network (3, 8). Internalized SERINC5 is ultimately directed to lysosomes, where it is subsequently degraded (9). Due to its removal from the cell surface, SERINC5 is no longer incorporated into virions during egress, effectively restoring HIV-1 particle infectivity (3, 4). To link SERINC5 to the endolysosomal network, Nef engages the adaptor protein 2 (AP-2) complex via the highly conserved Nef dileucine motif ([D/E]xxxL[L/I]165) (9–11). The [D/E]xxxL[L/I]165 motif, where x represents any amino acid, engages AP-2 by interacting with a hydrophobic pocket formed by the AP-2 σ subunit (12, 13). The AP-2 α subunit also contacts Nef, further stabilizing the Nef–AP-2 protein-protein interaction (12).
Nef also utilizes its dileucine motif to interact with AP-2 to functionally downregulate CD4 from the cell surface (12–14). Indeed, mutating the two leucine residues in the Nef dileucine motif to alanine (LL165AA) severs the Nef–AP-2 interaction, thereby impeding Nef’s ability to downregulate both CD4 (14, 15) and SERINC5 (16). Several Nef motifs required to downregulate cell surface CD4 are also required to downregulate SERINC5. These include the G2 motif (3, 9, 16), required for Nef myristoylation, the Nef CAW57 motif (9, 16), and the Nef C-terminal DD175 motif (3, 9, 16). However, the complete molecular determinants underlying Nef-mediated SERINC5 antagonism are still incompletely understood. For instance, Nef can indirectly counter the antiviral functions of SERINC5 without disrupting SERINC5 incorporation into egressing virions (16). Thus, the molecular determinants underlying this alternative antagonistic pathway requires further investigation.
Although Nef motifs utilized to downregulate CD4 and SERINC5 may overlap, genetic diversity within primary Nef protein sequences outside the conserved motifs may result in variations of specific Nef functions, including CD4 and major histocompatibility complex class I (MHC-I) downregulation (17–19). Furthermore, primary subtype B HIV-1 Nef clones isolated from elite controllers displayed reduced ability to antagonize in vitro SERINC5 compared to chronic progressors, suggesting that the extent to which Nef downregulates SERINC5 contributes to reduced viremia and concomitant slower disease progression (20).
Here, we identified naturally occurring polymorphisms found within primary nef sequences in regions responsible for interacting with the membrane trafficking machinery and characterized how these polymorphisms mechanistically link to subsequent Nef function(s). We demonstrate divergence between Nef isolates and their ability to downregulate cell surface SERINC5 and CD4. Using flow cytometry-based assays, we identified a naturally occurring ND164 polymorphism within the Nef dileucine [D/E]xxxL[L/I]165 motif, which uncoupled the Nef-mediated CD4 and SERINC5 antagonistic pathways. Furthermore, this polymorphism resulted in a severe reduction in infectious virus yield when produced in the presence of SERINC5, which may explain differential rates of plasma CD4+ T cell decline observed within the infected patients.
RESULTS
Primary HIV-1 nef sequences.
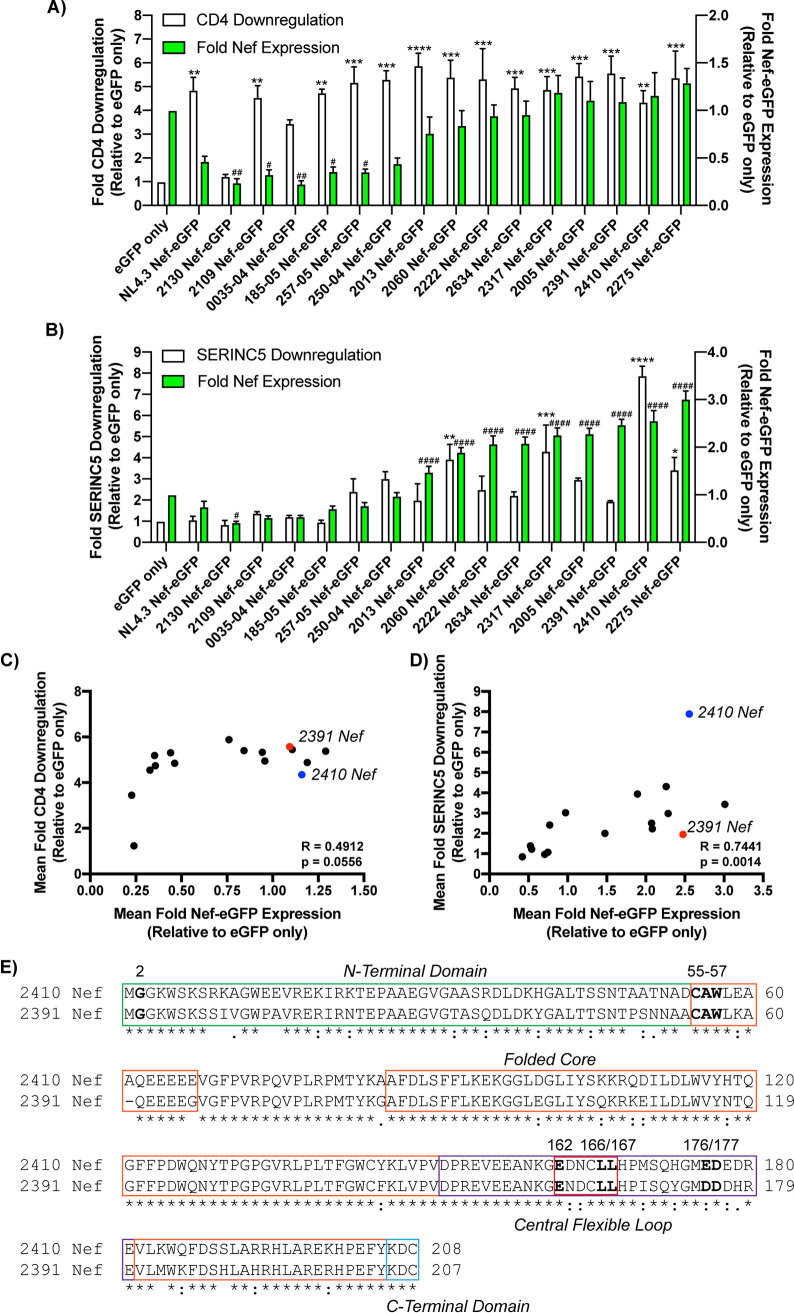
We initially sought to determine whether nef sequences obtained from a patient cohort displayed differential abilities to downregulate cell surface SERINC5. As such, we isolated a panel of 15 Nef isolates acquired from newly HIV-1-infected Ugandan and Zimbabwean women that were subsequently monitored during disease progression as part of the Hormonal Contraception and HIV-1 Genital Shedding and Disease Progression among Women with Primary HIV-1 Infection (GS) study (21, 22). Accordingly, nef sequences were isolated from blood samples acquired at the first visit postseroconversion and cloned into enhanced green fluorescent protein (eGFP) expression vectors to allow for flow cytometry-based assessment of Nef-eGFP fusion protein expression. These Nef-eGFP vectors were subsequently expressed in CD4+ HeLa cells in the absence or presence of SERINC5.intHA (here referred to as SERINC5) to assess cell surface CD4 (Fig. 1A) and SERINC5 (Fig. 1B) downregulation. Downregulation of cell surface CD4 was initially assessed to confirm whether the Nef-eGFP isolates were functional, as downregulation of CD4 is a highly conserved Nef function (23, 24). Our flow cytometry analysis revealed the various Nef isolates generally downregulated CD4 from the cell surface efficiently, despite variable Nef-eGFP expression (Fig. 1C). Conversely, the primary Nef isolates drastically differed in their ability to downregulate cell surface SERINC5, with the level of Nef-eGFP expression correlating to the extent by which Nef downregulates cell surface SERINC5 (Fig. 1D). Despite this correlation between Nef-eGFP expression and SERINC5 downregulation, we identified variable SERINC5 downregulation, including Nef isolates capable of downregulating SERINC5 robustly (Fig. 1B, 2410 Nef) as well as isolates demonstrating a low degree of SERINC5 downregulation ability (Fig. 1B, 2391 Nef). Interestingly, isolates 2410 and 2391 Nef were expressed to similar extents, suggesting the differences observed in cell surface SERINC5 downregulation were not due to variable levels of Nef-eGFP expression but rather due to the presence of polymorphisms between these two Nef isolates. Analysis of the amino acid sequences of the two Nef isolates revealed that both retain the known shared functional motifs required to downregulate both cell surface CD4 and enhance virion infectivity in the presence of SERINC5 (Fig. 1E, boldfaced). However, both of the Nef isolates had notable variations within the amino (N)-terminal domain and carboxyl (C)-terminal folded core of the protein, suggesting these polymorphisms are responsible for the impairments in SERINC5 downregulation comparing 2391 to 2410 Nef (Fig. 1E).
FIG 1.
Primary Nef isolates displayed diverging Nef functionality. (A) CD4+ HeLa cells expressing Nef isolates were stained for cell surface CD4 and analyzed by flow cytometry. Shown is a summary of the fold CD4 downregulation ability (±SE) on the left y axis and fold Nef expression (±SE) on the right y axis for each Nef isolate (n = 3). (B) CD4+ HeLa cells coexpressing each Nef isolate and SERINC5.intHA were stained for cell surface SERINC5 and analyzed by flow cytometry. Shown is a summary of the fold SERINC5 downregulation ability (±SE) on the left y axis, and fold Nef-expression (±SE) on the right y axis for each Nef isolate (n = 3). Significance of Nef-eGFP expression and CD4/SERINC5 cell surface downregulation are denoted by # and ∗, respectively, upon comparison to the eGFP only control. Correlation analysis between the mean fold Nef-eGFP expression (x axis) and mean fold CD4 downregulation (C) and mean fold SERINC5 downregulation (y axis) (D) are shown for the 15 described primary Nef isolates and NL4.3 Nef. The blue and red dots indicate the 2410 Nef-eGFP and 2391 Nef-eGFP samples, respectively. Statistical analysis was conducted using the Spearman rank correlation test. (E) Amino acid sequence alignment of 2410 and 2391 Nef. Boldfaced Nef residues denote shared residues/motifs involved in Nef-mediated CD4 downregulation or enhancing virion infectivity in the presence of SERINC5. The location of the specific Nef domains, including the N-terminal domain (green), folded core (orange), central flexible loop (purple), Nef dileucine motif (red), and the C-terminal domain (blue), are highlighted in the appropriate boxes. An asterisk indicates positions with a fully conserved residue between the isolates. A colon indicates conservation of residues with similar properties (scoring >0.5 in the Gonnet PAM 250 matrix). A period indicates conservation of residues with less similar properties (scoring ≤0.5 and >0 in the Gonnet PAM 250 matrix). eGFP, enhanced green fluorescent protein; SE, standard error; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; #, P ≤ 0.05; ##, P ≤ 0.01; ####, P ≤ 0.0001.
