ABSTRACT
Chikungunya virus (CHIKV) is one of the most pathogenic members of the Alphavirus genus in the Togaviridae family. Within the last 2 decades, CHIKV has expanded its presence to both hemispheres and is currently circulating in both Old and New Worlds. Despite the severity and persistence of the arthritis it causes in humans, no approved vaccines or therapeutic means have been developed for CHIKV infection. Replication of alphaviruses, including CHIKV, is determined not only by their nonstructural proteins but also by a wide range of host factors, which are indispensable components of viral replication complexes (vRCs). Alphavirus nsP3s contain hypervariable domains (HVDs), which encode multiple motifs that drive recruitment of cell- and virus-specific host proteins into vRCs. Our previous data suggested that NAP1 family members are a group of host factors that may interact with CHIKV nsP3 HVD. In this study, we performed a detailed investigation of the NAP1 function in CHIKV replication in vertebrate cells. Our data demonstrate that (i) the NAP1-HVD interactions have strong stimulatory effects on CHIKV replication, (ii) both NAP1L1 and NAP1L4 interact with the CHIKV HVD, (iii) NAP1 family members interact with two motifs, which are located upstream and downstream of the G3BP-binding motifs of CHIKV HVD, (iv) NAP1 proteins interact only with a phosphorylated form of CHIKV HVD, and HVD phosphorylation is mediated by CK2 kinase, and (v) NAP1 and other families of host factors redundantly promote CHIKV replication and their bindings have additive stimulatory effects on viral replication.
IMPORTANCE Cellular proteins play critical roles in the assembly of alphavirus replication complexes (vRCs). Their recruitment is determined by the viral nonstructural protein 3 (nsP3). This protein contains a long, disordered hypervariable domain (HVD), which encodes virus-specific combinations of short linear motifs interacting with host factors during vRC assembly. Our study defined the binding mechanism of NAP1 family members to CHIKV HVD and demonstrated a stimulatory effect of this interaction on viral replication. We show that interaction with NAP1L1 is mediated by two HVD motifs and requires phosphorylation of HVD by CK2 kinase. Based on the accumulated data, we present a map of the binding motifs of the critical host factors currently known to interact with CHIKV HVD. It can be used to manipulate cell specificity of viral replication and pathogenesis, and to develop a new generation of vaccine candidates.
KEYWORDS: CK2 kinase, NAP1L1, NAP1L4, alphavirus, chikungunya virus, intrinsically disordered proteins, nsP3, protein phosphorylation, viral pathogenesis, viral replication
INTRODUCTION
Members of the Alphavirus genus in the Togaviridae family are circulating on all continents (1). Most of them are transmitted by mosquito vectors between vertebrate hosts, in which they cause diseases of different severities. Based on their geographical distribution, alphaviruses are referred to as the New World (NW) and the Old World (OW) alphaviruses. However, recently, chikungunya virus (CHIKV) became an exception, since within the last 2 decades it has dramatically expanded its circulation area and is currently present in both the New and the Old Worlds (2, 3). In humans, CHIKV causes a highly debilitating disease characterized by rash, fever and, most importantly, severe joint pain that can persist for years (4). Thus, CHIKV represents an unquestionable public health threat, but to date, no licensed vaccines or therapeutic means have been developed for this infection. As for other alphaviruses, the molecular mechanisms of CHIKV replication, virus-host interactions, and pathogenesis remain to be better understood.
CHIKV genome is represented by a single-stranded RNA (G RNA) of positive polarity (5–7). Similar to cellular mRNAs, it contains a cap and a poly(A) tail at the 5′ and 3′ termini, respectively. Upon delivery into the cytoplasm by viral particles or RNA transfection, the genome is translated into nonstructural (ns) polyproteins P123 and P1234. They are targeted to the plasma membrane and sequentially processed into individual viral nonstructural proteins (nsP1 to nsP4) by the nsP2-encoded protease (8). nsPs serve as viral components of the replication complexes (vRCs) that synthesize dsRNA replication intermediates on the G RNA template, and new G and subgenomic (SG) RNAs. Processing of ns polyproteins regulates the template specificities of vRCs at different steps of viral replication. The partially processed P123+nsP4 complex functions in the negative-strand RNA synthesis and the completely processed nsPs mediate synthesis of the positive-strand G and SG RNAs but cannot synthesize the negative-strand RNAs.
The enzymatic functions of nsP1, nsP2, and nsP4 in the synthesis of virus-specific RNAs and in the unique cascade of capping reactions are relatively well understood (7, 9). However, to date, no direct enzymatic activities in G RNA replication and transcription of SG RNA have been ascribed for nsP3 protein. Nevertheless, nsP3 is indispensable for viral RNA synthesis, and mutations in this protein have deleterious effects on viral replication (10–13). nsP3 contains two N-terminal, conserved, structured domains (the macro domain and the alphavirus unique domain [AUD]), whose functions remain to be determined, and the C-terminal hypervariable domain (HVD) (14–16). Our previous studies and those of other groups demonstrated that alphavirus HVDs encode a variety of short motifs, which recruit specific combinations of host factors into vRCs, and alterations of HVD interactions with host proteins either have strong negative effects on the rates of viral replication or can make alphaviruses nonviable (13, 17–23).
The CHIKV nsP3 HVD is intrinsically disordered and was shown to interact with cell-specific sets of host proteins (17, 19, 21, 24, 25). The accumulated data about its interactions with host factors can be summarized as follows. (i) G3BP1 and G3BP2 are the critical host factors for CHIKV replication in vertebrate cells (17, 26). In mosquito cells, the insect-specific homolog RIN1 appears to have the same proviral function as G3BPs of vertebrates (27). Mutations in G3BP-binding sites of nsP3 HVD or knockout (KO) of both G3BP1/2 genes in mammalian cells make CHIKV not viable (17). (ii) Binding of only G3BPs to CHIKV HVD is insufficient for vRC formation and function, and the variants that have only G3BP-binding sites in their HVDs remain nonviable (19). Interactions with other families of host proteins have stimulatory effects on viral replication. These additional interacting partners include the SH3 domain-containing proteins (BIN1, CD2AP, and SH3KBP1), members of the FHL family (FHL1 to FLH3) and NAP1 (nucleosome assembly protein 1) family members (19, 23). However, this list probably remains incomplete. (iii) The distinguishing characteristic of the HVD-binding factors is the high redundancy of their function. It allows CHIKV to utilize multiple members of the families and different protein families to facilitate vRC formation and function. The use of more than one family member likely promotes viral replication in a wider range of tissues and host species, in which the presence of particular protein factors may vary. (iv) We have recently characterized by nuclear magnetic resonance (NMR) spectroscopy and genetic approaches the binding sites of the members of G3BP and FHL families, and the SH3 domain-containing proteins. The additive proviral effects of these interactions were demonstrated in biological studies (23, 24). (v) Binding of NAP1 proteins to CHIKV HVD and its biological significance remains poorly explored, although this interaction appears to be CHIKV specific and is required for efficient viral replication in vertebrate cells. NAP1 proteins are not extensively characterized but are known to be essential for cell growth. NAP1L1 is also believed to be involved in DNA replication with higher expression in rapidly proliferating cells and in tumors. NAP1L1 and NAP1L4 were also implicated in regulation of cellular transcription (28–30).
In this new study, we used a wide variety of approaches to identify the binding sites of NAP1L1 and NAP1L4 in CHIKV HVD and demonstrated the proviral role of this interaction in CHIKV replication. Our data reveal a unique mechanism of NAP1L1 binding to HVD. It is determined by two HVD-encoded motifs located upstream and downstream of the G3BP-binding sites. However, NAP1-HVD interaction remains G3BP independent. Importantly, interaction of NAP1 proteins with CHIKV nsP3 is dependent on HVD phosphorylation by CK2 kinase The presence of only NAP1-binding sites in CHIKV HVD in addition to the G3BP-binding motifs is sufficient for CHIKV viability, but binding of other host factors has strong stimulatory effect on viral replication. These results additionally support our overall hypothesis regarding redundant functions of alphavirus HVD-interacting partners.
RESULTS
NAP1L1 interacts with the C-terminal fragment of CHIKV nsP3 HVD both in the presence or absence of G3BP.
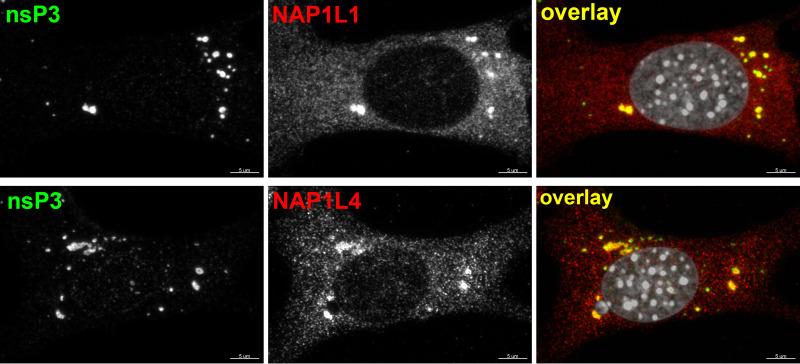
In previous studies, we and other groups have identified sets of host factors that interact with CHIKV nsP3 HVD in human and mouse cells (18–23). These HVD-interacting partners include the members of G3BP family (G3BP1 and G3BP2), the SH3 domain-containing proteins (CD2AP, BIN1, and SH3KBP1), FHL family members (FHL1, FHL2, and FHL3), and proteins of the NAP1 family (NAP1L1 and NAP1L4). To further understand the mechanism of NAP1L1 and NAP1L4 interactions with CHIKV HVD and their roles in viral replication, we first confirmed that in CHIKV-infected cells NAP1L1 and NAP1L4 are present in the nsP3 complexes. NIH 3T3 cells were infected with CHIKV 181/25 and stained with nsP3-, NAP1L1-, and NAP1L4-specific antibodies (Abs). As demonstrated in Fig. 1, both NAP1 proteins clearly accumulated in large nsP3-containing cytoplasmic complexes.
FIG 1.
NAP1L1 and NAP1L4 accumulate in CHIKV nsP3 complexes. NIH 3T3 cells were infected with CHIKV 181/25 at an MOI of 10 PFU/cell in 8-well Ibidi plates. At 7 h p.i., the cells were fixed and stained with CHIKV nsP3- and NAP1-specific Abs and DAPI as described in Materials and Methods. Images were acquired on a Zeiss LSM 800 confocal microscope in Airyscan mode with a 63 × 1.4 NA PlanApochromat oil objective.
