ABSTRACT
NKG2C is an activating NK cell receptor encoded by a gene having an unexpressed deletion variant. Cytomegalovirus (CMV) infection expands a population of NKG2C+ NK cells with adaptive-like properties. Previous reports found that carriage of the deleted NKG2C− variant was more frequent in people living with HIV (PLWH) than in HIV− controls unexposed to HIV. The frequency of NKG2C+ NK cells positively correlated with HIV viral load (VL) in some studies and negatively correlated with VL in others. Here, we investigated the link between NKG2C genotype and HIV susceptibility and VL set point in PLWH. NKG2C genotyping was performed on 434 PLWH and 157 HIV-exposed seronegative (HESN) subjects. Comparison of the distributions of the three possible NKG2C genotypes in these populations revealed that the frequencies of NKG2C+/+ and NKG2C+/− carriers did not differ significantly between PLWH and HESN subjects, while that of NKG2C−/− carriers was higher in PLWH than in HESN subjects, in which none were found (P = 0.03, χ2 test). We were unable to replicate that carriage of at least 1 NKG2C− allele was more frequent in PLWH. Information on the pretreatment VL set point was available for 160 NKG2C+/+, 83 NKG2C+/−, and 6 NKG2C−/− PLWH. HIV VL set points were similar between NKG2C genotypes. The frequency of NKG2C+ CD3− CD14− CD19− CD56dim NK cells and the mean fluorescence intensity (MFI) of NKG2C expression on NK cells were higher on cells from CMV+ PLWH who carried 2, versus 1, NKG2C+ alleles. We observed no correlations between VL set point and either the frequency or the MFI of NKG2C expression.
IMPORTANCE We compared NKG2C allele and genotype distributions in subjects who remained HIV uninfected despite multiple HIV exposures (HESN subjects) with those in the group PLWH. This allowed us to determine whether NKG2C genotype influenced susceptibility to HIV infection. The absence of the NKG2C−/− genotype among HESN subjects but not PLWH suggested that carriage of this genotype was associated with HIV susceptibility. We calculated the VL set point in a subset of 252 NKG2C-genotyped PLWH. We observed no between-group differences in the VL set point in carriers of the three possible NKG2C genotypes. No significant correlations were seen between the frequency or MFI of NKG2C expression on NK cells and VL set point in cytomegalovirus-coinfected PLWH. These findings suggested that adaptive NK cells played no role in establishing the in VL set point, a parameter that is a predictor of the rate of treatment-naive HIV disease progression.
KEYWORDS: adaptive NK cells, HIV exposed seronegative, HIV load set point, injection drug users, men who have sex with men, people living with HIV, human immunodeficiency virus
INTRODUCTION
Natural killer (NK) cells are cytotoxic lymphocytes that generate early immune responses to virus-infected and cancer cells (1). The activation state of NK cells is determined by the integration of signals received from activating and inhibitory receptors (2, 3). Among the types of receptors present on NK cells are the NKG2 receptors, which belong to the C-type lectin family. The genes encoding these receptors are located in the 12p13 region of chromosome 12, within the NK receptor complex (4, 5). The NKG2C activating receptor, like its inhibitory counterpart NKG2A, is expressed as a heterodimer with CD94 (6). The ligand for NKG2C and NKG2A is HLA-E, a nonclassical major histocompatibility complex class Ib (MHC-Ib) molecule, stabilized by peptides derived from classical MHC-I antigens and HLA-G (7, 8). HLA-E molecules complexed with epitopes from the human cytomegalovirus (CMV)-encoded viral protein UL40 leader sequences are ligands for NKG2C (9–12). Among CD56dim NK cells, NKG2C+ NK cells are typically NKG2A− (13, 14). The interaction of NKG2C with its ligands transmits signals that activate cells bearing this receptor (7, 15).
Although NK cells are traditionally thought to be part of the innate immune system, NKG2C+ NK cells, which often coexpress CD57, can undergo clonal expansion in response to CMV infection (9, 13, 16, 17). Because the expansion of NKG2C+ cells resembles that seen in adaptive immune responses, these NK cells are called adaptive NK cells. Expanded adaptive NK cells frequently lack the signaling proteins Ewing’s sarcoma’s/FLI-1 activated transcript-2 (EAT-2), spleen tyrosine kinase (SYK), and FcεRγ, as well as the transcription factor promyelocytic leukemia zinc finger (PLZF) (18, 19). This is due to DNA methylation-dependent epigenetic modifications, which distinguish adaptive from conventional NK cells. Adaptive NKG2C+ cells exhibit enhanced CD16-dependent cytokine secretion due to epigenetic remodeling of the gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) promoter regions (20–22).
Some individuals do not express NKG2C at the NK cell surface due to homozygous deletion of an ∼16-kb genomic region that includes the nkg2c gene (also called klrc2), which encodes NKG2C (23, 24). In several Caucasian populations and one Japanese and Tanzanian population each, the frequency (percentage) of the NKG2C deletion haplotype is close to 20%, with a homozygous deletion frequency of approximately 4% (22, 24–26). However, frequencies of the NKG2C deletion haplotype were found to be as low as 10.3% in Mexican mestizos and as high as 36.8% in West African populations from the Gambia and Guinea-Bissau (26–28). NK cells expressing NKG2C have been shown to play a role in the immune surveillance of CMV (17).
CMV infection also drives the expansion of NKG2C− NK cells in people who are NKG2C−/− (29). NKG2C− NK cells having an epigenetic footprint characteristic of NKG2C+ adaptive NK cells are observed in NKG2C−/− carriers (22). Comparisons of the phenotypes and functions of adaptive NK cells from NKG2C−/− with those from NKG2C+/+ and NKG2C+/− carriers found few differences, suggesting that the contribution of NKG2C to NK cell adaptation to CMV infection can be compensated for in NK cells from NKG2C−/− carriers in CMV-monoinfected as well as in HIV-CMV-coinfected subjects (22, 30). In adaptive NK cells from NKG2C−/− carriers, CD2 costimulation plays an important role in compensating for the absence of NKG2C in antibody-dependent responses (22).
HIV-CMV coinfection has been reported by many to drive the expansion of NKG2C+ NK cells over that seen in CMV-monoinfected persons (31, 32). Several studies have questioned whether NKG2C+ cells play a role in protection from HIV infection or in a slower disease course in those infected. Supporting a role for the NKG2C− variant in susceptibility to HIV infection was the observation that the percentage of carriers of an nkg2c− allele in either the homozygous or heterozygous form was higher in HIV-infected individuals than in HIV-uninfected individuals with no history of HIV exposure (33). Whether NKG2C+ NK cells play a role in HIV control is unclear. Thomas et al. showed that among HIV-infected persons, the proportion of individuals with a pretreatment plasma viral load (VL) of <30,000 copies/ml was higher in carriers of the NKG2C+/+ genotype than among those carrying an nkg2c− allele (33). Contrasting with the notion that the NKG2C+/+ genotype was associated with lower VL control was the finding that the percentage of NKG2C+ NK cells from seven NKG2C+/+ carriers was positively correlated with a single pretreatment HIV VL (33). However, in two other studies, the percentage of NKG2C+ NK cells was negatively correlated with VL in early infection (34, 35).
Here, we compared NKG2C genotypes in people living with HIV (PLWH) enrolled in the Montreal Primary HIV infection (PI) cohort with HIV-exposed seronegative (HESN) subjects who remained HIV uninfected despite multiple high-risk HIV exposures. We found that carriage of the NKG2C−/− genotype was associated with increased HIV susceptibility. However, neither the NKG2C+/+ nor NKG2C+/− genotype alone nor the combination of both NKG2C+/− and NKG2C−/− genotypes was associated with changes in HIV susceptibility. We observed no differences in VL set points between HIV-infected carriers of the three possible NKG2C genotypes. We also observed no correlation between VL set point and the percentage of NKG2C+ NK cells or the intensity of NKG2C expression. Thus, carriage of an nkg2c− allele does not appear to affect HIV VL set point, which is a determinant of the rate of HIV disease progression.
RESULTS
PLWH and HESN populations differ in NKG2C−/− genotype frequencies.
Table 1 provides information on the racial/ethnic composition of the study population. Both populations were composed mainly of Caucasians (92.9 and 88.5% for PLWH and HESN participants, respectively) living in the same geographical region (Montreal, QC, Canada). There were no significant between-group differences in their ethnic/racial compositions (P > 0.11 for comparisons of Caucasians, Asians, Latinos, and American/African Blacks by two-tailed χ2 tests).
TABLE 1.