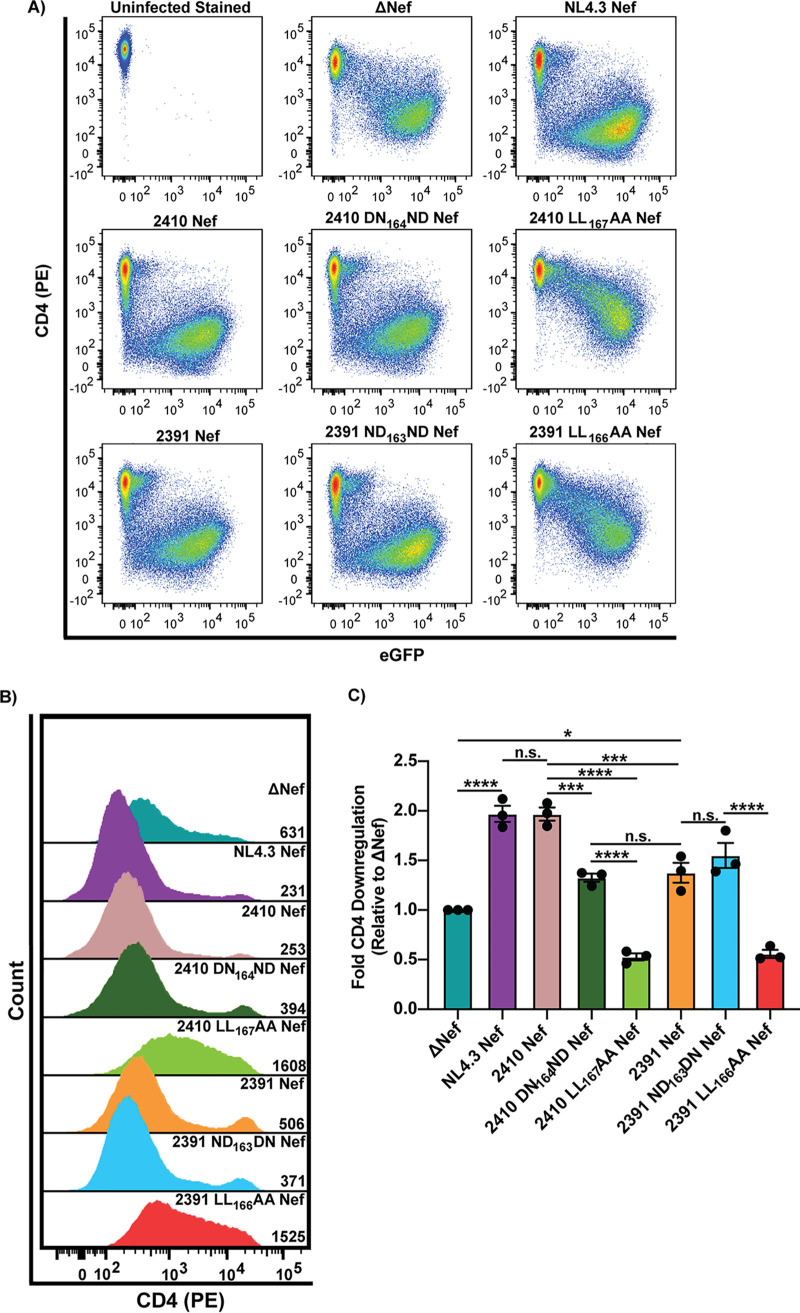
Nef isolates 2410 and 2391 demonstrate diverging abilities to downregulate SERINC5.
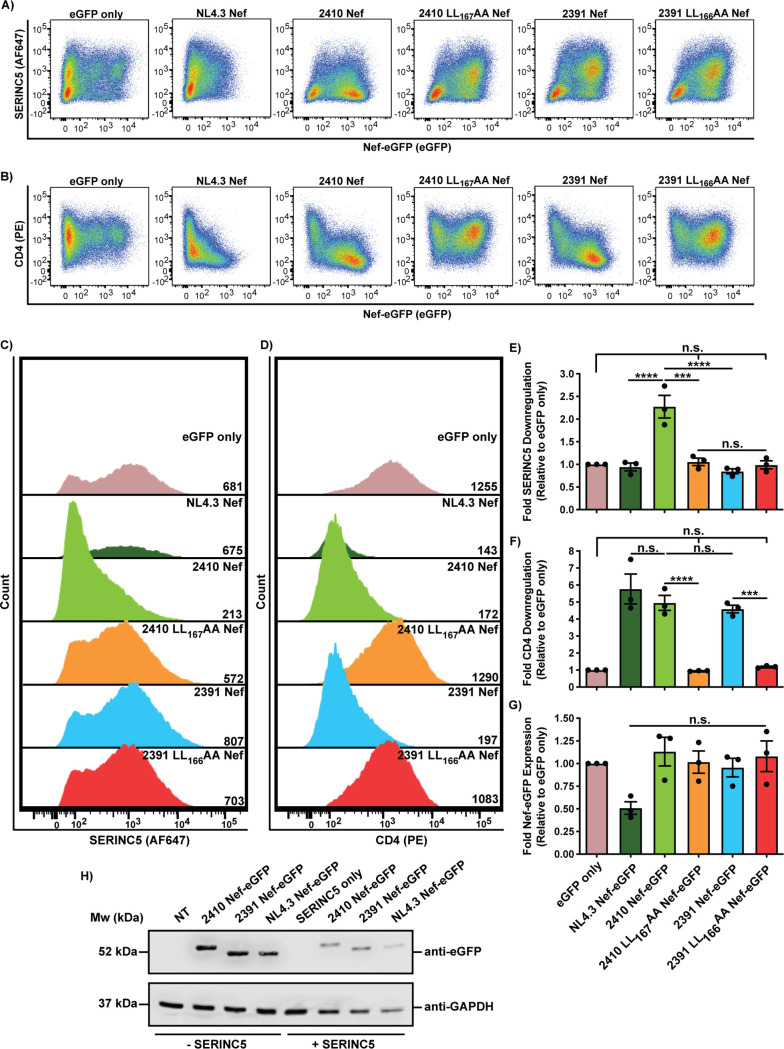
We next analyzed the ability of 2410 and 2391 Nef to mediate cell surface SERINC5 and CD4 downregulation concurrently and compared these levels to a laboratory-adapted wild-type HIV-1 NL4.3 Nef. In addition, the leucine residues within the [D/E]xxxL[L/I]165 motif were mutated in 2410 and 2391 Nef (2410 LL167AA and 2391 LL166AA Nef), thereby disrupting the Nef–AP-2 interaction. As such, the eGFP-tagged Nef isolates were coexpressed with SERINC5 in CD4+ HeLa cells, and 24 h later, the cells were stained for cell surface CD4 and SERINC5 and analyzed by flow cytometry. Compared to cells expressing eGFP only, cells expressing eGFP-tagged isolates 2410 and 2391 Nef, along with NL4.3 Nef, demonstrated efficient CD4 cell surface downregulation (Fig. 2B, D, and F). As expected, the 2410 LL167AA and 2391 LL166AA Nef dileucine mutants were unable to downregulate cell surface CD4 and SERINC5, as the cell surface levels of both proteins were not significantly different than those of cells expressing eGFP only (Fig. 2A to F). As in Fig. 1, isolate 2410 Nef, but not 2391 Nef, additionally retained the ability to downregulate cell surface SERINC5 (Fig. 2A, C, and E), despite being expressed at equivalent levels (Fig. 2G). Moreover, NL4.3 Nef downregulated cell surface SERINC5 to the same level as 2391 Nef (Fig. 2A, C, and E). This may be due to the low expression level of NL4.3 Nef relative to 2410 and 2391 Nef in the presence of SERINC5 (Fig. 2G and H) and potentially differing SERINC5 levels in CD4+ HeLa cells compared to endogenous SERINC5 expression in Jurkat E6.1/TAg cell lines where NL4.3 Nef downregulated SERINC5 (3, 4). To confirm the eGFP fluorescence detected via flow cytometry represented Nef-eGFP fusion protein expression, we conducted a Western blot analysis of cells expressing the Nef-eGFP isolates and determined that 2410 and 2391 Nef-eGFP fusion proteins were expressed to similar levels, both in the presence and absence of SERINC5 (Fig. 2H). 2391 Nef was able to downregulate cell surface CD4 efficiently, but not SERINC5, which suggests 2391 Nef can encode polymorphisms negatively impacting SERINC5 but not CD4 cell surface downregulation, thereby effectively uncoupling the two antagonistic pathways.
FIG 2.
Nef isolates 2410 and 2391 both downregulate cell surface CD4 but differentially downregulate cell surface SERINC5. CD4+ HeLa cells expressing 2410 or 2391 Nef-eGFP and SERINC5.intHA were stained for cell surface CD4 and SERINC5 and analyzed by flow cytometry. CD4+ HeLa cell populations were determined by gating on eGFP+ cells using an FMO control. (A and B) Representative pseudocolor plots illustrating cell surface SERINC5 (Alexa Fluor 647) (A) and cell surface CD4 (PE) (B) levels of Nef+ cell populations (eGFP+). (C and D) Representative histograms illustrating cell surface SERINC5 (C) or CD4 (D) levels on CD4+ HeLa cells after gating on single and Nef+ (eGFP+) cells. Geometric mean fluorescence intensities (MFIs) of the cell surface proteins are indicated. (E to G) Summary of the fold downregulation ability (±SE) for cell surface SERINC5 (E), cell surface CD4 (F), and fold Nef-eGFP (G) expression (n = 3). (H) Western blot illustrating the expression of Nef-eGFP fusion proteins in the absence and presence of SERINC5 in CD4+ HeLa cells. eGFP, enhanced green fluorescent protein; AF647, Alexa Fluor 647; PE, phycoerythrin; SE, standard error; Mw, molecular weight; kDa, kilodaltons; NT, nontransfected; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., nonsignificant (P > 0.05).
The N-terminal region of 2391 Nef does not mediate the uncoupling of SERINC5 and CD4 cell surface downregulation.
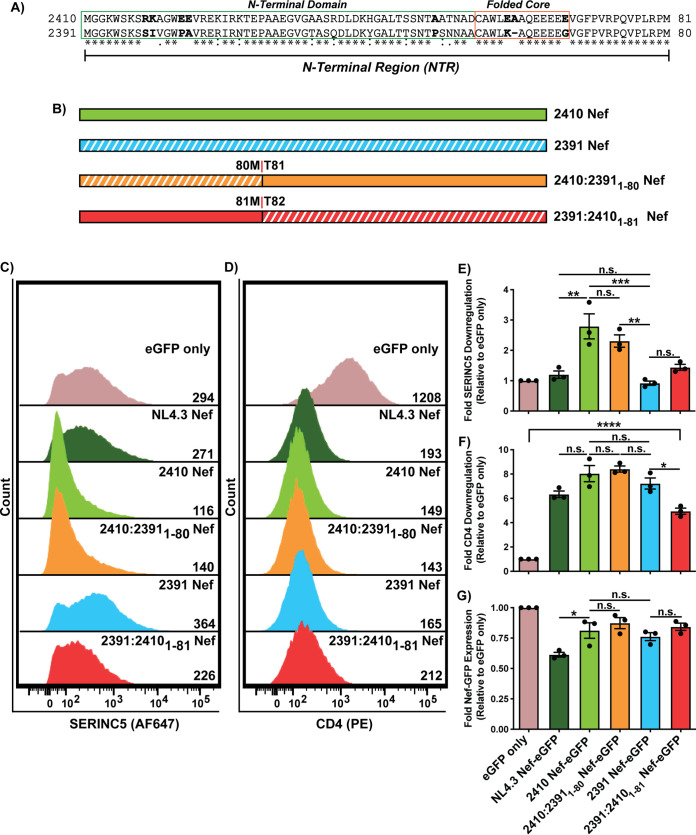
To determine the region of 2391 Nef responsible for impaired cell surface SERINC5 downregulation, we created Nef chimeras where segments of 2410 Nef were swapped with the corresponding region of 2391 Nef and subsequently determined their respective abilities to downregulate cell surface CD4 and SERINC5. While isolates 2410 and 2391 are conserved in much of the core region of Nef, we identified numerous polymorphisms within the first 81 amino acids, here referred to as the Nef N-terminal region (NTR) (Fig. 3A, residues 1 to 81; numbering based on 2410 Nef). Accordingly, the NTR of 2410 Nef was swapped with the NTR of 2391 Nef (2410:23911-80 Nef) and vice versa (2391:24101-81 Nef) (Fig. 3B). Flow cytometry analysis demonstrated that the 2410:23911-80 Nef chimera suffered a modest, but statistically insignificant, impairment in cell surface SERINC5 downregulation compared to 2410 Nef (Fig. 3C and E). Consistent with this finding, the 2391:24101-81 Nef chimera also demonstrated a modest, but insignificant, improvement in cell surface SERINC5 downregulation ability (Fig. 3C and E). Interestingly, 2391:24101-81 Nef suffered statistically significant decreases in cell surface CD4 downregulation compared to 2391 Nef, suggesting polymorphisms within the NTR of 2410 Nef negatively affect CD4 downregulation (Fig. 3D and F). However, this 2391:24101-81 Nef chimera retained considerable cell surface CD4 downregulation compared to eGFP only. Considering the 2410:23911-80 Nef chimera did not suffer significant impairments in cell surface SERINC5 downregulation, we next investigated the carboxyl (C)-terminal portion of Nef.