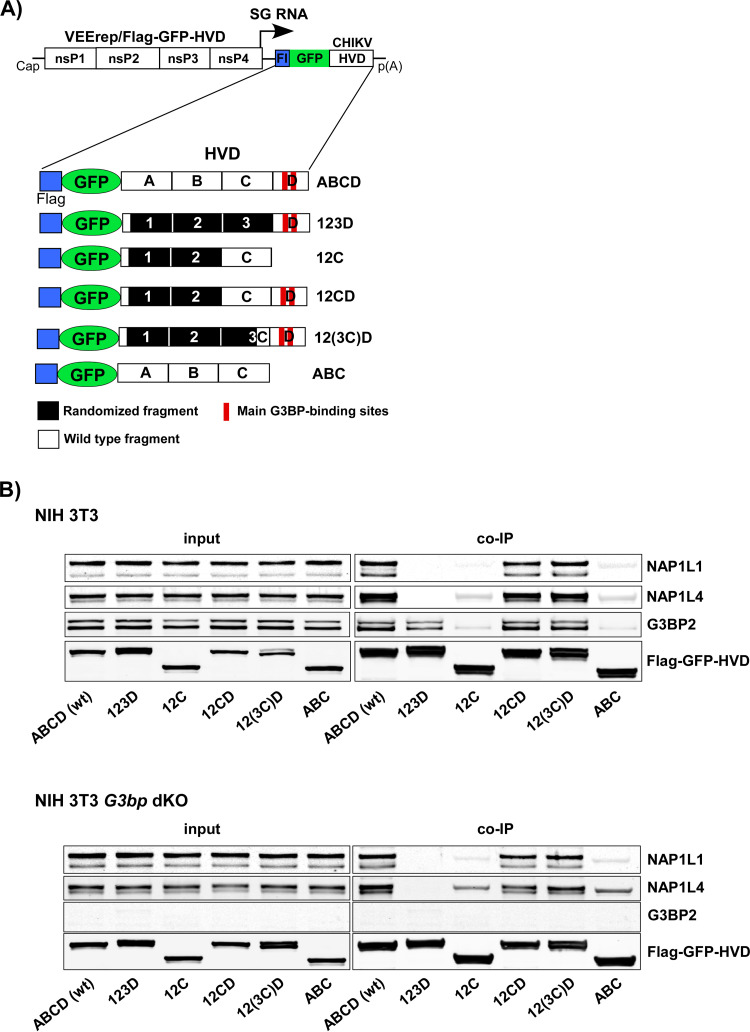
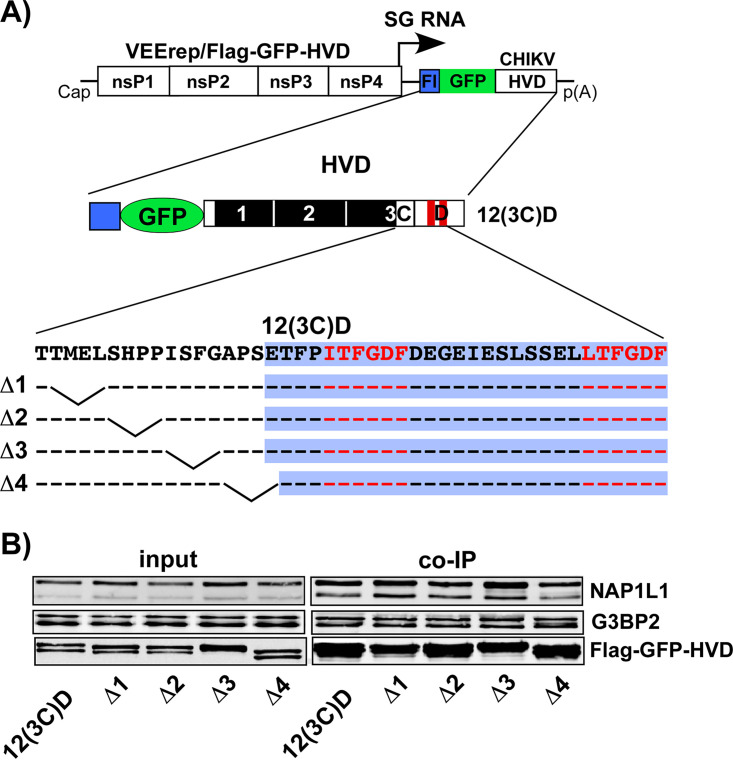
Next, we intended to identify the NAP1L1-binding motif in CHIKV nsP3 HVD. As in our recent studies (19), we utilized a set of Venezuelan equine encephalitis virus (VEEV) replicons (VEErep) to express different combinations of CHIKV HVD fragments fused with Flag-tagged green fluorescent protein (Flag-GFP) (Fig. 2A). The HVD was divided into four fragments either having a wild-type (wt) amino acid (aa) sequence (fragments A, B, C, and D) or having aa sequences randomized by changing the positions of closely located aa (fragments 1, 2, and 3). These modifications were aimed at eliminating the known and other potential binding sites of host proteins in the individual fragments. In the “(3C)” fragment, only the C-terminal 16-aa peptide of fragment C was left intact, and the remaining two thirds of the fragment were randomized. The designed replicons were packaged into infectious viral particles using VEEV TC-83-based helper RNA (see Materials and Methods for details). Then, NIH 3T3 cells and their G3bp dKO derivatives (17) were infected with packaged replicons, and protein complexes were isolated using magnetic beads loaded with Flag-specific monoclonal Abs (MAbs). Only the ABCD-, 12CD- and 12(3C)d-encoding constructs demonstrated the ability to efficiently coimmunoprecipitate NAP1L1 and NAP1L4 (Fig. 2B). Thus, interaction of NAP1 family members with CHIKV HVD was determined by the C-terminal HVD fragment (3C)D, which contained binding sites of the G3BP1/2 (fragment D), and the last 16 aa of the upstream fragment C. Since the results generated on G3bp dKO and parental NIH 3T3 cells were similar (Fig. 2B), we concluded that both NAP1 proteins can bind to CHIKV HVD in the absence of the G3BP interaction with CHIKV HVD. However, since G3BP- and NAP1-binding sites are so closely located, the possibility of cooperativity in interactions of these two protein families with CHIKV HVD could not be completely ruled out.
FIG 2.
NAP1 family members are capable of interacting with the C-terminal fragment of CHIKV HVD independently of G3BP. (A) The schematic presentation of VEEV replicons encoding different variants of CHIKV HVD fused with Flag-GFP. Labels A, B, C, and D indicate wt aa sequences. Fragments 1, 2, and 3 have the aa sequences of the corresponding A, B, and C fragments randomized. In fragment (3C), only the C-terminal 16-aa peptide had the wt sequence. wt and mutated fragments are additionally indicated by open and black boxes, respectively. (B) NIH 3T3 cells and their G3bp dKO derivatives were infected at an MOI of 10 inf.u/cell with the packaged VEEV replicons expressing the indicated fragments of CHIKV HVD. Cells were collected at 4 h p.i. Protein complexes were isolated on magnetic beads as described in Materials and Methods and analyzed by Western blotting with NAP1L1-, NAP1L4-, G3BP2-, and Flag-specific Abs and corresponding secondary Abs. Membranes were scanned on a Li-Cor imager. The experiment was repeated three times with similar results.
Mutations in the (3C) fragment of CHIKV nsP3 HVD affect viral replication.
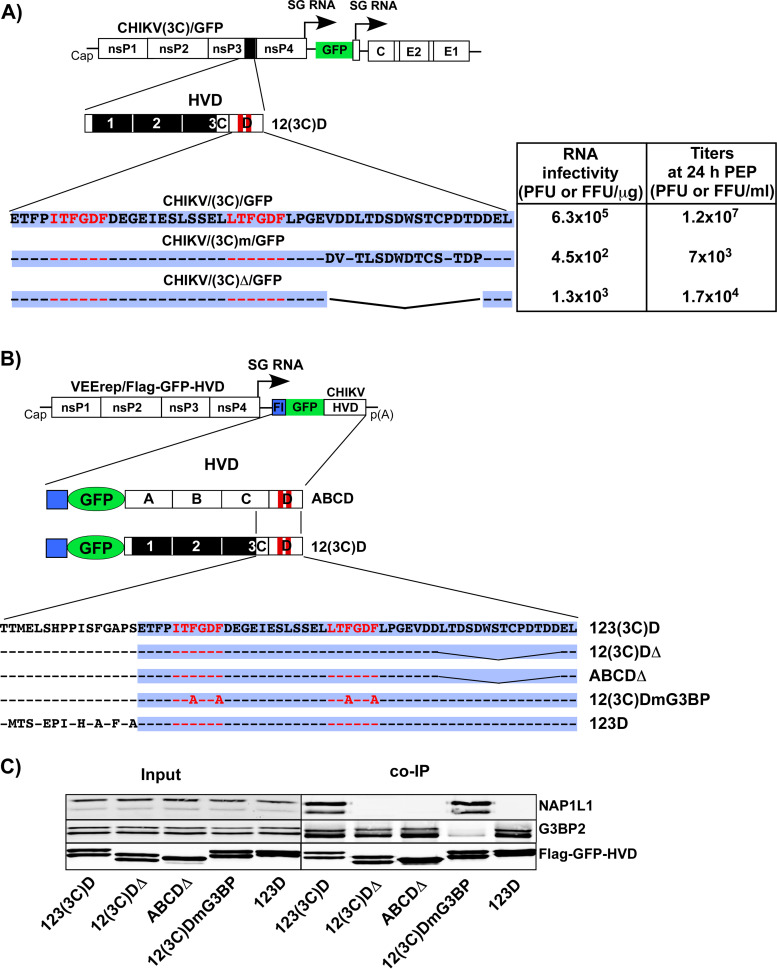
Next, we further characterized function of the identified (3C) fragment in CHIKV replication. For this analysis, we applied a CHIKV 181/25 variant encoding 123(3C)D HVD, termed CHIKV/(3C)/GFP (Fig. 3A). The nsP3 HVD of this virus contained no binding motifs of the FHL or the SH3 domain-containing proteins. Its replication was expected to rely only on the interactions of mutated nsP3 HVD with host G3BP and NAP1 protein families. Thus, in this study, it was used as one of the experimental systems to further dissect the HVD sites that mediate interaction with NAP1 proteins. CHIKV/(3C)/GFP and the designed derivatives, which were used in these and following experiments, also contained GFP gene under the control of a viral subgenomic promoter. Its expression simplified evaluation of the spread of the mutants in cultured cells, because some of them were expected to be unable to form distinct plaques.
FIG 3.
Mutations in NAP1-binding site affect CHIKV replication. (A) Schematic presentation of the CHIKV/(3C)/GFP genome, deletions introduced into (3C) fragment, and infectivities of the in vitro-synthesized RNA in the ICA (see Materials and Methods for details). Dashes indicate aa identical to those in HVD of the parental virus. Fragment D is additionally indicated by blue background. (B) BHK-21 cells were electroporated with 3 µg of the in vitro-synthesized RNAs of parental CHIKV/(3C)/GFP and its deletion mutants. Samples of the media were collected at the indicated times postelectroporation, and titers were determined by plaque assay on BHK-21 cells. The ICA and titers of the released viruses were assessed in the same experiment, which was reproducibly repeated three times. Means and standard deviations (SD) are indicated. The significance of differences was determined by one-way analysis of variance (ANOVA) with the Tukey test (***, P < 0.001; ****, P < 0.0001; n = 3).