Study population demographicsa
| Population | No. (%) in population |
P value | |
|---|---|---|---|
| PLWH (n = 434) | HIV− (n = 157) | ||
| Sex | |||
| Males | 408 (94.0) | 141 (89.8) | |
| Females | 25 (5.8) | 23 (14.6) | |
| HIV exposure risk group | |||
| Sexually exposed | 371 | 78 | |
| MSM | 337 | 67b | |
| Heterosexually exposed | |||
| Men | 9 | 3 | |
| Women | 25 | 8 | |
| IDUs | 63 | 79 | |
| Low-risk control | 11 | ||
| Ethnicity | |||
| Caucasian | 384 (88.5) | 146 (92.9) | 0.11 |
| American/African black | 19 (4.5) | 4 (2.5) | 0.25 |
| Latino | 27 (6.2) | 5 (3.2) | 0.19 |
| Asian | 4 (0.9) | 2 (1.3) | 0.76 |
Abbreviations: PLWH, people living with HIV; MSM, men who have sex with men; IDUs, injection drug users.
At risk for sexual exposure.
The numbers and percentages of PLWH and HESN subjects carrying the wild-type (nkg2c+) and deletion (nkg2c−) alleles and the three NKG2C genotypes are shown in Table 2. The allele percentages were similar in both populations. The distribution of the three NKG2C genotypes NKG2C+/+, NKG2C+/−, and NKG2C−/− at this locus did not deviate statistically from the Hardy-Weinberg equilibrium (HWE) in PLWH (P = 0.09 by χ2 test), while it did in the HESN subjects (P = 0.003 by χ2 test). When the proportions of NKG2C+/+, NKG2C+/−, and NKG2C−/− genotypes were compared in PLWH and HESN subjects, there was a significantly higher frequency of NKG2C−/− individuals among PLWH than HESN subjects (odds ratio [OR], 8.60; 95% confidence interval [CI], 0.50 to 146; P = 0.04 by two-tailed Fisher’s exact test), while the proportions of NKG2C+/+ and NKG2C+/− genotypes in these two populations did not differ significantly (Table 2). Thomas et al. previously reported that HIV-uninfected persons at low risk for infection were significantly more likely than PLWH to carry the NKG2C+/+ genotype, suggesting that carriage of at least 1 NKG2C− variant was associated with higher HIV susceptibility (33). Comparisons of the PLWH and HESN subjects revealed no between-population differences for either the NKG2C+/+ or combined NKG2C+/− NKG2C−/− genotypes. In summary, carriage of the NKG2C−/− genotype was associated with higher susceptibility to HIV infection.
TABLE 2.
NKG2C allele and genotype frequencies in people living with HIV and HIV-exposed seronegative subjectsa
| NKG2C allotype/genotype | Frequency of allele or genotype in population |
OR | 95% CI | P value | |
|---|---|---|---|---|---|
| PLWH | HESN | ||||
| All, n | 434 | 157 | |||
| Allele frequency, % | |||||
| nkg2c+ | 80.5 | 80.9 | 1.0 | 0.5–2.1 | 1.00 |
| nkg2c− | 19.5 | 19.1 | |||
| Genotype frequency, no. (%) | |||||
| NKG2C+/+ | 276 (63.6) | 97 (61.8) | 1.1 | 0.7–1.6 | 0.70 |
| NKG2C+/− | 147 (33.9) | 60 (38.2) | 0.8 | 0.6–1.2 | 0.38 |
| NKG2C−/− | 11 (2.5) | 0 | 8.6** | 0.5–146.0** | 0.04* |
| NKG2C+/− + NKG2C−/− | 158 (36.4) | 60 (38.2) | 0.9 | 0.6–1.5 | 0.70 |
| Sexually exposed, n | 371 | 78 | |||
| Allele frequency, % | |||||
| nkg2c+ | 81.4 | 78.2 | 1.21 | 0.6–2.4 | 0.72 |
| nkg2c− | 18.6 | 21.9 | |||
| Genotype frequency, no. (%) | |||||
| NKG2C+/+ | 240 (64.7) | 44 (56.4) | 1.4 | 0.9–2.3 | 0.20 |
| NKG2C+/− | 124 (33.4) | 34 (43.6) | 0.6 | 0.4–1.1 | 0.09 |
| NKG2C−/− | 7 (1.9) | 0 | 3.2** | 0.2–57.2** | 0.61* |
| IDUs, n | 63 | 79 | |||
| Allele frequency, % | |||||
| nkg2c+ | 75.4 | 83.5 | 0.6 | 0.3–1.2 | 0.21 |
| nkg2c− | 24.6 | 16.5 | |||
| Genotype frequency, no. (%) | |||||
| NKG2C+/+ | 36 (57.1) | 53 (67.1) | 0.6 | 0.3–1.3 | 0.29 |
| NKG2C+/− | 23 (36.5) | 26 (32.9) | 1.2 | 0.6–2.3 | 0.72 |
| NKG2C−/− | 4 (6.3) | 0 | 12.0** | 0.6–277.7** | 0.04* |
Abbreviations: PLWH, people living with HIV; HESN, HIV-exposed seronegative; OR, odds ratio; 95% CI, 95% confidence interval; IDUs, injection drug users. Asterisks indicate statistical significance measured by Fisher’s exact test (*) with Haldane’s correction (**).
Risks for HIV transmission include sexual exposure to and needle sharing with PLWH. As the PLWH and HESN populations included individuals who were at risk for sexual exposure, as well as injection drug users (IDUs), we investigated whether there was evidence that carriage of NKG2C−/− genotype was linked to HIV susceptibility by mucosal or parenteral exposure. Of the sexually exposed (SE) subjects, 371 were PLWH and 78 were HESN, with genotype distributions shown in Table 2. The NKG2C genotype distributions diverged from HWE in both the SE PLWH and HESN subjects (P = 0.045 and P = 0.001, respectively, by χ2 tests). When the proportions of NKG2C+/+, NKG2C+/−, and NKG2C−/− genotypes were compared in SE PLWH and HESN subjects, no significant between-group differences were observed. Of the 63 PLWH and 79 HESN IDUs evaluated for NKG2C genotypes, the genotype distribution in both populations was in HWE (P = 0.9 and P = 0.07, respectively, by χ2 tests). The proportion of NKG2C−/− genotypes was significantly higher among injection drug-using PLWH than HESN subjects (OR, 12; 95% CI, 0.6 to 277.7; P = 0.04 by Fisher’s exact test); the proportions of NKG2C+/+ and NKG2C+/− genotypes in the IDU PLWH and HESN subpopulations did not differ significantly from each other (Table 2). In summary, carriage of the NKG2C−/− genotype was significantly associated with higher HIV susceptibility in IDUs but not in SE subjects.
NKG2C cell surface expression is genotype dependent.
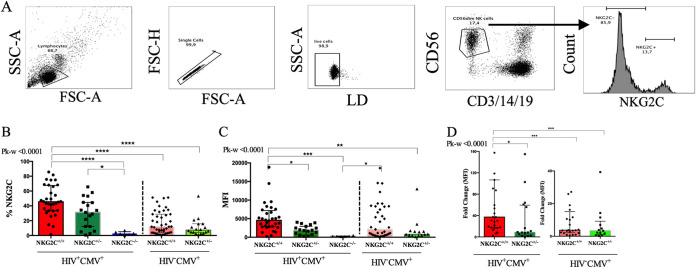
We next compared the percentages of NKG2C+ cells and the intensities of NKG2C expression on CD56dim NK cells from carriers of the three NKG2C genotypes. As CMV infection drives the expansion of NKG2C+ NK cells (9, 13, 31), for this analysis, we included only subjects who were CMV+ from whom cells were available for staining. Cells from 32 NKG2C+/+, 19 NKG2C+/−, and 6 NKG2C−/− PLWH and 43 NKG2C+/+and 18 NKG2C+/− HIV− subjects were tested. Figure 1A shows the strategy used to gate on live singlet CD3− CD14− CD19− CD56dim NK cells, which is the predominant population expressing NKG2C (36). From these, NKG2C+ cells were gated on. Figure 1B shows that CMV+ PLWH who were NKG2C+/+ and NKG2C+/− had a higher percentage of NKG2C+ NK cells than did NKG2C−/− carriers, with medians of 45.5% (interquartile range [IQR], 33.5 to 67.5%) and 30.1% (IQR, 7.41 to 44.63%) for NKG2C+/+ and NKG2C+/− carriers, respectively, and background levels of 2.7% (IQR, 1.04 to 5.14%) for NKG2C−/− carriers (P < 0.001 and P < 0.05 for comparisons of NKG2C+/+ and NKG2C+/− carriers with NKG2C−/− carriers by Dunn’s posttests). In a subanalysis comparing NKG2C+/+ with NKG2C+/− carriers, we found that the percentage of NKG2C+ CD56dim NK cells was significantly higher in CMV+ PLWH who were NKG2C+/+ than in NKG2C+/− carriers (P < 0.05 by Mann-Whitney test). For intensity measurements, we examined the mean fluorescence intensity (MFI), the median fluorescence intensity, and the fold change over background in the MFI of NKG2C staining. The latter measure controls for between-experiment variations in MFI. Since values for mean and median fluorescence intensities did not differ substantially, we only report MFI values here. The MFI of NKG2C expression from NKG2C+/+, NKG2C+/−, and NKG2C−/− carriers were 4,562 (IQR, 2,813 to 7,175), 1,870 (IQR, 671 to 3,061), and 269.6 (IQR, 212.8 to 325.3), respectively (Fig. 1C). NKG2C expression was higher on CD56dim NK cells from NKG2C+/+ carriers than on those from NKG2C−/− carriers (P < 0.001 by Dunn’s posttests). NK cells from NKG2C+/+ carriers expressed higher levels of NKG2C than those from NKG2C+/− carriers (P < 0.05 by Dunn’s posttest). The fold change in MFI over background for NKG2C expression intensity was also significantly higher on NK cells from NKG2C+/+ than NKG2C+/− CMV+ PLWH carriers (Fig. 1D). We also investigated the percentage of NKG2C+ CD56dim NK cells and the intensity of NKG2C expression on these cells from CMV-monoinfected subjects. Although the percentage of NKG2C+ NK cells was higher on cells from NKG2C+/+ than NKG2C+/− carriers (11.8% [IQR, 3.77 to 28.7%] and 7.23% [IQR, 4.08 to 15.75%], respectively), as were the MFI and fold change in MFI intensity of NKG2C expression on CD56dim NK cells, the difference did not achieve significance (Fig. 1B to D). Figure 1B D to D also show that the percentage of cells, MFI, and fold change in MFI over background of NKG2C expression were significantly higher among CD56dim NK cells from NKG2C+/+ carriers who were CMV+ PLWH than among NKG2C+/+ and NKG2C+/− cells from CMV-monoinfected persons (P < 0.006 for all by Mann-Whitney tests).