FIG 3.
Polymorphisms within the N-terminal domain of 2391 Nef do not impair its ability to downregulate cell surface SERINC5. CD4+ HeLa cells expressing 2410 and 2391 Nef or N-terminal Nef chimeras with SERINC5.intHA were stained for cell surface CD4 and SERINC5 and analyzed by flow cytometry. Nef+ CD4+ HeLa cells were determined by gating on eGFP+ cells using an FMO control. (A) Amino acid sequence alignment comparing the N-terminal region (residues 1 to 81; numbering based on the 2410 Nef sequence) of isolate 2410 and 2391 Nef. Boldfaced residues indicate polymorphisms of interest within the N-terminal region. The N-terminal domain and folded core region are denoted in green and orange boxes, respectively. (B) Schematic illustrating the Nef isolates and N-terminal Nef chimeras. The chimera breakpoint for each isolate is indicated. Subscripted residues indicate swapping with the other Nef isolate at the corresponding position. Regions belonging to 2391 Nef are indicated with white diagonal stripes, while regions belonging to 2410 Nef are indicated with solid colors. (C and D) Representative histograms illustrating cell surface SERINC5 (C) or CD4 levels (D) on CD4+ HeLa cells after gating on single and transfected (eGFP+) cells. Geometric mean fluorescence intensities (MFIs) of the respective cell surface proteins are indicated. (E to G) Summary of the fold-downregulation ability (±SE) for cell surface SERINC5 (E), cell surface CD4 (F), and fold Nef-eGFP (G) expression for each Nef isolate (n = 3). eGFP, enhanced green fluorescent protein; AF647, Alexa Fluor 647; PE, phycoerythrin; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., nonsignificant (P > 0.05).
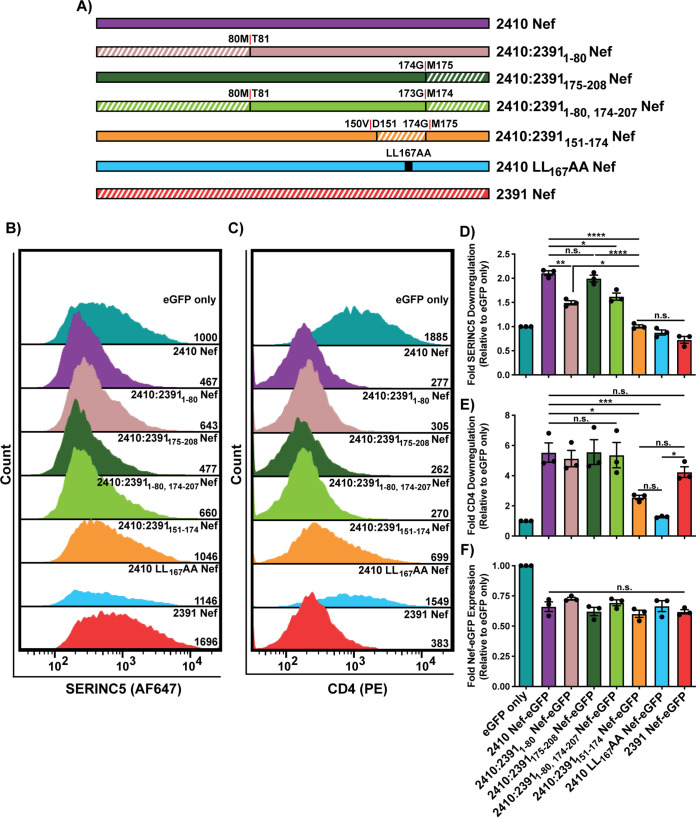
A C-terminal region within the flexible loop of 2391 Nef severely impairs cell surface SERINC5 and partially impairs cell surface CD4 downregulation.
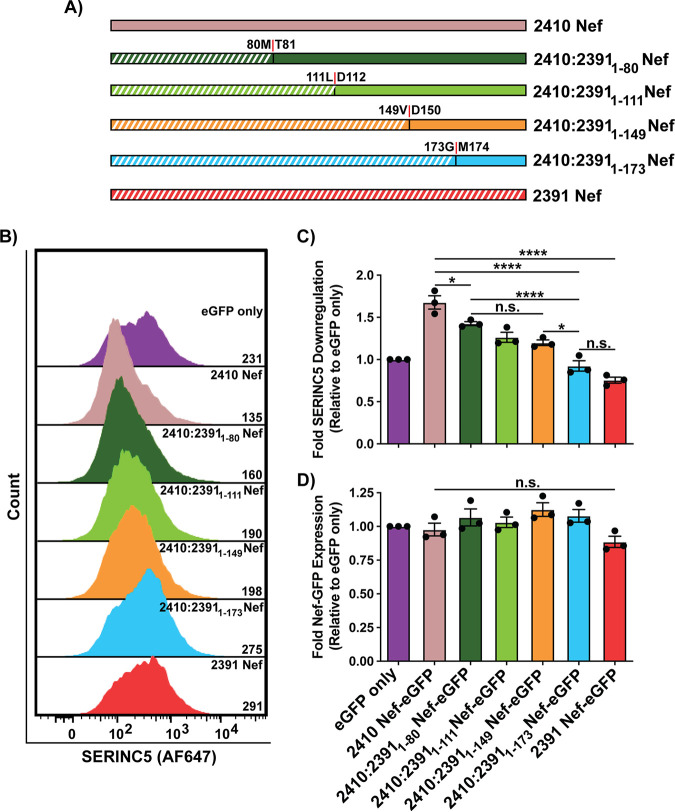
Next, we synthesized several Nef chimeras in which incrementally larger C-terminal regions of 2410 Nef were swapped with the corresponding regions of 2391 Nef (Fig. 4A). Consistent with Fig. 3, the 2410:23911-80 Nef chimera again suffered modest impairments in cell surface SERINC5 downregulation compared to 2410 Nef (Fig. 4B and C). However, the ability of 2410:23911-80 Nef to downregulate cell surface SERINC5 was more efficient than that of 2391 Nef, further suggesting the implicated region/polymorphism(s) resides outside the Nef NTR (Fig. 4B and C). When the first 111 and 149 residues of 2410 were replaced with 2391 Nef (2410:23911-111 Nef and 2410:23911-149 Nef respectively), no further decreases were observed in SERINC5 downregulation relative to 2410:23911-80 Nef (Fig. 4B and C). However, when the first 173 residues of 2410 Nef were replaced with the corresponding region of 2391 Nef (2410:23911-173 Nef), there was a significant reduction in cell surface SERINC5 downregulation relative to all other chimeras as well as 2410 Nef (Fig. 4B and C). Importantly, the ability of 2410:23911-173 Nef and 2391 Nef to downregulate cell surface SERINC5 was equivalent (Fig. 4B and C). Taken together, these results suggest the Nef region underlying the impairment in SERINC5 downregulation resides between residues 150 to 173. Given that 2410:23911-149 Nef did not alter cell surface SERINC5 downregulation relative to 2410:23911-80 Nef while 2410:23911-173 Nef robustly decreased SERINC5 downregulation, 2410 Nef chimeras were subsequently designed in which residues 151 to 174 of 2410 Nef (residue numbering shifted, since 2410 Nef is one amino acid longer than 2391 Nef) were replaced with the corresponding region of 2391 Nef (2410:2391151-174 Nef) and a 2410 chimera in which the C-terminal 175 to 208 residues were replaced with the corresponding region of 2391 Nef (2410:2391175-208 Nef) (Fig. 5A). Accordingly, the 2410:2391151-174 Nef chimera was impaired in cell surface SERINC5 downregulation relative to 2410 Nef and 2410:23911-80 Nef, but, importantly, equivalent to 2391 Nef, suggesting the implicated Nef residues reside within this region (Fig. 5B and D). Furthermore, the 2410:2391175-208 Nef and 2410:23911-80, 174-207 Nef chimeras did not suffer significant impairments in SERINC5 downregulation, confirming the implicated region does not reside within the C-terminal 174 to 207 residues (Fig. 5B and D). Conversely, the 2410:2391151-174 Nef chimera suffered an ∼2-fold reduction in cell surface CD4 downregulation ability relative to 2410 Nef and associated chimeras (Fig. 5C and E). While this Nef chimera did suffer impairments in downregulating cell surface CD4, it was not completely impaired in its ability to downregulate cell surface CD4, as evidenced upon comparison to eGFP only and the 2410 LL167AA Nef dileucine mutant (Fig. 5C and E). Taken together, these results suggest cell surface SERINC5 downregulation is severely impaired and cell surface CD4 downregulation is partially impaired when residues 151 to 174 of 2410 Nef are swapped with the corresponding region of 2391 Nef.
FIG 4.
C-terminal region of 2391 Nef mediates severe impairments in downregulating cell surface SERINC5. CD4+ HeLa cells expressing various 2410 and 2391 Nefs with SERINC5.intHA were stained for cell surface SERINC5 and analyzed by flow cytometry. Nef+ CD4+ HeLa cells were determined by gating on eGFP+ cells using an FMO control. (A) Schematic illustrating Nef isolates and C-terminal 2410 Nef chimeras. Chimera break points for each isolate are indicated. Regions belonging to 2391 Nef are indicated with white diagonal stripes, while regions belonging to 2410 Nef are indicated with solid colors. (B) Representative histogram illustrating the cell surface SERINC5 levels on CD4+ HeLa cells after gating on single and transfected (eGFP+) cells. Geometric mean fluorescence intensities (MFIs) of cell surface SERINC5 are indicated. (C) Summary of the fold downregulation ability (±SE) for cell surface SERINC5. (D) Fold Nef-eGFP expression for each Nef isolate (n = 3). eGFP, enhanced green fluorescent protein; AF647, Alexa Fluor 647; SE, standard error; *, P ≤ 0.05; ****, P ≤ 0.0001; n.s., nonsignificant (P > 0.05).
FIG 5.
C-terminal region mediating SERINC5 downregulation impairments is located between residues 151 and 174. CD4+ HeLa cells expressing 2410 and 2391 Nef as well as Nef chimeras in which residues 151 to 174 and residues 175 to 208 of 2410 Nef were replaced with the corresponding region of 2391 Nef (2410:2391151-174 Nef and 2410:2391175-208 Nef, respectively) and with SERINC5.intHA were stained for cell surface CD4 and SERINC5 and analyzed by flow cytometry. Nef+ CD4+ HeLa cells were determined by gating on eGFP+ cells using an FMO control. (A) Schematic illustrating Nef isolates and 2410 Nef chimeras. Chimera break points for each isolate are indicated. Regions belonging to 2391 Nef are indicated with white diagonal stripes, while regions belonging to 2410 Nef are indicated with solid colors. (B and C) Representative histograms illustrating cell surface SERINC5 (B) or CD4 (C) levels on CD4+ HeLa cells after gating on single and transfected (eGFP+) cells. Geometric mean fluorescence intensities (MFIs) of the respective cell surface proteins are indicated. (D to F) Summary of the fold downregulation ability (±SE) for cell surface SERINC5 (D), cell surface CD4 (E), and fold Nef-eGFP (F) expression for each Nef isolate (n = 3). eGFP, enhanced green fluorescent protein; AF647, Alexa Fluor 647; PE, phycoerythrin; SE, standard error; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., nonsignificant (P > 0.05).