To map the aa in CHIKV HVD, which determine the proviral effect of NAP1, we introduced sequential 4-aa deletions into the (3C) fragment of CHIKV/(3C)/GFP HVD (Fig. 3A). Equal amounts of the in vitro-synthesized RNAs were electroporated into BHK-21 cells, and we evaluated effects of the introduced deletions on RNA infectivity using the infectious center assay (ICA) (Fig. 3A) and on the rates of viral replication (Fig. 3B). Deletion Δ1 had no effect on viral replication. Δ2 deletion detectably affected the replication rates, but its effect was not as prominent as those of Δ3 and Δ4. The latter deletions reduced the efficiency of plaque formation in ICA by more than 100-fold (Fig. 3A), and the replication rates of these mutants were a few orders of magnitude lower than that of the parental CHKV/(3C)/GFP (Fig. 3B). A few orders of magnitude lower RNA infectivity was an indication that the detected plaques were likely formed by pseudorevertants, which evolved to a more efficiently replicating phenotype. They could accumulate adaptive mutations either in nsPs or in the cis-acting RNA elements of the viral genome. We randomly selected directly in the ICA four plaques formed by CHIKV/(3C)Δ3/GFP and CHIKV/(3C)Δ4/GFP. Sequencing the genomes of these plaque-purified viruses revealed the presence of mutations in the structured, N-terminal domains of nsP3. Three isolated pseudorevertants contained a D31A mutation in the macro domain, and another one had a K258R substitution in the AUD. To confirm the positive effects of these mutations on CHIKV replication, they were introduced into cDNAs of the parental CHIKV/(3C)Δ3/GFP and CHIKV/(3C)Δ4/GFP genomes (Fig. 4A and B). The mutations had a profound positive effect on infectivities of the in vitro-synthesized RNAs in the ICA (Fig. 4B) and increased viral replication rates by a few orders of magnitude (Fig. 4C), indicating their true compensatory functions.
FIG 4.
Adaptive mutations in the macro domain and AUD compensate negative effects of the deletions in the potential NAP1-binding site. (A) The schematic presentation of CHIKV/(3C)/GFP genome and deletions made in potential NAP1-binding site. (B) Schematic presentation of the domain structure of CHIKV nsP3 in the 12(3C)D variant, positions of the deletions introduced upstream of the G3BP-binding site, and adaptive mutations in the macro domain and AUD (indicated by red arrows). Infectivities of the in vitro-synthesized RNAs of the parental deletion mutants and those with adaptive mutations in the ICA (see Materials and Methods for details). The experiment was reproducibly repeated twice, and the results of one experiment are presented. (C) Replication of the variants with adaptive mutations in structured domains and the parental deletion mutants in BHK-21 cells after electroporation of the in vitro-synthesized RNAs. Samples of the media were collected at the indicated times postelectroporation, and titers were determined by plaque assay on BHK-21 cells. The experiment was reproducibly repeated three times. Means and SD are indicated. The significance of differences was determined by one-way ANOVA with the Tukey test (****, P < 0.0001; n = 3).
NAP1L1 interaction with CHIKV HVD is determined by more than one HVD motif.
The above data (Fig. 2 and 3) strongly suggested that the binding motif of NAP1L1 is located right upstream of the first G3BP-binding site in CHIKV HVD. To confirm this, we introduced the above deletions Δ1, Δ2, Δ3, and Δ4 into the Flag-GFP-12(3C)D fusion encoded by VEEV replicon (Fig. 5A). The designed replicons were packaged into viral particles and used for infection of the NIH 3T3 cells. As described in Materials and Methods, the protein complexes were isolated from the cell lysates on anti-Flag MAb magnetic beads and analyzed by Western blotting with NAP1L1-specific Abs. Surprisingly, the mutated HVDs were still able to bind NAP1L1 and, as expected, the deletions had no profound effect on the ability of HVD to interact with G3BP (Fig. 5B). In contrast, 123D HVD encoding the randomized (3C) peptide was incapable of interacting with NAP1L1 (Fig. 2). CHIKV encoding the latter HVD (CHIKV/123D/GFP) was also nonviable (19). However, smaller modifications, such as 4-aa deletions (Fig. 5), did not completely abrogate the ability of CHIKV HVD to coprecipitate NAP1L1. Moreover, the corresponding mutant viruses remained capable of at least very inefficient replication, which ultimately led to their evolution to a more efficiently replicating phenotype (Fig. 3 and 4). Thus, small deletions affected the function of NAP1L1 in replication of viral RNA but did not completely eliminate NAP1L1-HVD interaction. Taken together, these results suggested the possibility that NAP1L1 protein may have an additional binding site in CHIKV HVD. This second interaction could stabilize NAP1L1-HVD complex but appeared to be insufficient for promoting viral replication in the absence of the wt peptide (3C peptide) upstream of the G3BP-binding site.
FIG 5.
Deletions in putative NAP1-binding site do not abrogate 12(3C)D HVD interaction with NAP1L1. (A) Schematic presentation of VEEV replicon encoding Flag-GFP-12(3C)D HVD fusion with the indicated deletions in the motif located upstream of the G3BP-binding site. (B) NIH 3T3 cells were infected with packaged replicons at an MOI of 20 inf.u/cell. Cells were harvested at ∼3 h p.i. Protein complexes were isolated on magnetic beads as described in Materials and Methods. They were analyzed by Western blotting with NAP1L1- and Flag-specific Abs and corresponding secondary Abs. Membranes were scanned on a Li-Cor imager. The experiment was repeated twice with identical results. The results of one experiment are presented.
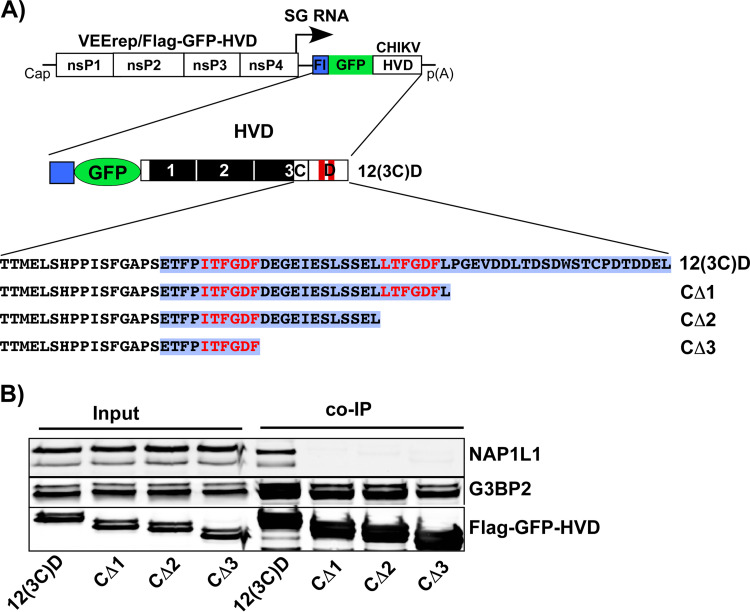
The C-terminal fragment of CHIKV nsP3 HVD is required for interaction with NAP1L1.
To examine the possibility that NAP1L1-HVD interaction is mediated by more than one motif in CHIKV HVD, we designed a set of VEEV replicons encoding Flag-GFP fused with 12(3C)D, which contained additional modifications (Fig. 6A). Since the C-terminal D fragment was the only one left intact in the previous constructs (Fig. 5), we deleted the aa sequences in the very C-terminal peptide located downstream of the first G3BP-binding motif (Fig. 6A). Next, NIH 3T3 cells were infected with these packaged replicons, and the HVD-specific protein complexes were isolated using anti-Flag MAb magnetic beads. Further analyses by Western blotting detected no NAP1L1 in the isolated complexes (Fig. 6B), but the mutated HVDs remained capable of interacting with G3BP. This was a strong indication that CHIKV HVD has at least two motifs that determine its interaction with NAP1L1. One of them is located upstream of the first G3BP-binding site and is required for the stimulatory effect of NAP1L1 on viral replication. The second motif is positioned downstream of the G3BP-binding repeat, at the very C terminus of nsP3 HVD, and its presence alone, without the first upstream peptide, is insufficient for driving CHIKV replication.
FIG 6.
Deletions in the C-terminal fragment of 12(3C)D HVD abrogate its binding of NAP1L1, but not of G3BP. (A) Schematic presentation of VEEV replicons encoding deletion variants of CHIKV 12(3C)D HVD fused with Flag-GFP. Fragment D is additionally indicated by blue background. (B) NIH 3T3 cells were infected with packaged replicons at an MOI of 20 inf.u/cell. Cells were harvested at 3 h p.i. Protein complexes were isolated on magnetic beads and analyzed by Western blotting with NAP1L1-, G3BP2-, and Flag-specific Abs and corresponding secondary Abs. Membranes were scanned on a Li-Cor imager. The experiment was repeated twice with identical results. The results of one experiment are presented.
Modifications of the C-terminal fragment of CHIKV HVD result in negative effects on viral replication and presence of NAP1L1 in HVD complexes.
NAP1 family members are a group of cell proteins that are essential for cell growth, and double knockout (KO) of these genes will likely be lethal for mammalian cells (31–33). Therefore, to demonstrate biological significance of the above finding, we modified the C-terminal fragment of HVD in CHIKV/(3C)/GFP genome (Fig. 7A). The C-terminal fragment was either randomized by changing the positions of the aa or contained a deletion of the 16 aa [CHIKV/(3C)m/GFP and CHIKV/(3C)Δ/GFP, respectively]. The in vitro-synthesized viral RNAs were electroporated into the cells, and their infectivities and levels of infectious virus release were evaluated in the ICA and by assessing titers in samples harvested at 24 h posttransfection. Modifications introduced into the C terminus of the HVD had deleterious effects on viral replication (Fig. 7A). The latter variants became almost nonviable. Synthesized RNAs exhibited very low efficiencies in the ICA with only pinpoint foci, but not plaques, being detected. Infection spread was almost undetectable, and most of electroporated cells remained GFP negative at 2 days postelectroporation. A few orders of magnitude decrease in RNA infectivity indicated that even the detected pinpoint GFP-positive foci were most likely the result of viral evolution and accumulation of adaptive mutations. However, since the infectious titers were very low, this adaptation was not further investigated. Taken together, these data supported the hypothesis that the C terminus of CHIKV nsP3 HVD is required for the proviral function of NAP1 family members.
FIG 7.
Modifications in the C-terminal fragment of CHIKV/(3C)/GFP have deleterious effects on viral replication. (A) Schematic presentation of the genomes and HVDs of the designed viruses and aa sequences of their D fragments. Dashes indicate aa identical to those in the wt sequence. The table presents the infectivities of the in vitro-synthesized RNAs in the ICA and viral titers at 24 h postelectroporation. RNA infectivities and titers were measured in PFU and GFP-positive focus-forming units (FFUs) for CHIKV/(3C)/GFP and its derivatives, respectively. The experiment was repeated twice with reproducible results. The data from one experiment are presented. (B) Schematic presentation of VEEV replicons encoding different variants of CHIKV HVD fused with Flag-GFP. Fragment D is additionally indicated by blue background. (C) NIH 3T3 cells were infected with packaged replicons at an MOI of 20 inf.u/cell. Cells were harvested at 3 h p.i. Protein complexes were isolated on magnetic beads and analyzed by Western blotting with NAP1L1-, G3BP2-, and Flag-specific Abs and corresponding secondary Abs. Membranes were scanned on a Li-Cor imager.