FIG 1.
Evaluation of the frequency of NKG2C+ NK cells and mean fluorescence intensity (MFI) of NKG2C expression. (A) Shown is the gating strategy used to detect the frequency and MFI of NKG2C expression. Peripheral blood mononuclear cells were stained for viability and cell surface CD3, CD56, CD14, CD19, and NKG2C. CD3− CD14− CD19− CD56dim NK cells were gated on from the live singlet lymphocyte gate. From these, we determined the frequencies of NKG2C+ CD56dim NK cells and MFI of NKG2C expression on NK cells. The y axes show the frequency (B), MFI (C), and fold change over background in the MFI (D) of NKG2C expression on CD56dim NK cells from CMV+ people living with HIV (HIV+ CMV+) carrying the NKG2C+/+ (n = 32), NKG2C+/− (n = 19), and NKG2C−/− (n = 6) genotypes and from CMV-monoinfected (HIV− CMV+) individuals carrying the NKG2C+/+ (n = 43) and NKG2C+/− (n = 18) genotypes. Each point represents a single individual. Bar graph heights and error bars represent medians and interquartile ranges for the group. FSC-A, forward scatter area; SSC-A, side scatter area; LD, live/dead; FSC-H, forward scatter height; Pk-w, P value for the Kruskal-Wallis test used to analyze the significance of differences between groups: *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
NKG2C genotypes and HIV VL set point.
VL set points in pretreatment PLWH are measures of HIV progression associated with time to AIDS, CD4 counts of <200 copies/ml of plasma, and death (37, 38). When all NKG2C-genotyped PLWH for whom information on the VL set point was available were included, we found no significant differences between NKG2C genotypes in the VL set points (P = 0.26 by Kruskal-Wallis test) (Fig. 2).
FIG 2.
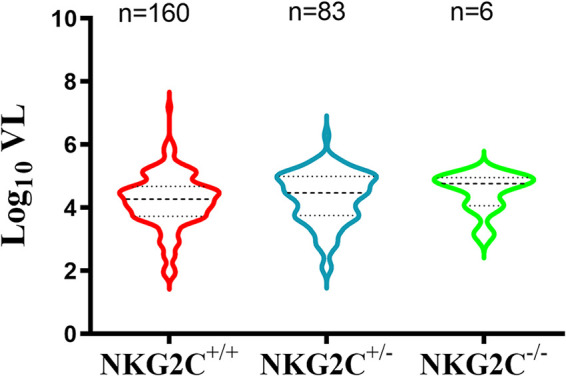
Log10 viral load (VL) set points in people living with HIV (PLWH) carriers of the NKG2C+/+, NKG2C+/−, and NKG2C−/− genotypes. Shown are violin plots of the median and interquartile range of the treatment-naive log10 VL set point in each NKG2C genotype group. The number of subjects included in each group is shown above each data set. A Kruskal-Wallis test was used to assess the significance of between-group differences in log10 VL set point.
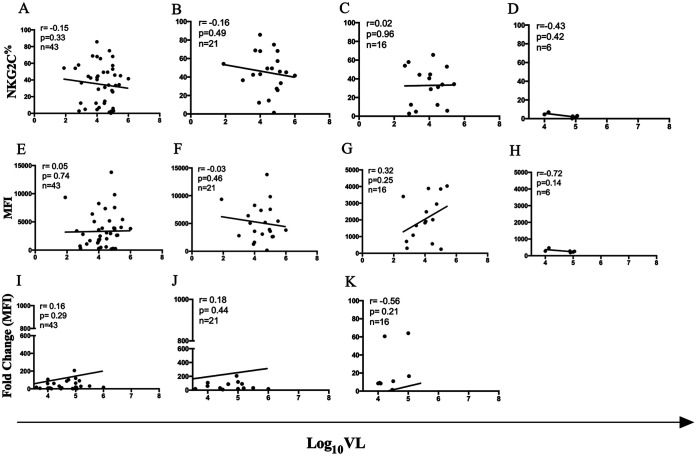
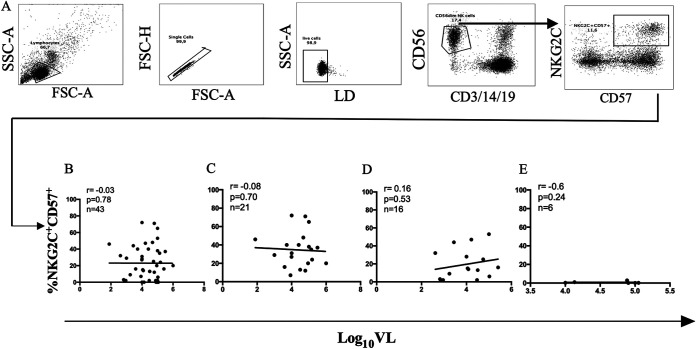
We next investigated whether the percentage of NKG2C+ CD56dim NK cells and/or the intensity of NKG2C expression correlated with the pretreatment VL set point. Forty-three NKG2C-genotyped CMV+ PLWH with a known HIV VL set point were included in this analysis: 21 NKG2C+/+, 16 NKG2C+/−, and 6 NKG2C−/− HIV+ CMV+ subjects. Neither the percentage nor the intensity of NKG2C expression (MFI or fold change over background in NKG2C MFI) was significantly correlated with VL set point when all observations were considered together or when results were stratified according to NKG2C genotype (Spearman’s correlation tests) (Fig. 3A to L) As adaptive NK cells are typically also CD57+, we also tested whether there was a correlation between the percentage of NKG2C+ CD57+ CD56dim NK cells and HIV VL set point. Figure 4A shows the strategy used to gate on NKG2C+ CD57+ CD56dim NK cells. No significant correlation was observed between these parameters for all NKG2C genotypes or for results stratified by NKG2C genotype (Spearman’s test) (Fig. 4B to E).
FIG 3.
Correlation between log10 VL set point and frequency of NKG2C+ NK cells, mean fluorescence intensity (MFI) of NKG2C expression and fold change in NKG2C MFI over background in cells from HIV+ CMV+ NKG2C+/+, NKG2C+/−, and NKG2C−/− carriers. Correlations between the frequency (A to D) MFI (E to H) of NKG2C expression and fold change in NKG2C MFI over background (I to K) on NK cells from CMV+ PLWH with log10 VL for carriers of all NKG2C genotypes tested (A, E, and I) and stratified by NKG2C+/+ (B, F, and J), NKG2C+/− (C, G, and K), and NKG2C−/− (D and H) genotypes. The number of subjects tested, the correlation coefficients (r), and the P values for each correlation are shown in the top left corner of the graphs.
FIG 4.
Correlation between log10 (VL) viral load set point and frequency of NKG2C+ CD57+ NK cells from CMV+ PLWH carrying the three possible NKG2C genotypes. (A) From the live singlet lymphocyte gate, CD56dim CD3− CD14− CD19− NK cells were gated on. From these NKG2C+ CD57+ NK cells were gated onto assess the frequency of these cells among CD56dim NK cells. Correlations between the frequency of NKG2C+ CD57+ (B to E) CD56dim NK cells with log10 viral load set point from for CMV+ PLWH carrying all NKG2C (B), NKG2C+/+ (C), NKG2C+/− (D), and NKG2C−/− (E) genotypes. The number of subjects tested, the correlation coefficients (r), and the P values for each correlation are shown in the top left corner of the graphs.