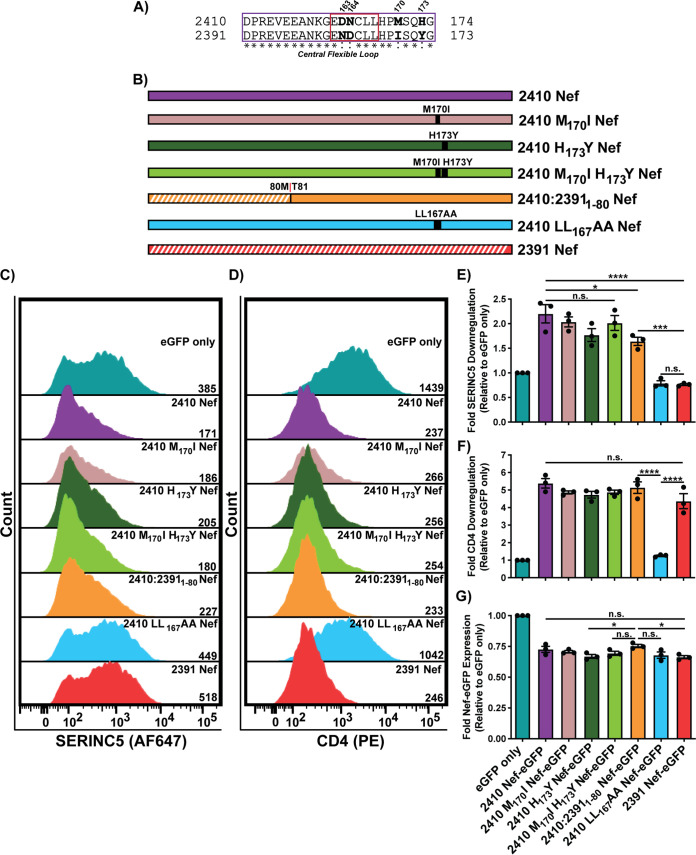
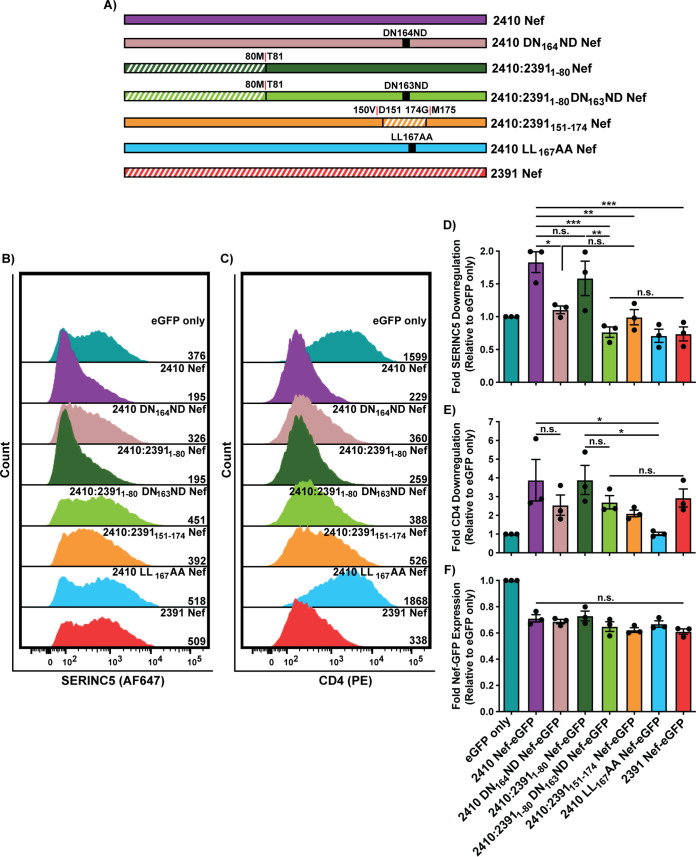
The Nef ND164 polymorphism underlies severe impairments in cell surface SERINC5 downregulation and partial impairments in CD4 cell surface downregulation.
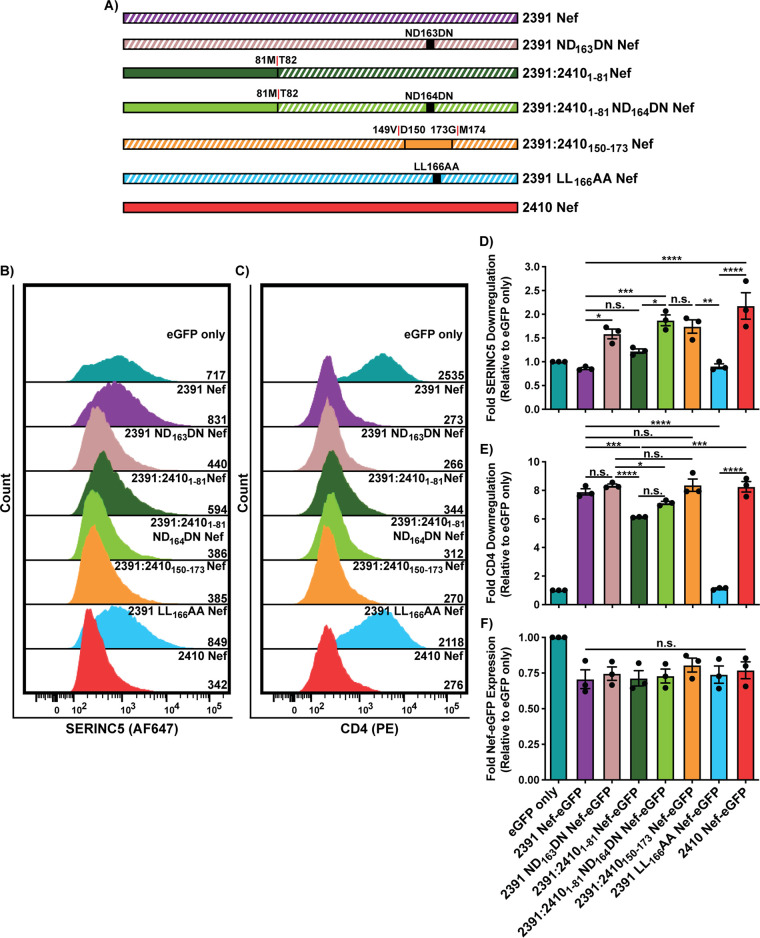
Nef residues 151 to 174 reside within the C-terminal central flexible loop that harbors the [D/E]xxxL[L/I]167 dileucine motif (Fig. 1E). Four polymorphisms exist within this region between the two Nef isolates; 2410 Nef encodes DPREVEEANKGEDNCLLHPMSQHG174, and 2391 Nef encodes DPREVEEANKGENDCLLHPISQYG173, with boldfaced residues indicating the polymorphisms and underlined residues identifying those comprising the Nef dileucine motif (Fig. 6A). Since the Nef dileucine motif has been defined as [D/E]xxxL[L/I], where x can represent any amino acid, we initially postulated that the latter two polymorphisms downstream from the dileucine motif at positions 170 and 173 (residue numbering based on 2410 Nef) result in the observed functional impairments in cell surface SERINC5 downregulation. As such, we generated single 2410 M170I and 2410 H173Y Nef mutants as well as a double 2410 M170I H173Y Nef mutant and tested their ability to downregulate cell surface SERINC5 and CD4 (Fig. 6B). We noticed no significant impairments in the single mutant or the double mutant to downregulate cell surface SERINC5 or CD4 (Fig. 6C to F). With no observable reductions in cell surface SERINC5 downregulation, we next sought to address whether the position of the asparagine and aspartic acid (ND163) within the 2391 Nef dileucine motif plays a role. Thus, we swapped the positions of the aspartic acid and asparagine residues within 2410 Nef (2410 DN164ND Nef) or within the NTR chimera (2410:23911-80 DN163ND) (Fig. 7A). Importantly, the 2410 DN164ND Nef mutant suffered a significant and severe impairment in cell surface SERINC5 downregulation, which was further accentuated in the 2410:23911-80 DN163ND Nef chimera (Fig. 7B and D). Interestingly, there was a trend in which 2410 DN164ND Nef suffered impairments in cell surface CD4 downregulation, consistent with our results with the 2410:2391151-174 Nef chimera (Fig. 7C and E). Taken together, these results suggest that the inversion of DN164 to ND164 is a major factor capable of severely impairing cell surface SERINC5 downregulation, as observed with 2391 Nef. Furthermore, the ND164 polymorphism is sufficient to partially impair cell surface CD4 downregulation, thereby affecting both antagonistic functions of Nef and uncoupling the two pathways. With the prior results suggesting the ND164 mutation within 2410 Nef severely impairs cell surface SERINC5 downregulation and 2391 Nef exhibits impaired cell surface SERINC5 downregulation, we next tested whether the reverse mutation, a DN163 mutation in 2391 Nef (2391 ND163DN Nef), would rescue cell surface SERINC5 downregulation (Fig. 8A). Indeed, we found there was a significant improvement in cell surface SERINC5 downregulation observed with 2391 ND163DN Nef compared to 2391 Nef (Fig. 8B and D). An enhanced rescue in SERINC5 downregulation was further achieved with the 2391:24101-81 Nef mutant bearing the DN164 mutation (2391:24101-81 ND164DN Nef) (Fig. 8B and D). Importantly, the ability of this chimera to downregulate cell surface SERINC5 was similar to that of 2410 Nef, demonstrating cell surface SERINC5 downregulation could be rescued with the reverse mutation in this system.
FIG 6.
Mutations of 2410 Nef M170 or H173 do not impact SERINC5 downregulation. CD4+ HeLa cells expressing 2410 and 2391 Nef as well as the respective 2410 Nef mutants with SERINC5.intHA were stained for cell surface CD4 and SERINC5 and analyzed by flow cytometry. Nef+-transfected CD4+ HeLa cells were determined by gating on eGFP+ cells using an unstained control. (A) Amino acid sequence alignment comparing residues 151 to 174 of 2410 and 2391 Nef. Boldfaced residues indicate polymorphisms between the two Nef isolates within this region. The central flexible loop and Nef dileucine motif are denoted by purple and red boxes, respectively. (B) Schematic illustrating the Nef isolates and 2410 Nef chimeras/mutants utilized. Chimera break points for each isolate are indicated accordingly. Regions belonging to 2391 Nef are indicated with white diagonal stripes, while regions belonging to 2410 Nef are indicated with solid colors. (C and D) Representative histograms illustrating cell surface SERINC5 (C) and CD4 (D) levels on CD4+ HeLa cells after gating on single and transfected (eGFP+) cells. Geometric mean fluorescence intensities (MFIs) of the respective cell surface proteins are indicated. (E to G) Summary of the fold downregulation ability (±SE) for cell surface SERINC5 (E), cell surface CD4 (F), and fold Nef-eGFP (G) expression for each Nef isolate (n = 3). eGFP, enhanced green fluorescent protein; AF647, Alexa Fluor 647; PE, phycoerythrin; SE, standard error; *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., nonsignificant (P > 0.05).
FIG 7.
Mutations of 2410 Nef residues D163 and N164 (DN164) severely impairs SERINC5 downregulation. CD4+ HeLa cells expressing 2410 and 2391 as well as mutants with SERINC5.intHA were stained for cell surface SERINC5 and CD4 and analyzed by flow cytometry. Single transfected CD4+ HeLa cells were determined by gating on eGFP+ cells using an unstained control. (A) Schematic of Nef isolates and chimeras/mutants utilized. The chimera break points for the respective chimera isolates are indicated. Regions belonging to 2391 Nef are indicated with white diagonal stripes, while regions belonging to 2410 Nef are indicated with solid colors. (B and C) Representative histograms illustrating cell surface (B) SERINC5 or (C) CD4 levels on CD4+ HeLa cells after gating on single and transfected (eGFP+) cells. Geometric mean fluorescence intensities (MFIs) of the respective cell surface proteins are indicated. (D to F) Summary of the fold downregulation ability (±SE) for (D) cell surface SERINC5, (E) cell surface CD4, and (F) fold Nef-eGFP expression for each Nef isolate (n = 3). eGFP, enhanced green fluorescent protein; AF647, Alexa Fluor 647; PE, phycoerythrin; SE, standard error; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., nonsignificant (P > 0.05).
FIG 8.