To experimentally support these data, we expressed additional mutated HVDs as Flag-GFP fusions from VEEV replicon (Fig. 7B). As in the experiments described above, HVD complexes were isolated from the NIH 3T3 cells infected with packaged replicons and analyzed by Western blotting with NAP1L1-, G3BP2-, and Flag-specific Abs (Fig. 7C). Mutations in G3BP-binding repeat abrogated binding of this protein to 12(3C)DmG3BP but did not affect coimmunoprecipitation (co-IP) of NAPL1L1. The very low level of residual G3BP was likely a result of its binding to the cryptic site of HVD that we had described in our previous studies (19). The deletions of the C-terminal aa sequences in both 12(3C)D and natural ABCD HVDs [12(3C)DΔ and ABCDΔ, respectively] reduced the presence of NAP1L1 in the isolated complexes to undetectable levels but did not noticeably affect the presence of G3BP. Thus, the results of the co-IP experiments correlated with the inability of CHIKV/(3C)Δ/GFP and CHIKV/(3C)m/GFP mutants to develop spreading infection even in BHK-21 cells (Fig. 7A), which are highly susceptible to CHIKV.
NAP1L1 is one of the host factors with additive stimulatory effects on CHIKV replication.
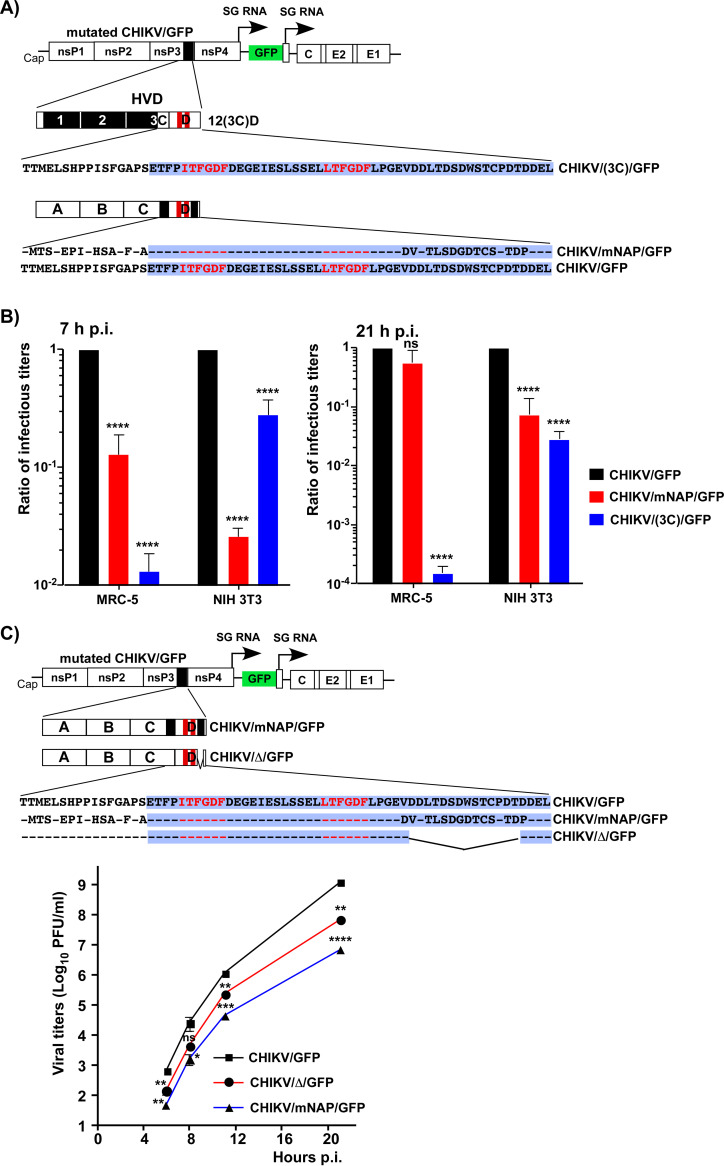
In the next set of experiments, we compared replication of additional CHIKV variants with modified nsP3 HVD (Fig. 8A). One of them, CHIKV/mNAP/GFP, had the NAP1-binding site randomized and thus was dependent on interactions with all of the CHIKV HVD-specific host factors, except NAP1 family members. Replication of the second one, CHIKV/(3C)/GFP, depended only on binding of G3BPs and NAP1 proteins. Both these mutants and the parental CHIKV/GFP were rescued in BHK-21 cells from in vitro-synthesized RNAs. The human and mouse fibroblasts (MRC-5 and NIH 3T3 cells, respectively) were then infected at the same multiplicity of infection (MOI). The modifications introduced into the NAP1-binding site had a negative effect on replication of CHIKV/mNAP/GFP in both cell lines, which was better detectable at early times postinfection (p.i.). Interestingly, the CHIKV/(3C)/GFP mutant replicated less efficiently in MRC-5 cells than in the NIH 3T3 cell line (Fig. 8B). This was an additional indication that HVD-binding factors function in species- or cell-specific modes, with NAP1 proteins playing a more significant role in CHIKV replication in mouse than in human cells.
FIG 8.
Interaction of CHIKV HVD with NAP1 stimulates viral replication. (A) Schematic presentation of CHIKV/GFP genome, its HVD and the HVDs of the designed mutants, and corresponding aa sequences. Dashes indicate aa identical to those in the wt sequence. The sequence of fragment D is additionally indicated by a blue background. (B) NIH 3T3 and MRC-5 cells were infected with the indicated viruses at an MOI of 0.1 PFU/cell, and media were harvested at the indicated times p.i. Infectious titers were determined by plaque assay on BHK-21 cells. Titers were normalized to those of the parental CHIKV/GFP. The experiment was repeated three times; means and SD are indicated. The significance of differences was determined by two-way ANOVA with the Fisher LSD test (****, P < 0.0001; n = 3). (C) Schematic presentation of mutated HVDs in the designed CHIKV variants and corresponding aa sequences of the modified fragments. NIH 3T3 cells were infected with the indicated mutants and the parental CHIKV/GFP at an MOI of 0.01 PFU/cell. Media were collected at the indicated times p.i., and infectious titers were determined by plaque assay on BHK-21 cells. The experiment was reproducibly repeated three times; means and SD are indicated. The significances of the differences were determined by two-way ANOVA with the Fisher LSD test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n = 3).
Next, NIH 3T3 cells were infected with the rescued viruses, which encoded HVD with either the C-terminal NAP1-interacting motif deleted (CHIKV/Δ/GFP) or both motifs located upstream and downstream of G3BP-binding sites were randomized (CHIKV/mNAP/GFP) (Fig. 8C). The rest of their HVDs had wt aa sequences. Both mutants demonstrated lower growth rates in the NIH 3T3 cells (Fig. 8C), but the negative effect of the mutations in both NAP1-specific motifs (CHIKV/mNAP/GFP) was more prominent than that of the deletion of one of them in CHIKV/Δ/GFP. CHIKV/mNAP/GFP replicated to almost 2 orders of magnitude lower titers than the parental CHIKV/GFP.
NAP1L1 binding requires phosphorylation of CHIKV nsP3 HVD.
To further understand NAP1 interaction with CHIKV HVD, we produced NAP1L1 and NAP1L4 proteins in Escherichia coli and purified them to homogeneity. Surprisingly, in the in vitro binding assay, we did not detect any interaction between CHIKV HVD and NAP1 proteins by size exclusion chromatography and by NMR titration (data not shown), despite these proteins being able to form complexes in vertebrate cells (Fig. 1 and 2). However, alphavirus HVDs are known to be highly phosphorylated (11, 13, 34–41), and proteins produced in E. coli lack this posttranslational modification. Moreover, it was previously suggested that NAP1L1 interaction with HCV NS5 protein is dependent on phosphorylation (42).
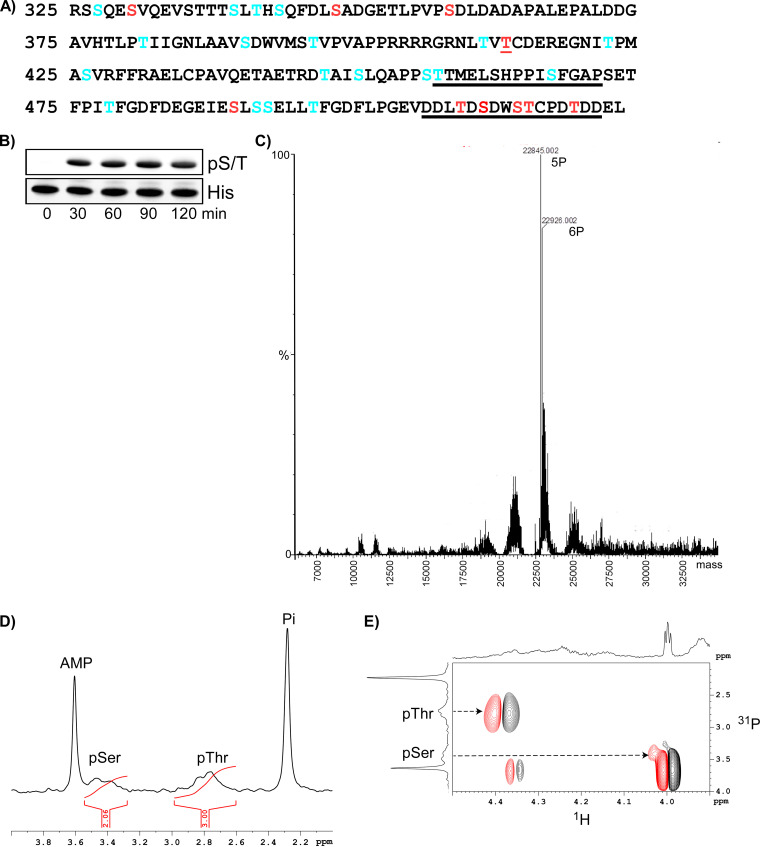
We analyzed the potential phosphorylation sites in CHIKV HVD using NetPhos 3.1 prediction server (43). It predicted that 16 serines and 9 threonines can potentially be phosphorylated (Fig. 9A). Interestingly the C-terminal NAP1-binding motif was predicted to contain five phosphorylated aa, all of which could potentially be phosphorylated by CK2 kinase. The recombinant CK2α was found to efficiently phosphorylate the recombinant CHIKV HVD in vitro (Fig. 9B). Mass spectrometry analysis of the in vitro phosphorylated protein suggested that the major phosphorylated species contains five phosphates, and a minor one has six phosphates (Fig. 9C). In good correlation with these data, 31P NMR suggested that both serines and threonines were phosphorylated and that the ratio of pSer to pThr was 2 to 3 (Fig. 9D and E). Proteomics analysis of phosphorylated HVD detected clearly only a single threonine, T412, phosphorylated at 20% level. However, the C-terminal protein fragment, containing the 16-aa peptide with the second NAP1-binding site, could not be detected at all. The most plausible explanation for this was that a high degree of phosphorylation was likely the reason that this peptide could not be identified by conventional mass spectrometry. Combined, these data strongly suggested that the second, C-terminal NAP1-binding site contains five phosphorylated Ser and Thr.
FIG 9.