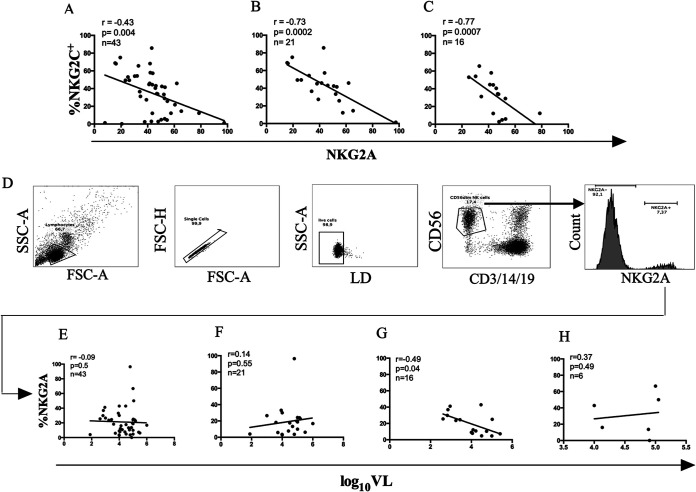
As others have shown, the percentage of NKG2C+ NK cells was significantly negatively correlated with the percentage of NKG2A+ NK cells. This was the case for all genotypes together and for the NKG2C+/+ and NKG2C+/− genotypes specifically (Fig. 5A). Figure 5B shows the strategy used to gate on NKG2A+ NKG2C− CD56dim NK cells. As for NKG2C+ and NKG2C+ CD57+ CD56dim NK cells, the percentage of NKG2A+ NKG2C− CD56dim NK cells did not correlate with the VL set point when results from all subjects were examined together or when results from NKG2C+/+ and NKG2C−/− carriers were examined separately. For NKG2C+/− carriers, a negative correlation was observed (r = −0.49, P = 0.04) (Fig. 5D). However, application of a Bonferroni correction for multiple correlations reduced the significance of the correlation below the level of significance.
FIG 5.
Correlation between log10 VL set point and frequency of NKG2A+ NKG2C− CD56dim NK cells from CMV+ PLWH carrying the three possible NKG2C genotypes. Correlations between the frequency of NKG2C+ CD56dim and NKG2A+ CD56dim NK cells from CMV+ PLWH for carriers of all NKG2C (A), NKG2C+/+ (B), and NKG2C+/− (C) genotypes. The number of subjects tested, the correlation coefficients (r) and the P values for each correlation are shown in the inset at the top left corner of each graph. (D) Shown is the strategy used to gate on CD56dim CD3− CD14− CD19− NK cells, from which NKG2A+ NKG2C− cells were gated onto assess their frequency among CD56dim NK cells. Correlations between the frequency of CD56dim NKG2A+ NKG2C− NK cells with log10 VL set point from for CMV+ PLWH carrying all NKG2C (E), NKG2C+/+ (F), NKG2C+/− (G), and NKG2C−/− (H) genotypes. The numbers of subjects tested, the correlation coefficients (r), and the P values for each correlation are shown in the top left corner of the graphs.
DISCUSSION
In this report, we assessed whether the NKG2C genotype distributions differed in a population of recently HIV-infected individuals compared to subjects who remained uninfected despite multiple documented exposures to HIV. We found that the NKG2C−/− genotype was more frequent among PLWH than HESN subjects. None of the 157 HESN subjects tested carried this genotype, which was present in 11 of 434 (2.53%) of PLWH. The distributions of NKG2C genotypes did not differ in the PLWH and HESN subpopulations who were exposed to HIV mucosally, while the NKG2C−/− genotype was more frequent in parenterally exposed PLWH than in HESN individuals. These findings suggest that the NKG2C−/− genotype is associated with a higher risk of HIV infection. The PLWH population included individuals who remained treatment naive long enough to calculate a post-acute infection, pretreatment plasma VL set point. When this parameter was compared in carriers of the three NKG2C genotypes, we found no between-genotype differences in VL set point. Furthermore, neither the percentage of NKG2C+ NK cells, MFI, nor fold change over background of the MFI of NKG2C expression on these cells correlated with VL set point in the CMV+ PLWH.
There exists a variation in chromosome 12 where a 16-kb genomic region that includes the nkg2c gene is either present or entirely absent (23, 24). Genotyping of the mainly Caucasian study population described in this article found that the frequency of the nkg2c− variant was close to 20% in both the PLWH and HESN populations and the frequency of the homozygous NKG2C−/− genotype was 2.53% in PLWH. The nkg2c− allele frequency and the distribution of NKG2C genotypes in the PLWH are in line with those reported for several populations of European extraction, as well as in a Japanese population and an East African Tanzanian population (22, 24–26, 33). The allele frequency of nkg2c− was lower (10.3%) in a population of Mexican mestizos and higher (29.3 to 36.7%) in West African populations from the Gambia and Guinea-Bissau (26–28) In contrast with what we found in PLWH, we observed no NKG2C−/− carriers among 157 HIV-uninfected persons at risk for HIV exposure, a difference that was statistically significant. The non-Caucasian ethnic composition of the study populations was balanced between PLWH and HESN subjects. However, if only Caucasians were included in the analysis, proportional between-group differences in the percentage of the NKG2C−/− genotype fell below the level of significance (P = 0.1). This may be due to the smaller sample sizes. It was not possible to compare the proportional between-group differences in the percentages of the NKG2C−/− genotype for the other ethnicities included in the study populations due to the small numbers of subjects in these subgroups.
The NKG2C genotype distributions in the PLWH and the uninfected population described here differed from those reported by Thomas et al. (33). They compared the NKG2C genotype distribution in 433 PLWH with that in 280 controls who had no history of HIV exposure (33). They found NKG2C−/− subjects among their HIV-uninfected population, while we did not. They reported a significant association between carriage of an nkg2c− allele (i.e., combined NKG2C+/− and NKG2C−/− carriers) with HIV infection and that there was a higher proportion of NKG2C+/+ carriers among uninfected controls than among PLWH. The main difference between the population reported by Thomas et al. and the one described here was the composition of the HIV-uninfected population. In the study by Thomas at al., the control population was not HIV exposed and thus was at a low risk for HIV infection. While it is possible that some of the people at high risk for HIV exposure we included remained HIV uninfected by chance, they represent a group that is likely to have a higher level of resistance to HIV infection than the HIV-uninfected population described by Thomas et al. The inclusion of HESN participants allowed us to explore more directly whether NKG2C genotypes were associated with HIV susceptibility. This may account for the discrepancy between our results and those reported by Thomas et al. regarding which NKG2C genotypes were associated with HIV susceptibility.
We stratified both PLWH and HESN subjects into those whose route of HIV infection/exposure was mucosal (SE) versus parenteral (IDU). When SE and IDU PLWH and HESN subjects were compared separately, we observed that the frequency of the NKG2C−/− genotype was significantly higher in the IDU PLWH than HESN subjects, while this frequency did not differ significantly between SE PLWH and those at risk for sexual exposure to HIV. Many factors influence the per-act risk of HIV transmission, including the VL of the transmitting partner, the route of exposure, the presence of genital ulcers, circumcision, and the frequency of exposure, among others (39, 40). The SE PLWH and high risk for HIV exposure subpopulations were mainly men who have sex with men (MSM). Of these, all reported unprotected receptive (where the receptive partner was HIV seronegative) anal intercourse. This route of exposure averages at least a 10-fold higher risk of transmission per act than unprotected insertive anal or vaginal intercourse and a per-act risk that is close to that of injection drug use (41–44).