Reversing the polymorphism in 2391 Nef (ND163) rescues the ability of 2391 Nef to downregulate cell surface SERINC5. CD4+ HeLa cells expressing 2410 and 2391 Nef as well as 2391 Nef mutants with SERINC5.intHA. were stained for cell surface SERINC5 and CD4 and analyzed by flow cytometry. Nef+ CD4+ HeLa cells were determined by gating on eGFP+ cells using an unstained control. (A) Schematic of the Nef isolates and associated mutants/chimeras utilized. The chimera break points for the Nef chimeras are indicated. Regions belonging to 2391 Nef are indicated with white diagonal stripes, while regions belonging to 2410 Nef are indicated with solid colors. (B and C) Representative histograms illustrating cell surface (B) SERINC5 or (C) CD4 levels of CD4+ HeLa cells after gating on single and transfected (eGFP+) cells. Geometric mean fluorescence intensities (MFIs) of the respective cell surface proteins are indicated. (D to F) Summary of the fold downregulation ability (±SE) for (D) cell surface SERINC5, (E) cell surface CD4, and (F) fold Nef-eGFP expression for each Nef isolate (n = 3). eGFP, enhanced green fluorescent protein; AF647, Alexa Fluor 647; PE, phycoerythrin; SE, standard error; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., nonsignificant (P > 0.05).
The ND164 polymorphism modestly impairs cell surface CD4 downregulation in infected cells.
Next, we tested whether the partial impairments in cell surface CD4 downregulation observed with the ND164 polymorphism in transfected CD4+ HeLa cells could be translated to an infected T cell line. Accordingly, Sup-T1 cells were infected with vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped virions encoding the respective Nef isolates and cell surface CD4 levels were determined via flow cytometry. We observed that 2410 Nef downregulated cell surface CD4 significantly more efficiently than 2391 Nef within infected cells (Fig. 9A to C). Furthermore, the 2410 DN164ND Nef mutant was partially impaired in cell surface CD4 downregulation relative to 2410 Nef (Fig. 9A and B), consistent with our results in transfected CD4+ HeLa cells (Fig. 7C and E). Importantly, the ability of 2410 DN164ND Nef to downregulate cell surface CD4 was not significantly different from that of 2391 Nef, which both retain the ability to downregulate cell surface CD4 upon comparison to cells infected with virus lacking Nef expression (ΔNef) and the respective dileucine mutants (Fig. 9A to C). Taken together, these results suggest the ND164 polymorphism partially impairs the ability of Nef to downregulate cell surface CD4 within infected cells.
FIG 9.
ND164 polymorphism impairs the ability of 2410 Nef to downregulate cell surface CD4 within infected cells. Sup-T1 cells were infected with VSV-G pseudotyped virions encoding the respective Nef proteins. The 2410 DN164ND and 2391 ND163DN mutants were included in these experiments. Infected cells were stained for cell surface CD4 and analyzed by flow cytometry. Live infected Sup-T1 cells were analyzed by gating on Zombie NIR – using the Zombie FMO control, and then infected (eGFP+) cells were determined using an eGFP FMO control. (A) Representative pseudocolor plots illustrating cell surface CD4 (PE) and infection (eGFP+) of live (Zombie NIR–) Sup-T1 cells. (B) Representative histograms illustrating cell surface levels of CD4 (PE) on Sup-T1 cells after gating on live (Zombie NIR–) and infected (eGFP+) cells. CD4 (PE) geometric mean fluorescence intensities (MFIs) are indicated. (C) Summary of the fold-downregulation ability (±SE) of cell surface CD4 for each Nef isolate (n = 3). Fold downregulation of cell surface CD4 was calculated by comparing the geometric MFI of cell surface CD4 in live cells infected with the ΔNef negative control to cells infected with pseudoviruses encoding the respective Nef proteins. Zombie NIR, Zombie near-infrared; FMO, fluorescence minus one; eGFP, enhanced green fluorescent protein; PE, phycoerythrin; SE, standard error; *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., nonsignificant (P > 0.05).
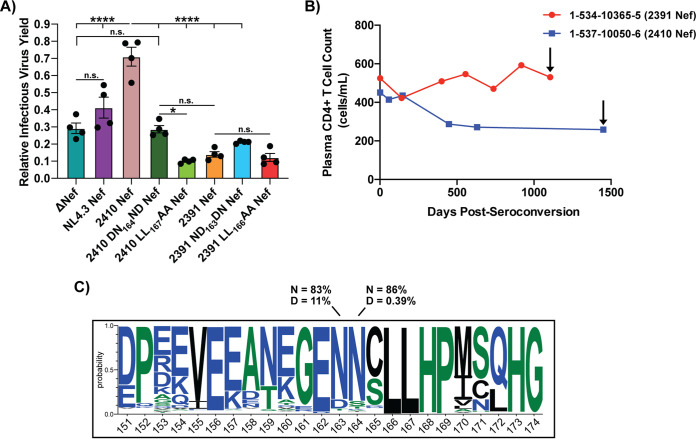
The ND164 polymorphism results in significantly lower infectious virus yield in the presence of SERINC5.
We next sought to determine whether infected cells expressing 2410 DN164ND Nef indeed produce lower infectious virus yield than 2410 Nef in the presence of SERINC5. To test this, HEK 293T cells were transfected with pNL4.3 eGFP proviral plasmids encoding the various Nef isolates along with a plasmid encoding Gag/Pol in either the presence or absence of SERINC5. TZM-bl target cells were then infected with purified virions produced in the presence or absence of SERINC5. We observed that virions produced from constructs encoding 2391 Nef yielded significantly lower infectious virus yield in the presence of SERINC5 than virions produced from constructs encoding 2410 Nef. Indeed, the infectious virus yield produced from constructs encoding 2391 Nef was equivalent to those encoding the 2410 LL167AA and 2391 LL166AA Nef dileucine mutants. These results effectively demonstrate that the ND164 polymorphism-containing isolate 2391 Nef lacks functionally significant cell surface SERINC5 downregulation, which therefore resulted in severe reductions in infectious virus yield (Fig. 10A). Importantly, virions produced from constructs encoding 2410 DN164ND Nef resulted in significantly lower infectious virus yield in the presence of SERINC5 compared to those encoding 2410 Nef and was equivalent to particles lacking Nef expression (ΔNef) (Fig. 10A). Furthermore, these results are consistent with the ability of Nef isolates to downregulate cell surface SERINC5 in CD4+ HeLa cells (Fig. 7B and D). While we observed the 2391 ND163DN Nef mutant resulted in a partial rescue in cell surface SERINC5 downregulation ability in the prior experiments (Fig. 8B and D), we only observed a modest, yet insignificant, rescue in infectious virus yield compared to virions expressing 2391 Nef (Fig. 10A). This suggests polymorphisms occurring outside the 2391 Nef dileucine motif are contributing in part to the impaired infectious virus yield in the presence of SERINC5, which is not being captured with the single ND163 polymorphism. However, in the context of 2410 Nef, the single ND164 polymorphism is sufficient to drastically impair infectious virus yield in the presence of SERINC5. Additionally, NL4.3 Nef resulted in a moderate enhancement in infectious virus yield upon comparison to cells infected with ΔNef (Fig. 10A). With the identification of functional consequences to the ND164 polymorphism in cell surface SERINC5 and CD4 downregulation ability, we considered whether there were any implications with respect to the rate of plasma CD4+ T cell decline observed within an infected individual, which we characterize as an indirect measure of disease progression to AIDS in this study. Indeed, the patient from which isolate 2391 Nef was derived (Fig. 10B, red) demonstrated a considerably slower loss of plasma CD4+ T cells than the patient from which isolate 2410 Nef was derived (Fig. 10B, blue), suggesting that this polymorphism plays a key role in pathogenesis. We next sought to determine whether the ND164 polymorphism is conserved among primary Nef isolates. Of the 7,475 mined Nef sequences of the Los Alamos HIV sequence database, asparagines encoded at positions 163 and 164 are conserved at a frequency of 83% and 86%, respectively (Fig. 10C). An aspartic acid encoded at position 163 in the context of 2410 Nef is present in 11% of the mined sequences and is the second most frequent residue at that position next to asparagine (Fig. 10C). Interestingly, an aspartic acid encoded at position 164 in the context of 2391 Nef is only present in 0.39% of the mined sequences, suggesting the ND164 in Nef is rare among circulating HIV-1 isolates (Fig. 10C). Taken together, these results suggest that the ND164 polymorphism in the context of 2410 Nef results in severe impairments in cell surface SERINC5 downregulation but only partial impairments in cell surface CD4 downregulation, thereby functionally uncoupling these two antagonistic pathways of Nef.
FIG 10.
ND164 polymorphism impairs the enhancement of infectious virus yield in the presence of SERINC5. (A) Virions encoding the respective Nef proteins were generated in the presence of SERINC5.intHA or an empty pBJ5 vector. Along with the primary Nef isolates, the 2410 DN164ND and 2391 ND163DN Nef mutants were included. TZM-bl cells were infected and relative infectious virus yield was determined by comparing the average relative luciferase units (RLUs) in the presence of SERINC5 to the absence of SERINC5. Results were obtained from four independent experiments (n = 4). (B) The patient from which 2391 Nef was derived (red) experienced a slower loss of plasma CD4+ T cells than the patient from which 2410 Nef was derived (blue). Black arrows indicate when the patients were placed on combined antiretroviral therapy (cART). (C) Amino acid frequencies of Nef residues 151 to 174, based on 7475 Nef sequences obtained from the Los Alamos HIV sequence database, are shown. Numbering is based on the 2410 Nef sequence. Listed percentages indicate the conservation of N (asparagine) or D (aspartic acid) at the indicated positions of Nef. *, P ≤ 0.05; ****, P ≤ 0.0001; n.s., nonsignificant (P > 0.05).
DISCUSSION
We identified that the Nef ND164 polymorphism, located in the first and second x residues of the Nef [D/E]xxxL[L/I]167 dileucine motif, severely impairs cell surface SERINC5 downregulation and partially impairs cell surface CD4 downregulation in the context of 2410 Nef. The ND164 polymorphism, which was only found in 2391 Nef out of the 15 Nef isolates tested (Fig. 1), resulted in a reduction of infectious virus yield, suggesting a functional consequence due to impaired cell surface SERINC5 downregulation.
As both CD4 and SERINC5 downregulation were affected by the ND164 polymorphism, and considering both pathways are AP-2 dependent, the polymorphism may alter the stability of the Nef–AP-2 interaction. We therefore speculate that Nef requires higher-affinity interactions with AP-2 to internalize cell surface SERINC5 compared to CD4. As SERINC5 harbors 10 transmembrane domains (25) and is more integral within the membrane than CD4, which only harbors a single transmembrane domain (26), variations in sequences that recognize AP-2 may ultimately uncouple the two antagonistic pathways. Ren et al. summarized known Nef residues responsible for stabilizing the interactions with the AP-2 α and σ subunits (12). Interestingly, within the [D/E]xxxL[L/I]167 dileucine motif, an asparagine residue at the first x residue and a serine residue at the third x residue contribute to AP-2 binding by forming weak van der Waals interactions and hydrogen bonds with the AP-2 σ subunit (12, 27). However, specific residues encoded at the second x position within the Nef dileucine motif were not identified as being responsible for stabilizing the interaction with AP-2 directly (12, 27). Importantly, the downstream Nef DD175 diacidic motif fails to contact AP-2 directly but rather stabilizes the conformation of the C-terminal flexible Nef loop by forming core-to-loop hydrogen bonds and salt bridges with upstream Nef residues (12). As the Nef diacidic DD175 motif is absolutely required to downregulate cell surface CD4 (27), mutations/polymorphisms altering the accessible conformations and dynamics of the flexible loop, which harbors motifs required to interact with AP-2, could indirectly modulate the stability of the Nef–AP-2 interaction, affecting AP-2-dependent Nef functions, including SERINC5 downregulation.
Nef forms a weaker interaction with AP-2 than with other adaptor protein complexes such as AP-1 and AP-3 (28, 29). As a result, a polymorphism occurring within the second x position of the Nef dileucine motif, such as the charged aspartic residue of 2391 Nef (D164), could alter the conformational stability of the C-terminal Nef flexible loop, thereby affecting the stability of the already weak Nef–AP-2 interaction. We speculate the presence of the negatively charged D164 residue could alter the mobility/dynamics of the flexible loop, possibly by disrupting hydrogen bond and salt bridge formation required by DD175 to stabilize the flexible Nef loop required to interact with AP-2 due to electrostatic repulsion. Indeed, the presence of an aspartic acid at the second x position within the Nef dileucine motif is not limited to just HIV-1 Nef. A histidine residue at the second x position of the dileucine motif of simian immunodeficiency virus Nef significantly impaired infectious virus yield in the presence of SERINC5 and impaired cell surface tetherin downregulation when mutated to an aspartic acid residue (H192D) (30). Of note, the H192D mutant did not suffer impairments in CD4 downregulation, supporting the notion that the ND164 polymorphism could be genetically and functionally uncoupling the two antagonistic pathways (30).