CHIKV HVD is phosphorylated by CK2α. (A) Potential phosphorylation sites in CHIKV HVD predicted by NetPhos 3.1. The CK2α-specific sites are indicated in red, and other potential sites are indicated in blue. The underlined aa represent the elements of the NAP1-binding sites upstream and downstream of the G3BP-binding motifs. (B) Western blot analysis of in vitro phosphorylation of CHIKV HVD-His tag by CK2α. (C) Charge-deconvoluted spectrum of phosphorylated HVD demonstrates the presence of two major species that contain five and six phosphates. (D) 1D 31P spectrum of the phosphorylated CHIKV HVD (pHVD) between 4.0 and 2.0 ppm. Two sharp peaks at 3.61 and 2.29 ppm correspond to AMP and free phosphate, respectively. The broad peaks at 3.48 and 2.76 ppm correspond to phosphorylated serine, labeled as pSer, and to phosphorylated threonine, labeled as pThr, respectively. Note that the ratio of the integrals of pSer to pThr is 2 to 3. (E) 2D 1H-31P correlation spectrum of the pHVD sample is shown. The arrows indicate the connection of the 31P resonance of pSer and pThr with corresponding cross peaks.
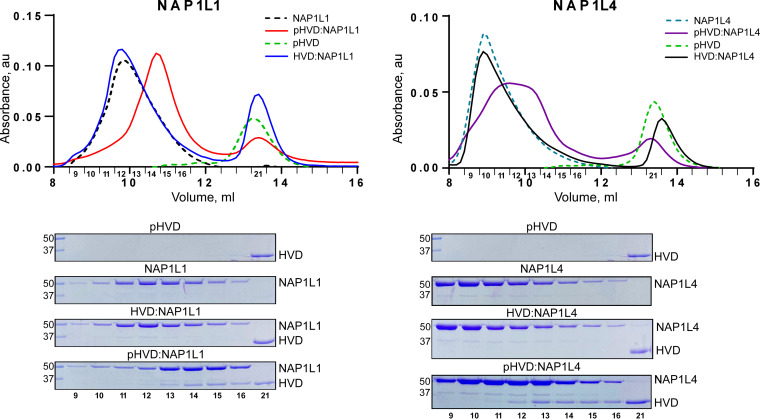
The in vitro-phosphorylated CHIKV HVD was used for the in vitro-binding experiments. Individual proteins or their complexes were analyzed by size exclusion chromatography (Fig. 10). HVD was eluted in a single peak. In correlation with the previously published data, both NAP1L1 and NAP1L4 eluted from the column as large aggregates (44, 45). A mixture of NAP1L1 or NAP1L4 with unphosphorylated HVD was eluted as two peaks, which corresponded to individual proteins. In the case of phosphorylated HVD, the peak height for HVD was reduced and formation of a new peak was detected for both NAP1L1 and NAP1L4. Gel electrophoresis demonstrated that the latter peak contained both NAP and HVD. Interestingly, the HVD/NAP1 complexes eluted later than aggregated NAP1 proteins, which suggested that binding to HVD led to a dissociation of NAP1 aggregates. Thus, nsP3 HVD phosphorylation is a prerequisite of its binding to NAP1 proteins.
FIG 10.
NAP1L1 and NAP1L4 bind in vitro only to phosphorylated CHIKV HVD. Analyses of binding were performed by using SEC on Superdex 200 increase 10/30 GL column, as described in Materials and Methods. Fractions corresponding to the peaks were analyzed by SDS-PAGE. Gels were stained with Coomassie blue.
Inhibition of CK2 kinases prevents NAP1 accumulation in nsP3 complexes and affects viral replication.
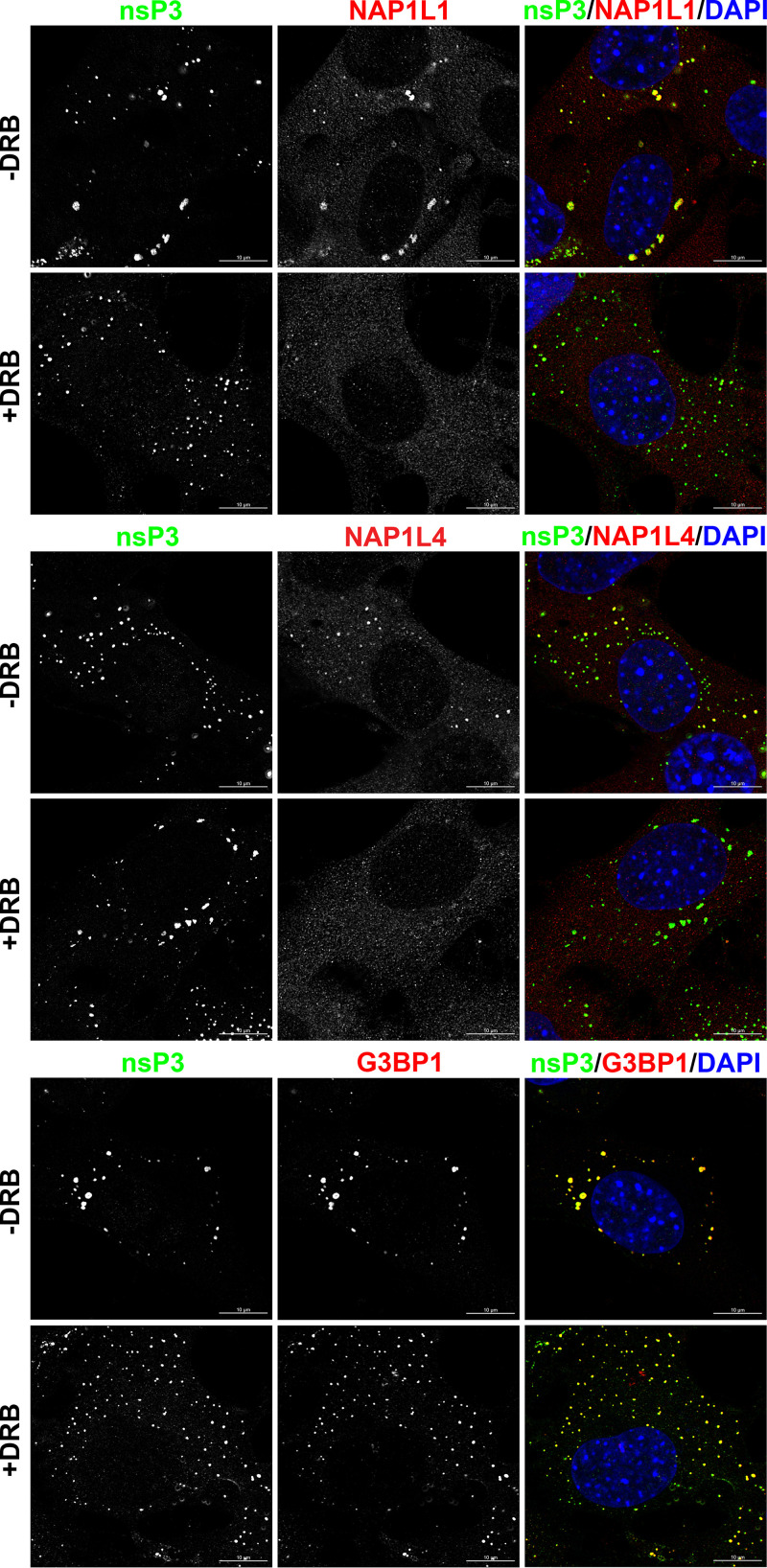
We next evaluated whether inhibition of CK2 kinase prevents interaction of NAP1 proteins with CHIKV HVD during viral infection. The infected cells were treated with CK2 kinase inhibitor 6-dichloro-1-(β-d-ribofuranosyl) benzimidazole (DRB) and processed for immunostaining with antibodies against nsP3, NAP1L1, NAP1L4, and G3BP1. In mock-treated cells, all three host proteins accumulated in the cytoplasmic nsP3 complexes (Fig. 11). In the presence of DRB, NAP1 proteins no longer colocalized with nsP3. As expected, the DRB treatment did not inhibit interaction between G3BP1 and nsP3 (Fig. 11). This suggested that CK2 kinase is responsible for phosphorylation of CHIKV HVD in vitro.
FIG 11.
Treatment with DRB prevents accumulation of NAP1 proteins, but not G3BPs, in the cytoplasmic nsP3 complexes. NIH 3T3 cells were infected with CHIKV/GFP at an MOI of 10 PFU/cell and incubated for 15 h in the presence (100 µM) or absence of CK2α inhibitor DRB. After fixing with PFA, cells were stained with antibodies against indicated proteins. Images were acquired on a Zeiss LSM 800 confocal microscope in Airyscan mode with a 63 × 1.4 NA PlanApochromat oil objective.
In the additional experiments, we evaluated the effect of DRB on CHIKV replication in the NIH 3T3 cells. The negative effect of this drug on viral replication was clearly detectable (Fig. 12). As expected, it was more prominent for CHIKV/(3C)/GFP mutant, whose replication was determined only by G3BPs and NAP1 family members. In case of CHIKV/GFP, other HVD-binding host factors were likely capable of mediating RNA replication, when interaction of HVD with NAP1 was affected by inhibition of the CK2-mediated HVD phosphorylation.
FIG 12.
Inhibition of phosphorylation has negative effect on CHIKV replication. NIH 3T3 cells were infected with the indicated viruses in the presence of 100 µM DRB or 0.2% dimethyl sulfoxide (DMSO) and after washing with PBS, incubated in media containing the same concentration of either drug or solvent. Samples of the media were collected at the indicated times p.i., and viral titers were determined by plaque assay on BHK-21 cells. Means and SD are indicated. Significance of differences was determined by two-way ANOVA with the Fisher LSD test (**, P < 0.01; ****, P < 0.0001; n = 3).
DISCUSSION
Within the last few years, intrinsically disordered proteins (IDPs) and protein domains have attracted a lot of attention, which they deserve. In addition to demonstrating unusual physical characteristics, IDPs can interact with multiple proteins and mediate the formation of large complexes and thus are critically involved in numerous cellular processes (46–49). HVDs of CHIKV and VEEV nsP3 and likely nsP3 proteins of other alphaviruses are intrinsically disordered and encode sets of motifs that bind specific host proteins in virus- and cell-specific manners (18, 19, 23, 24, 50–52). These interacting host factors are required for assembly of vRCs and recruitment of G RNA and also possibly for other functions that remain to be determined. The disordered structure of HVDs and lack of specific enzymatic activities promote their rapid evolution and adaptation for hijacking different host proteins into alphavirus-specific complexes. Some alphavirus-specific HVDs have even evolved an ability to interact with the same essential host factors by using HVD motifs having different aa sequences (18). For example, the G3BP-binding sites in the HVDs of CHIKV and eastern equine encephalitis virus have different sequences, but both of them recruit G3BP family members to function in viral RNA replication. Similarly, in the HVDs of Venezuelan and eastern equine encephalitis viruses, the critical FXR-binding motifs demonstrate very limited identity, but both function in recruitment of critically important FXR family members (18). The lack of HVD conservation between geographically isolated alphaviruses suggests that these domains may evolve to adapt viruses to mosquito species and vertebrate hosts that circulate in particular areas.