What accounts for the frequencies of the NKG2C−/− genotype not differing significantly between SE PLWH and those at risk for mucosal HIV exposure is unknown. The level of exposure to HIV may be a factor if a higher proportion of HIV-transmitting partners of SE than IDU HESN populations are on antiretroviral treatment (ART). In the context of NKG2C+ cells, the biology of HIV transmission by injection versus sexual exposure may be a factor. Parenteral exposure involves the introduction of needles contaminated with HIV-infected cells and/or virions into the circulation. Transmitted HIV-infected cells will express HLA-E, the ligand for NKG2C, and downmodulate HLA-A, -B, and -C, the ligands for inhibitory killer immunoglobulin-like receptors also present on NKG2C+ cells (2, 3, 7, 8, 36). The integration of these signals promotes NKG2C+ NK cell activation that may contribute to HIV clearance prior to the establishment of a productive infection. In this setting, the absence of NKG2C+ cells in NKG2C−/− carriers may be linked to heightened HIV susceptibility in those who became infected. In the case of sexual exposure, HIV-infected cells or virions must cross mucosal barriers to access the NKG2C+ NK cells in the circulation. Our knowledge of NKG2C+ NK cells at mucosal genital/anal sites is limited. NK, tissue-resident NK (TrNK), and NK-like innate lymphoid cells are present in tissues, including in the female genital tract (45, 46). The NK receptor profile of these cells differs from that of circulating NK cells, making it challenging to evaluate their stage of maturity, their ability to interact with HIV-infected cells, and the consequences of such an interaction in the context of what is known about circulating NK cells. Whether NK-like cells at portals of HIV entry express NKG2C is unknown. A study of the transcriptomic and protein expression patterns of TrNK cells in lung mucosal tissue did not report expression of NKG2C, while this receptor was shown to be expressed on adaptive NK cells in the liver, although these NK cells had distinct NK cell receptor profiles from those in the circulation (46, 47). If NKG2C+ NK cells were absent at the portals of HIV entry, it would reduce the relevance of NKG2C genotypes in modulating infection risk through a mucosal route. In sum, more information on the NK cell landscape at mucosal portals of HIV entry would aid in understanding the discrepancy between the percentage of NKG2C−/− carriers in SE versus IDU PLWH and HESN subjects.
The reason underlying why none of the 157 HESN subjects carried the NKG2C−/− genotype and how this may contribute to the maintenance of seronegative status despite multiple HIV exposures are unknown. CMV infection drives the expansion of adaptive NK cells (17, 29, 48). It is notable that NKG2C− adaptive NK cells also expand in CMV-infected NKG2C−/− carriers (22, 36, 49, 50). Adaptive NKG2C− and NKG2C+ NK cells are found at similar frequencies in those who do not and those who do carry an nkg2c+ allele, and these cells share phenotypic, epigenetic, and functional properties that distinguish them from conventional NK cells (22, 30, 36, 49, 50). One of the differences between adaptive and conventional NK cells is that the former are more likely to express CD2. CD2 is a major coactivating receptor found on NK cells and T cell subsets, whose ligand is CD58 (LFA-3), which is expressed on many tissues (22, 51). CD2 is present on a higher percentage of adaptive NK cells from NKG2C−/− than NKG2C+ carriers (51, 52). It compensates for the absence of NKG2C on adaptive NK cells from NKG2C−/− carriers in a manner that contributes to the activation of these cells. Although signaling through CD2 alone has little effect on adaptive NK cell activation, it synergizes with CD16 signaling, to potently activate NK cells to secrete IFN-γ and TNF-α (22). It is tempting to speculate that CMV infection provides the costimulatory signals (i.e., CD16 cross-linking by anti-CMV antibody Fc regions and CD2-CD58 interactions) to activate adaptive NK cells. CMV has tropism for epithelial cells, fibroblasts, myeloid cells, and endothelial cells, all of which express CD58 and thus have the potential to be adaptive NK cell-interacting partners (53). CMV infection is a common infection, with a prevalence close to 40% in HIV-uninfected Canadians that increases with age (54–56). In ART-naive PLWH enrolled in the Montreal PI cohort, the prevalence of CMV coinfection is 84% (57). It would be interesting to investigate whether the higher frequency of the NKG2C−/− genotype in PLWH than in HESN subjects is linked to differential activation of these NKG2C− adaptive NK cells in PLWH than in HESN subjects due to factors such as differential levels of CMV infection or other factors that affect NK cell activity in a manner that influences HIV susceptibility.
We observed that the percentages of NKG2C+ NK cells in CMV+ PLWH and in CMV-monoinfected persons differed according to NKG2C genotype. CMV infection drives the expansion of NKG2C+ NK cells (9, 13, 31). This was the rationale for confining this analysis to PLWH and HIV-uninfected subjects who were CMV seropositive. Cell surface NKG2C percentage, MFI, and intensity of fold change over background in the MFI of NKG2C expression results reported by others did not test for CMV serostatus, which if negative, would preclude the expansion of NKG2C+ NK cells (33). In CMV-monoinfected subjects, differences in the percentages and intensities of NKG2C expression between NKG2C+/+ and NKG2C+/− carriers were not significant. However, these values in CMV+ PLWH compared to CMV-monoinfected persons were higher for cells from carriers of both NKG2C+/+ and NKG2C+/− genotypes, as has been seen by others (31, 32, 58).
Treatment-naive VL set point is associated with the rate of HIV disease progression, as measured by time to CD4 counts of <200/mm3, AIDS, and death (37, 38). We found no significant correlations between either the percentage of NKG2C+ CD56dim NK cells or the intensity of NKG2C expression on NK cells and the VL set point. This was also the case for correlations between the percentage of NKG2C+ CD57+ and NKG2A+ CD56dim NK cells and the VL set point. These results differ from those of others who correlated the percentage of NKG2C+ cells with single VL measures in ART-naive individuals. Thomas et al. found a positive correlation between these parameters, although their analysis only included 7 untreated subjects in the chronic phase of infection (33). In contrast, Ma et al. found a negative correlation between the percentage of NKG2C+ NK cells and concurrent VL in 22 treatment-naive PLWH infected at least 120 days, which corresponded to the VL set point (34). Gondois-Rey et al. also found a negative correlation between the percentage of NKG2C+ NK cells and concurrent VL in 18 treatment-naive subjects tested at time points in acute/early infection (35). The analysis performed here was done on a larger group of 43 HIV+ and CMV+ individuals together and stratified by NKG2C genotype. To our knowledge this is the first report investigating correlations between the intensity of NKG2C expression on NK cells and VL set point. Overall, we found no evidence that NKG2C+ NK cell parameters influenced VL set point, which is a determinant of the rate of HIV disease progression.
γδ T cells also express NKG2C and have been shown to respond to HIV-infected cells (59, 60). Future studies should explore the link between NKG2C genotype, CMV infection, and frequency of NKG2C-expressing γδ T cells at the level of susceptibility/resistance to HIV infection and at the level of HIV control.
In summary, our results support that carriage of the NKG2C−/− genotype is associated with higher susceptibility to HIV infection, particularly by the parenteral infection route. Although, NKG2C copy number was associated with percentage and intensity of NKG2C expression on NK cells, these parameters did not correlate with HIV VL set point.
MATERIALS AND METHODS
Ethics statement.
This research study was approved by the Institutional Review Board of the Research Ethics Committee of the McGill University Health Centre (study identification code 2018-4501). It was conducted according to the principles expressed in the Declaration of Helsinki. Written informed consent for the collection of each individual’s specimens and subsequent analyses using these samples was obtained from all study subjects.
Study population.
The study population included 591 individuals: 434 were PLWH enrolled in the Montreal PI study, and 157 were HESN subjects (61). Persons at high risk of being sexually exposed to HIV, which we will designate here are sexually exposed (SE) HESN (n = 78), included HIV-uninfected men who have sex with men (MSM) recruited from the Clinique Médicale l’Actuel (n = 40) and subjects enrolled in the Ipergay Pre-Exposure Prophylaxis (PrEP) on-demand study followed in Montreal (n = 21) (62). These MSM SE HESN subjects answered “yes” to the question “Have you had unprotected receptive anal intercourse with a partner of unknown HIV serostatus or known to be HIV-infected, at least 5 times in the last 6 months or at least 50 times in your lifetime before starting PrEP?” An additional 17 SE HESN subjects were HIV-negative partners in HIV-discordant couples who remained HIV uninfected despite multiple exposures that occurred before the availability of antiretroviral treatment (ART). These included 9 men and 8 women; 6 of the men were MSM (63). We also recruited HIV-negative injection drug user (IDU) HESN subjects from the St. Luc cohort (n = 79) (64). All IDU HESN subjects answered “yes” to the question “Have you shared needles and/or injection equipment with partners known to be HIV-infected at least 5 times?” Clinic visits for St. Luc cohort participants occurred approximately every 6 months, at which time information was collected regarding the frequency of their at-risk behavior for HIV exposure. All HESN subjects provided a blood sample from which peripheral blood mononuclear cells (PBMCs) and plasma were isolated and stored frozen until use. HIV serostatus was assessed using HIV enzyme immunoassays (EIAs) (65). Subjects enrolled in the Montreal PI cohort included individuals recruited within the first 6 months of HIV infection, who were then followed an average of every 3 months for up to 4 years (65). At each clinic visit, CD4 and CD8 counts and plasma VL were measured, ART status was recorded, and blood was drawn for isolation of PBMCs and plasma, which was stored frozen until use. For one experiment comparing the expression of NKG2C on cells from HIV− CMV+ persons, 11 additional subjects who had minimal HIV exposure, were included.
NKG2C genotyping.