Other polymorphisms within the Nef flexible loop have previously been shown to functionally uncouple the CD4 and SERINC5 antagonistic pathways. Indeed, a G176R polymorphism, located within the conserved hydrophobic region immediately downstream the Nef dileucine motif, has been described within a cohort of primary nef sequences (31, 32). The G176R mutation impaired CD4 downregulation but not the ability of Nef to rescue particle infectivity in the presence of SERINC5 (31). As G176R is present in a Nef region required for stabilizing the AP-2 interaction, it was suggested this mutation may alter the specific Nef–AP-2 complex molecular organization required for CD4, but not SERINC5, downregulation (31). Thus, an aspartic acid at the second x position in the context of ND164 can be selectively disrupting the molecular organization of the Nef–AP-2 complex required for SERINC5 downregulation, thereby resulting in functional uncoupling. The finding that the reverse DN163 polymorphism in 2391 Nef (2391 ND163DN Nef) did not significantly rescue infectious virus yield in the presence of SERINC5 may be due to the polymorphic nature of the N-terminal region (NTR) of 2391 Nef. Indeed, swapping the NTR of 2410 Nef with the 2391 NTR modestly reduced SERINC5 downregulation (Fig. 3E and 4C). Recent evidence suggests the Nef NTR contains a SERINC5 antagonism motif (S5AM) represented by the VGAxS core sequence (33). Such a S5AM is found in 2410, EGVGAASR35, but not in 2391 Nef, EGVGTASQ35. Thus, the altered S5AM within the 2391 Nef NTR may disrupt SERINC5 antagonism, which could explain the lack of rescued infectious virus yield observed with the DN163 reverse mutation (2391 ND163DN Nef). Overall, our data suggest the ND164 polymorphism within the Nef dileucine motif differentially impairs the CD4 and SERINC5 antagonistic pathways of Nef, resulting in functional uncoupling when S5AM is preserved, as is the case within the 2410 NTR.
The ability of Nef to downregulate CD4 and SERINC5 has been primarily attributed to an interaction with AP-2 and not other adaptor protein complex family members such as AP-1 (34). However, recent studies have demonstrated that a variant AP-1 tetramer, which contains the variant γ2 subunit as opposed to the classically defined γ1 subunit (here referred to as AP-1 γ2), was involved in Nef-mediated CD4 downregulation (35) as well as MHC-I downregulation (36). Interestingly, Nef requires engagement with both γ1 and γ2 containing AP-1 tetramers to downregulate MHC-I, as depletion of either individually dampened the ability of Nef to downregulate MHC-I (36). This suggests that Nef requires an interaction with distinct AP complexes to completely downregulate a target protein, potentially providing spatiotemporal regulation to these antagonistic pathways. With respect to Nef-mediated CD4 downregulation, Nef requires engagement with AP-2 to trigger endocytosis of CD4 from the cell surface, which is eventually delivered to multivesicular bodies (MVBs) and lysosomes for degradation (37). A critical step in directing CD4 to the degradative route requires targeting of CD4 to the intraluminal vesicles (ILVs) of MVBs, which is dependent on the endosomal sorting complexes required for transport (ESCRT) machinery (37, 38). The ESCRT serves several crucial physiological roles, including mediating ILV formation and MVB biogenesis (38, 39). Additionally, Nef physically engages with the Alix adaptor, a protein associated with the ESCRT machinery, which is necessary for directing Nef-bound CD4 monomers in late endosomes to the degradative route (39). Furthermore, Nef binding to the AP-1 γ2 variant tetramer is required for directing endocytosed proteins such as CD4 to the degradative pathway, a step likely preceding Nef interactions with Alix (35, 36). Indeed, SERINC5 can be sorted to MVBs, likely due to an interaction between Nef and Alix, suggesting AP-1 γ2 is required in this antagonistic pathway (9, 36, 39). Interestingly, it is known that the sequence requirements for Nef physical interactions with AP-2 are considerably less strict than what is required for interacting with AP-1 (40). As such, we speculate the ND164 polymorphism is predominantly impacting the extent to which Nef engages AP-1 γ2, which would theoretically impair rerouting of both CD4 and SERINC5 to the degradative route. How Nef spatiotemporally regulates interactions with AP-2 and AP-1 γ2 to downregulate CD4 and SERINC5 requires further examination. While both AP-2 and AP-1 γ2 are required for CD4 downregulation, the spatiotemporal contributions of either adaptor in SERINC5 downregulation remains unknown. We postulate downregulating SERINC5 and eventual rerouting to the degradative pathway could require more comprehensive engagements with AP-1 γ2 as opposed to the CD4 downregulation pathway, which could explain the differential antagonistic functions observed in this study.
The finding that the patient from which the 2410 Nef isolate was derived demonstrated a more rapid loss of plasma CD4+ T cells over time compared to the patient from which 2391 Nef was derived supports previous observations that the extent to which Nef antagonizes SERINC5 could affect disease progression to AIDS (19, 20). The patients from which isolates 2410 and 2391 Nef were derived became HIV-1 positive via heterosexual male-to-female transmission (21). This form of transmission is associated with an extreme genetic bottleneck, in which one HIV-1 variant typically establishes infection (41, 42). As such, in the early stages of acute HIV-1 infection, the virus is extremely clonal, with minimal circulating variants (42). Since we acquired these samples at the first visit postseroconversion, the associated Nef isolates may be representative of the original transmitter/founder (T/F) strain or genetically similar; however, we cannot exclude the possibility some mutations have been introduced during the cloning process. Furthermore, the potential ability of these Nef isolates to downregulate SERINC3, a SERINC family member possessing antiviral effects on HIV-1 replication, albeit to a lesser extent than SERINC5 (3, 4), could also be a factor contributing to differential rates of plasma CD4+ T cell decline. Moreover, specific HIV-1 Env proteins can be susceptible to SERINC5 antagonism, while others remain resistant (43). As such, env or other HIV-1 viral genes may also factor into the observed differences in the rate of plasma CD4+ T cell decline observed within the implicated infected individuals.
Overall, our results demonstrate the ND164 polymorphism within the Nef dileucine uncouples the CD4 and SERINC5 antagonistic pathways and results in lower infectious virus yield in the presence of SERINC5. Our results support the role that Nef adaptor protein recognition sequences are required for both antagonistic pathways, and, as such, polymorphisms outside previously defined Nef functional motifs could positively or negatively impact these functional abilities.
MATERIALS AND METHODS
Cell culture.
HEK 293T (ATCC, Manassas, Virginia), CD4+ HeLa (NIH AIDS Reagent Program), and TZM-bl (NIH AIDS Reagent Program) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 4 mM l-glutamine (Cytiva Life Sciences, Vancouver, BC), 4.5 g/liter glucose (Cytiva Life Sciences) and supplemented with 10% fetal bovine serum (FBS; Wisent, St. Bruno, QC, Canada) and 1% penicillin and streptomycin (HyClone, Logan, UT). Sup-T1 cells (ATCC) were maintained in Roswell Park Memorial Institute (RPMI) medium 1640, which was supplemented with 100 μg/ml penicillin-streptomycin (HyClone), 1% sodium pyruvate (HyClone), 1% nonessential amino acids (HyClone), 2 mM l-glutamine (HyClone), and 10% FBS (Wisent). All cell lines were grown at 37°C in the presence of 5% CO2 and were subcultured in accordance with the manufacturer’s recommendations.
Ethics statement and patient DNA extraction.
Primary nef genes utilized in this study were acquired from the Hormonal Contraception and HIV-1 Genital Shedding and Disease Progression among Women with Primary HIV-1 Infection (GS) study. A complete description of this study has been previously described (21, 22). Ethics approval was obtained from the Institutional Review Boards (IRBs) from the Joint Clinical Research Center and UNST in Uganda, the University of Zimbabwe, the University Hospitals of Cleveland, and from Western University (21). Briefly, blood and cervical samples were collected every month for the first 6 months, every 3 months for the first 2 years, and then every 6 months for up to 9.5 years. Women who had plasma CD4+ T cell counts of 200 cells/ml and/or developed severe symptoms of HIV-1 infection (WHO clinical stage IV or advanced stage III disease) were offered combination ART (cART) and trimethoprim-sulfamethoxazole (21). CD4+ T cell counts were determined by standard flow cytometry using FACSCalibur (Becton, Dickinson, Sparks, MD) (21).
DNA constructs.
Primary nef genes utilized in this study were derived from Ugandan and Zimbabwean women enrolled in the GS study (21, 22). At the first visit postseroconversion, DNA was extracted from peripheral blood mononuclear cells (PBMCs) taken from plasma samples. Patient nef genes were first amplified from the purified DNA using a nested PCR. The subtype of each primary Nef isolate utilized in Fig. 1 was determined using the recombinant identification profram (RIP) tool from the Los Alamos HIV sequence database (44). Twelve of the Nef isolates were subtype C (2013, 2275, 2391, 2317, 2005, 2634, 2410, 2130, 2060, 2222, 2109, and 250-04 Nef) and three of the Nef isolates were subtype A1 (185-05, 257-05, and 0035-04 Nef). In the first reaction, multiple primers were designed to account for the genetic variability of the integrated provirus. Forward primers were designed ∼1,000 bp upstream of the nef open reading frame (ORF) within env, and reverse primers were designed in the 3′ long terminal repeat (LTR). Successful amplicons were PCR purified and subjected to the second round of nested PCR. The forward primers used in the second PCR were designed ∼200 bp upstream of the nef ORF within the rev-env overlap and reverse primers were designed to anneal within the 3′ LTR. Successful PCR products were purified and cloned into a pREC_nfl_HIV-1 Δnef/URA3 proviral vector using a yeast recombination system (45). Sanger sequencing was subsequently performed to confirm nef sequence identity.
The nef genes were cloned into the pN1 vector (Clontech) containing a 3′ egfp ORF. Forward primers specific to each nef sequences were designed to harbor a 5′ EcoRI restriction site, and reverse primers were designed to harbor a 3′ BamHI restriction site.
The various Nef chimeras and mutants were generated using overlapping PCR mutagenesis techniques. The breakpoint for each Nef chimera was selected in a region of nef that would not induce a frameshift mutation in either 2410 or 2391 nef. In a first PCR, the N-terminal nef region was amplified using a forward primer bearing the EcoRI restriction site, as described above, and a reverse primer including the appropriate mutation. The second PCR then amplified the C-terminal nef region of the corresponding nef gene using the forward primer complementary to the reverse mutagenic primer and a reverse primer which harbors the BamHI site. In the final PCR, both purified amplicons were mixed 1:1 and amplified using the 5′ forward primer with the EcoRI site and the 3′ reverse primer with the BamHI site to generate the full-length nef chimera. For the 2410:2391151-174 Nef and the 2391:2410150-173 Nef chimeras, an extra overlapping PCR mutagenesis reaction was required to generate an intermediate chimera nef product, which served as a template for subsequent cloning steps.
To assess Nef function, the respective nef genes were also cloned into the pNL4.3 ΔGag/Pol eGFP Nef replication-incompetent proviral plasmid, which has been described (46–48). Briefly, this proviral plasmid was engineered to readily clone nef using flanking 5′ XmaI and 3′ NotI sites. Forward and reverse primers were specifically designed to clone the respective Nef isolates into the proviral backbone by harboring XmaI and NotI restriction sites, respectively.
To exogenously express SERINC5 in trans, a pBJ5-SERINC5.intHA vector was utilized, which was a kind gift from Heinrich Göttlinger (UMass Medical School) (4). This SERINC5 harbors a hemagglutinin (HA) tag inserted between residues 290 and 291 of SERINC5, which is located on the fourth extracellular loop, connecting the 7th and 8th transmembrane helices.