An interesting characteristic of alphavirus HVDs is that in order to support viral RNA synthesis, they can redundantly interact with multiple members of host protein families. In the case of CHIKV, both members of the G3BP family (G3BP1 and G3BP2), three members of the FHL family (FHL1, FHL2, and FHL3) and at least three SH3 domain-containing proteins (CD2AP, BIN1, and SH3KBP1) can function in viral replication with comparable efficiencies (17, 23, 24). The NAP1 family is likely not an exception. Previously, both NAP1L1 and NAP1L4 were readily detectable in co-IP samples of CHIKV HVD-specific complexes (19, 22). Thus, both NAP1L1 and NAP1L4 may redundantly stimulate CHIKV replication.
Thus far, only the G3BP-HVD interaction was found to be indispensable for CHIKV infection (17). No virus replication or evolution to a more efficiently replicating phenotype was found when both G3BP-binding sites in CHIKV HVD were deleted or mutated or when G3bp dKO cells were infected with wt CHIKV. However, CHIKV HVD interaction with G3BPs only is insufficient for viral replication, and other above-described factors demonstrate redundant stimulatory, proviral effects. The presence of at least one more interacting motif, besides those specific to G3BP, makes CHIKV viable. Binding of the members of additional families, such as FHL, NAP1, or SH3 domain-containing proteins (BIN1, CD2AP, or SH3KBP1), to HVD additively stimulates CHIKV replication. Moreover, the results of our previous NMR-based studies also demonstrated that binding of either CD2AP or FHL1 induces allosteric changes in the distantly located C-terminal HVD peptide, which contains two G3BP-binding motifs (23, 24). These allosteric changes are still difficult to understand, but they may explain the cumulative stimulatory effects of host factor interactions with CHIKV HVD on G3BP-mediated viral replication.
NAP1 family (nucleosome assembly protein 1) contains five members both in humans and mice. NAP1L1 and NAP1L4 are the most ancient members and are proposed to form both mono- and heterodimers (28–30, 33). NAP1L1 is likely involved in DNA replication and is detectable in most human tissues and cell lines of human origin, with higher expression in rapidly proliferating cells and in tumors. NAP1L1 and NAP1L4 were also proposed to regulate cellular transcription. Both proteins are mostly present in the cytoplasm but are also detectable in the nucleus. Their shuttling appears to be determined by the encoded nuclear export and nuclear import signals (NES and NLS, respectively) and probably by their phosphorylation status. The mechanism of NAP1 function in viral replication remains incompletely understood, but CHIKV is not the only virus that hijacks NAP1 proteins for its replication. Previously, NAP1L1 was shown to bind to the disordered fragment of NS5A of HCV, but the biological significance of this interaction remains to be determined (42). In the case of CHIKV infection, NAP1L1/4-HVD interactions have proviral effects and likely act directly at the level of viral RNA replication. In our experiments, only fractions of cytoplasmic NAP1L1 and NAP1L4, but not their entire pools, were found in nsP3 complexes (Fig. 1). However, we cannot rule out a possibility that in the infected cells, NAP1 proteins may additionally interact with the fraction of nsP3 that is also diffusely distributed in the cytoplasm outside the large complexes (53).
An important characteristic of CHIKV HVD, and likely HVDs of other alphaviruses, is that it binds host factors (other than G3BPs and BIN1) with low affinities (21, 23–25). The binding sites are also either relatively long or more than one binding motif is required for interaction with either host protein. The identified FHL1-interacting peptide contains 47 aa (23). CD2AP binds simultaneously to two motifs through its SH3-binding domains (24). Moreover, the CD2AP- and FHL1-specific sites overlap, but at least in vitro, these proteins are capable of binding to CHIKV HVD simultaneously (23). In this new study, NAP1 function was found to require two HVD motifs, separated by G3BP-binding sites, to express its stimulatory function in viral replication. Moreover, in contrast to other HVD-interacting host factors, NAP1 proteins bind only to the phosphorylated form of the HVD. Phosphorylation of CHIKV HVD by CK2 kinase is required for its binding of NAP1 proteins in vitro, and inhibition of CK2 abrogates NAP1/HVD interaction and reduces viral replication. Importantly, it has been demonstrated for Mayaro virus that inhibition of CK2α could also strongly reduce viral replication (54). The requirement for HVD phosphorylation for interaction with other host partners is not so obvious but might also result in higher affinities of protein binding, and this provides a plausible explanation for the HVD phosphorylation that was described for all known alphaviruses (11, 13, 34–41). Taken together, the accumulating data strongly suggest an additional level of the viral life cycle regulation by phosphorylation of HVD, which leads to differential interactions with host proteins. It would be important to reevaluate the effect of HVD phosphorylation on interactions with other host factors and identify additional kinases involved in this posttranslational modification of HVD.
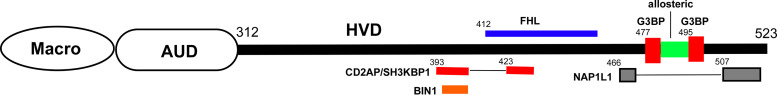
A map of the interacting motifs in CHIKV HVD that summarizes the currently available data is shown in Fig. 13. It is clear that the C-terminal half of HVD interacts with all of the host factors studied to date. Thus, after binding of such complex combination of cellular proteins, the C-terminal part of CHIKV HVD is likely not as flexible as it is in its free form and might adopt a new conformation. The function of the N-terminal fragment of CHIKV HVD remains unclear. No interacting partners have been determined for this fragment (fragment A), and it can be used for insertion of relatively large proteins, such as GFP or Cherry without strong negative effects on viral replication (16, 55). However, the 62-aa deletion in this fragment attenuates CHIKV replication both in vivo and in vitro (56). To date, the mechanism of attenuation remains unclear. It is possible that either the list of HVD-interacting partners remains incomplete or the distance between structured nsP3 domains and the above-described interacting motifs in HVD is important. However, other explanations, such as the essential role of phosphorylation of the latter fragment, are also possible. Taken together, the accumulated data suggest that we are only beginning to understand the complex structure and function(s) of HVD complexes. An additional complicating factor in understanding the HVD function comes from the analysis of adaptive mutations that are selected in response to strong negative effects of HVD-specific mutations on replication of CHIKV and other alphaviruses (13, 16, 51). In this study, the same adaptive mutation, D31A, was detected in in the macro domains of two different CHIKV HVD mutants (Fig. 4). It is in the same position as those we have previously described for the evolved VEEV mutants with strongly modified HVDs (13, 51). Function of the latter mutation in virus replication remains to be determined. However, since it also appears in response to modifications in the promoter elements in the 5′ untranslated region of the viral genome (57), it likely represents a universal response to dramatic decreases in viral RNA replication rather than a specific response to modifications in HVDs. In addition, D31 is located in the binding site of ADR-ribose in the alphavirus nsP3 macro domains (14, 58). Mutation of N24A in wt CHIKV nsP3, which inhibit ADR-ribose hydrolase activity of nsP3, requires additional mutation D31G for efficient viral replication (59). Thus, aa 31 appears to be involved in multiple functions of nsP3.
FIG 13.
Schematic presentation of the distribution of the binding sites of cellular factors on CHIKV HVD. Numbers indicate the positions of aa in CHIKV nsP3.
An additional adaptive mutation was identified in another structured domain of nsP3, the AUD (Fig. 4). Functions of AUD are even less understood, except that introduced mutations may affect RNA synthesis from the SG promoter (60). Both the macro domain- and AUD-specific mutations demonstrate plasticity of alphaviruses and their rapid evolution to higher replication rates and infection spread. However, they likely are of no benefit for the wt virus replication, which has already been optimized by previous evolution.
Taken together, our data demonstrate that (i) NAP1-HVD interaction has a strong stimulatory effect on CHIKV replication; (ii) both NAP1L1 and NAP1L4 interact with CHIKV HVD and likely have redundant functions; (iii) NAP1 family members interact with two peptides, which are located upstream and downstream of the G3BP-binding site of CHIKV HVD, but the NAP1-HVD interaction is independent of G3BP; (iv) NAP1 family members interact only with a phosphorylated form of CHIKV HVD, and this phosphorylation is mediated by CK2 kinase; (v) inhibitors of CK2 kinase could be explored as a therapeutic means against CHIK infection; and (vi) NAP1 and other families of host factors have additive stimulatory effects on CHIKV replication.
MATERIALS AND METHODS
Cell cultures.
The BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, MO). The NIH 3T3 and MRC-5 cells were obtained from the American Tissue Culture Collection (Manassas, VA). MRC-5 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS). Other cell lines were maintained in alpha minimum essential medium supplemented with 10% FBS and vitamins.
Plasmid constructs.
Plasmids encoding infectious cDNA of CHIKV 181/25 containing GFP under the control of viral subgenomic promoter (CHIKV/GFP) and its CHIKV/(3C)/GFP derivative have been described elsewhere (61). Their HVD-coding sequences were modified by using gene blocks from Integrated DNA Technologies (IDT) to replace the original nucleotide sequences. The presence of correct sets of mutations was verified by sequencing HVD-coding sequences in the final constructs. Plasmids encoding VEEV replicons with mutated CHIKV HVDs fused with Flag-GFP under the control of the subgenomic promoter have been previously described (17).
Rescuing of the viruses.
Plasmids encoding cDNAs of viral genomes with either wt or mutated HVDs (see the figures for details) were purified by ultracentrifugation in CsCl gradients. DNAs were linearized using NotI restriction site located downstream of the poly(A) tail. RNAs were synthesized in vitro by SP6 RNA polymerase (New England Biolabs) in the presence of a cap analog (New England Biolabs) under the conditions recommended by the manufacturer. The qualities of the RNAs were evaluated by electrophoresis in nondenaturing agarose gels, and equal amounts of RNAs were used for electroporation without additional purification. Electroporations of BHK-21 cells by the synthesized RNAs were performed under previously described conditions (62, 63). Viruses were harvested at 24 h postelectroporation, and titers were determined by a plaque assay on BHK-21 cells (64). To rule out a possibility that the designed CHIKV/GFP mutants require adaptive mutations for their viability, we used an infectious center assay (ICA) to evaluate RNA infectivity. The equal numbers of electroporated cells were seeded on the subconfluent monolayers of naive BHK-21 cells in 6-well Costar plates. After cell attachment, monolayers were covered by 0.5% agarose, supplemented with DMEM and 3% FBS. After ∼60 h of incubation at 37°C, the cells were fixed with 4% paraformaldehyde (PFA), and plaques were stained with crystal violet.
In some experiments, the release of infectious virus was analyzed directly after RNA electroporation. Aliquots of the media were harvested at the time points indicated in the figures, and infectious titers were determined by plaque assay on BHK-21 cells.
Packaging of VEEV replicons.
The in vitro-synthesized RNAs of VEEV replicons and helper, which encodes VEEV TC-83-specific structural genes under the control of the subgenomic promoter, were electroporated into BHK-21 cells as described above for viral genomes. Packaged replicons were harvested at ∼24 h postelectroporation. Titers were determined in infectious units (inf.u) per ml by infecting BHK-21 cells in 6-well Costar plates (5 × 105 per well) with different dilutions of the harvested stocks, and the numbers of GFP-positive cells were evaluated at 6 h p.i.