Genomic DNA was extracted from the PBMCs of all study subjects with the QIAamp DNA blood minikit (Qiagen, Inc., Toronto, ON, Canada) according to the manufacturer’s instructions. Full-length nkg2c (nkg2c+) and the deletion variant (nkg2c−) are alleles at the same locus (24). NKG2A is encoded at a separate locus. The presence of nkg2c+ or nkg2c− alleles and the nkg2a locus, as a positive control present in all subjects, was determined by sequence-specific PCR. Three sets of forward and reverse sequence-specific primers for nkg2c+, nkg2c−, and nkg2a were used to amplify the allele groups at the nkg2c and nkg2a loci. The forward and reverse primers for amplification of the nkg2c+ allele were NKG2CT/F (5′-ATCAATTATTGAAATAGGATGC-3′) and NKG2CT/R (5′-CGCAAAGTTACAACCATCACCAT-3′) (24). Those amplifying the nkg2c− allele were BREAK-F (5′-ACTCGGATTTCTATTTGATGC-3′) and BREAK-R (5′-ACAAGTGATGTATAAGAAAAAG-3′) (24). Those amplifying the nkg2a internal control were NKG2A3F (5′-TGTATCCACCTCTCCTTTCG-3′) and NKG2A4R (5′-TTTGTACAGCCTAAGATCAAG-3′) (24). Twenty-five nanograms per microliter of genomic DNA from each participant was amplified with Platinum Taq (Thermo Fisher Scientific, Burlington, ON, Canada) in a T100 Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA) using the following conditions: denaturation at 95°C for 2 min, then 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, followed by a 5-min extension at 72°C. Amplicons were visualized by gel electrophoresis on a 2% agarose gel in 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA) run at 125 V for 30 min in Fluo-DNA loading buffer (6×; Zmtech Scientifique, Montreal, QC, Canada) and imaged with an Omega Lum C imaging system (Gel Company, Inc., San Francisco, CA). Band sizes of 300 bp corresponded to nkg2c+ alleles, 400 bp to nkg2c− alleles, and 800 bp to nkg2a (29). Samples were classified as homozygous for the presence of the nkg2c+ allele when only the 300-bp band was present (NKG2C+/+), homozygous for nkg2c− (NKG2C−/−) when only the 400-bp band was present, and heterozygous for nkg2c+ and nkg2c− when bands of both sizes (NKG2C+/−) were present (29).
Flow cytometry analysis of the frequency of NKG2C+ cells and the intensity of NKG2C expression.
PBMCs from 32 NKG2C+/+, 19 NKG2C+/−, and 6 NKG2C−/− HIV+ CMV+ subjects were stained with an antibody cocktail that allowed for gating on live NK cells as CD3− CD14− CD19− CD56dim lymphocytes. We also stained PBMCs from 43 NKG2C+/+ and 18 NKG2C+/− HIV− CMV+ subjects with this antibody cocktail; all belonged to the HESN group, except for 11 HIV− CMV+ low-risk controls. These were examined for differences in the percentage of NKG2C+ CD56dim, NKG2C+ CD57+ CD56dim, and NKG2A+ CD56dim NK cells and the intensity of NKG2C expression on CD56dim NK cells from subjects carrying each NKG2C genotype. The intensity of NKG2C staining was assessed by measuring the mean fluorescence intensity (MFI), the median fluorescence intensity, and the fold change over background in the MFI of NKG2C staining. Cryopreserved PBMCs were thawed and resuspended in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 IU/ml penicillin, and 50 mg/ml streptomycin (R10) (all from Wisent, St Jean Baptiste, QC, Canada). PBMCs (106 in 100 μl of R10) were cell surface stained for 25 min at 4°C with previously optimized concentrations of fluorochrome-conjugated antibodies to the following cell surface markers: CD3-BV785 (clone OKT3), CD19-BV785 (HIB19), CD14-BV785 (M5E2), and CD56-BV605 (HCD56) from Biolegend, San Diego, CA; CD16-allophycocyanin (APC)-Cy7 (3G8) from BD Biosciences, Baltimore, MD; NKG2C-phycoerythrin (PE)-Cy7 (REA250) and NKG2A-APC (REA110) from Miltenyi Biotec, Auburn, CA; CD57-PE (TB01) from Life Technologies, Burlington, ON, Canada); and Indo-Violet LIVE/DEAD (L/D) stain from Fisher Scientific, Waltham, MA. Cells were then washed twice with fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline [PBS], 4% fetal bovine serum [FBS], 0.05% NaN3) and fixed in 2% paraformaldehyde (Santa Cruz Biotechnology, Santa Cruz, CA). Between 5 × 105 and 7 × 105 cells were acquired using an LSRFortessa X-20 flow cytometer (BD Biosciences, San Jose, CA). Results were analyzed using FlowJo v10.6.2 software (Tree Star, Ashland, OR).
VL set point determination.
VL set points were calculated for 160 NKG2C+/+, 83 NKG2C+/−, and 6 NKG2C−/− HIV+ carriers. The average of the VLs from all treatment-naive time points 6 months after the estimated date of infection to the end of their follow-up in the Montreal PI cohort were used to calculate the VL set point.
Statistical analysis.
Statistical analysis and graphical presentation of results were performed using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA) and Statistical Analysis System (SAS) software version 9.4 (SAS Institute, Cary, NC). The statistical significance of differences in the racial/ethnic composition of the HIV+ and HESN populations and deviations in the distributions of NKG2C genotype from Hardy-Weinberg equilibrium (HWE) was assessed using χ2 tests. Between-group differences in the frequency of NKG2C genotypes in PLWH and HESN populations were determined using two-tailed Fisher’s exact tests with Haldane’s correction. The statistical significance of between-genotype differences in the percentage of NKG2C+ NK cells, the intensity of NKG2C expression on CD56dim NK cells, and VL set point in ART-naive PLWH was assessed using Kruskal-Wallis tests with Dunn’s posttests. The significance of correlations between the percentages of NKG2C+, NKG2C+ CD57+, and NKG2A+ CD56dim NK cells and intensity of NKG2C expression and VL set point in ART-naive CMV+ PLWH was assessed using Spearman’s correlation tests. P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
We are thankful to the individuals who took part in this study over the years and their physicians for their collaboration: S. Vézina, L. Charest, C. Milne, S. Lavoie, J. Friedman, F. Asselin, M. Boissonnault, P.-J. Maziade, B. Deligne, V. To, J.-G. Baril, B. Lessard, M.-A. Charron, S. Dufresne, E. Huchet, S. Poulin, D. Longpré, M. Lonpré, R. Pilarski, E. Sasseville, A. Cloutier-Blais, F. Chano, L. Labrecque, C. Fortin, M. Munoz, V. Martel-Laferrière, B. Lebouché, A. de Pokomandy, J. Cox, L.-P. Haraoui, M. Potter, J. MacLeod, M. Klein, G. Smith, N. Gilmore, R. Lalonde, C. Frenette, D. Murphy, A. Talbot, H. Dion, C. M. Tsoukas, N. Lapointe, C. Olivier, and E. Lefebvre. We wish to acknowledge Olfa Debbeche for laboratory sample processing, handling, storage, and shipment of Montreal Primary HIV Infection cohort samples. We thank Mario Legault for administrative support and coordination of the Montreal Primary HIV Infection cohort and Rachel Bouchard for administrative support and coordination of the St. Luc cohort. We acknowledge the Immunophenotyping Technology Platform of the Research Institute of the McGill University Health Center and staff for their contribution to this publication.
We also acknowledge the financial support provided by the Fond de Recherche du Québec—Santé to support general expenditures incurred by the Research Institute of the McGill University Health Centre, of which N.F.B. is a member, to support indirect costs related to the implementation of research projects.
Contributor Information
Nicole F. Bernard, Email: nicole.bernard@mcgill.ca.
Guido Silvestri, Emory University.