For pseudovirion generation, the pCMV-DR8.2 plasmid (catalog number 12263; Addgene) encoding Gag/Pol and the pMD2.G plasmid (catalog number 12259; Addgene) encoding VSV-G were utilized.
Transfections and infections.
To determine the ability of each Nef isolate to downregulate cell surface SERINC5 and CD4 in CD4+ HeLa cells, 5 × 105 CD4+ HeLa cells were plated in 6-well plates 24 h prior to transfection. For cohort screening experiments, cell surface CD4 downregulation was determined by transfecting CD4+ HeLa cells with 1 μg of the respective cloned pN1-Nef-eGFP plasmids using PolyJet (FroggaBio, North York, ON). Cell surface SERINC5 downregulation ability was determined by cotransfecting CD4+ HeLa cells with 0.5 μg of the cloned pN1-Nef-eGFP plasmids along with 0.5 μg of pBJ5-SERINC5.intHA, also using PolyJet. To determine the ability of the respective Nef isolates to downregulate cell surface SERINC5 and CD4, CD4+ HeLa cells were cotransfected with 0.64 μg of the respective pN1-Nef-eGFP plasmids and 0.80 μg pBJ5-SERINC5.intHa plasmid using PolyJet. To determine the level of Nef-eGFP expression via Western blotting, CD4+ HeLa cells were transfected with 0.64 μg of the respective cloned pN1-Nef-eGFP plasmids in the absence or presence of 0.80 μg of pBJ5-SERINC5.intHA using PolyJet.
For VSV-G pseudotyped lentivirus production, 1 × 106 HEK 293T cells were plated in 6-well plates 24 h prior to transfection. HEK 293T cells were then triply transfected with 0.83 μg of the respective cloned pNL4.3 ΔGag/Pol eGFP Nef plasmids, 0.83 μg of the pCMV-DR8.2 Gag/Pol-encoding plasmid, and 0.40 μg of the VSV-G-encoding pMD2.G plasmid using PolyJet. Seventy-two hours posttransfection, virus-containing supernatants were collected, supplemented with 20% FBS, filtered using a 0.45-μm filter, and stored at −80°C. For infection of Sup-T1 cells with VSV-G pseudotyped viruses, 1 × 106 Sup-T1 cells were pelleted, resuspended in the appropriate amount of pseudovirus in 20% FBS, 3 μg/ml Polybrene (Sigma-Aldrich, St. Louis, MO), and brought to 700 μl with complete RPMI. After 8 h, the cells were pelleted, the virus-containing medium was removed, and the pelleted cells were resuspended in 1 ml of complete RPMI for an additional 40 h.
For generating lentivirus particles with or without SERINC5 incorporation as part of the infectivity assay, 3 × 105 HEK 293T cells were plated in 12-well dishes 1 day prior to transfection. HEK 293T cells were then triply transfected with 0.60 μg of the respective cloned pNL4.3 ΔGag/Pol eGFP Nef plasmids and 0.60 μg of the Gag/Pol-encoding pCMV-DR8.2 plasmid, along with either 0.24 μg of pBJ5-SERINC5.intHA or the empty pBJ5 vector using PolyJet. Forty-eight hours posttransfection, virus-containing supernatants were collected, supplemented with 20% FBS, filtered using a 0.45-μm filter, and stored at −80°C. For infection of TZM-bl cells to determine infectious virus yield, 5,000 TZM-bl cells were seeded per well in a 96-well plate. Prior to infection, the TZM-bl cells were incubated for 30 min with 1 μg/ml Polybrene at 37°C for 30 min. Following this incubation, the cell medium was aspirated and replaced with 100 μl of complete DMEM. The TZM-bl cells were infected with 100 μl of each virus condition (with and without SERINC5), with each condition being performed with 5 technical replicates for 72 h.
Flow cytometry analysis of cell surface receptors.
For cell surface staining of CD4+ HeLa cells, 24 h posttransfection, cells were washed twice with 1× phosphate-buffered saline (PBS; Wisent), displaced with 0.25% Trypsin (Life Technologies, Carlsbad, CA), and collected into fluorescence-activated cell sorting (FACS) tubes. Collected cells were spun at 1,500 rpm for 5 min at room temperature and were washed twice with 1 ml of 1× PBS. Pelleted cells were fixed with 1.5% paraformaldehyde (PFA) for 15 min in the dark and washed twice with 1 ml FACS buffer (3% FBS, 5 mM ethylenediaminetetraacetic acid [EDTA] in 1× PBS). Cells were subsequently stained with the appropriate antibodies for 50 min at room temperature, washed twice with FACS buffer, and resuspended in 1× PBS. For cell surface staining of CD4 only, as depicted in Fig. 1A, 1:100 allophycocyanin (APC)-conjugated anti-human CD4 antibody (clone OKT4; BioLegend) in FACS buffer was used. For cell surface staining of SERINC5.intHA, 1:500 Alexa Fluor 647-conjugated anti-HA.11 epitope tag antibody (clone 16B12; BioLegend) in FACS buffer was used. When staining for both cell surface CD4 and SERINC5, 1:100 phycoerythrin (PE)-conjugated anti-human CD4 antibody (clone OKT4; BioLegend) was used, along with the same concentration of the anti-HA.11 epitope tag antibody described above. The isotype control sample in these experiments was stained with 1:50 Alexa Fluor 647-conjugated mouse IgG1, κ isotype control antibody (clone MOPC-21; BioLegend), and 1:200 PE-conjugated mouse IgG2B, κ isotype control antibody (clone MPC-11; BioLegend) in FACS buffer was used. Cells were then washed twice with cell staining buffer and resuspended in 1× PBS prior to analysis. For flow cytometry analysis, cells were analyzed using a BD Biosciences FACS-Canto SORP (London Regional Flow Cytometry Facility, London, ON). To determine the transfected (eGFP+) cell population, an unstained cell control was used in the single CD4 and SERINC5 downregulation assays as depicted in Fig. 1, and a fluorescence minus one (FMO) control lacking eGFP fluorescence was used in the SERINC5 and CD4 downregulation assays depicted in subsequent experiments. The geometric mean fluorescence intensity (MFI) was then determined, where appropriate, with Alexa Fluor 647 (SERINC5), PE or APC (CD4), and eGFP (Nef-eGFP) within the eGFP+ cell population for each sample. Fold SERINC5 and CD4 downregulation was then determined by comparing the MFI of Alexa Fluor 647 (SERINC5) and PE or APC (CD4) in cells transfected with the eGFP only control to cells expressing the respective Nef-eGFP isolates. Fold Nef-eGFP expression was calculated in a similar manner.
For cell surface staining of Sup-T1 cells, 48 h postinfection, Sup-T1 cells were collected, washed twice with 1× PBS, and stained for 20 min in the dark at room temperature with 1:200 Zombie NIR viability dye (catalog number 423105; BioLegend) in 1× PBS. Cells were washed twice with FACS buffer and fixed with 1% PFA for 20 min at room temperature. Cells were subsequently washed twice with FACS buffer and stained with the appropriate antibodies by rocking for 50 min at room temperature. Cells were washed twice with FACS buffer and resuspended in 1× PBS. For cell surface CD4 staining, 1:20 PE-conjugated anti-human CD4 antibody (clone OKT4; BioLegend) in FACS buffer was utilized. For the isotype control, cells were stained with 1:40 PE-conjugated mouse IgG2B, k isotype control antibody (clone MPC-11; BioLegend). For flow cytometry analysis, cells were analyzed as described above. FMO controls lacking the Zombie NIR dye and eGFP fluorescence were utilized to determine live (Zombie NIR−) and then infected (eGFP+) cell populations. The geometric MFI of PE (cell surface CD4) was then determined within the Zombie NIR− eGFP+ cell populations for each sample. The MFI of PE (CD4) in cells infected with the ΔNef negative control was compared to the MFI of PE in cells infected with the respective Nef isolates to determine fold cell surface CD4 downregulation. Flow cytometry data were analyzed using FlowJo software (version 10.7.1; FlowJo LLC, Ashland, OR).
Infectivity assay.
After 72 h of infection, the medium was aspirated and replaced with 100 μl complete DMEM. This was then supplemented with 100 μl of the Steady-Glo reagent (catalog number E2520; Promega) and incubated in the dark for 15 min; 100 μl was then transferred to a black opaque 96-well plate, with the relative luciferase units (RLUs) of each condition being quantified by the Cytation5 microplate reader. The average number of RLUs in the presence and absence of SERINC5 was then determined for each technical replicate. The relative infectious virus yield was then calculated by comparing the average RLU in the presence of SERINC5 to the average number of RLU in the absence of SERINC5.
Fusion protein analysis.
Lysate collection for Western blot analysis of the CD4+ HeLa cells transfected with the respective pN1-eGFP Nef plasmids in the presence and absence of SERINC5.intHA has been described previously (46). The primary antibody utilized in this experiment to detect Nef-eGFP fusion proteins was a rabbit anti-GFP recombinant monoclonal antibody (Invitrogen) diluted to 1:200, and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control was detected using a primary mouse anti-GAPDH monoclonal antibody (Invitrogen) diluted to 1:3,000 in 5% nonfat skim milk in Tris-buffered saline with Tween 20 (TBST). Twenty-four hours later, membranes were washed three times with TBST and incubated for 1 h at room temperature with a secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L) antibody diluted to 1:1,000 (Invitrogen) and a secondary HRP-conjugated goat anti-mouse IgG (H+L) antibody diluted to 1:3,000 in 5% nonfat skim milk in TBST. Blots were subsequently washed and developed using the Crescendo HRP chemiluminescent detection (Millipore Sigma) for 2 min prior to imaging with a C-DiGit blot scanner (Li-Cor Biosciences, Lincoln, NE). Images were collected using Image Studio v 5.2 (Li-Cor).
Data and statistical analysis.
All statistical tests were completed using Graph Pad Prism 8 (Graph Pad Software Inc., La Jolla, CA). All experimental data were tested for normalcy using a Shapiro-Wilk normality test. Statistical significance was then determined using a nonmatching one-way analysis of variance (ANOVA) test, in which the means of each group were compared to the negative-control sample or compared to each other (where indicated), followed by a Tukey multiple-comparison test with a 95% confidence interval (α = 0.05).
Data availability.
The accession numbers of the primary nef sequences deposited in GenBank are MW892600–MW892614.
ACKNOWLEDGMENTS
This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR) to J.D.D. (CIHR project grant 389413) and by infrastructure grants from the Canadian Foundation for Innovation and The University of Western Ontario. M.J.M. is supported by an Ontario Graduate Scholarship from the Government of Ontario. C.R.E. is supported by a Graduate Studentship from CIHR.