Viral infections.
For analysis of viral replication, equal numbers of cells indicated in the figures were seeded into the wells of the 6-well Costar plates. After 4 h of attachment at 37°C, cells were infected at the indicated MOIs in phosphate-buffered saline (PBS) supplemented with 1% FBS. After incubation for 1 h at 37°C, the cells were washed with PBS and then incubated in corresponding complete media. At the times indicated in the figures, media were replaced, and viral titers were determined by plaque assay on BHK-21 cells.
Immunostaining.
Cells were seeded into µ-Slide 8-well plates (Ibidi) (2 × 104 cells/well) and infected at MOIs indicated in the figure legends. In some experiments infected cells were treated with CK2 inhibitor, DRB. At the indicated times p.i., cells were fixed with 4% PFA, permeabilized, and stained with specific primary Abs and corresponding fluorescent secondary Abs. Images were acquired on a Zeiss LSM 800 confocal microscope in Airyscan mode with a 63 × 1.4 NA PlanApochromat oil objective. The following antibodies were used: anti-NAP1L1 rabbit polyclonal antibodies (14898-1; Proteintech), anti-NAP1L4 rabbit polyclonal antibodies (16018-1; Proteintech), anti-G3BP1 rabbit polyclonal antibodies (a gift from Richard Lloyd), and anti-CHIKV nsP3 (MAB2.19; custom).
Immunoprecipitations.
NIH 3T3 cells and their G3bp dKO derivatives were infected with packaged VEEV replicons expressing Flag-GFP fused with different variants of CHIKV HVD at MOIs of 10 or 20 inf.u/cell. Cells were harvested at 3 to 4 h p.i., when GFP expression began to be detectable under fluorescence microscope. Protein complexes were isolated from the postnuclear fraction of the NP-40-lysed cells using magnetic beads loaded with Flag-specific MAb (Sigma, St. Louis, MO), as described elsewhere (17). Presence of proteins of interest in the isolated complexes was analyzed by Western blotting using specific Abs and corresponding secondary Abs labeled with infrared dyes. Membranes were scanned on a Li-Cor imager. The following antibodies were used: anti-NAP1L1 rabbit polyclonal antibodies (14898-1; Proteintech), anti-NAP1L4 rabbit polyclonal antibodies (16018-1; Proteintech), and anti-G3BP2 rabbit polyclonal antibodies (HPA018304; Sigma).
Purification of recombinant proteins.
Nucleotide sequence of CHIKV HVD (aa 325 to 523) was codon-optimized for expression in E. coli, and His tag coding sequence was added to the N terminus. The synthetic DNA fragment was obtained from Integrated DNA Technologies. It was cloned into pE-SUMOpro-3 plasmid (LifeSensors, Inc.) between NcoI and XhoI restriction sites. Plasmid encoding His-HVDchikv (termed HVD) was transformed into E. coli LOBSTR-BL21(DE3) RIL (Kerafast), and protein was produced in the M9 media. The expression was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) after the cells reached optical density at 600 nm (OD600) of ∼2. The cells then continued to grow at 37°C for 3 to 5 h. Freshly prepared or frozen cell pellets were lysed in Emulsiflex B15 (Avestin). The lysates were loaded on HisTrap HP column (GE Healthcare) and, after extensive washing, the recombinant protein was eluted by imidazole gradient. Fractions containing HVD were combined, diluted to contain 25 mM NaCl, and further purified on a Resource Q column (GE Healthcare). Size exclusion chromatography on a Superdex 75 10/300 column (GE Healthcare) was used as a final purification step.
Nucleotide sequences of NAP1L1 (aa 69 to 391 of isoform 1, NP_001317160.1) and NAP1L4 (aa 2 to 375 of isoform 1, NP_001356304.1) were codon optimized for the expression in E. coli. The synthetic DNA fragments were cloned into pE-SUMOpro-3 plasmid (LifeSensors Inc.) between KifI and XhoI sites. The resulting proteins contained His tags at the N termini (MGHHHHHHG for NAP1L1 and MGHHHHHHGSR for NAP1L4). These plasmids were transformed into E. coli BL21 Star(DE3) (Thermo Fisher), and proteins were produced in M9 media. Protein expression was induced by 1 mM IPTG after the cells reached an OD600 of 1 to 1.5. Then cells continued to grow at 18°C for 18 h. Freshly prepared or frozen pellets were lysed in Emulsiflex B15 (Avestin). The recombinant proteins were first purified on a HisTrap HP column (GE Healthcare), followed by buffer exchange using HiPrep 26/10 desalting column (GE Healthcare).
Protein purities and identities were confirmed by SDS-PAGE and mass spectrometry, respectively. In all proteins, the first methionine was present in less than 50% of purified products. Protein concentrations were determined at 280 nm using extinction coefficients, which were determined by ProteinCalculator v3.4 (http://protcalc.sourceforge.net/).
Analysis of protein interaction by size exclusion chromatography.
All size exclusion chromatography (SEC) experiments were performed on the Superdex 200 increase 10/300GL column (GE Healthcare) in 25 mM Na-phosphate buffer [pH 6.8 with 100 mM NaCl, 1 mM tris(hydroxypropyl)phosphine (TCEP)]. For complex formation samples were mixed in equal molar ratios and incubated for 15 to 20 min at room temperature. Then, 50-µl aliquots were injected into the SEC column, and 400-µl aliquots were collected and analyzed by 14% SDS-PAGE.
HVD phosphorylation.
Samples of purified CHIKV HVD were diluted to 100 µM in the buffer containing 25 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM ATP, and 1 mM TCEP. Reactions were initiated by adding the recombinant CK2α to ∼10 µg/ml. Aliquots were collected at the time points indicated in the figure. Samples were analyzed by Western blotting with Abs specific to His tag (catalog no. 66005-1-Ig; Proteintech) and Phospho-CK2 substrate [(pS/pT)DXE] MultiMab (catalog no. 8738; Cell Signaling technology) and corresponding secondary Abs labeled with infrared dyes. Membranes were scanned on a Li-Cor imager. The in vitro-phosphorylated CHIKV HVD was also submitted to MSBioworks (MSB-service) for identification of phosphorylation sites.
Analyses of CHIKV HVD phosphorylation by 31P NMR.
The 1D 31P experiment was performed on a Bruker Avance III spectrometer operating at 600.13 MHz for 1H and 242.94 MHz for 31P nuclei equipped with a SMART probe. The spectrum was recorded with acquisition times (AQ) of 1.35 s, a relaxation delay (D1) of 2 s, and 20,480 scans. 1H decoupling during D1 and AQ was applied. The sweep width was 50 ppm with the carrier at 0.0 ppm.
The 2D 1H-31P correlation spectrum was acquired on a Bruker Avance III spectrometer operating at 600.18 MHz for 1H and 242.96 MHz for 31P nuclei equipped with a cryo-enhanced QCI-P probe. The spectrum was recorded with acquisition times of 170.4 and 10.1 ms in (t2,t1) in 1,024 × 64 complex matrices. The sweep widths were 10 and 26 ppm, with the carriers at 4.68 and 0.0 ppm, respectively, for 1H and 31P. A D1 of 1.5 s was employed, and 256 scans were collected. DSS (0.1 mM) was added as a reference for chemical shift. The control 1H 1D spectrum was collected with an AQ of 1.70 s, a D1 of 1 s, and 16 scans. The sweep width was 16 ppm with the carrier at 4.68 ppm.
All pulse sequences were taken from the Bruker standard library: zgpg30, zgesgp, and na_xhcoctetf3gp for 1D 31P and 1H experiments and 2D 1H-31P experiments, respectively. Spectra were recorded at 25°C. All NMR spectra were processed by TopSpin 4.0.6 software.
Analyses of CHIV HVD phosphorylation by native mass spectrometry.
Masses of the intact unphosphorylated and phosphorylated CHIKV HVDs were determined using ESI-ToF mass spectrometry [Waters Synapt G2-S(i)]. A total of 60 pmol of the proteins were desalted using a Waters BEH SEC column (200 Å, 1.7 µm, 2.1 mm × 150 mm) equilibrated in 0.1% formic acid in H2O. Spectra were recorded using MassLynx 4.1 in positive-ion resolution mode using glu-fib as a lockmass standard. Masses were determined from the acquired spectra using the Waters MaxEnt1 algorithm of MassLynx.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grants R01AI133159 and R01AI118867 to E.I.F. and R21AI146969 to I.F. and by Swedish Foundation for Strategic Research grant ITM17-0218 to P.A.