REFERENCES
- 1.Lanier LL. 2005. NK cell recognition. Annu Rev Immunol 23:225–274. 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. 2008. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 9:495–502. 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raulet DH, Vance RE, McMahon CW. 2001. Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol 19:291–330. 10.1146/annurev.immunol.19.1.291. [DOI] [PubMed] [Google Scholar]
- 4.Houchins JP, Yabe T, McSherry C, Bach FH. 1991. DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. J Exp Med 173:1017–1020. 10.1084/jem.173.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yabe T, McSherry C, Bach FH, Fisch P, Schall RP, Sondel PM, Houchins JP. 1993. A multigene family on human chromosome 12 encodes natural killer-cell lectins. Immunogenetics 37:455–460. 10.1007/BF00222470. [DOI] [PubMed] [Google Scholar]
- 6.Chang C, Rodríguez A, Carretero M, López-Botet M, Phillips JH, Lanier LL. 1995. Molecular characterization of human CD94: a type II membrane glycoprotein related to the C-type lectin superfamily. Eur J Immunol 25:2433–2437. 10.1002/eji.1830250904. [DOI] [PubMed] [Google Scholar]
- 7.Braud VM, Allan DS, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391:795–799. 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 8.Llano M, Lee N, Navarro F, García P, Albar JP, Geraghty DE, López-Botet M. 1998. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur J Immunol 28:2854–2863. . [DOI] [PubMed] [Google Scholar]
- 9.Guma M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, López-Botet M. 2006. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis 194:38–41. 10.1086/504719. [DOI] [PubMed] [Google Scholar]
- 10.Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, Heinrich F, Gasparoni G, Babic M, Tomic A, Pietra G, Nienen M, Blau IW, Hofmann J, Na IK, Prinz I, Koenecke C, Hemmati P, Babel N, Arnold R, Walter J, Thurley K, Mashreghi MF, Messerle M, Romagnani C. 2018. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol 19:453–463. 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- 11.Rolle A, Meyer M, Calderazzo S, Jager D, Momburg F. 2018. Distinct HLA-E peptide complexes modify antibody-driven effector functions of adaptive NK cells. Cell Rep 24:1967–1976.e4. 10.1016/j.celrep.2018.07.069. [DOI] [PubMed] [Google Scholar]
- 12.Rolle A, Pollmann J, Ewen EM, Le VT, Halenius A, Hengel H, Cerwenka A. 2014. IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J Clin Invest 124:5305–5316. 10.1172/JCI77440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M, 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104:3664–3671. 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 14.Beziat V, Descours B, Parizot C, Debre P, Vieillard V. 2010. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS One 5:e11966. 10.1371/journal.pone.0011966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanier LL, Corliss B, Wu J, Phillips JH. 1998. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 8:693–701. 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, Lanier LL. 2010. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood 116:3865–3874. 10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, Nixon DF, Lanier LL. 2011. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A 108:14725–14732. 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SC, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg KJ, Ljunggren HG, Miller JS, Bryceson YT. 2015. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42:443–456. 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, Kim S. 2015. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42:431–442. 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luetke-Eversloh M, Hammer Q, Durek P, Nordstrom K, Gasparoni G, Pink M, Hamann A, Walter J, Chang HD, Dong J, Romagnani C. 2014. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog 10:e1004441. 10.1371/journal.ppat.1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luetke-Eversloh M, Cicek BB, Siracusa F, Thom JT, Hamann A, Frischbutter S, Baumgrass R, Chang HD, Thiel A, Dong J, Romagnani C. 2014. NK cells gain higher IFN-gamma competence during terminal differentiation. Eur J Immunol 44:2074–2084. 10.1002/eji.201344072. [DOI] [PubMed] [Google Scholar]
- 22.Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA, Hammer Q, Goodridge JP, Larsson S, Jayaraman J, Oei VYS, Schaffer M, Tasken K, Ljunggren HG, Romagnani C, Trowsdale J, Malmberg KJ, Beziat V. 2016. Critical role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep 15:1088–1099. 10.1016/j.celrep.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hikami K, Tsuchiya N, Yabe T, Tokunaga K. 2003. Variations of human killer cell lectin-like receptors: common occurrence of NKG2-C deletion in the general population. Genes Immun 4:160–167. 10.1038/sj.gene.6363940. [DOI] [PubMed] [Google Scholar]
- 24.Miyashita R, Tsuchiya N, Hikami K, Kuroki K, Fukazawa T, Bijl M, Kallenberg CG, Hashimoto H, Yabe T, Tokunaga K. 2004. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int Immunol 16:163–168. 10.1093/intimm/dxh013. [DOI] [PubMed] [Google Scholar]
- 25.Moraru M, Canizares M, Muntasell A, de Pablo R, Lopez-Botet M, Vilches C. 2012. Assessment of copy-number variation in the NKG2C receptor gene in a single-tube and characterization of a reference cell panel, using standard polymerase chain reaction. Tissue Antigens 80:184–187. 10.1111/j.1399-0039.2012.01911.x. [DOI] [PubMed] [Google Scholar]
- 26.Goncalves A, Makalo P, Joof H, Burr S, Ramadhani A, Massae P, Malisa A, Mtuy T, Derrick T, Last AR, Nabicassa M, Cassama E, Houghton J, Palmer CD, Pickering H, Burton MJ, Mabey DC, Bailey RL, Goodier MR, Holland MJ, Roberts CH. 2016. Differential frequency of NKG2C/KLRC2 deletion in distinct African populations and susceptibility to trachoma: a new method for imputation of KLRC2 genotypes from SNP genotyping data. Hum Genet 135:939–951. 10.1007/s00439-016-1694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangel-Ramirez VV, Garcia-Sepulveda CA, Escalante-Padron F, Perez-Gonzalez LF, Rangel-Castilla A, Aranda-Romo S, Noyola DE. 2014. NKG2C gene deletion in the Mexican population and lack of association to respiratory viral infections. Int J Immunogenet 41:126–130. 10.1111/iji.12104. [DOI] [PubMed] [Google Scholar]
- 28.Goodier MR, White MJ, Darboe A, Nielsen CM, Goncalves A, Bottomley C, Moore SE, Riley EM. 2014. Rapid NK cell differentiation in a population with near-universal human cytomegalovirus infection is attenuated by NKG2C deletions. Blood 124:2213–2222. 10.1182/blood-2014-05-576124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Della Chiesa M, Falco M, Bertaina A, Muccio L, Alicata C, Frassoni F, Locatelli F, Moretta L, Moretta A. 2014. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C−/− umbilical cord blood. J Immunol 192:1471–1479. 10.4049/jimmunol.1302053. [DOI] [PubMed] [Google Scholar]
- 30.Comeau EM, Holder KA, Fudge NJ, Grant MD. 2019. Cytomegalovirus-driven adaption of natural killer cells in NKG2Cnull human immunodeficiency virus-infected individuals. Viruses 11:239. 10.3390/v11030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mela CM, Goodier MR. 2007. The contribution of cytomegalovirus to changes in NK cell receptor expression in HIV-1-infected individuals. J Infect Dis 195:158–159. 10.1086/509811. [DOI] [PubMed] [Google Scholar]
- 32.Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E, Moretta A, Mavilio D. 2009. The decreased expression of Siglec-7 represents an early marker of dysfunctional natural killer-cell subsets associated with high levels of HIV-1 viremia. Blood 114:3822–3830. 10.1182/blood-2009-06-226332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas R, Low HZ, Kniesch K, Jacobs R, Schmidt RE, Witte T. 2012. NKG2C deletion is a risk factor of HIV infection. AIDS Res Hum Retroviruses 28:844–851. 10.1089/aid.2011.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma M, Wang Z, Chen X, Tao A, He L, Fu S, Zhang Z, Fu Y, Guo C, Liu J, Han X, Xu J, Chu Z, Ding H, Shang H, Jiang Y. 2017. NKG2C+ NKG2A− natural killer cells are associated with a lower viral set point and may predict disease progression in individuals with primary HIV infection. Front Immunol 8:1176. 10.3389/fimmu.2017.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gondois-Rey F, Cheret A, Granjeaud S, Mallet F, Bidaut G, Lecuroux C, Ploquin M, Muller-Trutwin M, Rouzioux C, Avettand-Fenoel V, Moretta A, Pialoux G, Goujard C, Meyer L, Olive D. 2017. NKG2C+ memory-like NK cells contribute to the control of HIV viremia during primary infection: Optiprim-ANRS 147. Clin Transl Immunol 6:e150. 10.1038/cti.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, Sverremark-Ekstrom E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaelsson J, Ljunggren HG, Malmberg KJ. 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688. 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellors JW, Muñoz A, Giorgi JV, Margolick JB, Tassoni CJ, Gupta P, Kingsley LA, Todd JA, Saah AJ, Detels R, Phair JP, Rinaldo CR, Jr.. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med 126:946–954. 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 38.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167–1170. 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 39.Vanhems P, Caillat-Vallet E, Hirschel B, Routy J-P, Carr A, Vizzard J, Cooper DA, Perrin L, Swiss HIV Cohort Study. 2002. The CD4 cell count 3 months after acute retroviral syndrome is associated with the presence of AIDS in the source individual. AIDS 16:2234–2236. 10.1097/00002030-200211080-00022. [DOI] [PubMed] [Google Scholar]