CD4+ HeLa cells, ARP-154, were obtained from the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH, contributed by Richard Axel. TZM-bl cells, ARP-8129, were obtained from the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH, contributed by John C. Kappes and Xiaoyun Wu.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Contributor Information
Jimmy D. Dikeakos, Email: Jimmy.Dikeakos@uwo.ca.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Cruz NVG, Amorim R, Oliveira FE, Speranza FAC, Costa LJ. 2013. Mutations in the nef and vif genes associated with progression to AIDS in elite controller and slow-progressor patients. J Med Virol 85:563–574. 10.1002/jmv.23512. [DOI] [PubMed] [Google Scholar]
- 2.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. 1995. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med 332:228–232. 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 3.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Usami Y, Wu Y, Göttlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inuzuka M, Hayakawa M, Ingi T. 2005. Serine, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 280:35776–35783. 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 6.Trautz B, Wiedemann H, Lüchtenborg C, Pierini V, Kranich J, Glass B, Kräusslich HG, Brocker T, Pizzato M, Ruggieri A, Brügger B, Fackler OT. 2017. The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J Biol Chem 292:13702–13713. 10.1074/jbc.M117.797332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. 2017. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 292:6014–6026. 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matheson NJ, Sumner J, Wals K, Rapiteanu R, Weekes MP, Vigan R, Weinelt J, Schindler M, Antrobus R, Costa ASH, Frezza C, Clish CB, Neil SJD, Lehner PJ. 2015. Cell surface proteomic map of HIV infection reveals antagonism of amino acid metabolism by Vpu and Nef. Cell Host Microbe 18:409–423. 10.1016/j.chom.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J, Xiong R, Zhou T, Su P, Zhang X, Qiu X, Li H, Li S, Yu C, Wang B, Ding C, Smithgall TE, Zheng Y-H. 2018. HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J Virol 92:e00196-18. 10.1128/JVI.00196-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staudt RP, Smithgall TE. 2020. Nef homodimers down-regulate SERINC5 by AP-2–mediated endocytosis to promote HIV-1 infectivity. J Biol Chem 295:15540–15552. 10.1074/jbc.RA120.014668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig HM, Pandori MW, Guatelli JC. 1998. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci U S A 95:11229–11234. 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren X, Park SY, Bonifacino JS, Hurley JH. 2014. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. Elife 3:e01754. 10.7554/eLife.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon Y, Kaake RM, Echeverria I, Suarez M, Karimian Shamsabadi M, Stoneham C, Ramirez PW, Kress J, Singh R, Sali A, Krogan N, Guatelli J, Jia X. 2020. Structural basis of CD4 downregulation by HIV-1 Nef. Nat Struct Mol Biol 27:822–828. 10.1038/s41594-020-0463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. 2007. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol 81:3877–3890. 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doray B, Lee I, Knisely J, Bu G, Kornfeld S. 2007. The γ/σ1 and α/σ2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell 18:1887–1896. 10.1091/mbc.e07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trautz B, Pierini V, Wombacher R, Stolp B, Chase AJ, Pizzato M, Fackler OT. 2016. The antagonism of HIV-1 Nef to SERINC5 particle infectivity restriction involves the counteraction of virion-associated pools of the restriction factor. J Virol 90:10915–10927. 10.1128/JVI.01246-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwimanzi P, Markle TJ, Ogata Y, Martin E, Tokunaga M, Mahiti M, Kuang XT, Walker BD, Brockman MA, Brumme ZL, Ueno T. 2013. Dynamic range of Nef functions in chronic HIV-1 infection. Virology 439:74–80. 10.1016/j.virol.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Cotton LA, Kuang XT, Le AQ, Carlson JM, Chan B, Chopera DR, Brumme CJ, Markle TJ, Martin E, Shahid A, Anmole G, Mwimanzi P, Nassab P, Penney KA, Rahman MA, Milloy MJ, Schechter MT, Markowitz M, Carrington M, Walker BD, Wagner T, Buchbinder S, Fuchs J, Koblin B, Mayer KH, Harrigan PR, Brockman MA, Poon AFY, Brumme ZL. 2014. Genotypic and functional impact of HIV-1 adaptation to its host population during the North American epidemic. PLoS Genet 10:e1004295. 10.1371/journal.pgen.1004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin SW, Mwimanzi FM, Mann JK, Bwana MB, Lee GQ, Brumme CJ, Hunt PW, Martin JN, Bangsberg DR, Ndung’u T, Brumme ZL, Brockman MA. 2020. Variation in HIV-1 nef function within and among viral subtypes reveals genetically separable antagonism of serinc3 and serinc5. PLoS Pathog 16:e1008813. 10.1371/journal.ppat.1008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin SW, Alsahafi N, Kuang XT, Swann SA, Toyoda M, Göttlinger H, Walker BD, Ueno T, Finzi A, Brumme ZL, Brockman MA. 2019. Natural HIV-1 Nef polymorphisms impair SERINC5 downregulation activity. Cell Rep 29:1449–1457. 10.1016/j.celrep.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venner CM, Nankya I, Kyeyune F, Demers K, Kwok C, Chen PL, Rwambuya S, Munjoma M, Chipato T, Byamugisha J, Van Der Pol B, Mugyenyi P, Salata RA, Morrison CS, Arts EJ. 2016. Infecting HIV-1 subtype predicts disease progression in women of sub-Saharan Africa. EBioMedicine 13:305–314. 10.1016/j.ebiom.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison CS, Chen PL, Nankya I, Rinaldi A, Van Der Pol B, Ma YR, Chipato T, Mugerwa R, Dunbar M, Arts E, Salata RA. 2011. Hormonal contraceptive use and HIV disease progression among women in Uganda and Zimbabwe. J Acquir Immune Defic Syndr 57:157–164. 10.1097/QAI.0b013e318214ba4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobiume M, Takahoko M, Yamada T, Tatsumi M, Iwamoto A, Matsuda M. 2002. Inefficient enhancement of viral infectivity and CD4 downregulation by human immunodeficiency virus type 1 Nef from Japanese long-term nonprogressors. J Virol 76:5959–5965. 10.1128/jvi.76.12.5959-5965.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casartelli N, Matteo GD, Potestà M, Rossi P, Doria M. 2003. CD4 and major histocompatibility complex class I downregulation by the human immunodeficiency virus type 1 Nef protein in pediatric AIDS progression. J Virol 77:11536–11545. 10.1128/jvi.77.21.11536-11545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pye VE, Rosa A, Bertelli C, Struwe WB, Maslen SL, Corey R, Liko I, Hassall M, Mattiuzzo G, Ballandras-Colas A, Nans A, Takeuchi Y, Stansfeld PJ, Skehel JM, Robinson CV, Pizzato M, Cherepanov P. 2020. A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Nat Struct Mol Biol 27:78–83. 10.1038/s41594-019-0357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parrish HL, Glassman CR, Keenen MM, Deshpande NR, Bronnimann MP, Kuhns MS. 2015. A transmembrane domain GGxxG motif in CD4 contributes to its lck-independent function but does not mediate CD4 dimerization. PLoS One 10:e0132333. 10.1371/journal.pone.0132333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindwasser OW, Smith WJ, Chaudhuri R, Yang P, Hurley JH, Bonifacino JS. 2008. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J Virol 82:1166–1174. 10.1128/JVI.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim B-Y, Venkatesan S, Bonifacino JS. 2003. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1–1 and AP-3–3 hemicomplexes. J Cell Biol 163:1281–1290. 10.1083/jcb.200307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craig HM, Reddy TR, Riggs NL, Dao PP, Guatelli JC. 2000. Interactions of HIV-1 Nef with the μ subunits of adaptor protein complexes 1, 2, and 3: role of the dileucine-based sorting motif. Virology 271:9–17. 10.1006/viro.2000.0277. [DOI] [PubMed] [Google Scholar]
- 30.Buffalo CZ, Stürzel CM, Heusinger E, Kmiec D, Kirchhoff F, Hurley JH, Ren X. 2019. Structural basis for tetherin antagonism as a barrier to zoonotic lentiviral transmission. Cell Host Microbe 26:359–368.e8. 10.1016/j.chom.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obermaier B, Ananth S, Tibroni N, Pierini V, Shytaj IL, Diaz RS, Lusic M, Fackler OT. 2020. Patient-derived HIV-1 Nef alleles reveal uncoupling of CD4 downregulation and SERINC5 antagonism functions of the viral pathogenesis factor. J Acquir Immune Defic Syndr 85:e23–e26. 10.1097/QAI.0000000000002418. [DOI] [PubMed] [Google Scholar]
- 32.Jin YJ, Cai CY, Mezei M, Ohlmeyer M, Sanchez R, Burakoff SJ. 2013. Identification of a novel binding site between HIV type 1 nef c-terminal flexible loop and AP2 required for nef-mediated CD4 downregulation. AIDS Res Hum Retroviruses 29:725–731. 10.1089/AID.2012.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ananth S, Morath K, Trautz B, Tibroni N, Shytaj IL, Obermaier B, Stolp B, Lusic M, Fackler OT. 2019. Multifunctional roles of the N-terminal region of HIV-1SF2Nef are mediated by three independent protein interaction sites. J Virol 94:e01398-19. 10.1128/JVI.01398-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gondim MV, Wiltzer-Bach L, Maurer B, Banning C, Arganaraz E, Schindler M. 2015. AP-2 is the crucial clathrin adaptor protein for CD4 downmodulation by HIV-1 Nef in infected primary CD4+ T cells. J Virol 89:12518–12524. 10.1128/JVI.01838-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavares LA, da Silva EML, da Silva-Januário ME, Januário YC, de Cavalho JV, Czernisz ÉS, Mardones GA, daSilva LLP. 2017. CD4 downregulation by the HIV-1 protein Nef reveals distinct roles for the γ1 and γ2 subunits of the AP-1 complex in protein trafficking. J Cell Sci 130:429–443. 10.1242/jcs.192104. [DOI] [PubMed] [Google Scholar]
- 36.Tavares LA, de Carvalho JV, Costa CS, Silveira RM, de Carvalho AN, Donadi EA, daSilva LLP. 2020. Two functional variants of AP-1 complexes composed of either γ2 or γ1 subunits are independently required for major histocompatibility complex class I downregulation by HIV-1 Nef. J Virol 94:e02039-19. 10.1128/JVI.02039-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.daSilva LLP, Sougrat R, Burgos PV, Janvier K, Mattera R, Bonifacino JS. 2009. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J Virol 83:6578–6590. 10.1128/JVI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katzmann DJ, Babst M, Emr SD. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155. 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 39.Amorim NA, da Silva EML, de Castro RO, da Silva-Januário ME, Mendonça LM, Bonifacino JS, da Costa LJ, daSilva LLP. 2014. Interaction of HIV-1 nef protein with the host protein Alix promotes lysosomal targeting of cd4 receptor. J Biol Chem 289:27744–27756. 10.1074/jbc.M114.560193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coleman SH, Madrid R, Van Damme N, Mitchell RS, Bouchet J, Servant C, Pillai S, Benichou S, Guatelli JC. 2006. Modulation of cellular protein trafficking by human immunodeficiency virus type 1 Nef: role of the acidic residue in the ExxxLL motif. J Virol 80:1837–1849. 10.1128/JVI.80.4.1837-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin T-H, Peng J, Seese AM, Shapiro R, Frater J, Ndung'u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJR, Allen TM, Allen S, Hunter E. 2014. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345:1254031. 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein K, Nickel G, Nankya I, Kyeyune F, Demers K, Ndashimye E, Kwok C, Chen PL, Rwambuya S, Poon A, Munjoma M, Chipato T, Byamugisha J, Mugyenyi P, Salata RA, Morrison CS, Arts EJ. 2018. Higher sequence diversity in the vaginal tract than in blood at early HIV-1 infection. PLoS Pathog 14:e1006754. 10.1371/journal.ppat.1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beitari S, Ding S, Pan Q, Finzi A, Liang C. 2017. Effect of HIV-1 Env on SERINC5 antagonism. J Virol 91:e02214-16. 10.1128/JVI.02214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siepel AC, Halpern AL, Macken C, Korber BT. 1995. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses 11:1413–1416. 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 45.Dudley DM, Gao Y, Nelson KN, Henry KR, Nankya I, Gibson RM, Arts EJ. 2009. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques 46:458–467. 10.2144/000113119. [DOI] [PubMed] [Google Scholar]
- 46.Pawlak EN, Dirk BS, Jacob RA, Johnson AL, Dikeakos JD. 2018. The HIV-1 accessory proteins Nef and Vpu downregulate total and cell surface CD28 in CD4+ T cells. Retrovirology 15:6. 10.1186/s12977-018-0388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacob RA, Johnson AL, Pawlak EN, Dirk BS, Van Nynatten LR, Mansour HS, Dikeakos JD. 2017. The interaction between HIV-1 Nef and adaptor protein-2 reduces Nef-mediated CD4+ T cell apoptosis. Virology 509:1–10. 10.1016/j.virol.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 48.Johnson AL, Dirk BS, Coutu M, Haeryfar SMM, Arts EJ, Finzi A, Dikeakos JD. 2016. A highly conserved residue in HIV-1 Nef alpha helix 2 modulates protein expression. mSphere 1:e00288-16. 10.1128/mSphere.00288-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The accession numbers of the primary nef sequences deposited in GenBank are MW892600–MW892614.