Contributor Information
Ilya Frolov, Email: ivfrolov@uab.edu.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol Rev 58:491–562. 10.1128/MR.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McSweegan E, Weaver SC, Lecuit M, Frieman M, Morrison TE, Hrynkow S. 2015. The Global Virus Network: challenging chikungunya. Antiviral Res 120:147–152. 10.1016/j.antiviral.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver SC, Forrester NL. 2015. Chikungunya: evolutionary history and recent epidemic spread. Antiviral Res 120:32–39. 10.1016/j.antiviral.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Weaver SC, Lecuit M. 2015. Chikungunya virus infections. N Engl J Med 373:94–95. 10.1056/NEJMc1505501. [DOI] [PubMed] [Google Scholar]
- 5.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3:e201. 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss EG, Rice CM, Strauss JH. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92–110. 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 7.Frolov I, Frolova EI. 2019. Molecular virology of chikungunya virus. Curr Top Microbiol Immunol 10.1007/82_2018_146. [DOI] [PubMed] [Google Scholar]
- 8.Lemm JA, Bergqvist A, Read CM, Rice CM. 1998. Template-dependent initiation of Sindbis virus replication in vitro. J Virol 72:6546–6553. 10.1128/JVI.72.8.6546-6553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and nonstructural protein functions. J Gen Virol 96:2483–2500. 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaStarza MW, Lemm JA, Rice CM. 1994. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J Virol 68:5781–5791. 10.1128/JVI.68.9.5781-5791.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lastarza MW, Grakoui A, Rice CM. 1994. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology 202:224–232. 10.1006/viro.1994.1338. [DOI] [PubMed] [Google Scholar]
- 12.Wang YF, Sawicki SG, Sawicki DL. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J Virol 68:6466–6475. 10.1128/JVI.68.10.6466-6475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foy NJ, Akhrymuk M, Shustov AV, Frolova EI, Frolov I. 2013. Hypervariable domain of nonstructural protein nsP3 of Venezuelan equine encephalitis virus determines cell-specific mode of virus replication. J Virol 87:7569–7584. 10.1128/JVI.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malet H, Coutard B, Jamal S, Dutartre H, Papageorgiou N, Neuvonen M, Ahola T, Forrester N, Gould EA, Lafitte D, Ferron F, Lescar J, Gorbalenya AE, de Lamballerie X, Canard B. 2009. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol 83:6534–6545. 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin G, Yost SA, Miller MT, Elrod EJ, Grakoui A, Marcotrigiano J. 2012. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci U S A 109:16534–16539. 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foy NJ, Akhrymuk M, Akhrymuk I, Atasheva S, Bopda-Waffo A, Frolov I, Frolova EI. 2013. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J Virol 87:1997–2010. 10.1128/JVI.02853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, Frolova EI. 2016. New World and Old World alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog 12:e1005810. 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolov I, Kim DY, Akhrymuk M, Mobley JA, Frolova EI. 2017. Hypervariable domain of eastern equine encephalitis virus nsP3 redundantly utilizes multiple cellular proteins for replication complex assembly. J Virol 91:e00371-17. 10.1128/JVI.00371-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meshram CD, Agback P, Shiliaev N, Urakova N, Mobley JA, Agback T, Frolova EI, Frolov I. 2018. Multiple host factors interact with the hypervariable domain of chikungunya virus nsP3 and determine viral replication in cell-specific mode. J Virol 92:e00838-18. 10.1128/JVI.00838-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panas MD, Ahola T, McInerney GM. 2014. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J Virol 88:5888–5893. 10.1128/JVI.00439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neuvonen M, Kazlauskas A, Martikainen M, Hinkkanen A, Ahola T, Saksela K. 2011. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog 7:e1002383. 10.1371/journal.ppat.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutso M, Morro AM, Smedberg C, Kasvandik S, Aquilimeba M, Teppor M, Tarve L, Lulla A, Lulla V, Saul S, Thaa B, McInerney GM, Merits A, Varjak M. 2018. Mutation of CD2AP and SH3KBP1 binding motif in alphavirus nsP3 hypervariable domain results in attenuated virus. Viruses 10:226. 10.3390/v10050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukash T, Agback T, Dominguez F, Shiliaev N, Meshram C, Frolova EI, Agback P, Frolov I. 2020. Structural and functional characterization of host FHL1 protein interaction with hypervariable domain of chikungunya virus nsP3 protein. J Virol 95:e01672-20. 10.1128/JVI.01672-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agback P, Dominguez F, Pustovalova Y, Lukash T, Shiliaev N, Orekhov VY, Frolov I, Agback T, Frolova EI. 2019. Structural characterization and biological function of bivalent binding of CD2AP to intrinsically disordered domain of chikungunya virus nsP3 protein. Virology 537:130–142. 10.1016/j.virol.2019.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte T, Liu L, Panas MD, Thaa B, Dickson N, Gotte B, Achour A, McInerney GM. 2016. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol 6:160078. 10.1098/rsob.160078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholte FE, Tas A, Albulescu IC, Zusinaite E, Merits A, Snijder EJ, van Hemert MJ. 2015. Stress granule components G3BP1 and G3BP2 play a proviral role early in chikungunya virus replication. J Virol 89:4457–4469. 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goertz GP, Lingemann M, Geertsema C, Abma-Henkens MHC, Vogels CBF, Koenraadt CJM, van Oers MM, Pijlman GP. 2018. Conserved motifs in the hypervariable domain of chikungunya virus nsP3 required for transmission by Aedes aegypti mosquitoes. PLoS Negl Trop Dis 12:e0006958. 10.1371/journal.pntd.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park YJ, Luger K. 2006. Structure and function of nucleosome assembly proteins. Biochem Cell Biol 84:549–558. 10.1139/o06-088. [DOI] [PubMed] [Google Scholar]
- 29.Attia M, Rachez C, Avner P, Rogner UC. 2013. Nucleosome assembly proteins and their interacting proteins in neuronal differentiation. Arch Biochem Biophys 534:20–26. 10.1016/j.abb.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Zlatanova J, Seebart C, Tomschik M. 2007. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J 21:1294–1310. 10.1096/fj.06-7199rev. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka T, Hozumi Y, Martelli AM, Iino M, Goto K. 2019. Nucleosome assembly proteins NAP1L1 and NAP1L4 modulate p53 acetylation to regulate cell fate. Biochim Biophys Acta Mol Cell Res 1866:118560. 10.1016/j.bbamcr.2019.118560. [DOI] [PubMed] [Google Scholar]
- 32.Lankenau S, Barnickel T, Marhold J, Lyko F, Mechler BM, Lankenau DH. 2003. Knockout targeting of the Drosophila nap1 gene and examination of DNA repair tracts in the recombination products. Genetics 163:611–623. 10.1093/genetics/163.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, Zhu Y, Dong A, Shen WH. 2015. Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development. Plant J 83:78–95. 10.1111/tpj.12830. [DOI] [PubMed] [Google Scholar]
- 34.Vihinen H, Ahola T, Tuittila M, Merits A, Kaariainen L. 2001. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J Biol Chem 276:5745–5752. 10.1074/jbc.M006077200. [DOI] [PubMed] [Google Scholar]
- 35.Mazzon M, Castro C, Thaa B, Liu L, Mutso M, Liu X, Mahalingam S, Griffin JL, Marsh M, McInerney GM. 2018. Alphavirus-induced hyperactivation of PI3K/AKT directs pro-viral metabolic changes. PLoS Pathog 14:e1006835. 10.1371/journal.ppat.1006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakovic A, Bhalla N, Kortchak S, Sun C, Zhou W, Ahmed A, Risner K, Klimstra WB, Narayanan A. 2020. Venezuelan equine encephalitis virus nsP3 phosphorylation can be mediated by IKKβ kinase activity and abrogation of phosphorylation inhibits negative-strand synthesis. Viruses 12:1021. 10.3390/v12091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teppor M, Zusinaite E, Merits A. 2021. Phosphorylation sites in the hypervariable domain in chikungunya virus nsP3 are crucial for viral replication. J Virol 95:e02276-20. 10.1128/JVI.02276-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li GP, La Starza MW, Hardy WR, Strauss JH, Rice CM. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416–427. 10.1016/0042-6822(90)90310-n. [DOI] [PubMed] [Google Scholar]
- 39.Peranen J, Takkinen K, Kalkkinen N, Kaariainen L. 1988. Semliki Forest virus-specific nonstructural protein nsP3 is a phosphoprotein. J Gen Virol 69:2165–2178. 10.1099/0022-1317-69-9-2165. [DOI] [PubMed] [Google Scholar]
- 40.De I, Fata-Hartley C, Sawicki SG, Sawicki DL. 2003. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J Virol 77:13106–13116. 10.1128/jvi.77.24.13106-13116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vihinen H, Saarinen J. 2000. Phosphorylation site analysis of Semliki Forest virus nonstructural protein 3. J Biol Chem 275:27775–27783. 10.1074/jbc.M002195200. [DOI] [PubMed] [Google Scholar]
- 42.Goonawardane N, Gebhardt A, Bartlett C, Pichlmair A, Harris M. 2017. Phosphorylation of Serine 225 in hepatitis C virus NS5A regulates protein-protein interactions. J Virol 91:e00805-17. 10.1128/JVI.00805-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. 2004. Prediction of posttranslational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649. 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 44.Park YJ, McBryant SJ, Luger K. 2008. A beta-hairpin comprising the nuclear localization sequence sustains the self-associated states of nucleosome assembly protein 1. J Mol Biol 375:1076–1085. 10.1016/j.jmb.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy D, Bhattacharya S, Jani V, Sonavane U, Joshi R, Gupta S. 2018. Biochemical and biophysical characterization of higher oligomeric structure of rat nucleosome assembly protein 1. Protein J 37:58–69. 10.1007/s10930-017-9751-9. [DOI] [PubMed] [Google Scholar]
- 46.Tsang B, Pritisanac I, Scherer SW, Moses AM, Forman-Kay JD. 2020. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 183:1742–1756. 10.1016/j.cell.2020.11.050. [DOI] [PubMed] [Google Scholar]
- 47.Bugge K, Brakti I, Fernandes CB, Dreier JE, Lundsgaard JE, Olsen JG, Skriver K, Kragelund BB. 2020. Interactions by disorder: a matter of context. Front Mol Biosci 7:110. 10.3389/fmolb.2020.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright PE, Dyson HJ. 2015. Intrinsically disordered proteins in cellular signaling and regulation. Nat Rev Mol Cell Biol 16:18–29. 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babu MM. 2016. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans 44:1185–1200. 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agback P, Shernyukov A, Dominguez F, Agback T, Frolova EI. 2020. Novel NMR assignment strategy reveals structural heterogeneity in solution of the nsP3 HVD domain of Venezuelan equine encephalitis virus. Molecules 25:5824. 10.3390/molecules25245824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meshram CD, Phillips AT, Lukash T, Shiliaev N, Frolova EI, Frolov I. 2020. Mutations in hypervariable domain of Venezuelan equine encephalitis virus nsP3 protein differentially affect viral replication. J Virol 94:e00617-20. 10.1128/JVI.00617-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meshram CD, Shiliaev N, Frolova EI, Frolov I. 2020. Hypervariable domain of nsP3 of eastern equine encephalitis virus is a critical determinant of viral virulence. J Virol 94:e00617-20. 10.1128/JVI.00617-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 82:10088–10101. 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barroso MM, Lima CS, Silva-Neto MA, Da Poian AT. 2002. Mayaro virus infection cycle relies on casein kinase 2 activity. Biochem Biophys Res Commun 296:1334–1339. 10.1016/s0006-291x(02)02093-4. [DOI] [PubMed] [Google Scholar]
- 55.Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol 80:4122–4134. 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallengard D, Kakoulidou M, Lulla A, Kummerer BM, Johansson DX, Mutso M, Lulla V, Fazakerley JK, Roques P, Le Grand R, Merits A, Liljestrom P. 2014. Novel attenuated chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol 88:2858–2866. 10.1128/JVI.03453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michel G, Petrakova O, Atasheva S, Frolov I. 2007. Adaptation of Venezuelan equine encephalitis virus lacking 51-nt conserved sequence element to replication in mammalian and mosquito cells. Virology 362:475–487. 10.1016/j.virol.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eckei L, Krieg S, Butepage M, Lehmann A, Gross A, Lippok B, Grimm AR, Kummerer BM, Rossetti G, Luscher B, Verheugd P. 2017. The conserved macrodomains of the nonstructural proteins of chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci Rep 7:41746. 10.1038/srep41746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meshram CD, Lukash T, Phillips AT, Akhrymuk I, Frolova EI, Frolov I. 2019. Lack of nsP2-specific nuclear functions attenuates chikungunya virus replication both in vitro and in vivo. Virology 534:14–24. 10.1016/j.virol.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao Y, Goonawardane N, Ward J, Tuplin A, Harris M. 2019. Multiple roles of the nonstructural protein 3 (nsP3) alphavirus unique domain (AUD) during chikungunya virus genome replication and transcription. PLoS Pathog 15:e1007239. 10.1371/journal.ppat.1007239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reynaud JM, Kim DY, Atasheva S, Rasalouskaya A, White JP, Diamond MS, Weaver SC, Frolova EI, Frolov I. 2015. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog 11:e1004863. 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liljeström P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol 65:4107–4113. 10.1128/JVI.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorchakov R, Hardy R, Rice CM, Frolov I. 2004. Selection of functional 5′ cis-acting elements promoting efficient Sindbis virus genome replication. J Virol 78:61–75. 10.1128/jvi.78.1.61-75.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemm JA, Durbin RK, Stollar V, Rice CM. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol 64:3001–3011. 10.1128/JVI.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]