- 40.Routy J-P, Danielle VP, Tsoukas C, Lefèbvre E, Côté P, LeBlanc R, Conway B, Alary M, Bruneau J, Sekaly R-P. 2000. Comparison of clinical features of acute HIV1 infection in patients infected sexually or through injection drug use. J Acquir Immune Defic Syndr 24:425–432. 10.1097/00042560-200008150-00004. [DOI] [PubMed] [Google Scholar]
- 41.Fox J, White PJ, Weber J, Garnett GP, Ward H, Fidler S. 2011. Quantifying sexual exposure to HIV within an HIV-serodiscordant relationship: development of an algorithm. AIDS 25:1065–1082. 10.1097/QAD.0b013e328344fe4a. [DOI] [PubMed] [Google Scholar]
- 42.Boily MC, Baggaley RF, Wang L, Masse B, White RG, Hayes RJ, Alary M. 2009. Heterosexual risk of HIV-1 infection per sexual act: systematic review and meta-analysis of observational studies. Lancet Infect Dis 9:118–129. 10.1016/S1473-3099(09)70021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baggaley RF, Boily MC, White RG, Alary M. 2006. Risk of HIV-1 transmission for parenteral exposure and blood transfusion: a systematic review and meta-analysis. AIDS 20:805–812. 10.1097/01.aids.0000218543.46963.6d. [DOI] [PubMed] [Google Scholar]
- 44.Vittinghoff E, Douglas J, Judson F, McKirnan D, MacQueen K, Buchbinder SP. 1999. Per-contact risk of human immunodeficiency virus transmission between male sexual partners. Am J Epidemiol 150:306–311. 10.1093/oxfordjournals.aje.a010003. [DOI] [PubMed] [Google Scholar]
- 45.Agostinis C, Mangogna A, Bossi F, Ricci G, Kishore U, Bulla R. 2019. Uterine immunity and microbiota: a shifting paradigm. Front Immunol 10:2387. 10.3389/fimmu.2019.02387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marquardt N, Kekalainen E, Chen P, Lourda M, Wilson JN, Scharenberg M, Bergman P, Al-Ameri M, Hard J, Mold JE, Ljunggren HG, Michaelsson J. 2019. Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat Commun 10:3841. 10.1038/s41467-019-11632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E, Johansson H, Mjosberg J, Westgren M, Lankisch TO, Wedemeyer H, Ellis EC, Ljunggren HG, Michaelsson J, Bjorkstrom NK. 2015. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol 194:2467–2471. 10.4049/jimmunol.1402756. [DOI] [PubMed] [Google Scholar]
- 48.Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Verges S, Lanier LL, Weisdorf D, Miller JS. 2012. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119:2665–2674. 10.1182/blood-2011-10-386995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beziat V, Traherne J, Malmberg JA, Ivarsson MA, Bjorkstrom NK, Retiere C, Ljunggren HG, Michaelsson J, Trowsdale J, Malmberg KJ. 2014. Tracing dynamic expansion of human NK-cell subsets by high-resolution analysis of KIR repertoires and cellular differentiation. Eur J Immunol 44:2192–2196. 10.1002/eji.201444464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA, Hammer Q, Goodridge JP, Larsson S, Jayaraman J, Oei VYS, Schaffer M, Taskén K, Ljunggren HG, Romagnani C, Trowsdale J, Malmberg KJ, Béziat V. 2016. Critical role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep 15:1088–1099. 10.1016/j.celrep.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith ME, Thomas JA. 1990. Cellular expression of lymphocyte function associated antigens and the intercellular adhesion molecule-1 in normal tissue. J Clin Pathol 43:893–900. 10.1136/jcp.43.11.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolle A, Halenius A, Ewen EM, Cerwenka A, Hengel H, Momburg F. 2016. CD2-CD58 interactions are pivotal for the activation and function of adaptive natural killer cells in human cytomegalovirus infection. Eur J Immunol 46:2420–2425. 10.1002/eji.201646492. [DOI] [PubMed] [Google Scholar]
- 53.Revello MG, Gerna G. 2010. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev Med Virol 20:136–155. 10.1002/rmv.645. [DOI] [PubMed] [Google Scholar]
- 54.Olsson J, Wikby A, Johansson B, Löfgren S, Nilsson BO, Ferguson FG. 2000. Age-related change in peripheral blood T-lymphocyte subpopulations and cytomegalovirus infection in the very old: the Swedish longitudinal OCTO immune study. Mech Ageing Dev 121:187–201. 10.1016/S0047-6374(00)00210-4. [DOI] [PubMed] [Google Scholar]
- 55.Lamarre V, Gilbert NL, Rousseau C, Gyorkos TW, Fraser WD. 2016. Seroconversion for cytomegalovirus infection in a cohort of pregnant women in Quebec, 2010–2013. Epidemiol Infect 144:1701–1709. 10.1017/S0950268815003167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuhair M, Smit GSA, Wallis G, Jabbar F, Smith C, Devleesschauwer B, Griffiths P. 2019. Estimation of the worldwide seroprevalence of cytomegalovirus: a systematic review and meta-analysis. Rev Med Virol 29:e2034. 10.1002/rmv.2034. [DOI] [PubMed] [Google Scholar]
- 57.Ramendra R, Isnard S, Lin J, Fombuena B, Ouyang J, Mehraj V, Zhang Y, Finkelman M, Costiniuk C, Lebouche B, Chartrand-Lefebvre C, Durand M, Tremblay C, Ancuta P, Boivin G, Routy JP. 2020. Cytomegalovirus seropositivity is associated with increased microbial translocation in people living with human immunodeficiency virus and uninfected controls. Clin Infect Dis 71:1438–1446. 10.1093/cid/ciz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muntasell A, Lopez-Montanes M, Vera A, Heredia G, Romo N, Penafiel J, Moraru M, Vila J, Vilches C, Lopez-Botet M. 2013. NKG2C zygosity influences CD94/NKG2C receptor function and the NK-cell compartment redistribution in response to human cytomegalovirus. Eur J Immunol 43:3268–3278. 10.1002/eji.201343773. [DOI] [PubMed] [Google Scholar]
- 59.Juno JA, Eriksson EM. 2019. gammadelta T-cell responses during HIV infection and antiretroviral therapy. Clin Transl Immunol 8:e01069. 10.1002/cti2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fausther-Bovendo H, Wauquier N, Cherfils-Vicini J, Cremer I, Debre P, Vieillard V. 2008. NKG2C is a major triggering receptor involved in the V[delta]1 T cell-mediated cytotoxicity against HIV-infected CD4 T cells. AIDS 22:217–226. 10.1097/QAD.0b013e3282f46e7c. [DOI] [PubMed] [Google Scholar]
- 61.Cao W, Mehraj V, Trottier B, Baril JG, Leblanc R, Lebouche B, Cox J, Tremblay C, Lu W, Singer J, Li T, Routy JP, Vezina S, Charest L, Milne M, Huchet E, Lavoie S, Friedman J, Duchastel M, Villielm F, Cote P, Potter M, Lessard B, Charron MA, Dufresne S, Turgeon ME, Rouleau D, Labrecque L, Fortin C, de Pokomandy A, Hal-Gagne V, Munoz M, Deligne B, Martel-Laferriere V, Gilmore N, Fletcher M, Szabo J, Montreal Primary HIV Infection Study Group. 2016. Early initiation rather than prolonged duration of antiretroviral therapy in HIV infection contributes to the normalization of CD8 T-cell counts. Clin Infect Dis 62:250–257. 10.1093/cid/civ809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molina JM, Capitant C, Spire B, Pialoux G, Cotte L, Charreau I, Tremblay C, Le Gall JM, Cua E, Pasquet A, Raffi F, Pintado C, Chidiac C, Chas J, Charbonneau P, Delaugerre C, Suzan-Monti M, Loze B, Fonsart J, Peytavin G, Cheret A, Timsit J, Girard G, Lorente N, Preau M, Rooney JF, Wainberg MA, Thompson D, Rozenbaum W, Dore V, Marchand L, Simon MC, Etien N, Aboulker JP, Meyer L, Delfraissy JF, ANRS IPERGAY Study Group. 2015. On-demand preexposure prophylaxis in men at high risk for HIV-1 infection. N Engl J Med 373:2237–2246. 10.1056/NEJMoa1506273. [DOI] [PubMed] [Google Scholar]
- 63.Makedonas G, Bruneau J, Alary M, Tsoukas CM, Lowndes CM, Lamothe F, Bernard NF. 2005. Comparison of HIV-specific CD8 T-cell responses among uninfected individuals exposed to HIV parenterally and mucosally. AIDS 19:251–259. [PubMed] [Google Scholar]
- 64.Bruneau J, Daniel M, Abrahamowicz M, Zang G, Lamothe F, Vincelette J. 2011. Trends in human immunodeficiency virus incidence and risk behavior among injection drug users in Montreal, Canada: a 16-year longitudinal study. Am J Epidemiol 173:1049–1058. 10.1093/aje/kwq479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mehraj V, Cox J, Lebouche B, Costiniuk C, Cao W, Li T, Ponte R, Thomas R, Szabo J, Baril JG, Trottier B, Cote P, LeBlanc R, Bruneau J, Tremblay C, Routy JP, Montreal Primary HIV Infection Study Group. 2018. Socio-economic status and time trends associated with early ART initiation following primary HIV infection in Montreal, Canada: 1996 to 2015. J Int AIDS Soc 21:e25034. 10.1002/jia2.25034. [DOI] [PMC free article] [PubMed] [Google Scholar]